Abstract
Mutation of the immunophilin-like FK506-binding protein TWISTED DWARF1 (FKBP42/TWD1) causes dwarf and twisted-organ phenotypes in Arabidopsis. However, the function of FKBP42 is not fully understood at the molecular level. Using genetic, physiological and immunological experiments, we show here that FKBP42 is necessary for brassinosteroid (BR) signal transduction. The twd1 mutant showed reduced BR sensitivity in growth responses and activation of the BZR1 transcription factor. However, twd1 showed normal responses to an inhibitor of BIN2/GSK3, suggesting that twd1 has defect upstream of BIN2 in the BR signaling pathway. In vitro and in vivo assays showed that TWD1 interacts physically with the kinase domains of the BR receptor kinases BRI1 and BAK1. TWD1 is not required for normal localization of BRI1-GFP to the plasma membrane or for activation of the flagellin receptor kinase FLS2. Our results suggest that FKBP42/TWD1 plays a specific role in the activation of BRI1 receptor kinase.
Introduction
Brassinosteroid (BR) is a plant steroid hormone that plays a major role in regulating a wide range of developmental and physiological processes throughout the life cycle of plants (Clouse and Sasse, 1998). BR-deficient and signaling mutants share characteristic phenotypes, including dwarf stature, small-round and dark green leaves, short hypocotyls and constitutive photomorphogenesis in the dark. BR binds to the extracellular domain of a leucine-rich repeat receptor-like kinase (LRR-RLK) BRI1, to activate a signal transduction cascade that regulates transcription activities of transcription factors BZR1 and BZR2/BES1 (Clouse, 2011; Kim and Wang, 2010; Wang et al., 2012).
When BR levels are low, BZR1 is inactivated due to phosphorylation by the GSK3-like kinase BIN2 (He et al., 2002) and subsequent cytoplasmic retention by the 14-3-3 proteins (Gampala et al., 2007). When BR levels are high, BR binding to the extracellular domain of BRI1 recruits its co-receptor kinase BAK1, which leads receptor kinase activation through transphosphorylation between their kinase domains. BRI1 and BAK1 are mainly localized at the plasma membrane, but also undergo endocytosis and cycle between the endosome and plasma membrane (Geldner et al., 2007; Russinova et al., 2004; Song et al., 2009). Activation of the BRI1/BAK1 receptor complex then leads to BRI1 phosphorylation of BR-signaling kinases (BSKs) (Tang et al., 2008), BSK1 activation of the PP1-like phosphatase BSU1, BSU1-mediated dephosphorylation and inactivation of BIN2 kinase (Kim et al., 2009), and PP2A-mediated dephosphorylation and activation of the BZR1 transcription factor (Tang et al., 2011). Unphosphorylated BZR1 accumulates in the nucleus and binds to genomic DNA to directly repress or activate thousands of BZR1 target genes (He et al., 2005; Oh et al., 2014; Sun et al., 2010; Yin et al., 2005).
TWISTED DWARF1 (TWD1) encodes an immunophilin-like FK506-binding protein 42 (FKBP42) with an in-plane membrane-anchored domain in the C terminus (Bailly et al., 2014; Scheidt et al., 2007). FKBP42 has been shown to be localized to several membranes, including the plasma membrane, tonoplast, and endoplasmic reticulum (ER) (Geisler et al., 2003; Wang et al., 2013; Wu et al., 2010). Loss-of-function mutations of TWD1, such as twd1-1 and ultracurvata2-1 (ucu2-1), cause dwarf and twisted-organ phenotypes in Arabidopsis (Bowman et al., 1994). The twd1 mutants share reduced cell elongation, helical organ twisting, and epinastic leaf phenotypes with mutants defective in polar auxin transport, such as the double abcb1/abcb19 mutant that lacks the ABC transporters class B (ABCB) 1 and 19 (Bouchard et al., 2006; Geisler et al., 2003). However, twd1/ucu2 mutants show more severe dwarfism than abcb1/abcb19 as well as dark green leaf phenotypes (Geisler et al., 2003; Pérez-Pérez et al., 2004), which are distinct from those of auxin transport mutants but resemble the characteristic phenotypes of the BR-signaling or biosynthesis mutants (Li et al., 2001; Pérez-Pérez et al., 2004). The ucu2-1 mutant also showed reduced BR sensitivity in root growth and expression of some BR-responsive genes (Pérez-Pérez et al., 2004). Hence, the phenotype of twd1 suggests that FKBP42/TWD1 might play a role in the BR signaling pathway to regulate plant growth and development. However, the molecular function of FKBP42/TWD1 in BR signaling remains unknown.
In this study, we showed that the dwarf phenotype of twd1 can be partially suppressed by the hypermorphic bzr1-1D mutation, suggesting a defect of twd1 upstream of bzr1-1D. The twd1 mutant was less sensitive to BR treatment but responded normally to an inhibitor of BIN2 kinase, which indicates that FKBP42/TWD1 is involved in a BR signaling step upstream of BIN2. Using in vitro and in vivo protein-protein interaction assays, we demonstrated that FKBP42/TWD1 functions in the BR signaling pathway by means of physical interaction with the kinase domains of BRI1 and BAK1 receptors.
Results
FKBP42/TWD1 regulates BR signaling upstream of BZR1
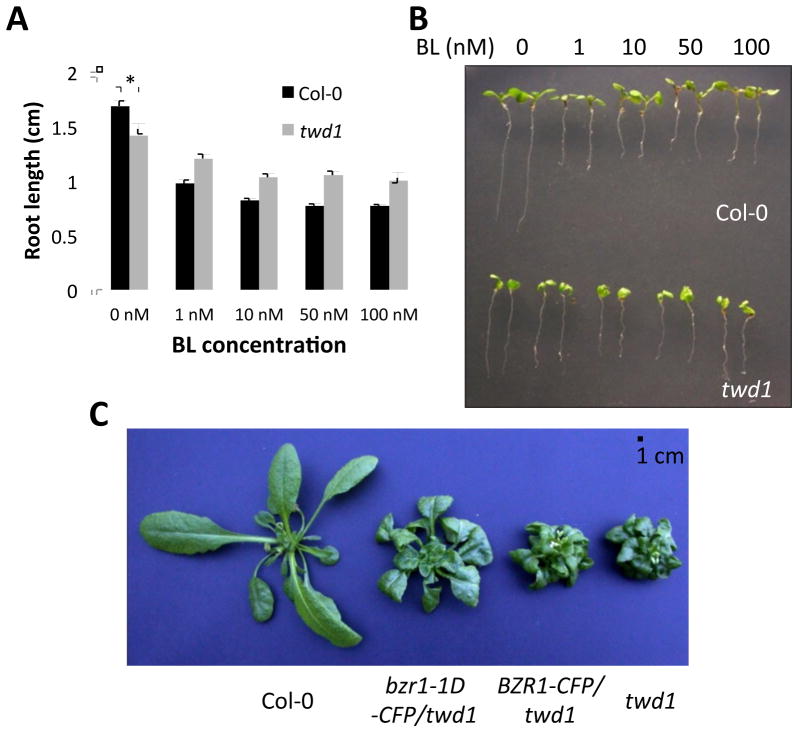
It has been reported that the ucu2 mutants (alleles of twd1) showed reduced responses to 24-epibrassinolide inhibition of root growth and expression of CPD and ROT3 (Pérez-Pérez et al., 2004). Consistent with the previous study, we found that twd1-2 (Salk_012836) mutant was less sensitive than wild type plants to brassinolide (BL, the most active form of brassinosteroids), in root growth and hypocotyl elongation (Figure 1A–B). Furthermore, high concentrations of BL (50–100 nM) did not promote hypocotyl elongation in twd1 seedlings, in contrast to wild-type Col-0 (Figure 1B). These results suggest that FKBP42/TWD1 is required for normal BR signal transduction.
Figure 1. FKBP42/TWD1 regulates BR signaling upstream of BZR1.
(A–B) Col-0 and twd1 seedlings were grown on media supplemented with various concentrations of BL. (A) Measurement of primary root growth (n>20, mean ± SD). *indicates T-test’s p-value<0.01. (B) Representative images of the seedlings showing reduced BR responses in root growth inhibition and hypocotyl elongation. Note that the seedlings were transferred to new plates for imaging and thus subtle morphology such as root twisting is not obvious.
(C) Gain-of-function bzr1-1D mutation partially suppressed growth phenotypes in twd1. Wild-type, pBZR1:bzr1-1D-CFP/twd1, pBZR1:BZR1-CFP/twd1 and twd1 plants were grown in the greenhouse for 3 weeks.
To determine genetic interaction between FKBP42/TWD1 and the BR signaling pathway, we crossed twd1 with transgenic plants expressing the gain-of-function bzr1-1D-CFP (pBZR1:bzr1-1D-CFP), in which the mutation leads to a constitutively active, hypophosphorylated form of BZR1 due to increased affinity to PP2A phosphatases (Tang et al., 2011). Phenotypic analyses of three-week old pBZR1:bzr1-1D-CFP/twd1 and pBZR1:BZR1-CFP/twd1 comparing to twd1 showed that the bzr1-1D mutation partially suppressed the dwarf phenotype of twd1 (Figure 1C), which is similar to bzr1-1D suppression of bri1 (Wang et al., 2002). The results suggest that FKBP42/TWD1 is involved in BR signaling upstream of BZR1.
FKBP42/TWD1 is involved in BR signaling upstream of BIN2 kinases
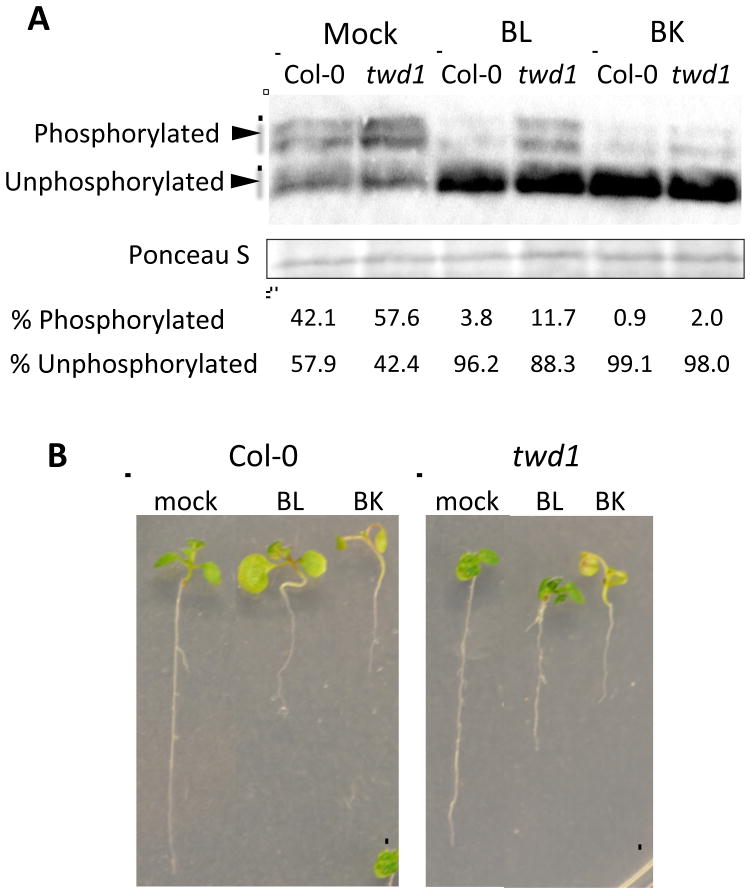
To determine if FKBP42/TWD1 regulates BZR1 activities by regulating BIN2, we treated seedlings with bikinin, an inhibitor of GSK3/BIN2 kinases (De Rybel et al., 2009), compared to BL and mock treatments. We determined BZR1 phosphorylation status by western blotting, using transgenic lines expressing pBZR1:BZR1-CFP in Col-0 and twd1. Western blot results demonstrated that, without BL treatment, twd1 accumulated a higher level of inactive, phosphorylated forms of BZR1-CFP protein (57.6% phosphorylated) than wild-type (42.1% phosphorylated) (Figure 2A). Furthermore, treatment of 100 nM brassinolide for 20 min only partially dephosphorylated BZR1-CFP in twd1 (88.3% dephosphorylated), whereas the same treatment almost fully dephosphorylated BZR1-CFP in wild-type (96.2% dephosphorylated) (Figure 2A). The reduced responsiveness to BR in terms of BZR1 dephosphorylation in twd1 is consistent with the reduced BR responses observed in root growth inhibition and hypocotyl elongation (Figure 1).
Figure 2. FKBP42/TWD1 regulates BZR1 dephosphorylation by regulating BR-signaling components upstream of BIN2.
(A) Western blot analysis of BZR1-CFP phosphorylation in Col-0 and twd1 background. Seven-day-old pBZR1:BZR1-CFP/Col and pBZR1:BZR1-CFP/twd1 seedlings were treated with mock, 100 nM BL, or 30μM Bikinin for 20 min. Phosphorylated and unphosphorylated BZR1-CFP were detected with anti-GFP antibodies, and the band intensities were quantified using ImageJ. Blot stained with Ponceau S is presented to show equal loading.
(B) Col-0 and twd1 were grown on mock, 100 nM BL, or 30 μM Bikinin for seven days. Scale bars, 5 mm.
However, unlike BR treatment, treatment with bikinin, an inhibitor of BIN2 kinase, induced similar dephosphorylation of BZR1 in wild-type and twd1 (99.1% and 98.0% dephosphorylated BZR1-CFP in wild-type and twd1, respectively) (Figure 2A). Bikinin also induced similar growth responses in wild-type and twd1 (Figure 2B). Thus, the results suggest that FKBP42/TWD1 is likely involved in the BR signaling pathway upstream of BIN2.
FKBP42/TWD1 physically interacts with BRI1 and BAK1
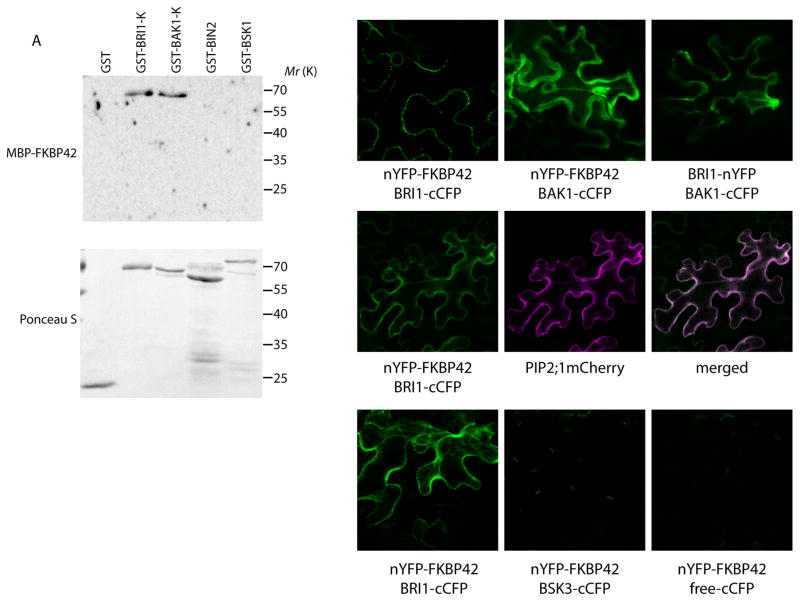
Next, we tested whether BR signaling components upstream of BIN2, such as BRI1, BAK1 and BSKs, interact with FKBP42/TWD1. First, protein-protein interactions were tested in vitro by gel blot overlay assays. The GST, GST-tagged kinase domains of BRI1 and BAK1 and GST-tagged BIN2 and BSK1 were gel blotted and probed with MBP-tagged FKBP42 protein. The results further indicated that FKBP42/TWD1 interacts with the kinase domains of BRI1 and BAK1, but not BIN2 or BSK1 (Figure 3A). This interaction was further confirmed in vivo by bimolecular fluorescence complementation (BiFC) assays. Tobacco epidermal cells co-transformed with the amino-terminal half of yellow fluorescent protein (YFP) fused to the amino terminus of FKBP42 (nYFP-FKBP42) and BRI1 or BAK1 fused to the carboxy-terminal half of cyan fluorescent protein (BRI1-cCFP and BAK1-cCFP) showed strong fluorescence signals, similar to the signals of BRI1-nYFP and BAK1-cCFP, which served as a positive control (Figure 3B). The BiFC signal of nYFP-FKBP42 and BRI1-cCFP was co-localized with fluorescence signal of plasma membrane intrinsic protein (PIP) aquaporin (PIP2;1-mCherry) (Figure 3C), which has been shown to be localized at the plasma membrane and endosomal membrane mediated through endocytosis (Wudick et al., 2015). Treatment of the leaf tissues with BL or propiconazole (PPZ), which blocks BR biosynthesis, did not obviously alter the interaction between nYFP-FKBP42 and BRI1-cCFP (Figure 3C–D), suggesting that FKBP42/TWD1 interacts with BRI1 in a BR-independent manner. Tobacco leaves co-transformed with nYFP-FKBP42 and BSK3-cCFP showed no fluorescence signal (Figure 3E–F). Thus, the results suggest that FKBP42/TWD1 affects BR signaling by binding to the kinase domains of the receptors BRI1 and BAK1.
Figure 3. FKBP42/TWD1 interacts with BRI1 and BAK1 kinase domains.
(A) The GST fusion proteins of BRI1 kinase domain (GST-BRI1-K), BAK1 kinase domain (GST-BAK1-K), BIN2 (GST-BIN2) and BSK1 (GST-BSK1), as well as GST itself, were separated by SDS-PAGE and blotted onto a nitrocellulose membrane. The blot was probed with the MBP fusion protein of FKBP42/TWD1 (MBP-FKBP42) and an anti-MBP antibody (top) and then stained with Ponceau S (bottom).
(B–F) Confocal fluorescence images showing BiFC fluorescence signals from tobacco leaf epidermal cells infiltrated with the indicated constructs. Scale bar, 20 μm. (B) BiFC assays show interaction between FKBP42/TWD1 and BRI1 or BAK1 in vivo. Positive interaction between BRI1-nYFP and BAK1-cCFP served as a positive control.
(C–D) The BiFC constructs nYFP-FKBP42 and BRI1-cCFP were co-infiltrated with 35S:PIP2;1-mCherry in the presence of 1 μM BL (similar patterns were also observed when BiFC and PIP2;1-mCherry were expressed in different cells) (C), or infiltrated in the presence of 2 μM PPZ (D).
(E–F) No interaction was observed in the nYFP-FKBP42/BSK3-cCFP (E) and nYFP-FKBP42/cCFP cells (F).
TWD1 mutation does not affect BRI1 localization to the plasma membrane
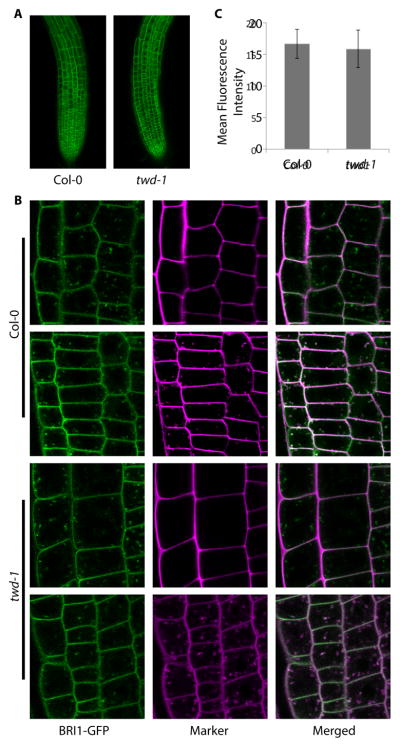
A previous study has demonstrated that FKBP42/TWD1 is required for localization of plasma membrane protein ABCB auxin transporters and mutations in TWD1 lead to mislocalization of ABCB1, ABCB4 and ABCB19 to the ER instead of the plasma membrane (Wang et al., 2013; Wu et al., 2010). To test whether FKBP42/TWD1 is also necessary for localization of the BRI1 receptor to the plasma membrane, we crossed transgenic plants expressing GFP-tagged BRI1 (pBRI1:BRI1-GFP) with the twd1 mutant. Confocal microscopic analysis demonstrated that the root tips of Col-0 wild-type and twd1 exhibited similar BRI1-GFP fluorescence intensities (Figure 4A). We detected BRI1-GFP signals at the plasma membrane and in the endocytic vesicles, which were labeled with propidium iodide (for plasma membrane) and FM4-64 (for plasma membrane and endosomal membrane) (Figure 4B), consistent with that reported in a previous study (Russinova et al., 2004). Our results showed that subcellular localization of BRI1-GFP was not affected in the twd1 mutant (Figure 4B). Quantification of GFP fluorescence intensities further supported that the twd1 mutation did not significantly alter BRI1-GFP levels (Figure 4C). Thus, unlike the ABCB proteins, BRI1 localization to the plasma membrane does not require TWD1.
Figure 4. The twd1 mutation does not affect BRI1 subcellular localization.
(A–B) Confocal images of root tips of pBRI1:BRI1-GFP transgenic Col-0 and twd1 mutant. (A) Maximum intensity projection view of the root tips (20X objective). Scale bar, 75 μm. (B) Zoom-in images of the epidermis (63X objective). BRI1-GFP signal was detected on the plasma membrane stained with propidium iodide (PI) and in the endocytic vesicles stained with FM4-64 in both twd1 and Col-0 backgrounds. Arrowheads indicate endocytic vesicles. Scale bar, 10 μm. (C) Quantification of BRI1-GFP fluorescence intensity. Areas of 20 μm × 20 μm (covered approximately 2–4 cells) were selected, and mean intensity values were calculated using ImageJ software (n = 20, mean ± SD).
FKBP42/TWD1 is not required for normal flagellin signaling
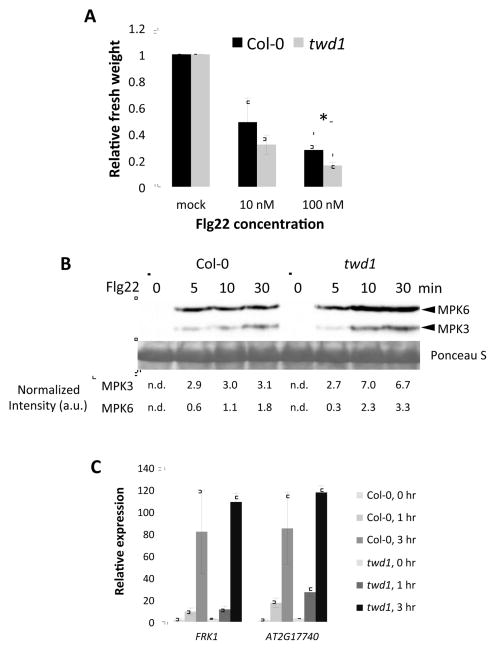
Our results suggest that FKBP42/TWD1 functions in the BR signaling pathway by means of protein-protein interaction with the BRI1 and BAK1 receptor kinases. To determine whether FKBP42/TWD1 plays a specific role in BR signaling or also modulates other receptor kinase pathways, we tested the effect of twd1 mutation on flagellin responses, which is mediated by the FLS2 and BAK1 receptor kinases (Chinchilla et al., 2007; Heese et al., 2007). Treatments with flagellin (flg22) for 7 days reduced growth (indicated by fresh weight) in twd1 more than wild type (Figure 5A). Time-course analysis of flg22-induced MAP kinase (MAPK) activation by western blotting indicated that twd1 responded to flg22 more strongly than wild type plants after 10 and 30 min of flg22 treatment (Figure 5B). Quantitative RT-PCR analysis of expression of flagellin-induced genes, FRK1 and AT2G17740, showed that twd1 was slightly more responsive than wild type to flg22 treatment (Figure 5C). These observations of enhanced flg22 responses in twd1 suggest that FKBP42/TWD1 is not required for FLS2 and BAK1 functions in mediating immunity signaling and that FKBP42/TWD1 might negatively regulate flg22 responses either directly through interaction with BAK1 or indirectly through reduced BR signaling, which is known to antagonize flagellin responses (Wang and Wang, 2014).
Figure 5. The twd1 mutation does not inhibit BAK1-mediated flg22 responses.
(A) Relative fresh weight of 12-day-old Col-0 and twd1 seedlings treated with mock or different concentrations of flg22 for 7 days (n = 20, mean±SD). * indicates T-test’s p-value < 0.05
(B) Activation of MAPKs in wild-type and twd1 in response to flg22 treatment. Twelve-day-old Col-0 and twd1 seedlings were treated with 100 nM flg22 for indicated time. Arrowheads indicated phosphorylated MPK3 and MPK6. Blot stained with Ponceau S is presented to show equal loading. The band intensities were quantified using ImageJ. Normalized intensity (a.u., arbitrary unit) was calculated from the intensity of MPK3 and MPK6 divided by that of Ponceau S staining (n.d., not determined).
(C) Quantitative RT-PCR analysis of flg22-responsive genes, FRK1 and AT2G17740. Twelve-day-old seedlings were treated with 100 nM flg22 for 0, 1 or 3 hr (n = 3 biological replicates, mean±SEM). Gene expression was normalized to PP2A.
Discussion
The BR signal transduction pathway has been characterized in detail. All major components of the pathway, from BR receptor BRI1 to transcription factor BZR1, seem to have been identified, and the protein-protein interactions that mediate each step of signal relay have been illustrated at the molecular level (Zhu et al., 2013). It has been unclear how twd1 modulates BR response. It is also intriguing that the twd1 mutant shows both BR hyposensitivity phenotypes and auxin response defects. A role for the TWD1/UCU2 gene has been proposed in the cross talk between the auxin and BR signal transduction pathways, which could be achieved by regulation of auxin homeostasis through activation of one or both of the ABCB1 and ABCB19 auxin transporters or by the regulation of the BRI1/BAK1 receptor complex (Pérez-Pérez et al., 2004). In this study, we provide evidence for a direct role of FKBP42/TWD1 in the normal function of BR signaling receptor kinases.
While twd1’s reduced BR sensitivity in growth and BR-responsive gene expression is consistent with defects in either BR signaling or auxin-BR synergism, the increased accumulation of phosphorylated form of BZR1 in twd1 supports a direct role of FKBP42/TWD1 in BR signaling. The increase of BZR1 phosphorylation is unlikely due to the defect in auxin transport because auxin has little effect on the phosphorylation status of BZR1 or BZR2/BES1 in seedlings (He et al., 2002; Yin et al., 2002). The increased BZR1 phosphorylation apparently contributed to the dwarf phenotype of twd1 because the bzr1-1D mutation, which increases BZR1 dephosphorylation by PP2A, partly suppressed the dwarf phenotype of twd1. The reduced responsiveness of twd1 to BR than to bikinin indicates that BR cannot effectively inactivate BIN2 in the twd1 mutant. Consistent with twd1 having a defect in BR signaling upstream of BIN2, we found FKBP42/TWD1 directly interacts with BRI1 and BAK1. Our results therefore demonstrate that FKBP42/TWD1 plays a direct role in BR response through physical interaction with the BRI1 and BAK1 receptor kinases.
FKBP42/TWD1 shares structural similarity with mammalian FKBP52, with an N-terminal peptidyl-prolyl isomerase (PPIase) domain and a C-terminal tetratricopeptide repeat (TPR) domain. Mammalian FKBP52 associates with heat shock protein (HSP90) and functions as co-chaperons of steroid receptors, which are ligand-dependent nuclear transcription factors (Sivils et al., 2011). It is intriguing that TWD1, which also associates with HSP90 (Kamphausen et al., 2002), also positively regulates plant steroid responses. However, the mechanisms appear to have diverged. While mammalian FKBP-HSP90 complex facilitates ligand binding and subcellular localization of the steroid hormone receptor transcription factors, FKBP42/TWD1 appears to act directly on the receptor kinases. Although BZR1 contains a proline residue that is crucial for its phosphorylation (Pro mutated to Leu in bzr1-1D and bes1-D) and thus potentially a target for regulation by PPIase, our BiFC assay did not detect interaction between BZR1 and FKBP42/TWD1 (data not shown). In contrast, results of bikinin treatment further support that FKBP42/TWD1 acts on BR signaling upstream of BIN2. Our results indicate that FKBP42/TWD1 positively regulates BR response by directly acting on or downstream of the BRI1 and BAK1 receptor kinases.
The mechanism by which FKBP42/TWD1 positively regulates BRI1/BAK1 function remains unclear. FKBP42/TWD1 has been shown to be localized to several membranes, including the plasma membrane, tonoplast, and ER (Geisler et al., 2003; Wang et al., 2013; Wu et al., 2010). While FKBP42/TWD1 is required for localization of ABCB transporters from the ER to the plasma membrane (Wu et al., 2010), our microscopic analysis of BRI1-GFP localization in the twd1 mutant suggests that FKBP42/TWD1 is not involved in the localization of BRI1. This suggests that localization of transmembrane proteins such as the ABCB transporters and the LRR-RLK BRI1 from the ER to the plasma membrane may involve different mechanisms. Thus, it is possible that FKBP42/TWD1 and HSP90 might facilitate folding of membrane proteins, and such correct folding might be required for ER to plasma membrane translocation of some proteins but for activity of other proteins.
Alternatively, FKBP42/TWD1 may facilitate BRI1 and BAK1 phosphorylation of each other or of downstream substrates, although FKBP42/TWD1 did not interact with BRI1’s substrate BSKs in our overlay and BiFC assays. Indeed, Zhao et al provided evidence that the twd1 mutant show reduced phosphorylation of BRI1 (Zhao et al., 2016). Interestingly, FKBP42/TWD1 has been shown to interact with the AGC kinase PINOID (PID), which phosphorylates the ABCB1 linker domain and regulates ABCB1 activity (Henrichs et al., 2012). Furthermore, FKBP binding alters the phoshorylation of glucocorticoid receptor in mouse embryonic fibroblast cells (Hinds et al., 2014). Therefore, FKBP42/TWD1 may play a role in kinase activation or kinase-substrate interaction.
FKBP42/TWD1 appears to be a positive regulator of BRI1-BAK1 signaling, but not for other receptor kinases, such as FLS2. While FKBP42/TWD1 positively regulates BRI1-BAK1 receptor function, it seems to have a negative effect on flagellin signaling mediated by FLS2, which, like BRI1, also uses BAK1 as a co-receptor kinase. The twd1 mutant displayed enhanced responses to flg22 treatments in growth inhibition, MAPK activation, and expression of pathogen-induced genes. It is possible that FKBP42/TWD1 facilitates BRI1 recruitment of BAK1 or stabilization of the BRI1-BAK1 complex, and thus the twd1 mutant has more BAK1 available for FLS2 activation. Alternatively, the enhanced flagellin responses could be an indirect result of reduced BR signaling, as BR and flagellin pathways are known to antagonize each other at multiple levels (Wang and Wang, 2014). Future studies will be required to understand the mechanism by which FKBP42/TWD1 positively regulates BRI1-BAK1 receptor kinase signaling.
Methods
Plant material
All wild-type, various mutants, and transgenic lines are in the Arabidopsis thaliana Columbia (Col-0) background. These include twd1-2 (Salk_012836), bzr1-1D (Wang et al., 2002), pBZR1:BZR1-CFP, pBZR1:bzr1-1D-CFP (He et al., 2002), and pBRI1:BRI1-GFP (Friedrichsen et al., 2000).
Phenotypic analysis
Seeds were surface-sterilized and plated on half-strength MS media containing 1% sucrose, 0.8% Phytoblend agar, and indicated chemicals, hormones or mock. Plates were placed vertically under constant light. Root lengths were measured from images using ImageJ.
Plasmids
The coding sequence of full-length FKBP42/TWD1 was first cloned into pENTRY/SD/D-TOPO vector (Invitrogen), and then subcloned into the gateway-compatible pnYFP (Gampala et al., 2007) and pMALc2 (New England Biolab) vectors to generate 35S:nYFP-FKBP42 for BiFC experiments and 35S:MBP-FKBP42 for overlay assays, respectively. The 35S:nYFP-FKBP42 construct was transformed into Nicotiana benthamiana via Agrobacterium tumefaciens GV3101.
Western blot analysis
For BZR1 and MAPK activation analyses, total protein samples were extracted from 7-day-old seedlings or 12-day-old seedlings using 2x SDS sample buffer and separated on SDS-PAGE gels. Proteins were transferred to a nitrocellulose membrane and probed with a monoclonal anti-GFP antibody (Clontech; 1:2000 dilution) or anti-phosphop44/42 mitogen-activated protein kinase (Erk1/2) (Thr202/Tyr204) antibody (Cell Signaling; 1:1000 dilution).
For overlay assays, GST-BRI1-K, GST-BAK1-K, GST-BIN2, GST-BSK1 (Tang et al., 2008) and MBP-FKBP42 were expressed in E. coli and purified using glutathione agarose beads (GE Healthcare) or amylase agarose beads (New England Biolabs) as previously described (Tang et al., 2008). To test the interaction of FKBP42 with BR signaling components in vitro, GST, GST-BRI1-K, GST-BAK1-K, GST-BIN2, and GST-BSK1 were separated by SDS-PAGE and blotted on nitrocellulose membrane. The blot was then incubated with MBP-FKBP42 (4 μg) in 5% non-fat dry milk/PBS buffer for one hour, washed three times, and then probed with a monoclonal anti-MBP antibody (New England Biolabs, 1:4000) for one hour. Overlay signal was detected using the SuperSignal West Dura chemiluminescence reagent (Pierce).
BiFC analysis
BiFC analysis was performed according to a previous study (Gampala et al., 2007). In brief, Agrobacterium cells containing BiFC expression vectors or 35S:PIP2;1-mCherry construct (Emami et al., 2013) were resuspended in the induction medium and infiltrated into young leaves of 4-week-old Nicotiana benthamiana (tobacco) plants, which were grown in green houses under 16 hr light/8 hr dark conditions. At 36 to 72 hr after infiltration, fluorescent signals in the infiltrated epidermal cells of tobacco were observed by using confocal microscopy.
Confocal microscopy and quantification of fluorescence signal
Laser scanning confocal microscopy was performed on either a Leica SP5 or SP8 system. BiFC YFP fluorescence was visualized by excitation at 514 nm and emission at 520–580 nm. In Figure 3C, YFP and mCherry fluorescence was visualized by excitation at 488 nm and emission at 500–550 nm and 575–625 nm, respectively. In Figure 4, fluorescence was visualized by excitation at 488 nm and emission at 500–550 nm (GFP), 650–700 nm (PI), and 700–725 nm (FM4-64). Quantification of BRI1-GFP intensities in root epidermal cells was performed in ImageJ. Areas of 20 μm × 20 μm were selected, and mean intensity value of each selection was calculated. Averages of the mean GFP intensity values were calculated with Microsoft Excel.
Seedling growth inhibition assay
Seedlings growth inhibition assay was carried out following a previous study (Belkhadir et al., 2012). Seedlings were grown on half-strength MS medium containing 1% Sucrose under constant light for 5 days, then transferred to liquid half-strength MS medium containing 1% Sucrose supplemented with the indicated concentrations of flg22 peptides. Seedlings were weighted 7 days after treatment.
Total RNA extraction and quantitative RT-PCR analysis
Whole seedlings were harvested for total RNA extraction using the Spectrum Plant Total RNA kit (Sigma). For quantitative RT-PCR, the total RNA was used to prepare the first-strand cDNA using RevertAid reverse transcriptase (Fermentas). Quantitative RT-PCR was performed on LightCycler 480 (Roche) using a SYBR Green reagent (Bioline) with gene-specific primers: FRK1-F 5′ATCTTCGCTTGGAGCTTCTC-3′; FRK1-R 5′-TGCAGCGCAAGGACTAGAG-3′; AT2G17740-F 5′-TGCTCCATCTCTCTTTGTGC-3′; AT2G17740-R 5′-ATGCGTTGCTGAAGAAGAGG-3′; PP2A-F 5′-TATCGGATGACGATTCTTCGTGCAG-3′; PP2A-R 5′-GCTTGGTCGACTATCGGAATGAGAG-3′. Three biological replicates were used in the analysis.
Acknowledgments
This research was supported by a grant from NIH (R01GM066258) to Z-Y. W. and by Ratchadapiseksomphot Endowment under Outstanding Research Performance Program (J.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailly A, Wang B, Zwiewka M, Pollmann S, Schenck D, Lüthen H, Schulz A, Friml J, Geisler M. Expression of TWISTED DWARF1 lacking its in-plane membrane anchor leads to increased cell elongation and hypermorphic growth. Plant J. 2014;77:108–118. doi: 10.1111/tpj.12369. [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemao-Pires E, Dangl JL, Chory J. Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci. 2012;109:297–302. doi: 10.1073/pnas.1112840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard R, Bailly A, Blakeslee JJ, Oehring SC, Vincenzetti V, Lee OR, Paponov I, Palme K, Mancuso S, Murphy AS, et al. Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J Biol Chem. 2006;281:30603–30612. doi: 10.1074/jbc.M604604200. [DOI] [PubMed] [Google Scholar]
- Bowman J, Callos JD, Behringer FJ, Vasinda J, Stewart D, Link BM, Medford JI, Griffith M, Pyke KA, Marrison JL. Arabidopsis. Springer; 1994. Vegetative development; pp. 1–89. [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Clouse SD. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23:1219–1230. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Emami S, Yee MC, Dinneny JR. A robust family of Golden Gate Agrobacterium vectors for plant synthetic biology. Front Plant Sci. 2013;4:339. doi: 10.3389/fpls.2013.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro Ca, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, Tang W, Deng Z, Bai MY, Guan S, Lalonde S, Sun Y, Gendron JM, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Bouchard R, Billion K, Berger J, Saal B, Frangne N, Koncz-ka Z, Koncz C, Dudler R, Blakeslee JJ, et al. TWISTED DWARF1 , a Unique Plasma Membrane-anchored Immunophilin-like Protein , Interacts with Arabidopsis Multidrug Resistance-like Transporters AtPGP1 and AtPGP19. Mol Biol Cell. 2003;14:4238–4249. doi: 10.1091/mbc.E02-10-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007;21:1598–1602. doi: 10.1101/gad.1561307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SSL, Gendron N, Sun CQ, Wang ZY. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci U S A. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichs S, Wang B, Fukao Y, Zhu J, Charrier L, Bailly A, Oehring SC, Linnert M, Weiwad M, Endler A, et al. Regulation of ABCB1/PGP1-catalysed auxin transport by linker phosphorylation. EMBO J. 2012;31:2965–2980. doi: 10.1038/emboj.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds TD, Stechschulte La, Elkhairi F, Sanchez ER. Analysis of FK506, timcodar (VX-853) and FKBP51 and FKBP52 chaperones in control of glucocorticoid receptor activity and phosphorylation. Pharmacol Res Perspect. 2014;2:e00076. doi: 10.1002/prp2.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphausen T, Fanghänel J, Neumann D, Schulz B, Rahfeld JU. Characterization of Arabidopsis thaliana AtFKBP42 that is membrane-bound and interacts with Hsp90. Plant J. 2002;32:263–276. doi: 10.1046/j.1365-313x.2002.01420.x. [DOI] [PubMed] [Google Scholar]
- Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart Ra, Sun Y, Wang ZY. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife. 2014;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Ponce MR, Micol JL. The ULTRACURVATA2 gene of Arabidopsis encodes an FK506-binding protein involved in auxin and brassinosteroid signaling. Plant Physiol. 2004;134:101–117. doi: 10.1104/pp.103.032524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Vert G, Rozhon W, Mayerhofer J, Peelman F, Coutuer S, Denayer T, Jansen L, Nguyen L, et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem Biol. 2009;16:594–604. doi: 10.1016/j.chembiol.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidt HA, Vogel A, Eckhov A, Koenig BW, Huster D. Solid-state NMR characterization of the putative membrane anchor of TWD1 from Arabidopsis thaliana. Eur Biophys J. 2007;36:393–404. doi: 10.1007/s00249-006-0094-2. [DOI] [PubMed] [Google Scholar]
- Sivils JC, Storer CL, Galigniana MD, Cox MB. Regulation of steroid hormone receptor function by the 52-kDa FK506-binding protein (FKBP52) Curr Opin Pharmacol. 2011;11:314–319. doi: 10.1016/j.coph.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Shi QM, Yang XH, Xu ZH, Xue HW. Membrane steroid-binding protein 1 (MSBP1) negatively regulates brassinosteroid signaling by enhancing the endocytosis of BAK1. Cell Res. 2009;19:864–876. doi: 10.1038/cr.2009.66. [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto Ja, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto Ja, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol. 2011;13:124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang ZY. At the intersection of plant growth and immunity. Cell Host Microbe. 2014;15:400–402. doi: 10.1016/j.chom.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Bailly A, Zwiewka M, Henrichs S, Azzarello E, Mancuso S, Maeshima M, Friml J, Schulz A, Geisler M. Arabidopsis TWISTED DWARF1 functionally interacts with auxin exporter ABCB1 on the root plasma membrane. Plant Cell. 2013;25:202–214. doi: 10.1105/tpc.112.105999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Wu G, Otegui MS, Spalding EP. The ER-Localized TWD1 Immunophilin Is Necessary for Localization of Multidrug Resistance-Like Proteins Required for Polar Auxin Transport in Arabidopsis Roots. Plant Cell. 2010;22:3295–3304. doi: 10.1105/tpc.110.078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudick MM, Li X, Valentini V, Geldner N, Chory J, Lin J, Maurel C, Luu DT. Subcellular Redistribution of Root Aquaporins Induced by Hydrogen Peroxide. Mol Plant. 2015;8:1103–1114. doi: 10.1016/j.molp.2015.02.017. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Zhao B, Lv M, Feng Z, Campbell T, Liscum E, Li J. TWISTED DWARF 1 associates with BRASSINOSTEROID INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Mol Plant xxx Companion paper. 2016 doi: 10.1016/j.molp.2016.01.007. [DOI] [PubMed] [Google Scholar]
- Zhu JY, Sae-Seaw J, Wang ZY. Brassinosteroid signalling. Development. 2013;140:1615–1620. doi: 10.1242/dev.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]