Abstract
Plants are equipped with the capacity to respond to a large number of diverse signals, both internal ones and those emanating from the environment, that are critical to their survival and adaption as sessile organisms. These signals need to be integrated through highly structured intracellular networks to ensure coherent cellular responses, and in addition, spatiotemporal actions of hormones and peptides both orchestrate local cell differentiation and coordinate growth and physiology over long distances. Further, signal interactions and signaling outputs vary significantly with developmental context. This review discusses our current understanding of the integrated intracellular and intercellular signaling networks that control plant growth.
Introduction
Cell-to-cell communication is essential for the life of multicellular organisms, in which growth and development requires coordination of cell proliferation and differentiation between cells. Survival also requires an organism to respond properly to a wide range of environmental signals, and such adaptive responses require both intracellular signal transduction and information flow from cells receiving the signal to the rest of the body. In animals, cell-to-cell communication is facilitated by both hormones and the neuronal systems. Plants lack neuronal systems and rely largely on hormones and secreted small peptides for communication. Further, plants are sessile and must adapt to the environment by altering growth, development, and metabolism. Consequently, plants have evolved robust intracellular information processing systems and sophisticated intercellular signaling networks.
At least nine groups of plant hormones have been studied extensively. Auxin, cytokinin, brassinosteroid (BR), gibberellin (GA), and strigolactone (SL) play essential roles in normal growth and development. Abscisic acid (ABA) and ethylene mediate responses to abiotic stresses. Jasmonic acid (JA) is required for defense responses to herbivore wounding and anther development, whereas salicylic acid (SA) activates immune responses to pathogen infection (Larrieu and Vernoux, 2015). In addition, many secreted peptides have been shown to have hormone-like functions as mobile signals (Tavormina et al., 2015). While different hormones play predominant roles in growth promotion or stress responses, each hormone affects a wide range of developmental and physiological processes, and every developmental process is co-regulated by multiple hormones. Plant development is also highly sensitive to many environmental factors, such as light, temperature, pathogens, and herbivores. Extensive studies have elucidated the molecular pathways that transduce these signals and revealed many connections between these pathways. Further, recent studies have revealed a central growth-regulation module that controls cell elongation in shoot organs and different signaling outputs and hormone interactions between shoot and root. These studies shed light on important general questions of how a cell processes complex signals into coherent responses and growth decisions, how a hormone induces cell-type-specific responses, and how hormone signaling and crosstalk are rewired in different developmental context. Here, we provide an overview of the intracellular circuits that integrate multiple signals into cellular decisions, as well as intercellular signal circuits that program development locally and globally. We cover classic phytohormones and peptide signals, and their interactions with environmental signals in regulating shoot and root growth. Proteins and RNA molecules that move between cells through the plasmodesmata also play important roles in communication; these topics have been covered in recent reviews (Otero et al., 2016), and will not be discussed here. Given the broad scope of the topic and high complexity of the system, we will use selected key examples to illustrate principles rather than giving a comprehensive coverage of the literature.
Regulation of Shoot Cell Elongation by Integration of Environmental and Hormonal Signals
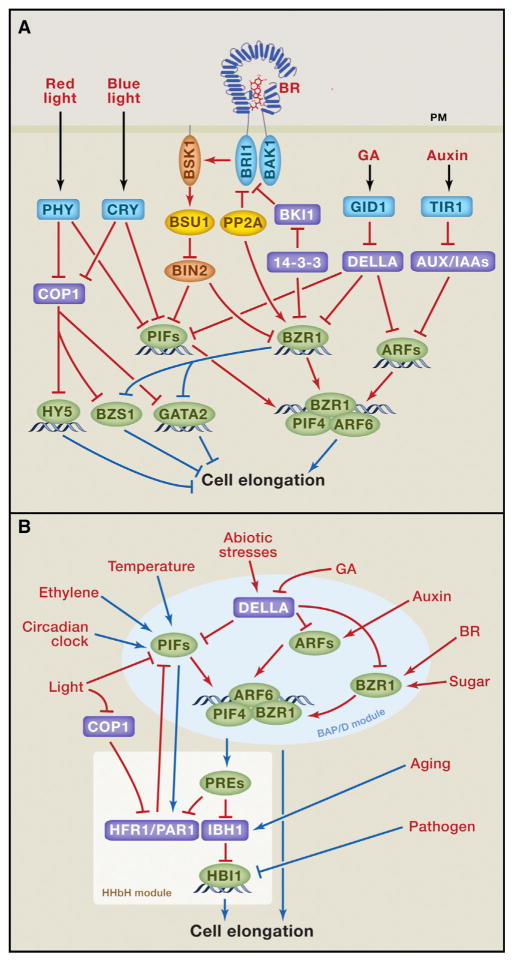
Growth in plants is driven by cell division in the stem cell populations maintained at the shoot apical meristem (SAM) and root apical meristem (RAM), followed by cell elongation. The balance between stem cell division and differentiation is crucial for maintaining the continuous growth (Sparks et al., 2013). However, cell elongation contributes to the majority of growth of shoot and root length and is controlled tightly by key environmental signals such as light and temperature, as well as major growth promoting hormones including auxin, BR, and GA (Figure 1).
Figure 1. Integration of Light and Hormone Signaling Pathways Regulates Hypocotyl Elongation.
(A) Light and hormonal signals (red text) are perceived by cell-surface or intracellular receptors (blue), which regulate transcription factors (green) through signaling/posttranslational mechanisms (red lines), whereas the transcription factors transcriptionally regulate (blue lines) downstream responses and components of other pathways. Orange: kinases; yellow: phosphatases; purple: inhibitors of transcription factors.
(B) Transcriptional integration by the BAP/D-HHbH circuit. Red and blue lines show regulation at the protein and RNA (transcriptional) levels, respectively.
The light-mediated morphological changes in Arabidopsis seedling, so called photomorphogenesis, has been a model system for studying the interactions between light and hormones and the regulation of cell elongation. Upon seed germination in the dark (under soil in nature), an Arabidopsis seedling undergoes skotomorphogenesis (also called etiolation), which is characterized by maximum hypocotyl elongation, limited root growth, closed cotyledons with an apical hook, and suppression of chloroplast development. Upon exposure to light, seedling development switches to photomorphogenesis (also called de-etiolation), which is characterized by inhibition of hypocotyl elongation, opening/expanding and greening of cotyledons and leaves, and acceleration of root growth. Deficiency in synthesis or signal transduction of BR, auxin, and GA all cause photomorphogenesis in the dark, suggesting intimate relationships between light and these growth hormones in co-regulating cell elongation and seedling photomorphogenesis. While some previous studies proposed hierarchical relationships of light regulating hormone levels, or one hormone regulating another, in order to control cell elongation, recent studies have revealed that these light and hormonal signals also converge at a central module of interacting transcription factors/regulators to co-regulate overlapping sets of genes. Furthermore, additional environmental and endogenous signals impinge on this central module to control shoot cell elongation (Figure 1).
For developmental regulation, plants detect light of different wavelength using several classes of photoreceptors (Galvão and Fankhauser, 2015), among which the red/far-red photoreceptor phytochromes and blue light receptor cryptochromes play major roles in seedling morphogenesis (Chory, 2010; Liu et al., 2011). Phytochromes exist in two photo-switchable forms: the red-absorbing Pr form, which is biologically inactive, and the active far-red-absorbing Pfr form. Light absorption switches phytochromes between Pr and Pfr forms. Such photo-reversibility allows phytochromes to measure not only light intensity but also wavelength, which changes with season and canopy (Chen and Chory, 2011).
Phytochromes and cryptochromes regulate cell elongation primarily through two classes of transcription factors that have opposite functions. The phytochrome interacting factors (PIFs), a class of basic helix-loop-helix (bHLH) factors, are major positive regulators of shoot cell elongation. Photo-activated phytochromes inactivate PIFs by inhibiting their DNA-binding activities and promoting their phosphorylation, ubiquitination, and degradation (de Lucas and Prat, 2014; Ni et al., 2014). Several PIFs are also inactivated by direct interaction with cryptochromes (Ma et al., 2016; Pedmale et al., 2016). Elongated hypocotyl5 (HY5), GATA2/4, and B-box factors including BZS1 are negative regulators of cell elongation, and they are degraded in the dark through the E3 ubiquitin ligase constitutive photomorphogenic1 (COP1), which is inactivated by both phytochromes and cryptochromes (Lau and Deng, 2012; Liu et al., 2011; Wang et al., 2012).
Auxin (primarily indole-3-acetic acid, IAA) regulates gene expression and promotes shoot cell elongation by activating the auxin response factor (ARF) family of transcription factors. ARFs are repressed by the Aux/IAA proteins, which are removed by auxin-induced degradation. Auxin binds to the TIR1/AFB family of F-box proteins, which recruit the Aux/IAA proteins to the SCFTIR1/AFB complex for ubiquitination and proteasome-mediated degradation, leading to de-repression of ARFs (Salehin et al., 2015).
BR acts through the cell surface receptor kinase brassinosteroid-insensitive1 (BRI1). BRI1 represents one of over 220 leucine-rich repeat receptor kinases (LRR-RKs) in Arabidopsis and is the best characterized among them (Belkhadir et al., 2014). BR binding to the extracellular domain of BRI1 induces its association with the co-receptor kinase BRI1-associated kinase1 (BAK1), which activates BRI1 by trans-phosphorylation (Belkhadir et al., 2014). Activated BRI1 phosphorylates the BR-signaling kinase1 (BSK1) and the constitutive differential growth1 (CDG1) kinases, which in turn phosphorylate the PP1-like phosphatase BRI1-suppressor1 (BSU1), leading to BSU1 dephosphorylation and inactivation of the GSK3-like kinase BR-insensitive2 (BIN2) (Wang et al., 2012). When BR levels are low, BRI1 is inactivated by the BRI1 kinase inhibitor1 (BKI1) and protein phosphatase 2A (PP2A), while BIN2 phosphorylates members of the brassinazole resistant1 (BZR1) family transcription factors, to both promote their cytoplasmic retention by the 14-3-3 proteins and inhibit their DNA-binding activity (Kim and Wang, 2010). Upon BIN2 inactivation by upstream BR signaling, BZR1 is dephosphorylated by PP2A and then moves into the nucleus to alter the expression of thousands of target genes (Sun et al., 2010; Wang et al., 2012).
GA, similar to auxin, acts through an intracellular receptor to promote ubiquitination and degradation of key repressor proteins, named DELLA proteins for containing a conserved Asp-Glu-Leu-Leu-Ala amino acid sequence (Sun, 2010). GA binding to its receptor gibberellin-insensitive 1 (GID1) causes DELLA interaction with the SCFSLY1 E3 ubiquitin ligase complex, leading to ubiquitination and degradation of DELLAs (Sun, 2010). When GA levels are low, DELLAs accumulate to high levels. DELLAs were initially found to interact with PIFs and inhibit their DNA-binding activity (de Lucas et al., 2008; Feng et al., 2008) but have since been found to inhibit DNA-binding activities of many transcription factors, including BZR1 and ARF6 of the BR and auxin pathways, respectively (Bai et al., 2012b; Locascio et al., 2013; Oh et al., 2014). DELLAs also function as transcriptional co-activators through interaction with several classes of DNA-binding proteins, including ARR1, which is a component of the cytokinin pathway that promotes photomorphogenesis (Marínde la Rosa et al., 2015; Yoshida et al., 2014). The ability of DELLAs to modulate DNA binding and transcriptional activities of many transcription factors allows GA to effectively control diverse developmental processes.
Signal Integration by the BAP/D-HHbH Circuit
Recent studies demonstrate that the BR, auxin, GA, and phytochrome pathways converge through direct interactions among their transcription factors/regulators. BZR1, PIF4, and ARF6 interact with each other, and they share a large number of common target genes (Oh et al., 2014; Oh et al., 2012). These three transcription factors enhance each other’s target binding and transcriptional activation activities, and their functions in activating many shared target genes and promoting hypocotyl elongation are genetically interdependent on each other. For example, increasing either BZR1 or PIF4 levels enhances ARF6 binding to several shared target gene promoters in vivo (Oh et al., 2014). The shared target genes activated by PIF4, BR, auxin, and GA are enriched with functions in cell elongation (such as cell wall synthesis and loosening), consistent with the roles of these transcription factors in promoting cell elongation (Bai et al., 2012b). Such cooperative interaction among BZR1, ARF6, and PIF4, as well as their inhibition by DELLAs, is named BZR-ARF-PIF/DELLA (BAP/D) module. BAP/D elegantly explains the genetic requirement of BR, auxin, GA, and PIF activities for skotomorphogenesis, as well as the synergistic interactions among BR, auxin, GA, and dark/shade in promoting shoot cell elongation (Wang et al., 2014).
The BAP/D module is flexible and its components also function independently on certain target genes. Based on ChIP-seq and RNA-seq data, BZR1, ARF6, and PIF4 activate a large number of shared target genes, but each of them also regulates subsets of genes uniquely or only together with one of the two partners (Oh et al., 2014; Oh et al., 2012). Among these unique functions are feedback inhibition of its own signaling pathway but cross activation of partner pathways. For example, BR, auxin, and GA each negatively feedback regulates its own biosynthesis. Light signaling is feedback attenuated through PIF activation of its inhibitors PAR1 and HFR1 (de Lucas and Prat, 2014). Such feedback inhibition presumably maintains signal homeostasis. In contrast to feedback inhibition within each pathway, cross-regulation between the pathways tend to be mostly positive. For example, BR increases auxin transport, auxin activates expression of the BR biosynthesis gene DWF4, both BR and auxin increase GA biosynthesis, and PIFs activate auxin biosynthesis and increase GA levels (de Lucas and Prat, 2014; Wang et al., 2014). Furthermore, BIN2 phosphorylates and inactivates both PIF4 and ARF2, and thus BR inactivation of BIN2 potentially also activates these transcription factors (Wang et al., 2012; Bernardo-García et al., 2014). BZR1 also directly regulates the expression of a large number of components of the light-signaling pathways, including phytochrome B, COP1, GATA2, and BZS1/BBX20, in a manner that is consistent with promoting cell elongation (Wang et al., 2012). Such positive cross-regulation apparently generates synergy and ensures cooperative system-wide responses. Positive cross-regulation may also mediate inter-organ communication. For example, shade activation of PIFs in leaves increases the level of auxin, which is transported to stem to promote stem elongation (Casal, 2013) (see more details below).
The BAP module is coupled with a branched tripartite module of helix-loop-helix (HLH) and basic helix-loop-helix (bHLH) factors. Among a large number of transcription regulators downstream of the BAP module is the paclobutrazole resistant1 (PRE1) family of non-DNA binding HLH factors, which positively regulate cell elongation. Members of the PRE family interact with and sequester another class of HLH factors, including PAR1, HFR1, IBH1, and AIFs, which are negative growth regulators. PAR1 and HFR1 bind PIFs and inhibit their DNA-binding activities, whereas PRE1 sequesters PAR1 and HFR1 from PIF4, forming a positive feedback loop of HLH-HLH-bHLH (HHbH) cascade (Wang et al., 2014). IBH1 and AIFs, on the other hand, inhibit members of another family of DNA-binding bHLH factors, including HBI1, ACEs, CIB5, and BEE2, which are positive regulators of cell elongation (Bai et al., 2012a; Ikeda et al., 2012).
In addition to sequestration by PREs, the levels of HFR1 and PAR1 are also controlled by COP1-mediated ubiquitination/degradation and PIF-mediated transcriptional activation (Figure 1B). Such complex regulation of PAR1 and HFR1 potentially provides the appropriate light responsiveness under a wide dynamic range of light/dark conditions. For example, seedlings grown in the dark would have a very low level of PAR1/HFR1 activities due to both degradation by COP1 and inactivation by PREs, and the low PAR1/HFR1 activities ensure full activity of PIF proteins (and the BAP module) and a high level of PRE expression. Light exposure triggers not only degradation of PIFs but also accumulation of their inhibitors, PAR1 and HFR1, ensuring rapid decrease of PIF activities. The PIF dependence of PAR1/HFR1 transcription would provide a delayed attenuation of the initial change of PIF levels (Hornitschek et al., 2009), whereas the hormone-dependent PIF activation of PREs would cancel some of the effects of PAR1/HFR1, providing additional mechanism of hormonal modulation of light response. As such, the coupling of the BAP/D and HHbH modules creates a highly robust central command system, which integrates light and hormonal signals into coherent and dynamic cell elongation responses (Figure 1B).
The BAP/D-HHbH circuit mediates growth regulation by many additional endogenous and environmental cues (Wang et al., 2014). For example, the circadian clock controls rhythmic growth by regulating the expression levels of PIF4 and PIF5 (de Lucas and Prat, 2014). Warm temperature activates PIF4 expression to induce thermo-responsive growth (Quint et al., 2016). Abiotic stresses cause accumulation of DELLAs to slow down growth and improve survival of adverse conditions (Achard et al., 2006). The expression of IBH1 increases upon organ maturation and thus functions as a break in developmental progression (Zhang et al., 2009; Ikeda et al., 2012; Wang et al., 2014) (Figure 1B). Sugar-regulated degradation of BZR1 contributes to growth arrest under starvation conditions (Zhang et al., 2015). In light-grown Arabidopsis seedlings, ethylene induces hypocotyl elongation through ethylene-insensitive 3-mediated activation of PIF3 expression (Zhong et al., 2012). Furthermore, pathogen-triggered signaling represses the RNA level of HBI1 to inhibit growth and activate immunity (Fan et al., 2014: Malinovsky et al., 2014) (more details below). Therefore, the BAP/D-HHbH circuit seems to be the central, and perhaps evolutionarily ancient, mechanism of cell elongation regulation, upon with various signaling pathways converge.
The Tradeoffs between Growth and Defense
In nature, plants constantly encounter a variety of pathogens and insect herbivores. To grow or defend is a life-death decision, as both growth and defense are energy demanding. Therefore, defense programs are turned off during normal growth. However, when attacked by pathogens or herbivores, the immune or wounding responses, respectively, must be turned on, whereas growth programs must be turned down to prioritize resource and energy for defense. Some microbial pathogens synthesize growth hormones to suppress host’s immune responses (Robert-Seilaniantz et al., 2011). Thus, balancing the growth-defense tradeoff is crucial for growth and survival, and involves complex interactions between signaling pathways, including transcriptional cross regulation mediated by the BAP/D-HHbH circuit (Wang and Wang, 2014; Huot et al., 2014; Belkhadir et al., 2014; Lozano-Durán and Zipfel, 2015).
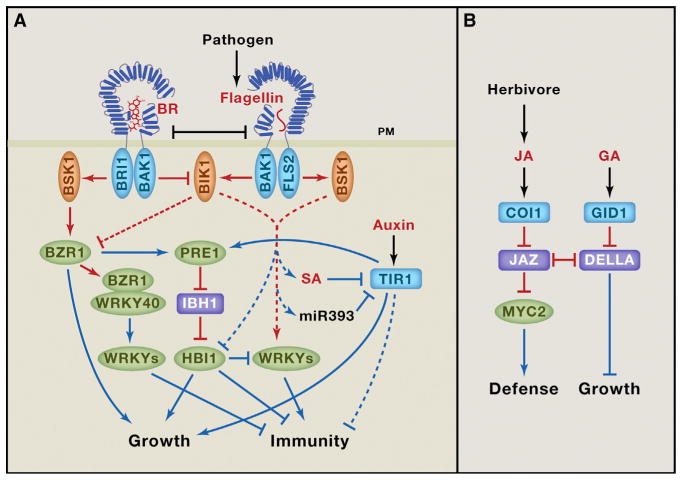
One example of such growth-defense trade-off is between the BR and the flagellin-signaling pathways (Belkhadir et al., 2014; Lozano-Durán and Zipfel, 2015) (Figure 2A). A peptide (flg22) from bacterial flagellin protein is perceived as a pathogen-associated molecular pattern (PAMP) by the LRR-RK named FLS2 (flagellin-sensitive2), which has an overall similar structure as the BR receptor BRI1 (Belkhadir et al., 2014). FLS2 and BRI1 use the same co-receptor kinase, BAK1, for ligand-induced activation. They further share some of their substrates, such as BSK1 and BIK1 kinases. BR and flagellin co-treatment experiments showed that BR signaling reduced flagellin-induced responses (Albrecht et al., 2012; Belkhadir et al., 2012). One hypothesis is that, when fully activated, BRI1 sequesters or phospho-codes BAK1 to prevent its activation of FLS2. However, experimental evidence has been inconsistent (Belkhadir et al., 2014; Lozano-Durán and Zipfel, 2015). On the other hand, genetic evidence supports a role for botrytis-induced kinase1 (BIK1) in the growth-immunity tradeoff. BIK1 is a substrate of both FLS2 and BRI1 kinase and it positively regulates the defense pathway but negatively regulates the BR pathway (Lin et al., 2013). In addition, BZR1 has been shown to directly interact with WRKY40 to activate several additional WRKY factors that negatively regulate immunity (Lozano-Durán and Zipfel, 2015). Furthermore, flagellin signaling represses the transcription level of HBI1, and this contributes to both inhibition of growth and activation of immunity (Fan et al., 2014; Malinovsky et al., 2014). Overexpression of HBI1 strongly suppresses the immunity to pathogen and growth inhibition caused by flagellin treatment, supporting a key role for HBI1 in balancing the tradeoff between innate immunity and growth. The aging-induced expression of IBH1 leads to inhibition of HBI1 (Ikeda et al., 2012; Zhang et al., 2009), which is likely to contribute to not only growth arrest but also the enhanced defense response in mature shoots and leaves (Carella et al., 2014; Fan et al., 2014) (Figure 2).
Figure 2. Mechanisms that Regulate the Tradeoff between Growth and Defense.
(A) Mechanisms of crosstalks of FLS2-mediated flagellin signaling with the BR and auxin pathways. (B) Growth regulation in response to herbivore attack, mediated by crosstalk between the GA and JA pathways. Red and blue lines show regulation at the protein and RNA (transcriptional) levels, respectively. Dashed lines indicate unknown mechanisms.
Flagellin signaling also inhibits auxin signaling to prioritize defense over growth (Figure 2A). Flg22 induces microRNA miR393, which targets the mRNAs of auxin receptor TIR1, AFB2, and AFB3 (Navarro et al., 2006). Flg22 also induces SA accumulation, and SA treatment stabilizes the Aux/IAA proteins, which inhibit auxin responsive gene expression (Wang et al., 2007). Auxin-deficient or -insensitive mutants display enhanced resistance to pathogens such as Pseudomonas syringae, indicating that auxin signaling inhibits immunity and that PAMP and SA signals enhance defense against pathogens by suppressing auxin signaling (Wang et al., 2007). The molecular mechanism by which auxin inhibits immunity, and particularly whether the HHbH module is involved, remains to be elucidated.
Wounding by herbivores induces production of JA, which acts as a mobile signal to induce systemic defense responses and inhibit vegetative growth (Song et al., 2014; Huot et al., 2014). For example, when some leaves of poplar trees are attacked by herbivores, plants not only turn on defense gene expression throughout the plants, but also increase sugar transport from leaves to roots, presumably to hide the food away from herbivore for later recovery. Such carbon relocation can be triggered by treating some leaves with JA (Babst et al., 2005). A recent study, using a fluorescent JA biosensor, showed that wounding of Arabidopsis leaves induces rapid increase of JA level in the root (Larrieu et al., 2015).
Similar to auxin and GA, JA induces degradation of the repressors of transcription factors (Song et al., 2014). JA-responsive genes are controlled by several transcription factors including MYC2, which, in the absence of JA, is inactivated by the JA ZIM-domain (JAZ) proteins (Figure 2B). JA binding to its receptor coronatine insensitive1 (COI1), which is an E3 ubiquitin ligase, induces its interaction with and ubiquitination of JAZ proteins, leading to de-repression of MYC2 (Song et al., 2014). Interestingly, several JAZ proteins also interact with the DELLA proteins, and their association prevents the DELLA interaction with PIFs and the JAZ interaction with MYC2 (Hou et al., 2010; Yang et al., 2012). This antagonistic interaction allows JA to prioritize defense over growth under normal light conditions, as JA-induced JAZ removal frees up DELLAs for inhibiting the growth-promoting factors (Yang et al., 2012), but it also explains why defense responses are compromised under shade conditions: shade increases GA level, which causes DELLAs removal, freeing up JAZ for inhibiting MYC2 (Leone et al., 2014). As such, the DELLA-JAZ interaction ensures optimal decisions to grow or to defend based on priority of environmental challenges (Figure 2B).
Shade Avoidance Syndrome: A Case of Inter-organ Growth Coordination
In nature, successful competition with neighbors for sunlight is crucial for plant survival, and thus canopy shade induces a variety of morphological changes that are collectively called shade avoidance syndrome (SAS). These include elongation of stem and petiole, leaf hyponasty (upward bending of the leaves caused by growth of the lower side), reduced shoot branching and root growth, and decreased seed and fruit production (Casal, 2013). SAS is a major cause of yield limitation in agricultural production, as growing crops at high densities stimulates SAS (Casal, 2013).
Shading by neighboring plants causes several unique features in the light environment, these include reductions of the red light to far-red light ratio (R:FR) and the intensities of blue and UV light, which are perceived by phytochromes, cryptochromes, and UVR8, respectively (Galvão and Fankhauser, 2015). All these photoreceptors act through members of the PIF family transcription factors to control SAS. Reduced phytochrome activity by low R:FR leads to increased accumulation of PIF3, PIF4, and PIF5 (Leivar et al., 2012), and enhanced binding of the light-stable PIF7 to target promoters (Li et al., 2012). Cryptochromes directly interact with PIF4 and PIF5 to control their transcriptional activities in response to low blue light (Ma et al., 2016; Pedmale et al., 2016), whereas UV light-activated UVR8 promotes the degradation of PIF4 and PIF5 through an unknown mechanism (Hayes et al., 2014).
Shade perceived by leaves lead to growth response in stems and roots through hormone-mediated inter-organ communication (Casal, 2013). Spotlight far-red light irradiation of leaf blades but not that of petioles altered petiole elongation (Kozuka et al., 2010), indicating that low R:FR is detected by (upper) leaves and the signal is communicated to the petiole and the rest of the plan body through mobile signals (Casal, 2013) (Figure 3A). Such shade perception by leaves rather than stem itself potentially prevents responses to shading of the stems by its own leaves.
Figure 3. Hormone-Mediated Growth Responses to Shade.
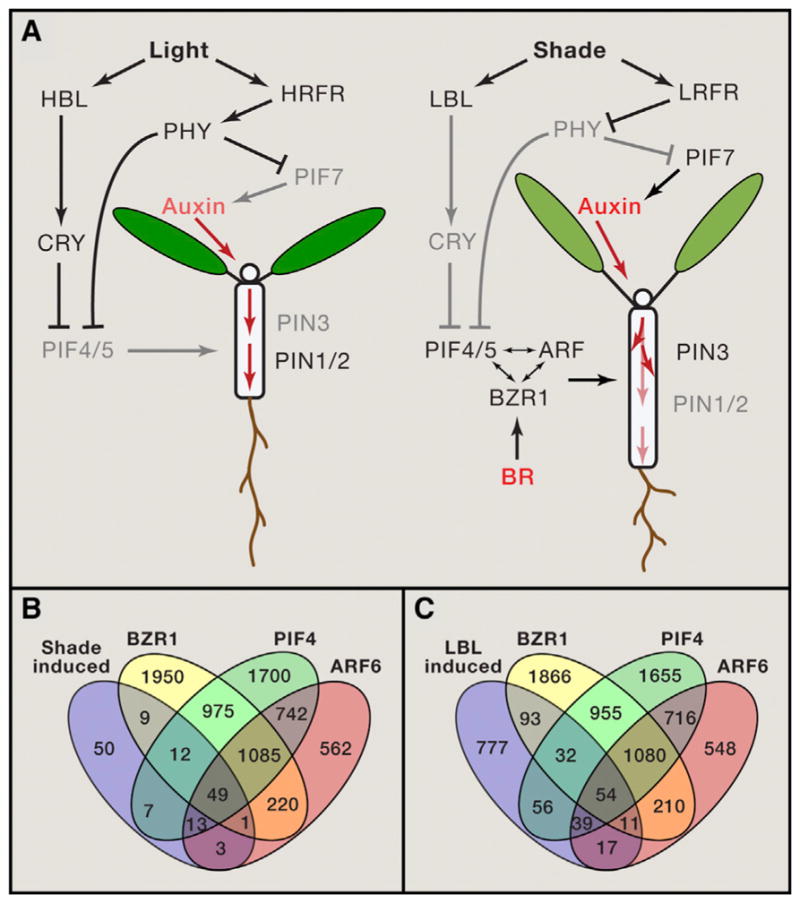
(A) Diagram of light-hormone interactions in growth regulation under full light (left) and shade (right) conditions. HBL: high blue light, LBL: low blue light, HRFR: high red:far-red ratio, LRFR: low red:far-red ratio. Dark text and arrows show active components and their activities, and dimmed text and arrows indicated inactivated components and activities. Red arrows show the flow of auxin.
(B) Venn diagram shows overlaps between genes induced by 1 hr low R:FR treatment (Li et al., 2012), or by 6 hr low blue light treatment of light-grown seedlings (Pedmale et al., 2016), and the target genes of BZR1, PIF4, and ARF6 identified by ChIP-seq in dark-grown seedlings (Oh et al., 2012, 2014).
The long-distance communication of shade signal is mediated mainly by auxin. Upon activation by shade condition, PIF7 activates the expression of the YUCCA family of auxin biosynthetic genes in leaves and cotyledons, and the increased auxin is transported to petioles and hypocotyls to promote their elongation (Li et al., 2012; Nito et al., 2015; Tao et al., 2008). Low R:FR also changes auxin distribution by altering the abundance and subcellular localization of the auxin efflux carrier PIN-formed3 (PIN3) in the endodermal cells and hence redirecting the auxin flow to the growth-limiting epidermal cell of stems/hypocotyls (Keuskamp et al., 2010), as well as by decreasing level of PIN1 auxin efflux carrier in hypocotyls to reduce the flow of auxin toward the roots (Sassi et al., 2012). Accumulation of PIF4 and PIF5 under low R:FR and low UV conditions increases both auxin synthesis and auxin sensitivity of the hypocotyls (de Wit et al., 2015; de Wit et al., 2014; Hayes et al., 2014; Hornitschek et al., 2012). Intriguingly, the cryptochrome-PIF4/5-mediated responses to blue light depletion appear to involve direct regulation of genes encoding cell wall synthesis and loosening enzymes (in hypocotyl cells, presumably), without changing auxin levels or sensitivity (Pedmale et al., 2016). However, genetic evidence supports that both auxin and BR play essential roles in SAS induced by blue light depletion (Keller et al., 2011; Keuskamp et al., 2011).
Full SAS also requires GA and BR, as defects in synthesis or signaling of these hormones compromise SAS (Casal, 2013). Shade induces GA synthesis and promotes degradation of the DELLAs repressor (Djakovic-Petrovic et al., 2007; Hayes et al., 2014). There is no evidence for increase of BR level under shade. In contrast, BZR1 is degraded and elongation response is abolished when light-grown plants are shifted into dark for extended period without exogenous sugar supplement (Zhang et al., 2015). This suggests that BR/BZR1 is responsive to endogenous sugar availability, and the requirement of BZR1 for PIFs and ARFs to promote cell elongation would ensure that the growth response to shade, which reduces the rate of photosynthesis, is within the limit of sugar availability.
The requirement of auxin, BR and GA for full SAS is consistent with the central role of the BAP/D module in shoot organ elongation. Further, genes induced by low R:FR include similarly large numbers of target genes of PIF4, ARF6, and BZR1 (Figure 3B). The genes induced by blue light depletion (in light-grown seedling), however, showed a smaller overlap with ARF6 targets than with BZR1 and PIF4 targets (identified in the dark-grown seedlings) (Figure 3C), consistent with the lack of overrepresentation of auxin responsive genes among the blue light-responsive genes (Pedmale et al., 2016). On the other hand, both hypocotyl elongation and gene expression responses to blue light depletion were suppressed by inhibitors of BR and auxin, suggesting that both BR and auxin are required for the responses to blue light depletion (Keuskamp et al., 2011). Since the overlaps between the target genes of BZR1, ARF6, and PIF4 are partial, it is conceivable that each pathway can have integrated as well as independent outputs, which will likely be dependent on the developmental and physiological contexts.
Root Growth Regulation: A Control Circuit of Both Local Orchestration and Long-Distance Coordination
A plant’s root system provides water and nutrients for the above-ground organs. Long-distance communication is important for balancing growth and resource allocation between shoot and root according to the environment and physiology. Plants rely on xylem and phloem vasculature system to transport mobile signals, such as plant hormones, nutrients, and secreted peptides, over long distances. Since roots and shoots compete for resources to growth and also need to respond oppositely to certain environmental cues, root growth involves different hormone interaction relationships and hormone signaling outputs compared to shoots.
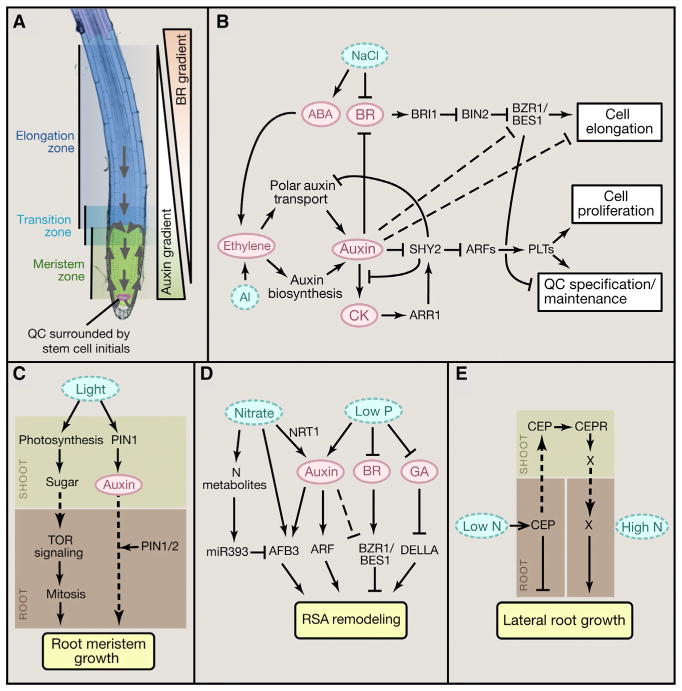
At the growing root tip, root apical meristem (RAM) contains a highly organized stem cell population (Figure 4A). The quiescent center (QC) at the apex contains cells that rarely divide and act to maintain the adjacent initial cells. Cell division maintains the stem cell population in the central meristem zone, whereas cells at the distal end of the meristem exit mitosis, enter the transition zone, and subsequently elongate rapidly and dramatically in the elongation zone, driving root tip growth (Figure 4A). The spatiotemporal balance of these stem cell activities determines root growth rate, and is maintained by patterned activities of several plant hormones (Pacifici et al., 2015). In particular, opposite gradient patterns of auxin and BR play prominent roles in enforcing the division and elongation zones and maintaining their balance (Chaiwanon and Wang, 2015), and therefore are also target for modulation by various endogenous and environmental signals that control root growth (Figure 4).
Figure 4. Signaling Networks Regulating Root Growth.
(A) Image of Arabidopsis root tip showing meristem zone, transition zone, and elongation zone. Arrows indicate the auxin reflux loop. PIN1 mediates polar auxin transport from shoot to the root tip. Auxin is then transported shootward by PIN2, creating auxin maximum in the QC and auxin gradient along the root developmental zones. BR distribution shows an opposite gradient with high BR levels in the elongation zone and low levels in the QC.
(B–E) Diagrams summarizing cross-regulation of root growth and development by hormones and environmental stimuli.
Arrows indicate positive regulation; bars indicate negative regulation; solid lines indicate direct regulation, dashed lines indicate indirect regulation; dotted arrows indicate movement of signals.
Balancing Division and Elongation by Two Opposite Hormonal Gradients
Auxin synthesized in the shoot is transported to the root via phloem transport and polar auxin transport mediated by the auxin efflux carrier PIN1 (Petrásek and Friml, 2009). The shoot-derived auxin, together with locally synthesized auxin, is then redistributed in the root tip and transported in a shootward direction by PIN2 (Petrásek and Friml, 2009). In addition to PIN-mediated polar transport, recent studies have shown that the nonpolar AUX1/LAX auxin influx carriers determine the locations of auxin accumulation, and play crucial roles in patterning auxin distribution (Band et al., 2014). Together these auxin transport mechanisms establish an auxin maximum at the QC and a gradient distribution of auxin in the meristem zone proximal to the QC (Figure 4A).
The auxin gradient contributes to patterned expression of large number of genes along the developmental gradient. Detailed transcriptome analysis of fine sections along root tip has identified large number of genes expressed specifically or preferentially at different developmental zones and cell types (Brady et al., 2007). Profiling of auxin responsive genes revealed a consistent pattern in which genes expressed in the QC and meristem zone, where endogenous auxin levels are high, are activated by auxin treatment, whereas genes expressed in the elongation zone are mostly repressed by auxin (Bargmann et al., 2013; Chaiwanon and Wang, 2015). Such correlation suggests that the gradient of endogenous auxin contributes to a large portion of the gene expression pattern along the auxin (and developmental) gradient (Chaiwanon and Wang, 2015).
A major role for BR as a patterning signal in root growth has emerged in recent studies. BR-deficient mutants have short roots due to insufficient cell elongation at maturity, while long-term treatment of roots with high concentrations of BR also inhibits root growth due to a reduced meristem size caused by acceleration of cell elongation (Chaiwanon and Wang, 2015; González-García et al., 2011; Hacham et al., 2011). BR mainly promotes root cell elongation through activation of BZR1 in the elongation zone, and endogenous BR is required for the balance between the meristem zone and the elongation zone. Under normal conditions, maximum BZR1 level is observed in the nucleus of the epidermal cells in the elongation zone, whereas BZR1 is mostly cytoplasmic in the QC and stem cell niche (Chaiwanon and Wang, 2015). This pattern of nuclear BZR1 depends on endogenous BR, which is likely patterned at least in part by localized BR catabolism. A low level of exogenous BR can recover the BZR1 pattern in a BR-deficient mutant, but high concentrations of BR cause rapid nuclear localization of BZR1 in all cells in root tip. The normal BZR1 gradient also requires the auxin gradient, as auxin treatment increases slowly the cytoplasmic BZR1 level in the elongation zone, whereas inhibiting auxin synthesis causes nuclear BZR1 accumulation in the cells of QC and stem cell niche (Chaiwanon and Wang, 2015). Thus, the gradient of BR/BZR1 appears to be established in part by the auxin gradient. BR may also modulate auxin level and distribution, as BR regulates expression of auxin transport (PIN2) and auxin biosynthesis genes (Vragović et al., 2015).
The gradient pattern of BR/BZR1 contributes to the pattern of gene expression and cell behavior along the gradient of stem cell quiescence, proliferation, and cell elongation. BR responsive genes identified by RNA-sequencing in root tip show that BR, acting through BZR1, represses large portion of genes expressed normally in the QC and meristem zone but activates genes expressed in the elongation zone. Such correlation suggests that the low BR/BZR1 levels in the QC and meristem zone allow expression of BR-repressed genes, and the high BR/BZR1 levels in the elongation zone contribute to the expression of BR-induced genes in this developmental zone (Chaiwanon and Wang, 2015; Vragović et al., 2015). The pattern of BR/BZR1 effects on gene expression is consistent with BZR1’s distinct functions of promoting cell elongation in epidermal cells of the elongation zone but promoting division of the QC cells (Chaiwanon and Wang, 2015; González-García et al., 2011).
In contrast to their synergistic interaction in shoot organs, auxin and BR show antagonistic interaction in regulating root cells. Auxin and BR are not only distributed with overall opposite gradients, but also have opposite effects on most of the genes they co-regulate and on cell elongation. In contrast to promoting cell elongation in stems and petioles, auxin inhibits root cell elongation and represses a large number of genes involved in cell elongation, which are activated by BR (Chaiwanon and Wang, 2015). Such opposite effects of auxin on the elongation of shoot and root cells are consistent with the facts that gravity, or light, induces similar redistribution of auxin but opposite bending response in shoot and root. On the other hand, BR appears to have the same signaling output (promoting cell elongation) in shoot and root.
The antagonism between BR and auxin is not limited to cell elongation, as they also regulated many key developmental factors in opposite ways. For example, the PLETHORA (PLT), and BRAVO/MYB56 genes are activated by auxin but repressed by BR. PLTs encode AP2-domain transcription factors. The highest levels of PLT proteins specify the position of the QC and promote stem cell identity and maintenance at the stem cell niche. Reduced PLT levels in the transition zone allow cell differentiation, thus determining root meristem size (Figure 4B) (Mähönen et al., 2014). Similarly, BR represses BRAVO expression to promote the division of QC cells (Vilarrasa-Blasi et al., 2014), but auxin positively regulates BRAVO expression (Chaiwanon and Wang, 2015). Therefore, the spatiotemporal antagonism between auxin and BR enforces the spatial domains of cell quiescence, division, and differentiation/elongation; the balance between auxin and BR controls the balance of these stem cell activities and hence the root growth rate (Chaiwanon and Wang, 2015).
Additional Hormones Act through Auxin to Control Root Growth
Many environmental and endogenous cues alter root growth rate through auxin and BR (Figure 4B). For example, cytokinin reduces cell division in the meristem zone. This is mediated by the cytokinin-activated transcription factor ARR1, which promotes the expression of SHY2/IAA3, an inhibitor of ARF in the auxin signaling pathway (Dello Ioio et al., 2008). This function of ARR1 requires its interaction with DELLAs, which act as its transcriptional co-activators (Marínde la Rosa et al., 2015). Furthermore, cytokinin signaling inhibits the expression of several auxin transporters in the root tip, leading to decreased auxin levels (Dello Ioio et al., 2008; Ruzicka et al., 2009; Zhang et al., 2013). On the other hand, auxin increases the expression of negative regulators of cytokinin signaling to establish the root meristem in embryo (Müller and Sheen, 2008), but activates a cytokinin biosynthesis gene to presumably balance meristem size during post-embryonic root development (Dello Ioio et al., 2008). Thus, cytokinin appears to regulate root growth mainly through altering auxin distribution and sensitivity, while auxin regulates cytokinin to maintain an appropriate balance.
Ethylene inhibits root cell elongation in the elongation zone by stimulating local expression of auxin biosynthesis enzymes in the root apex (Stepanova et al., 2008). In addition, ethylene also upregulates expression of PIN2 and AUX1, which promotes basipetal auxin transport toward the elongation zone, where auxin inhibits elongation. In turn, auxin also promotes ethylene biosynthesis in the elongation zone to enhance the inhibition of elongation caused by auxin (Stepanova et al., 2008). Recently, ethylene has been reported to also inhibit cell proliferation in the root meristem by promoting SHY2 expression (Street et al., 2015). Genetic screen for genes required for ABA inhibition of root growth identified components of the ethylene and auxin pathways (Thole et al., 2014). Thus, several phytohormones affect root growth through modulating auxin level, distribution, or signaling.
Environmental Cues Alter Root Growth through Hormones
Light exposure of shoots and leaves has major effects on root growth. When plants are grown in the dark or under shade, limited carbon resource is prioritized for growing stems and petioles in order to improve light exposure, and thus root growth is inhibited. Such root inhibition involves both direct effect of sugar availability to the root meristem, and regulatory effect of auxin. Without photosynthesis or exogenous sugar, primary root meristem is mitotically arrested after depletion of carbon supplies, and treatment of any growth hormones cannot promote root growth (Xiong et al., 2013). The shoot photosynthesis-derived glucose activates target-of-rapamycin (TOR) signaling to control metabolic networks and promote cell proliferation in the root meristem (Figure 4C). Glucose also promotes root growth through auxin and BR, and glucose promotion of lateral root formation is compromised in auxin and BR mutants (Gupta et al., 2015).
In addition to sugar, light signaling also directly alters auxin transport from shoot to root. In the dark, high activity of COP1 leads to repression of PIN1 expression in the hypocotyl and intracellular localization of PIN1 and PIN2 in the root, thus inhibiting shoot-to-root auxin transport and reducing auxin levels in the root (Sassi et al., 2012). On the other hand, light perception and photosynthesis in the shoot promotes root growth through shoot-derived sugars and auxin. Shade signal changes PIN3 subcellular localization to redirect auxin away from root-ward flux toward the epidermis of hypocotyls, and also decreases the level of PIN1 to reduce root-ward transport (Keuskamp et al., 2010; Sassi et al., 2012). Light is also perceived directly by photoreceptors in the root to promote root elongation (Dyachok et al., 2011). In the dark, COP1 regulates degradation of the SCAR complex, which organizes actin filaments required for root elongation (Dyachok et al., 2011). However, how this organ-autonomous light effect is relevant to roots covered by the soil remains unclear.
To navigate in the heterogeneous soil, plants’ root systems must respond appropriately to local environment, such as soil salinity, metal toxicity, and nutrient deficiency. High level of sodium chloride activates ABA signaling in the primary root and induces growth quiescence partly through downregulation of BR and GA signaling activities (Geng et al., 2013). Salt treatments induce immediate accumulation of DELLA proteins but reduce nuclear accumulation of BZR1, consistent with temporary reduction of primary root elongation induced by salt stress (Geng et al., 2013). It has been shown recently that auxin acts downstream of ethylene in mediating ABA-induced root elongation inhibition (Thole et al., 2014). In addition to ABA, aluminum toxicity induces ethylene-dependent local upregulation of the auxin biosynthesis gene TAA1 in the root transition zone, which leads to root growth inhibition in response to aluminum stress (Figure 4B) (Yang et al., 2014).
Arabidopsis respond to phosphate deficiency by inhibiting primary root growth and promoting the growth of lateral root and root hair in order to maximize phosphate uptake (Zhang et al., 2014) (Figure 4D). Such changes of root system architecture (RSA) are mainly mediated by auxin, as auxin treatment causes similar RSA changes, and phosphate deficiency increases auxin levels in both primary root tip and lateral root primordia (Zhang et al., 2014). In contrast, phosphate deficiency reduces BR biosynthesis and causes cytoplasmic accumulation of BZR1 and BES1/BZR2 in the root elongation zone (Singh et al., 2014). According to the BR-auxin antagonism model (Chaiwanon and Wang, 2015), such simultaneous increase of auxin level and decrease of BR level would enforce the inhibition of primary root growth. Furthermore, phosphate deficiency also promotes accumulation of DELLAs by reducing GA biosynthesis (Zhang et al., 2014) (Figure 4D). DELLAs inhibit root growth through interaction with ARR1 (Marínde la Rosa et al., 2015), and possibly also by modulating the activities of BZR1 and ARFs; however, the interactions between BZR1, ARFs, and DELLAs in root growth require further investigation.
Similar to the effect of phosphate deficiency, plant roots respond to nitrogen supply in the soil by stimulating lateral root initiation and elongation and inhibiting primary root growth. The N-induced RSA remodeling has been shown to involve cross regulation by nitrogen-signaling and auxin-signaling pathways (Figure 4D). NRT1.1 nitrate transporter has been shown to facilitate basipetal auxin transport in lateral root tips. High nitrate inhibits NRT1.1, thus leading to accumulation of auxin in lateral root tips to promote growth (Krouk et al., 2010). Furthermore, nitrate induces expression of AFB3, a member of the TIR1/AFB auxin receptor family, resulting in increased auxin signaling. However, the stimulation of AFB3 is feedback regulated by N metabolites, which induce expression of miR393 to target AFB3 transcript for degradation (Vidal et al., 2010). As such, lateral root growth is promoted when nitrate level is high in the soil and endogenous nitrogen supply is low, but inhibited when endogenous nitrogen supply is sufficient.
When grown in heterogeneous soil, a plant is able to inhibit root growth in nitrogen-poor area but enhance root growth in nitrogen-rich soil. Such foraging behavior of roots toward soil patches with high nitrate relies on highly elaborate signaling mechanisms that involve specialized peptide signals. Local nitrogen deficiency in soil induces the expression of C-terminally encoded peptide 1 (CEP1), which inhibits root growth locally (Tabata et al., 2014). However, CEP1 also acts as a mobile signal transmitted from the root to the shoot through the xylem. In the shoot, CEP1 interacts with the receptor kinases, CEPR1 and CEPR2, which induce production of an unknown signal that moves to the roots and promotes growth of the lateral roots in nitrogen-rich patches of soil (Figure 4E). A similar mechanism has been shown to regulate nodulation in legume roots, where the CLE peptides are induced by nitrogen-fixing rhizobia in root cells but transmitted to the shoot to interact with the LRR-RK HAR1 (Okamoto et al., 2013). An unknown signal induced by HAR1 is then transmitted from the shoot back to the root to control the quantity of nodulation (Soyano et al., 2014). Such root-to-shoot-to-root signaling mechanism allows plants to determine RSA by integrating information about both local soil environment and nutrient demand of shoots.
Conclusions
Plants are expected to have evolved highly sophisticated intracellular information processing systems to regulate development, defense, and immunity according to a plethora of environmental and endogenous cues. Research in the Arabidopsis model system has elucidated many individual pathways that transduce specific signals. Recent studies have started to address the more challenging questions of how signaling pathways are integrated with each other and with developmental programs to process complex information into coherent and orchestrated molecular responses within the cells as well as coordinated developmental responses between cells and organs. It has become clear that signaling pathways are highly integrated within the cells through many modes of molecular interaction and cross-regulation. In particular, the BAP/D-HHbH circuit explains how a large number of environmental and hormonal signals converge at a central command system to control the fundamental process of shoot cell elongation. In comparison, the regulation of root tip growth illustrates how interactions between hormones control the spatiotemporal dynamics of cell division and differentiation, as well as how signaling pathways can be rewired by developmental programs to enables tissue- and cell type-specific signaling outputs. One emerging theme is that signal integration and signaling outputs can vary significantly in different development contexts, and thus must be studied and interpreted in specific developmental and physiological context.
For response to light and hormones, plants make robust use of a small number of sensors by creating complex yet logical and modifiable downstream circuits. On the other hand, evolution has also expanded the repertoire of cell surface sensors: there are about 400 RKs in Arabidopsis and 600 in rice. BRI1, FLS2, and CEPR, examples of a few that have been studies, illustrate how powerful and elaborate regulatory systems RKs can enable. Understanding the functions of all these RKs has been one of the major challenges in plant biology, but the advent of CRISPR technology and improvement of mass spectrometry will likely to accelerate progress in near future.
Given the large number of sensors and the high degree of connectivity and integration between signaling pathways, the complexity of the information processing system in plant may be beyond anybody’s imagination. To fully understand the regulatory system of Arabidopsis will require not only continued genetic and molecular dissection, but also enhanced efforts in single cell analysis, high-throughput analysis of protein-protein interactions and protein modifications, as well as computation and modeling. Perhaps the biggest challenge—and opportunity—facing plant biologists is to apply the knowledge and experience gained in Arabidopsis research to the improvement of crop production and environmental protection.
Acknowledgments
This work was supported by grants from the National Institutes of Health (5 R01GM066258) and the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy (DE-FG02-08ER15973) to Z-Y.W, Fujian Agriculture and Forestry University (J.C. and W.W), and Ratchadaphiseksomphot Endowment Fund (J.C.). The authors thank Dr. Joanne Chory for comments on the manuscript. We apologize to the scientists whose important works are not cited here due to space limitation.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- Albrecht C, Boutrot F, Segonzac C, Schwessinger B, Gimenez-Ibanez S, Chinchilla D, Rathjen JP, de Vries SC, Zipfel C. Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc Natl Acad Sci USA. 2012;109:303–308. doi: 10.1073/pnas.1109921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst BA, Ferrieri RA, Gray DW, Lerdau M, Schlyer DJ, Schueller M, Thorpe MR, Orians CM. Jasmonic acid induces rapid changes in carbon transport and partitioning in Populus. New Phytol. 2005;167:63–72. doi: 10.1111/j.1469-8137.2005.01388.x. [DOI] [PubMed] [Google Scholar]
- Bai MY, Fan M, Oh E, Wang ZY. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell. 2012a;24:4917–4929. doi: 10.1105/tpc.112.105163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol. 2012b;14:810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Wells DM, Fozard JA, Ghetiu T, French AP, Pound MP, Wilson MH, Yu L, Li W, Hijazi HI, et al. Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell. 2014;26:862–875. doi: 10.1105/tpc.113.119495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BO, Vanneste S, Krouk G, Nawy T, Efroni I, Shani E, Choe G, Friml J, Bergmann DC, Estelle M, Birnbaum KD. A map of cell type-specific auxin responses. Mol Syst Biol. 2013;9:688. doi: 10.1038/msb.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão-Pires E, Dangl JL, Chory J. Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc Natl Acad Sci USA. 2012;109:297–302. doi: 10.1073/pnas.1112840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Yang L, Hetzel J, Dangl JL, Chory J. The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem Sci. 2014;39:447–456. doi: 10.1016/j.tibs.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo-García S, de Lucas M, Martínez C, Espinosa-Ruiz A, Davière JM, Prat S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014;28:1681–1694. doi: 10.1101/gad.243675.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Carella P, Wilson DC, Cameron RK. Some things get better with age: differences in salicylic acid accumulation and defense signaling in young and mature Arabidopsis. Front Plant Sci. 2014;5:775. doi: 10.3389/fpls.2014.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol. 2013;64:403–427. doi: 10.1146/annurev-arplant-050312-120221. [DOI] [PubMed] [Google Scholar]
- Chaiwanon J, Wang ZY. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr Biol. 2015;25:1031–1042. doi: 10.1016/j.cub.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011;21:664–671. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J. Light signal transduction: an infinite spectrum of possibilities. Plant J. 2010;61:982–991. doi: 10.1111/j.1365-313X.2009.04105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Prat S. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 2014;202:1126–1141. doi: 10.1111/nph.12725. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- de Wit M, Lorrain S, Fankhauser C. Auxin-mediated plant architectural changes in response to shade and high temperature. Physiol Plant. 2014;151:13–24. doi: 10.1111/ppl.12099. [DOI] [PubMed] [Google Scholar]
- de Wit M, Ljung K, Fankhauser C. Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 2015;208:198–209. doi: 10.1111/nph.13449. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- Djakovic-Petrovic T, de Wit M, Voesenek LA, Pierik R. DELLA protein function in growth responses to canopy signals. Plant J. 2007;51:117–126. doi: 10.1111/j.1365-313X.2007.03122.x. [DOI] [PubMed] [Google Scholar]
- Dyachok J, Zhu L, Liao F, He J, Huq E, Blancaflor EB. SCAR mediates light-induced root elongation in Arabidopsis through photoreceptors and proteasomes. Plant Cell. 2011;23:3610–3626. doi: 10.1105/tpc.111.088823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, Wang T, Oh E, Chen L, Park CH, Son SH, Kim SK, Mudgett MB, Wang ZY. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. Plant Cell. 2014;26:828–841. doi: 10.1105/tpc.113.121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C. Sensing the light environment in plants: photoreceptors and early signaling steps. Curr Opin Neurobiol. 2015;34:46–53. doi: 10.1016/j.conb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PMY, Tham C, Duan L, Dinneny JR. A spatiotemporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25:2132–2154. doi: 10.1105/tpc.113.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García MP, Vilarrasa-Blasi J, Zhiponova M, Divol F, Mora-García S, Russinova E, Caño-Delgado AI. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development. 2011;138:849–859. doi: 10.1242/dev.057331. [DOI] [PubMed] [Google Scholar]
- Gupta A, Singh M, Laxmi A. Interaction between glucose and brassinosteroid during the regulation of lateral root development in Arabidopsis. Plant Physiol. 2015;168:307–320. doi: 10.1104/pp.114.256313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S. Brassinosteroid perception in the epidermis controls root meristem size. Development. 2011;138:839–848. doi: 10.1242/dev.061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Velanis CN, Jenkins GI, Franklin KA. UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc Natl Acad Sci USA. 2014;111:11894–11899. doi: 10.1073/pnas.1403052111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71:699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- Hou X, Lee LY, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010;19:884–894. doi: 10.1016/j.devcel.2010.10.024. [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell. 2012;24:4483–4497. doi: 10.1105/tpc.112.105023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MM, Jaillais Y, Pedmale UV, Moreno JE, Chory J, Ballaré CL. Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 2011;67:195–207. doi: 10.1111/j.1365-313X.2011.04598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA. 2010;107:22740–22744. doi: 10.1073/pnas.1013457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Sasidharan R, Vos I, Peeters AJ, Voesenek LA, Pierik R. Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J. 2011;67:208–217. doi: 10.1111/j.1365-313X.2011.04597.x. [DOI] [PubMed] [Google Scholar]
- Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- Kozuka T, Kobayashi J, Horiguchi G, Demura T, Sakakibara H, Tsukaya H, Nagatani A. Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 2010;153:1608–1618. doi: 10.1104/pp.110.156802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 2010;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Larrieu A, Vernoux T. Comparison of plant hormone signalling systems. Essays Biochem. 2015;58:165–181. doi: 10.1042/bse0580165. [DOI] [PubMed] [Google Scholar]
- Larrieu A, Champion A, Legrand J, Lavenus J, Mast D, Brunoud G, Oh J, Guyomarc’h S, Pizot M, Farmer EE, et al. A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nat Commun. 2015;6:6043. doi: 10.1038/ncomms7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell. 2012;24:1398–1419. doi: 10.1105/tpc.112.095711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M, Keller MM, Cerrudo I, Ballaré CL. To grow or defend? Low red : far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytol. 2014;204:355–367. doi: 10.1111/nph.12971. [DOI] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012;26:785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Lu D, Gao X, Jiang S, Ma X, Wang Z, Mengiste T, He P, Shan L. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc Natl Acad Sci USA. 2013;110:12114–12119. doi: 10.1073/pnas.1302154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011;16:684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Blázquez MA, Alabadí D. Genomic analysis of DELLA protein activity. Plant Cell Physiol. 2013;54:1229–1237. doi: 10.1093/pcp/pct082. [DOI] [PubMed] [Google Scholar]
- Lozano-Durán R, Zipfel C. Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 2015;20:12–19. doi: 10.1016/j.tplants.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA. 2016;113:224–229. doi: 10.1073/pnas.1511437113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. PLETHORA gradient formation mechanism separates auxin responses. Nature. 2014;515:125–129. doi: 10.1038/nature13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky FG, Batoux M, Schwessinger B, Youn JH, Stransfeld L, Win J, Kim SK, Zipfel C. Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1. Plant Physiol. 2014;164:1443–1455. doi: 10.1104/pp.113.234625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-de la Rosa N, Pfeiffer A, Hill K, Locascio A, Bhalerao RP, Miskolczi P, Grønlund AL, Wanchoo-Kohli A, Thomas SG, Bennett MJ, et al. Genome wide Binding Site Analysis Reveals Transcriptional Co-activation of Cytokinin-Responsive Genes by DELLA Proteins. PLoS Genet. 2015;11:e1005337. doi: 10.1371/journal.pgen.1005337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–1164. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K, Kajiyama T, Unten-Kobayashi J, Fujii A, Mochizuki N, Kambara H, Nagatani A. Spatial Regulation of the Gene Expression Response to Shade in Arabidopsis Seedlings. Plant Cell Physiol. 2015;56:1306–1319. doi: 10.1093/pcp/pcv057. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife. 2014;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat Commun. 2013;4:2191. doi: 10.1038/ncomms3191. [DOI] [PubMed] [Google Scholar]
- Otero S, Helariutta Y, Benitez-Alfonso Y. Symplastic communication in organ formation and tissue patterning. Curr Opin Plant Biol. 2016;29:21–28. doi: 10.1016/j.pbi.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Pacifici E, Polverari L, Sabatini S. Plant hormone cross-talk: the pivot of root growth. J Exp Bot. 2015;66:1113–1121. doi: 10.1093/jxb/eru534. [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Huang SS, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PA, Sridevi P, Nito K, Nery JR, et al. Cryptochromes Interact Directly with PIFs to Control Plant Growth in Limiting Blue Light. Cell. 2016;164:233–245. doi: 10.1016/j.cell.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J, Friml J. Auxin transport routes in plant development. Development. 2009;136:2675–2688. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M. Molecular and genetic control of plant thermomorphogenesis. Nature Plants. 2016;2 doi: 10.1038/nplants.2015.190. Published online January 6, 2016 http://dx.doi.org/10.1038/nplants.2015.190. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- Ruzicka K, Simásková M, Duclercq J, Petrásek J, Zazímalová E, Simon S, Friml J, Van Montagu MCE, Benková E. Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA. 2009;106:4284–4289. doi: 10.1073/pnas.0900060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell. 2015;27:9–19. doi: 10.1105/tpc.114.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi M, Lu Y, Zhang Y, Wang J, Dhonukshe P, Blilou I, Dai M, Li J, Gong X, Jaillais Y, et al. COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development. 2012;139:3402–3412. doi: 10.1242/dev.078212. [DOI] [PubMed] [Google Scholar]
- Singh AP, Fridman Y, Friedlander-Shani L, Tarkowska D, Strnad M, Savaldi-Goldstein S. Activity of the brassinosteroid transcription factors BRASSINAZOLE RESISTANT1 and BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1/BRASSINAZOLE RESISTANT2 blocks developmental reprogramming in response to low phosphate availability. Plant Physiol. 2014;166:678–688. doi: 10.1104/pp.114.245019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Wasternack C, Xie D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr Opin Plant Biol. 2014;21:112–119. doi: 10.1016/j.pbi.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Soyano T, Hirakawa H, Sato S, Hayashi M, Kawaguchi M. Nodule Inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc Natl Acad Sci USA. 2014;111:14607–14612. doi: 10.1073/pnas.1412716111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks E, Wachsman G, Benfey PN. Spatiotemporal signalling in plant development. Nat Rev Genet. 2013;14:631–644. doi: 10.1038/nrg3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Street IH, Aman S, Zubo Y, Ramzan A, Wang X, Shakeel SN, Kieber JJ, Schaller GE. Ethylene Inhibits Cell Proliferation of the Arabidopsis Root Meristem. Plant Physiol. 2015;169:338–350. doi: 10.1104/pp.15.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol. 2010;154:567–570. doi: 10.1104/pp.110.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science. 2014;346:343–346. doi: 10.1126/science.1257800. [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BP. The Plant Peptidome: An Expanding Repertoire of Structural Features and Biological Functions. Plant Cell. 2015;27:2095–2118. doi: 10.1105/tpc.15.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole JM, Beisner ER, Liu J, Venkova SV, Strader LC. Abscisic acid regulates root elongation through the activities of auxin and ethylene in Arabidopsis thaliana. G3 (Bethesda) 2014;4:1259–1274. doi: 10.1534/g3.114.011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:4477–4482. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarrasa-Blasi J, González-García MP, Frigola D, Fàbregas N, Alexiou KG, López-Bigas N, Rivas S, Jauneau A, Lohmann JU, Benfey PN, et al. Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev Cell. 2014;30:36–47. doi: 10.1016/j.devcel.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Vragović K, Sela A, Friedlander-Shani L, Fridman Y, Hacham Y, Holland N, Bartom E, Mockler TC, Savaldi-Goldstein S. Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc Natl Acad Sci USA. 2015;112:923–928. doi: 10.1073/pnas.1417947112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang ZY. At the intersection of plant growth and immunity. Cell Host Microbe. 2014;15:400–402. doi: 10.1016/j.chom.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol. 2007;17:1784–1790. doi: 10.1016/j.cub.2007.09.025. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- Wang W, Bai MY, Wang ZY. The brassinosteroid signaling network-a paradigm of signal integration. Curr Opin Plant Biol. 2014;21:147–153. doi: 10.1016/j.pbi.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496:181–186. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZB, Geng X, He C, Zhang F, Wang R, Horst WJ, Ding Z. TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis. Plant Cell. 2014;26:2889–2904. doi: 10.1105/tpc.114.127993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Hirano K, Sato T, Mitsuda N, Nomoto M, Maeo K, Koketsu E, Mitani R, Kawamura M, Ishiguro S, et al. DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc Natl Acad Sci USA. 2014;111:7861–7866. doi: 10.1073/pnas.1321669111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Swarup R, Bennett M, Schaller GE, Kieber JJ. Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr Biol. 2013;23:1979–1989. doi: 10.1016/j.cub.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liao H, Lucas WJ. Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J Integr Plant Biol. 2014;56:192–220. doi: 10.1111/jipb.12163. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu Z, Wang J, Chen Y, Bi Y, He J. Brassinosteroid is required for sugar promotion of hypocotyl elongation in Arabidopsis in darkness. Planta. 2015;242:881–893. doi: 10.1007/s00425-015-2328-y. [DOI] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wang L, Xi Y, Li J, Quail PH, Deng XW, Guo H. A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol. 2012;22:1530–1535. doi: 10.1016/j.cub.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]