Abstract
Gallbladder carcinoma (GBC) is the most common biliary tract malignancy with high mortality. The median survival time is 6 months, and the 5-year survival rate less than 5% for GBC patients. Thus, it is imperative to investigate the molecular mechanisms underlying the pathogenesis of GBC. miR-133a may exert anti-tumor effects on a variety of cancers. However, the role of miR-133a in the pathogenesis of GBC remains unclear. In this study, quantitative real-time polymerase chain reaction (qRT-PCR) showed the miR-133a-3p expression markedly decreased in GBC as compared to adjacent normal tissues. Transient over-expression of miR-133a-3p inhibited the proliferation, migration and invasion abilities of GBC cells. Luciferase activity assay indicated that miR-133a-3p negatively regulated the expression of recombination signal-binding protein Jκ (RBPJ) directly, which is a key downstream transcription factor in the Notch signaling pathway. Moreover, PBPJ expression was up-regulated and negatively related to miR-133a-3p expression in GBC, and silencing of RBPJ achieved the effects as after miR-133a-3p over-expression. RBPJ over-expression could markedly reverse the inhibitory effects of miR-133a-3p on the proliferation, migration and invasion of GBC cells. Our findings indicate that miR-133a-3p acts as a tumor suppressor through directly targeting RBPJ in GBC.
Keywords: Gallbladder carcinoma, miR-133a-3p, recombination signal-binding protein Jκ, molecular mechanism
Introduction
Gallbladder cancer (GBC), which is a common malignancy with high mortality, accounting for 80%-90% of biliary tract cancers [1], represents the fifth most common cancer among gastrointestinal cancers [2]. The Surveillance, Epidemiology, and End Results (SEER) program estimates that the incidence of GBC is 2.5 cases per 1×105 people [3]. There are many risk factors of GBC, such as gallstones, chronic cholecystitis, gallbladder polyps, and primary sclerosing cholangitis (PSC) [4]. However, despite most risk factors have been confirmed, only a third of GBC can be recognized preoperatively, and the resectable GBC accounts for only 15%-47% [1,5]. In addition, the tumor progression is frequently rapid and silent, making the prognosis of GBC often unsatisfactory. According to the statistics, the median survival time is only 6 months, and the 5-year survival rate is less than 5% for GBC patients [6]. Therefore, it is important to investigate the molecular mechanisms underlying the pathogenesis of GBC and to develop early diagnostic methods or effective therapies for GBC.
Micro-RNAs (miRNAs) are endogenous short non-coding RNAs with ~24 nucleotides in length, and mediate post-transcriptional repression via binding to the 3’-untranslated region (3’UTR) of target messenger RNA transcripts. In some cases, they also mediate the transcription of target mRNA, leading the destruction of mRNA [7]. miRNAs play pivotal roles in various biological processes, including metabolism, cell proliferation, apoptosis, differentiation, transdifferentiation and cell signaling [8-10]. Numerous studies have suggested that some miRNAs are aberrantly expressed and function as oncogenes in many human cancers, since them actively participate in several important processes related to carcinogenesis [11-13]. A further understanding of miRNAs expression and function in tumorigenesis may improve the understanding of the initiation and development of human cancers, which may improve the diagnosis and therapy of cancers.
Recently, studies indicate that several miRNAs have aberrant expression and play a crucial role in the pathogenesis of GBC. For example, miR-101, miR-122 and miR-138 frequently display down-regulated expression and exert anti-tumor effect [14-16]. miR-182 and miR-155 show up-regulated expression and function as oncogenes [17,18]. miR-133a may exert anti-tumor effects on a variety of cancers. However, the role of miR-133a in the pathogenesis of GBC remains unclear. Our results showed that the expression of miR-133a-3p significantly reduced in GBC as compared to normal gallbladder tissues. In human GBC-SD and EHGB-1 cell lines, the role of miR-133a-3p in the biobehaviors of GBC cells as well as the potential mechanism was further investigated in vitro. Results showed the gain-function of miRNA-133a-3p in GBC cells could induce the inhibition of cell proliferation, cause G1/S phase arrest, and inhibit their migration and invasion in vitro. In addition, recombination signal-binding protein Jκ (RBPJ), a key downstream transcription factor of the Notch signaling pathway, was identified as a direct target gene of miRNA-133a-3p, and miRNA-133a-3p could exert its inhibitory effects on GBC cells via negatively regulating RBPJ.
Materials and methods
Tissue collection
GBC tissues and adjacent normal gallbladder tissues were obtained from 23 patients who underwent surgery between 2015 and 2016 in the Department of General Surgery in Xinhua Hospital, Zhongshan Hospital and People’s Hospital of Shanghai. The fresh GBC tissues and adjacent normal tissues were stored in liquid nitrogen immediately after collection until RNA extraction.
Cell culture and transfection
The human GBC cell lines (NOZ, SGC-996, OCUG, GBC-SD and EHGB-1 cells) were obtained from the Shanghai Life Science Laboratory of Chinese Academy of Sciences. NOZ cells were maintained in Williams medium (Gibico, USA), SGC-996 cells in RPMI 1640 (Gibico, USA), GBC-SD cells in DMEM (Gibico, USA) supplemented with 10% fetal bovine serum (FBS; Evergreen), and OCUG and EHGB-1 cells in DMEM containing 15% FBS. The medium was supplemented with penicillin (100 U/m) and streptomycin (100 U/ml), and cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. Prior to transfection, cells were cultured in complete medium without antibiotics for at least 24 h. When the cell confluence reached 30%-50%, cells were washed with 1×PBS once, the GBC-SD and EHGB-1 cells were transfected with 20 nM and 50 nM Hsa-miR-133a-3p mimic or mimic negative control, respectively, in the presence of Lipofectamine™2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
RNA extraction, reverse transcription and qRT-PCR
Total RNAs were extracted from tissues and cells using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RNA (1 µg) was reversely transcribed into cDNA using the miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen Beijing, China) according to the manufacturer’s instructions. qRT-PCR was performed using the miRcute miRNA q-PCR Detection Kit (SYBR GREEN) (Tiangen Beijing, China) in the ABI Step-one real-time PCR system (Applied Biosystems, Foster City, CA, USA). cDNA for miRNA and total cDNA were synthesized using the specific miRNA primers (Tiangen) and random hexamer (Shanghai Sangong), respectively. U6 or GAPDH was used as an internal control. The relative expression was calculated using the 2-ΔΔCt method.
CCK8 assay
The cell proliferation was measured using the WST-8 kit (YiSheng, ShangHai, China). The transfected cells were plated into 96-well plates (Corning Costar, Corning, NY) at a density of 1.0×103/well/100 µL, and then 10 µL of CCK8 solution was added to each well, followed by incubation for 2 h. The cell viability was determined by measuring the absorbance at 450 nm. Measurement of cell proliferation was done once every.
Colony formation assay
Cell transfection was performed 48 h later as above mentioned. The transfected cells were harvested after trypsinization, counted and plated at a density of 500 cells/6-cm dish. The medium was refreshed once every 3 days. Ten days later, cells were washed in 1×PBS twice, fixed in 3.7% methanol, stained with 0.1% crystal violet and then counted. The cell colony was counted (each colony contains at least 50 cells).
Transwell migration/invasion assay
Transwell chamber was used to measure cell migration and invasion abilities. In brief, culture inserts with 8-mm pore size (Transwell; Corning, NY) were placed into 24-well plates. Before the measurement of invasion ability, the plates were pre-coated with matrigel. 2 h before the addition of matrigel, 500 µL of serum-free medium was independently added to the upper and lower chambers, followed by incubation at 37°C for hydration. Cells were digested by typsin, and resuspended in serum-free medium. The cell density was adjusted to 1×105/mL. Then, 200 µL of cell suspension was added into the upper chamber, and 500 µL of DMEM containing 10% FBS into the lower chamber. After incubation at 37°C with 5% CO2 for 24 h, the Transwell chamber was removed, cells were washed with 1×PBS, fixed in paraformaldehyde for 20 min, and then stained with 0.1% crystal violet for 20 min. The cotton swab was used to clean the non-migrated cells in the upper chamber, cells migrating through the membrane were counted in 5 randomly selected fields under a microscope (Nikon) at a magnification of ×100.
Flow cytometry for cell-cycle assay
48 h after transfection, cells were trypsinized, washed with cold 1×PBS thrice, and fixed in cold 70% ethanol over night at 4°C. After washing with cold 1×PBS, cells were stained with propidium iodide (PI) containing RNase A in dark for 30 min. A total of 50,000 cells were counted for each sample and cell-cycle profiles were analyzed with ModFit 3.0 software (BD Biosciences). All the experiments were repeated in triplicate.
Western blotting
For the extraction of total cell protein, transfected cells were washed with ice-cold PBS, and then lysed in cell lysis buffer (50 mM Tris [pH 8.0], 120 mM NaCl, 0.5% NP-40, 50 mM NaF and, 1 mM phenylmethylsul-fonyl fluoride [Beyotime Institute of Biotechnology, Haimen, China]) on ice for 30 min. The homogenate was centrifuged at 14,000×g for 15 min at 4°C. The protein concentration was measured with the BCA Protein Assay Kit (Beyotime Institute of Biotechnology). Then, the bromophenol blue (0.01%) was added into the protein lysates, and proteins were separated on 10% sodium dodecyl sulfate-poly-acrylamide gel at 120 V for 1 h, followed by transferring onto polyvinylidene difluoride membranes. The membranes were blocked with 5% non-fat milk in Tris-buffered saline (pH7.4) containing 0.1% Tween 20 (TBST) for 1 h at room temperature, and then incubated with primary antibodies (anti-CyclinD1 [1:1,000, rabbit], anti-P53 [1:1,000; rabbit], anti-P21 [1:1,000; rabbit], anti-RBPJ [1:1,000; rabbit] and anti-β-actin [1:1,000, mouse] overnight at 4°C. After washing thrice in TBST for 5 min, the membranes were then incubated with HRP conjugated goat anti-rabbit IgG or anti-mouse IgG secondary antibody. Chemiluminescence detection was performed using AP buffer. The densitometry was performed using Gel Doc 2000 (BioRad. USA).
Dual-luciferase reporter assay
A dual luciferase reporter assay was carried out to investigate whether RBPJ expression is directly regulated by miR-133a-3p. The wild type and mutated type 3’UTR of RBPJ, which contains miR-133a-3p binding sequences, were synthesized and cloned into the pmirGLO luciferase reporter vector (Longqian, Shanghai, China). Using Lipofectamine 2000, the wild/mutant type RBPJ luciferase reporter vector with miR-133a-3p mimic was transfected into GBC-SD cells for 48 h. Then, the luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega). The relative expression of luciferase activity was normalized to Renilla luciferase activity.
Transfection of siRNA and construction of plasmids
The RBPJ-siRNA and negative control siRNA were designed and synthesized in TuoRan Company (Shanghai, China) and then independently transfected into GBC-SD cells and EHGB-1 cells for 48 h using Lipofectamine 2000. The pCMVPuro01 expression vector containing full-length RBPJ cDNA (Longqian, Shanghai, China) was independently transfected into GBC and EHGB-1 cells with miR-133a-3p mimic. The expression of RBPJ was detected by Western blotting. Empty vector was transfected into cells with miR-133a-3p mimic or mimic negative control as controls.
Statistical analysis
Data are expressed as mean ± standard deviation (SD). Unpaired t test and One-Way Analysis of Variance (ANOVA) were employed for comparisons using GraphPad Prism6.0. A value of P<0.05 was considered statistically significant. All experiments were repeated at least three times.
Results
miRNA-133a-3p expression is generally down-regulated in GBC tissues and cell lines
In order to ascertain the miR-133a-3p expression in human GBC tissues, RT-qPCR was performed in GBC tissues from 23 patients. As shown in Figure 1A, 69.6% (n=16) of GBC tissues showed reduced miR-133a-3p expression. The miR-133a-3p expression significantly reduced in GBC tissues as compared to adjacent normal gallbladder tissues (Figure 1B). Furthermore, the miR-133a-3p expression was detected in NOZ, SGC-996, OUCG-1, EHGB-1 and GBC-SD cells. As shown in Figure 1C, when compared with normal gallbladder tissue (n=3), miR-133a-3p expression significantly decreased in GBC cell lines. These findings indicate that miR-133a-3p expression is down-regulated in GBC tissues and cell lines. In the following experiments, GBC-SD and EHGB-1 cells were used since the miR-133a-3p expression was lower in these cell lines than in other cell lines.
Figure 1.
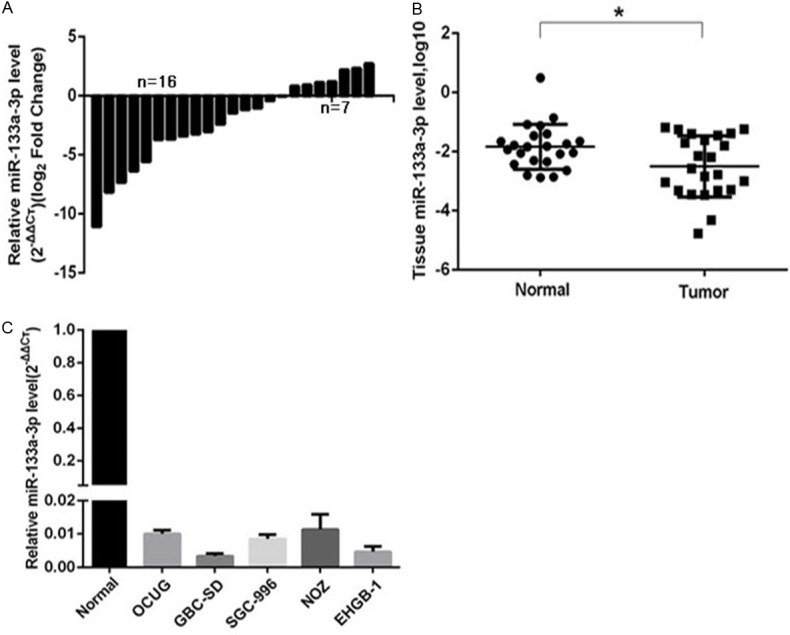
miR-133a-3p expression in human GBC tissues and cell lines. A and B: miR-133a-3p expression in GBC tissues and adjacent normal tissues from 23 patients (RT-qPCR; U6 as an internal control; Wilcoxon matched-pairs test) *P=0.0101 (<0.05) vs. normal tissues. C: miR-133a-3p expression in human GBC OCUG, GBC-SD, SGC-996, NOZ and EHGB-1 cell lines. **P<0.01 vs. normal tissues (n=3).
miR-133a-3p over-expression inhibits tumorigenic properties of GBC cells
The significant reduction in miR-133a-3p expression in GBC tissues and cell lines indicates that miR-133a-3p acts as a tumor suppressor in GBC. In order to verify this hypothesis, CCK8 assay and colony formation assay were performed in vitro. The expression of miR-133a-3p in GBC-SD and EHGB-1 cells were up-regulated via transfection with miR-133a-3p mimics, and the transfection efficiency was detected by RT-qPCR (Figure 2A). Subsequently, CCK8 assay was employed to detect the cell viability of GBC-SD and EHGB-1 cells. As shown in Figure 2B, miR-133a-3p over-expression inhibited the cell proliferation of both GBC-SD and EHGB-1 cells, and the longer the time of transfection, the more evident the inhibitory effect was. Furthermore, the over-expression of miR-133a significantly suppressed the colony formation by 86% in GBC-SD cells and 27% in EHGB-1 cells (P<0.01) (Figure 2C).
Figure 2.
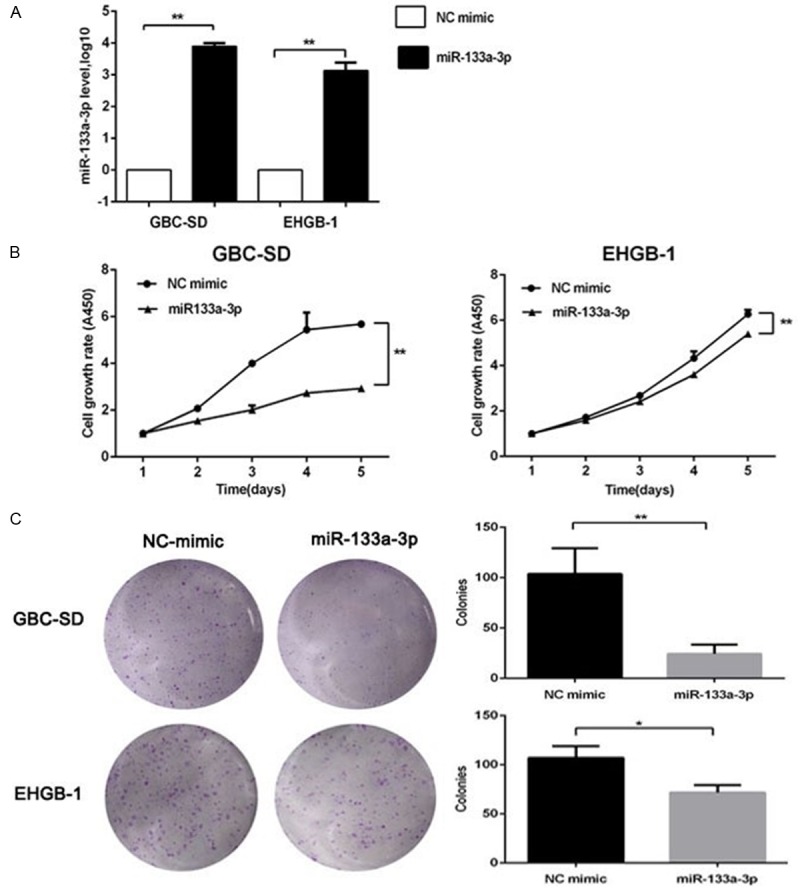
Effects of miR-133a-3p over-expression on GBC cell proliferation. A: miR-133a-3p mimic and NC mimic were transfected into GBC cells, and the transfection efficiency was determined by qRT-PCR. B: CCK8 assay showed that miR-133a-3p over-expression inhibited the proliferation of GBC-SD and EHGB-1 cells (vs miRNA-NC group). C: miR-133a-3p inhibited the colony formation of GBC-SD and EHGB-1 cells. *P<0.05, **P<0.01.
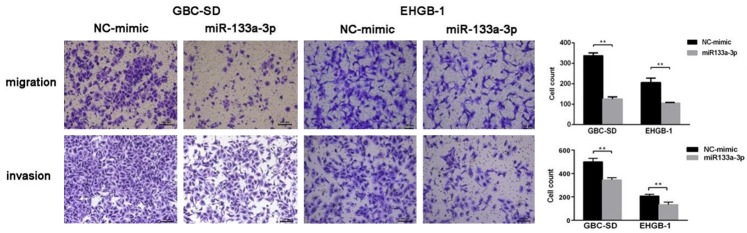
miR-133a-3p over-expression inhibits GBC cell migration and invasion
To investigate the effects of miR-133a-3p on the GBC cell migration and invasion, Transwell migration assay were performed in which matrigel gel was used to mimick the in vivo extracellular matrix. Using the transwell chamber containing matrigel gel, the invasion ability of GBC cells was evaluated. As shown in Figure 3, the number of migrated cells significantly decreased in both GBC-SD and EHGB-1 cells after transfection with miR-133a-3p mimic, as compared to miR-NC transfected cells. These indicate that miR-133a-3p over-expression inhibits the migration and invasion abilities of GBC cells in vitro.
Figure 3.
Effects of mir-133a-3p over-expression on GBC cell migration and invasion. miR-133a-3p over-expression inhibited the migration of GBC-SD and EHGB-1 cells as compared to miRNA-NC group. miR-133a-3p over-expression inhibited the invasion of GBC-SD and EHGB-1 cells. **P<0.01.
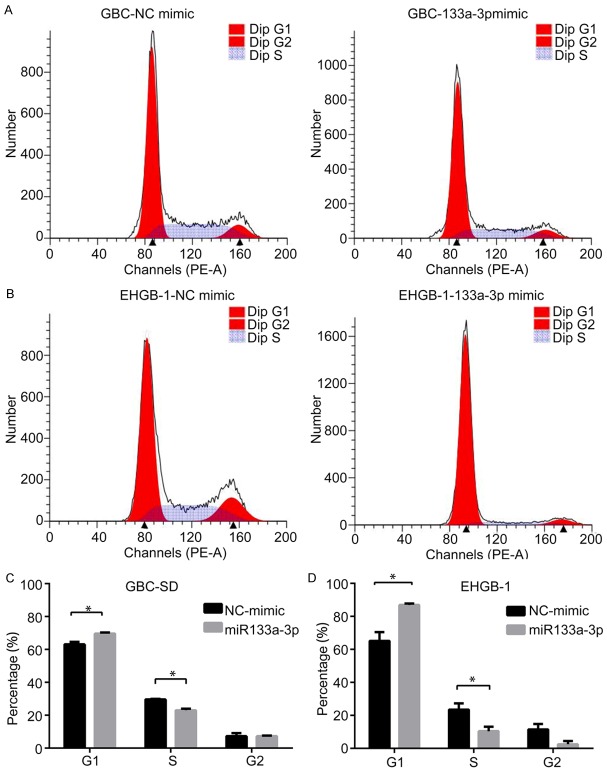
miR-133a-3p over-expression induces G1/S phase arrest in GBC cells
Above findings showed miR-133a-3p was able to inhibit the growth of GBC cells, and then impact of miR-133a-3p over-expression on cell cycle was further assessed by flow cytometry. As shown in Figure 4, miR-133a-3p over-expression remarkably increased the proportion of cells in G1-phase, and the proportion of cells in S phase significantly decreased as compared to miR-NC transfected GBC-SD and EHGB-1 cells (Figure 4C and 4D). These indicate that miRNA-133a-3p over-expression effectively induces G1/S phase arrest in GBC cells.
Figure 4.
Effects of mir-133a-3p over-expression on GBC cell cycle. The proportion of cells in G1-phase increased remarkably in GBC-SD cells (A, C) and EHGB-1 cells (B, D) compared with miRNA-NC group, while the proportion of cells in S phase significantly reduced as compared to miRNA-NC group (*P<0.05).
miR-133a-3p over-expression induces cell cycle arrest by up-regulating p53/p21 pathway and down-regulating cyclinD1 expression
To further explore the potential mechanism of G1/S phase arrest, the expression of cell cycle relative proteins was detected by Western blotting. The effect of miR-133a-3p on the expression of p53 was detected because p53 is a well-established tumor suppressor which is often dysfunctional in cancers, and p53 over-expression is closely related to cell cycle arrest. Our results indicated that the protein expression of p53 increased significantly in GBC-SD and EHGB-1 cells after transfection with miR-133a-3p (Figure 5A). As one of p53-inducible gene products, the expression of cyclin-dependent kinase inhibitor p21 led to cell cycle arrest in G1/S. Our results indicated that the p21 protein expression was up-regulated after miR-133a-3p over-expression in GBC cell lines (Figure 5A). Cyclin D1 is a critical mediator of cell cycle progression and can activate cyclin-dependent kinases (CDKs). Cyclin D1 over-expression has a pro-proliferative effect on cancer cells. In our study, the protein expression of cyclinD1 significantly decreased in GBC-SD and EHGB-1 cells after transfection with miR-133a-3p (Figure 5B). These suggest that miR-133a-3p restrains cell growth by up-regulating p53 and p21 expression and inhibiting cyclinD1 expression.
Figure 5.
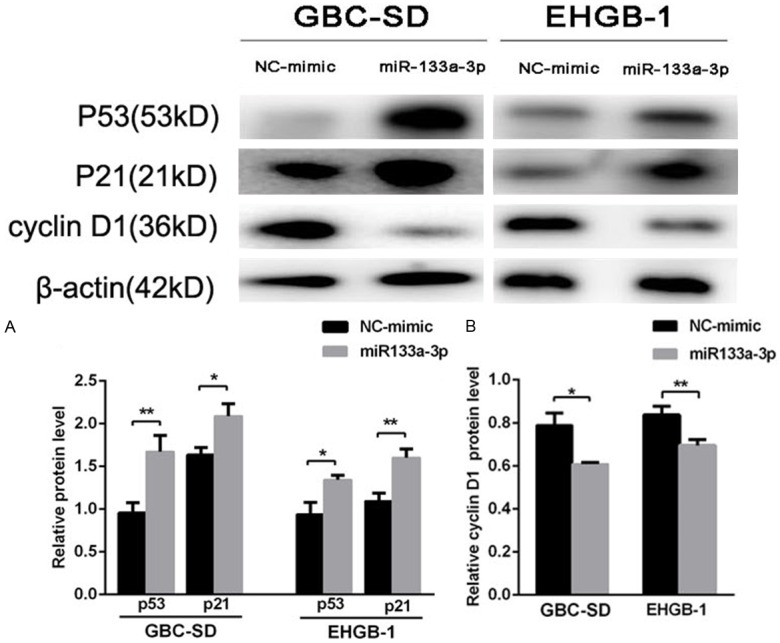
mir-133a-3p over-expression induced cell cycle arrest by up-regulating p53 and p21 expression and inhibiting cyclinD1 expression. miR-133a-3p over-expression increased the protein expression of p53 and p21 in GBC-SD and EHGB-1 cells (A). Cyclin D1 protein expression decreased after transfection with miR-133a in both cell lines as compared to miRNA-NC group (B). (*P<0.05, **P<0.01).
RBPJ is a direct target of miR-133a-3p
The molecular mechanism underlying the anti-tumor effect of miR-133a-3p was further investigated in GBC. Above findings indicated miR-133a-3p had anti-tumor activity, and thus its potential targets might have oncogenic properties. Bioinformatics prediction system (miRDB, PicTar AND Target Scan) showed RBPJ might be a target gene of miR-133a-3p. RBPJ has a conserved binding site for miR-133a-3p in the 372-378 of 3’-UTR (Figure 6A). To test whether RBPJ expression is direct regulated by miR-133a-3p, the 3’UTR reporter plasmids containing wild type or mutant RBPJ 3’UTR were constructed, and co-transfected into GBC-SD cells with miR-133a-3p mimic or negative control mimic. The luciferase reporter assay indicated that the luciferase activity of wild type RBPJ 3’UTR was significantly suppressed in miR-133a-3p mimic transfected cells compared with miR-NC transfected cells; while, there was no significant difference in the luciferase activity when the Mut-RBPJ 3’UTR was transfected along with miR-133a-3p mimic or negative control mimic (Figure 6B). Subsequently, the effect of miR-133a-3p on RBPJ expression was also investigated in GBC cell lines. Results showed that miR-138 mimic significantly reduced RBPJ protein expression in GBC and EHGB-1 cells (Figure 6C).
Figure 6.
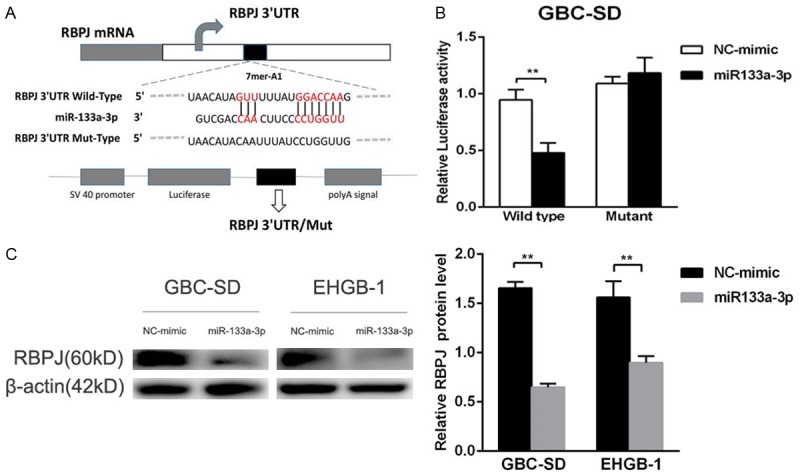
RBPJ was a direct target of miR-133a-3p. A: The wild type or mutant type of RBPJ 3’UTR for miR-133a-3p. B: The relative luciferase activity of wild type or mutant RBPJ 3’UTR in GBC-SD cells after transfection with miR-133a-3p mimic or NC-mimic. C: Over-expression of miR-133a-3p attenuated RBPJ protein expression in GBC-SD and EHGB-1 cells compared with miRNA-NC group. **P<0.01
PBPJ expression is up-regulated and negatively related to miR-133a-3p expression in GBC
To assess the correlation between RBPJ and miR-133a-3p in GBC tissues, qRT-PCR was performed to detect the expression of RBPJ and miR-133a-3p in GBC tissues from 23 patients. As shown in Figure 7A, 60.9% (n=14) of GBC tissues showed an increased RBPJ expression. The RBPJ expression was significantly higher in GBC tissues than in adjacent normal gallbladder tissues (Figure 7B). Furthermore, the correlation between miR-133a-3p and RBPJ in GBC tissues was evaluated with Spearman linear correlation analysis. As showed in Figure 7C, PBPJ expression was negatively related to miR-133a-3p expression at mRNA level in GBC tissues.
Figure 7.
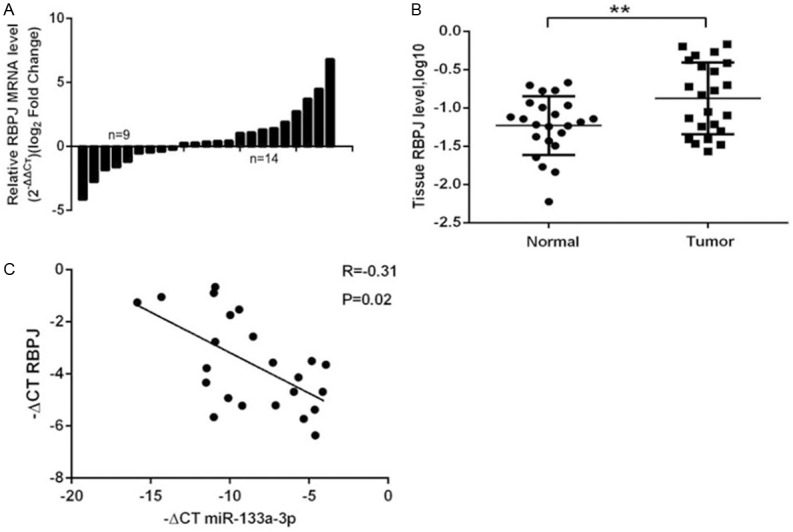
RBPJ expression in human GBC tissues. A and B: RBPJ expression in GBC tissues and adjacent normal tissues (n=23; RT-qPCR; GAPDH as an internal control; Wilcoxon matched-pairs test) **P=0.0074 vs. normal tissues. C: RBPJ mRNA expression reversely correlated with miR-133a-3p expression in GBC tissues.
Down-regulating RBPJ expression inhibits GBC cells proliferation, migration and invasion
To determine whether RBPJ acts as an oncogene in GBC, RBPJ siRNA was employed to down-regulate RBPJ expression in GBC-SD and EHGB-1 cells (Figure 8A). CCK8 and colony formation assay showed reducing RBPJ expression restrained cell proliferation (Figure 8B, 8C). Transwell assays indicated that decreasing RBPJ expression was able to inhibit the migration and invasion of GBC cells (Figure 8D). These results confirm that RBPJ has a strong tumorigenicity in GBC, which reinforces the finding that RBPJ is one of the functional target of miR-133a-3p in GBC.
Figure 8.
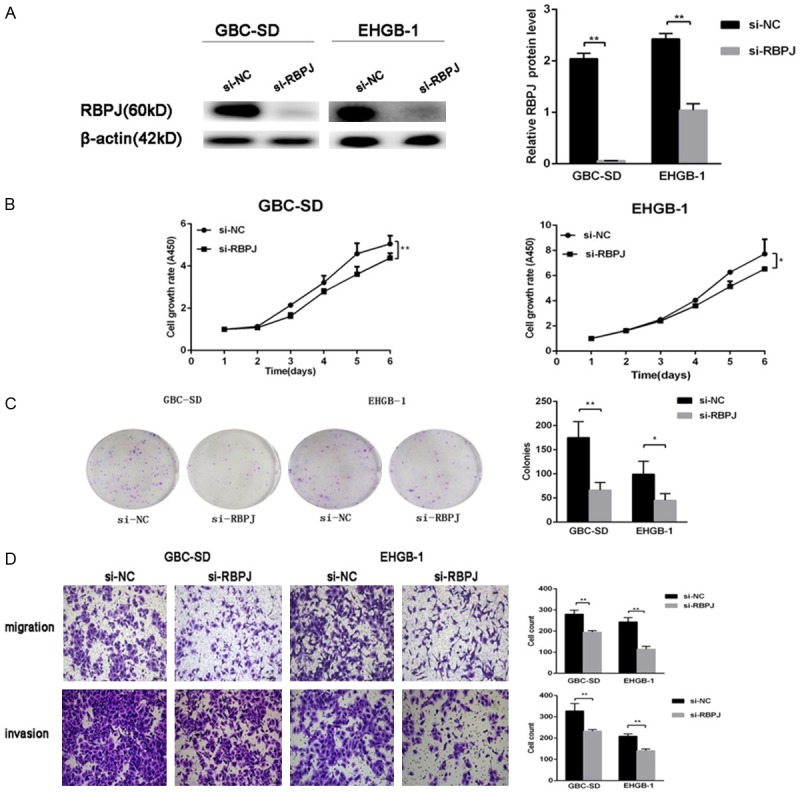
Effects of RBPJ down-regulation on GBC cells. A: RBPJ protein expression after transfection with siRNA-RBPJ. B and C: CCK8 assay and colony formation assay showed that down-regulation of RBPJ inhibited the proliferation and colony formation of GBC-SD and EHGB-1 cells compared with si-NC groups. D: Down-expression of RBPJ inhibited the migration and invasion of GBC-SD and EHGB-1 cells compared with si-NC groups. *P<0.05, **P<0.01.
RBPJ attenuates the inhibitory effects of miR-133a-3p on GBC cells
To investigate whether miR-133a-3p affects the proliferation, cell-cycle arrest, migration and invasion of human GBC cells through RBPJ, RBPJ over-expressing or empty vector was co-transfected with miR-133a-3p mimic or NC-mimic into GBC-SD cells. As shown in Figure 9A, the protein expression of RBPJ increased in miR-133a-3p+RBPJ cells compare with miR-133a-3p+MOCK cells. Meanwhile, cell proliferation and migration capabilities were also detected. As shown Figure 9B, 9C, the miR-133a-3p-mediated anti-tumor phenotypes of GBC-SD cells was weakened after transfection with RBPJ over-expressing vector.
Figure 9.
Over-expression of RBPJ attenuated the inhibitory effects of miR-133a-3p on GBC cells. A: Protein expression of RBPJ after transfection with NC-mimic/miR-133a-3p mimic, and RBPJ-vector/empty vector. Lane 1, NC mimic+Vector; lane 2, miR-133a-3p+Vector; lane 3, miR-133a-3p+RBPJ. B and C: CCK8 assay and migration assay showed that up-regulation of RBPJ partly reversed the effects of miR-133a-3p on the proliferation and migration of GBC-SD cells compared with miR-133a-3p+Vector. *P<0.05, **P<0.01.
Discussion
Studies have demonstrated that the reduced expression of miRNAs is associated with the development of various types of cancers. Microarray hybridization and qRT-PCR have shown that numerous miRNAs are aberrantly expressed, and several miRNAs may function as oncogenes or tumor-suppressors in GBC [14-18]. Nevertheless, the role of miRNAs in GBC remains largely unclear. Letelier et al. [19] found that miR-133a expression was down-regulated in GBC, suggesting that miR-133a plays a vital role in the development of GBC. In the present study, the expression of miR-133a-3p was detected in GBC tissues and cell lines, and its effects on the biobehaviors of GBC cells were further investigated.
miR-133a has two genes: miR-133a-1 and miR-133a-2, locating in 18q11.2 and 20q13.33 respectively. In most studies, miR-133a has been found to act as a myogenic miRNAs, and can regulate myoblast proliferation, differentiation and apoptosis via targeting special mRNAs [20-22]. However, on the basis of the reduced miR-133a expression in cancer, several studies indicate that miR-133a functions as a tumor suppressor in cancers [23,24]. In the present study, results showed that miR-133a-3p expression significantly decreased in GBC tissues and GBC cell lines, which was consistent with previous findings [19]. Therefore, gain-of-function experiments were conducted in vitro to investigate the role of miR-133a-3p in the pathogenesis of GBC. Our results showed that miR-133a-3p inhibited the proliferation, migration and invasion of GBC cells, suggesting that miR-133a-3p acts as a tumor suppressor in GBC.
Then, the mechanism underlying the inhibitory effects of miR-133a-3p on GBC cells was further investigated. miR-133a has been reported to induce cell cycle arrest in several types of cancer cells, including breast cancer cells, renal cell carcinoma cells and some cancer cells of the digestive system [25-27]. In our study, over-expression of miR-133a-3p resulted in an increased proportion of cells in G0/G1 phase, leading to G1/S phase arrest. The p53/p21 signal pathway is involved in the cell cycle progression in cancers [28], and activation of p53/p21 signal pathway is able to induce cell-cycle arrest and inhibit tumor cell growth. Our study revealed that over-expression of miR-133a-3p was associated with the up-regulation of p53 and p21 protein expression in GBC cells. Cyclin D1 is an oncoprotein that can be dimerized with cyclin-dependent kinase 4/6 to regulate G1/S phase transition [29,30]. Our results showed that the increased miR-133a-3p expression was related to the decreased cyclin D1 expression. These findings further indicate the growth-inhibiting capacity of miR-133a-3p.
The oncogenic or anti-oncogenic roles of miRNAs in cancers are tightly associated with their target mRNA [24]. Several candidate target genes of miR-133a-3p have been identified by bioinformatics analysis. Here, our study focused on RBPJ for its role as a vital regulator in the Notch pathway [31]. Aberrant Notch activity is implicated in the pathogenesis of several malignancies through mediating tumor proliferation, epithelial-mesenchymal transition (EMT) and differentiation [32-34]. Yoon et al. [35] reported that the Notch signaling pathway was tightly related to the clinical pathology of GBC in patients.
RBPJ gene locates in 4p15.2 and encoded RBPJ protein which is a transcriptional regulator important for the Notch signaling pathway. After binding to specific ligand, the intracellular domain of Notch receptor (NICD) is cleaved and translocates into the nucleus, where it binds to the transcription factor RBPJ to generate a Notch-RBPJ complex as an activator [31]. In the present study, luciferase reporter assay confirmed RBPJ as a novel direct target of miR-133a-3p. Furthermore, the RBPJ expression was negatively regulated by miR-133a-3p at post-transcriptional level in GBC cells.
Then, the RBPJ expression was detected in GBC tissues from 23 patients. Results showed that there was an inverse correlation between miR-133a-3p expression and RBPJ expression. To clarify the role of RBPJ in the development of GBC, loss-of-function experiments were performed in vitro. Results showed that the silencing of RBPJ expression inhibited the proliferation, migration and invasion of GBC cells, which revealed that RBPJ functions as an oncogene in GBC. This was consistent with the finding from the study of Yong et al. [36]. Moreover, RBPJ over-expression could markedly reverse the inhibitory effects of miR-133a-3p on the proliferation and migration of GBC cells. These findings suggest that miR-133a-3p exerts its effect on GBC cells, at least partially, by down-regulating the expression of downstream effector RBPJ.
In summary, our study expands the understanding of the regulatory effects of miR-133a-3p on GBC cells and for the first time characterizes a novel mechanism underlying the effects of miR-133a-3p on GBC cells: miR-133a-3p behaves as a tumor suppressor via directly repressing the Notch pathway protein RBPJ. Our findings may not only improve the understanding of molecular mechanism underlying the epigenetic regulation of miR-133a-3p in GBC, but also provide a novel therapeutic target for the treatment of GBC.
Disclosure of conflict of interest
None.
References
- 1.Lazcano-Ponce EC, Miquel JF, Munoz N, Herrero R, Ferrecio C, Wistuba II, Alonso de Ruiz P, Aristi Urista G, Nervi F. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin. 2001;51:349–364. doi: 10.3322/canjclin.51.6.349. [DOI] [PubMed] [Google Scholar]
- 2.Varshney S, Butturini G, Gupta R. Incidental carcinoma of the gallbladder. Eur J Surg Oncol. 2002;28:4–10. doi: 10.1053/ejso.2001.1175. [DOI] [PubMed] [Google Scholar]
- 3.Shu YJ, Weng H, Ye YY, Hu YP, Bao RF, Cao Y, Wang XA, Zhang F, Xiang SS, Li HF, Wu XS, Li ML, Jiang L, Lu W, Han BS, Jie ZG, Liu YB. SPOCK1 as a potential cancer prognostic marker promotes the proliferation and metastasis of gallbladder cancer cells by activating the PI3K/AKT pathway. Mol Cancer. 2015;14:12. doi: 10.1186/s12943-014-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanthan R, Senger JL, Ahmed S, Kanthan SC. Gallbladder Cancer in the 21st Century. J Oncol. 2015;2015:967472. doi: 10.1155/2015/967472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mekeel KL, Hemming AW. Surgical management of gallbladder carcinoma: a review. J Gastrointest Surg. 2007;11:1188–1193. doi: 10.1007/s11605-007-0115-1. [DOI] [PubMed] [Google Scholar]
- 6.Levy AD, Murakata LA, Rohrmann CA Jr. Gallbladder carcinoma: radiologic-pathologic correlation. Radiographics. 2001;21:295–314. doi: 10.1148/radiographics.21.2.g01mr16295. questionnaire, 549-255. [DOI] [PubMed] [Google Scholar]
- 7.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Shenoy A, Blelloch R. microRNA induced transdifferentiation. F1000 Biol Rep. 2012;4:3. doi: 10.3410/B4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 11.Riquelme I, Tapia O, Leal P, Sandoval A, Varga MG, Letelier P, Buchegger K, Bizama C, Espinoza JA, Peek RM, Araya JC, Roa JC. miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in gastric cancer through regulation of the PI3K/AKT/mTOR pathway. Cell Oncol (Dordr) 2016;39:23–33. doi: 10.1007/s13402-015-0247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutlu M, Raza U, Saatci O, Eyupoglu E, Yurdusev E, Sahin O. miR-200c: a versatile watchdog in cancer progression, EMT, and drug resistance. J Mol Med (Berl) 2016;94:629–644. doi: 10.1007/s00109-016-1420-5. [DOI] [PubMed] [Google Scholar]
- 13.Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang Y, Liu Z, Cao X, Chen P, Liu Z, Chen X, Tao Y, Xu C, Mao J, Cheng C, Li C, Hu Y, Wang L, Chin YE, Shi Y, Siebenlist U, Zhang X. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFkappaB and STAT3 in colorectal cancer cells. Gastroenterology. 2014;147:847–859. e811. doi: 10.1053/j.gastro.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao RF, Shu YJ, Hu YP, Wang XA, Zhang F, Liang HB, Ye YY, Li HF, Xiang SS, Weng H, Cao Y, Wu XS, Li ML, Wu WG, Zhang YJ, Jiang L, Dong Q, Liu YB. miR-101 targeting ZFX suppresses tumor proliferation and metastasis by regulating the MAPK/Erk and Smad pathways in gallbladder carcinoma. Oncotarget. 2016;7:22339–22354. doi: 10.18632/oncotarget.7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu W, Zhang Y, Zhou L, Wang X, Mu J, Jiang L, Hu Y, Dong P, Liu Y. miR-122 inhibits cancer cell malignancy by targeting PKM2 in gallbladder carcinoma. Tumour Biol. 2015 doi: 10.1007/s13277-015-4308-z. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.Ma F, Zhang M, Gong W, Weng M, Quan Z. MiR-138 Suppresses Cell Proliferation by Targeting Bag-1 in Gallbladder Carcinoma. PLoS One. 2015;10:e0126499. doi: 10.1371/journal.pone.0126499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu Y, Luo X, Kan T, Zhang Y, Yu W, Wei Y, Shen N, Yi B, Jiang X. TGF-beta upregulates miR-182 expression to promote gallbladder cancer metastasis by targeting CADM1. Mol Biosyst. 2014;10:679–685. doi: 10.1039/c3mb70479c. [DOI] [PubMed] [Google Scholar]
- 18.Kono H, Nakamura M, Ohtsuka T, Nagayoshi Y, Mori Y, Takahata S, Aishima S, Tanaka M. High expression of microRNA-155 is associated with the aggressive malignant behavior of gallbladder carcinoma. Oncol Rep. 2013;30:17–24. doi: 10.3892/or.2013.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letelier P, Garcia P, Leal P, Alvarez H, Ili C, Lopez J, Castillo J, Brebi P, Roa JC. miR-1 and miR-145 act as tumor suppressor microRNAs in gallbladder cancer. Int J Clin Exp Pathol. 2014;7:1849–1867. [PMC free article] [PubMed] [Google Scholar]
- 20.He B, Xiao J, Ren AJ, Zhang YF, Zhang H, Chen M, Xie B, Gao XG, Wang YW. Role of miR-1 and miR-133a in myocardial ischemic postconditioning. J Biomed Sci. 2011;18:22. doi: 10.1186/1423-0127-18-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Wang K, Chen J, Guo J, Yin Y, Cai X, Guo X, Wang G, Yang R, Zhu L, Zhang Y, Wang J, Xiang Y, Weng C, Zen K, Zhang J, Zhang CY. In vitro evidence suggests that miR-133a-mediated regulation of uncoupling protein 2 (UCP2) is an indispensable step in myogenic differentiation. J Biol Chem. 2009;284:5362–5369. doi: 10.1074/jbc.M807523200. [DOI] [PubMed] [Google Scholar]
- 22.Ge Y, Chen J. MicroRNAs in skeletal myogenesis. Cell Cycle. 2011;10:441–448. doi: 10.4161/cc.10.3.14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang LK, Hsiao TH, Hong TM, Chen HY, Kao SH, Wang WL, Yu SL, Lin CW, Yang PC. MicroRNA-133a suppresses multiple oncogenic membrane receptors and cell invasion in non-small cell lung carcinoma. PLoS One. 2014;9:e96765. doi: 10.1371/journal.pone.0096765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo J, Xia B, Meng F, Lou G. miR-133a suppresses ovarian cancer cell proliferation by directly targeting insulin-like growth factor 1 receptor. Tumour Biol. 2014;35:1557–1564. doi: 10.1007/s13277-013-1215-z. [DOI] [PubMed] [Google Scholar]
- 25.Cui W, Zhang S, Shan C, Zhou L, Zhou Z. microRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J. 2013;280:3962–3974. doi: 10.1111/febs.12398. [DOI] [PubMed] [Google Scholar]
- 26.Kawakami K, Enokida H, Chiyomaru T, Tatarano S, Yoshino H, Kagara I, Gotanda T, Tachiwada T, Nishiyama K, Nohata N, Seki N, Nakagawa M. The functional significance of miR-1 and miR-133a in renal cell carcinoma. Eur J Cancer. 2012;48:827–836. doi: 10.1016/j.ejca.2011.06.030. [DOI] [PubMed] [Google Scholar]
- 27.Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang Z, Zhu W, Shu Y, Liu P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588:1168–1177. doi: 10.1016/j.febslet.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 28.Lee EW, Lee MS, Camus S, Ghim J, Yang MR, Oh W, Ha NC, Lane DP, Song J. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 2009;28:2100–2113. doi: 10.1038/emboj.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 30.Alteri A, De Vito F, Messina G, Pompili M, Calconi A, Visca P, Mottolese M, Presutti C, Grossi M. Cyclin D1 is a major target of miR-206 in cell differentiation and transformation. Cell Cycle. 2013;12:3781–3790. doi: 10.4161/cc.26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64:2746–2762. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capaccione KM, Pine SR. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis. 2013;34:1420–1430. doi: 10.1093/carcin/bgt127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pannuti A, Foreman K, Rizzo P, Osipo C, Golde T, Osborne B, Miele L. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010;16:3141–3152. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon HA, Noh MH, Kim BG, Han JS, Jang JS, Choi SR, Jeong JS, Chun JH. Clinicopathological significance of altered Notch signaling in extrahepatic cholangiocarcinoma and gallbladder carcinoma. World J Gastroenterol. 2011;17:4023–4030. doi: 10.3748/wjg.v17.i35.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yong T, Sun A, Henry MD, Meyers S, Davis JN. Down regulation of CSL activity inhibits cell proliferation in prostate and breast cancer cells. J Cell Biochem. 2011;112:2340–2351. doi: 10.1002/jcb.23157. [DOI] [PubMed] [Google Scholar]