Abstract
Accumulating evidence suggests that aberrantly expressed microRNAs (miRNAs) contribute to the initiation and progression of human cancers. However, the underlying function of miR-193b in colorectal cancer (CRC) remains largely unexplored. Herein, we demonstrate that miR-193b is significantly down-regulated in CRC tissues compared with their normal counterparts. Kaplan-Meier analysis revealed that decreased miR-193b expression was closely associated with the shorter overall survival of patients with CRC. Through gain-and loss-of-function studies, we showed that miR-193b significantly suppressed CRC cell proliferation and invasion. In addition, bioinformatics analyses and luciferase reporter assays identified Stathmin 1 (STMN1) as the direct functional target of miR-193b in CRC. Furthermore, silencing of STMN1 resulted in a phenotype similar to that observed for overexpression of miR-193b, and restoration of STMN1 expression completely rescued the inhibitory effect of miR-193b in CRC cells. Taken together, our study implies the essential role of miR-193b in negatively regulating CRC progression, and a novel link between miR-193b and STMN1 in CRC.
Keywords: Colorectal cancer, miR-193b, STMN1, prognosis
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide [1]. Over the years, the incidence of CRC has become the fifth most common among cancer-related deaths in China [2]. Although patients with early-stage colorectal cancer can be cured by surgery, more than half of patients are diagnosed at advanced stages and endure a high risk of tumor recurrence [3,4]. Therefore, it is vital to clarify the molecular mechanism of colorectal cancer development to reveal novel diagnostic markers and therapeutic targets.
microRNAs (miRNAs) are a small class of nucleic acids that function in the transcriptional and post-transcriptional regulation of gene expression [5]. Deregulation of miRNAs is implicated in the initiation and progression of many diseases [6-8]. Growing evidence suggests that some miRNAs, similar to protein-coding genes, may mediate oncogenic or tumor-suppressing effects and could become a new class of cancer biomarkers and therapeutic targets [9-11]. A recent study showed that miR-193b is significantly downregulated in many types of human malignancies. For example, in ovarian cancer, miR-193b leads to a more aggressive, invasive phenotype and acts as an independent predictor of outcome [12,13]. In prostate cancer, miR-193b regulates tumor progression through targeting cyclin D1, which results in the inhibition of cell proliferation [14]. In esophageal cancer, miR-193b can induce autophagic flux and non-apoptotic cell death [15]. However, the possible functions and underlying mechanisms of miR-193b in CRC have not been reported.
In the current study, we revealed significant downregulation of miR-193b in CRC tissues. Overexpression of miR-193b suppressed cell proliferation and invasion. Inversely, knockdown of miR-193b significantly facilitated the malignant phenotype of CRC cells. Furthermore, we identified Stathmin 1 (STMN1) as a direct and functional target of miR-193b. Therefore, our results suggest that miR-193b may play a crucial role in the development and progression of CRC by targeting STMN1.
Materials and methods
Clinical samples
A total of 106 pairs of CRC tumor and matched, non-tumor tissues were collected at the Department of Gastrointestinal Surgery, Ren Ji Hospital between January 2010 and December 2014. None of the patients had received neoadjuvant chemotherapy before their operation. The presence of tumor in the tissue core was confirmed by H&E staining. The follow-up time was calculated from the date of surgery to the date of death, or the last known follow-up. All patients were well informed and the process was approved by the Ethics Committee of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, and written informed consent was obtained from each patient.
Cell culture and transfection
Human CRC cell lines (SW1116, SW480, HCT116, and HT29) and the normal colonic epithelial cell line (NCM460) were purchased from American Type Culture Collection. All of the cells lines were cultured in specific medium supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% antibiotics at 37°C in a humidified incubator under a 5% CO2 condition.
The transfections were performed using Lipofectamine 2000 (Invitrogen, USA), according to the manufacturer’s instructions. The miR-193b mimics, miR-193b inhibitor, and miRNA control were obtained from GenePharma Technology (Shanghai, China). Small interfering RNAs (siRNAs) targeting STMN1 and a negative control were also obtained from GenePharma Technology (Shanghai, China). The transfection was performed according to the manufacturer’s protocol.
Total RNA extraction and real-time quantitative PCR
Total RNA was extracted from tissues and cells using TRIzol reagent (Takara, Japan). The cDNA was synthesized using a microRNA Reverse Transcription Kit (Promega, USA) or a PrimeScript RT-PCR kit (Takara, Japan). RT-qPCR was performed using the StepOne Real-Time PCR System (Applied Biosystems, USA). The primers used in this study are shown in Table 1. The relative expression levels were determined by normalizing each Ct value to U6 or GAPDH Ct values, and the data were analyzed according to the 2-ΔΔCt formula.
Table 1.
Quantitative Real-time PCR primers used in this study
| Gene name | Primer sequence (5’ to 3’) |
|---|---|
| MiR-193b | Forward: CGGGGTTTTGAGGGCGAGAT |
| Reverse: ATGACCCCAAAAGCGGGACT | |
| STMN1 | Forward: GTACTTCTGGACTCACGGGC |
| Reverse: AAGGCAAGAGTGGTCTGCTC | |
| CADM1 | Forward: AGGCAAATCGGAGGTGGAAG |
| Reverse: ACTGTCCAGTTCTTCTGCGG | |
| IGFBP5 | Forward: GCCCTCCACCTCTCTCTACA |
| Reverse: TCGCGGTAGCTCTTTTCGTT | |
| U6 | Forward: CTCGCTTCGGCAGCACA |
| Reverse: AACGCTTCACGAATTTGCGT | |
| GAPDH | Forward: TGAAGGTCGGAGTCAACGGA |
| Reverse: CCTGGAAGATGGTGATGGGAT |
Cell proliferation assay
Following different treatments, CRC cells were seeded into a 96-well cell culture plate at a density of 3,000 cells/well, and grown at 37°C overnight. The cell viability was quantified by Cell Counting Kit-8 (CCK-8, Japan). Briefly, on the day that the growth rate of treated cells was measured, 100 μl of spent medium was replaced with an equal volume of fresh medium containing 10% CCK-8. Then, the cells continued to be incubated at 37°C for 1 h, and the absorbance was detected at 450 nm with a microplate reader.
Cell invasion assay
The invasive ability of CRC cells was detected by Transwell model (Corning, USA), according to the manufacturer’s instructions. Briefly, 5×104 cells suspended in serum-free medium were plated on the top of each chamber, while medium containing 20% FBS was put into the lower chamber. After incubating for 48 h, the chambers were disassembled. The non-invaded cells that remained on the upper chamber were removed, and the membranes were stained with a 2% crystal violet solution for 30 min and placed on a glass slide. Then, the cells that migrated across the membrane were counted in five random visual fields using a light microscope.
Western blotting
All proteins were resolved on a 10% SDS-PAGE, and then transferred onto a PVDF membrane. The membranes were incubated with blocking buffer for 90 min at room temperature, and then incubated with an antibody against STMN1 (1:1000, Abcam, UK) or β-actin (1:1000, Abcam, UK) overnight at 4°C. The membranes were washed and incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody. Protein expression was assessed by enhanced chemiluminescence and exposure to chemiluminescent film. The LabWorks image acquisition and analysis software was used to quantify the band intensities.
Luciferase reporter assays
The wild-type STMN1 3’UTR was cloned into the pMIR-REPORT luciferase vector (Genearray Biotechnology, Shanghai, China). The mutant STMN1 3’UTR was generated based on the pMIR-STMN1-3’UTR by mutating 3 nucleotides that are recognized by miR-193b. The reporter plasmid was transiently transfected into HT29 and SW1116 cells in the presence of either miR-193b or control mimics. After 48 h, the cells were harvested and lysed. The luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, USA), and Renilla-luciferase was used for normalization.
Tumorigenesis in nude mice
Xenograft tumors were generated by subcutaneous injection of 4×106 cells into the hind limbs of each 4-to 6-week-old BALB/C athymic nude mouse (nu/nu). The mice were obtained from the Animal Center of East China Normal University, Shanghai, China. All of the mice were housed and maintained under specific pathogen-free conditions, and used in accordance with institutional guidelines approved by the Use Committee for Animal Care. When the average tumor size reached 100 mm3, the mice were randomly separated into 2 groups, one with subcutaneous injection of miR-193b (Agomir) at different sites, and the other with the control (Agomir). Injections were performed twice a week. All of the mice were euthanized 16 days after the initial injection, and the tumors were excised.
Statistical analyses
The data were expressed as the mean ± SEM of at least three independent experiments. All of the statistical analyses were performed using the SPSS 19.0 software. The overall survival rate was calculated according to the Kaplan-Meier method, and the differences in the survival curves were evaluated by the log-rank test. Student’s t-test was used to analyze differences between two groups, and one-way ANOVA was used to determine the significant differences among multiple groups. P values less than 0.05 were considered statistically significant.
Results
miR-193b expression is significantly upregulated in CRC
To elucidate the expression pattern of miR-193b in CRC, we first detected the expression level of miR-193b in 106 matched CRC tumor and non-tumor tissues by quantitative reverse transcription PCR (RT-qPCR). The results showed that the expression level of miR-193b was significantly reduced in colorectal cancer tissues compared with their corresponding normal counterparts (Figure 1A). Furthermore, we measured the expression level of miR-193b in 5 CRC cell lines and the normal colonic epithelial cell line, NCM460 (Figure 1D). The highest miR-193b level was detected in the NCM460 cells. These results indicated that miR-193b expression was significantly reduced in colorectal cancer.
Figure 1.
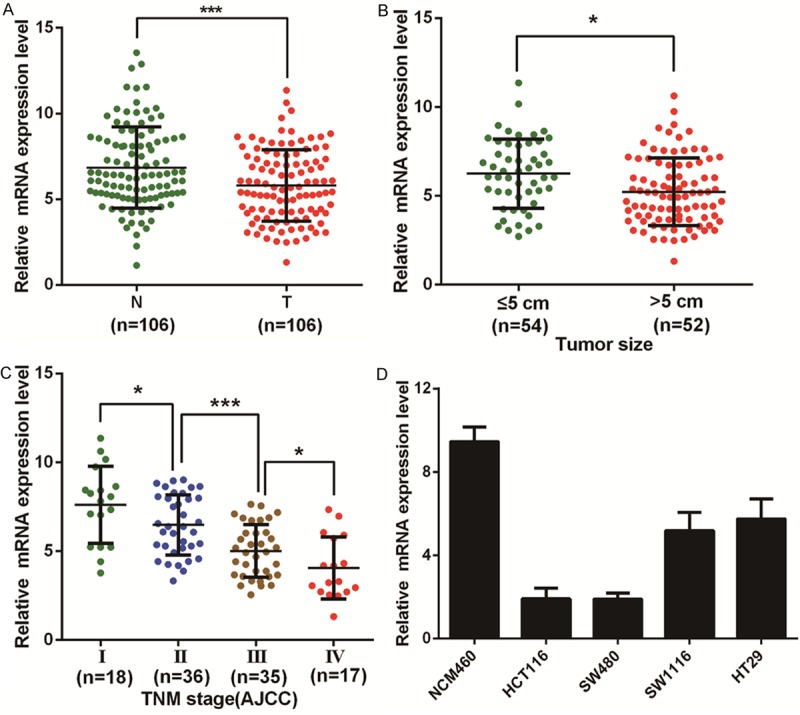
Expression of miR-193b in CRC tissues and cell lines. (A) The transcription level of miR-193b in 106 matched CRC tissues (T) and adjacent normal tissues (N) as measured by RT-qPCR. B, C. The relationship between miR-193b and clinicopathological characteristics, such as tumor size (B) and TNM stage (C). (D) Histograms of the transcription level of miR-193b in CRC cell lines and normal colonic epithelial cells (NCM460). The results are shown as the mean ± SEM (*P<0.05, ***P<0.001) of triplicate determination from three independent experiments.
Elevated miR-193b expression predicts a poor prognosis in patients with CRC
To determine the clinical significance of miR-193b, we next analyzed the association between miR-193b expression and the clinicopathological characteristics of CRC. As shown in Table 2, the level of miR-193b was negatively associated with tumor size (P=0.020), carcinoembryonic antigen (CEA) level (P=0.006), tumor (T) classification (P=0.020), lymph node metastasis (P=0.003), distant metastasis (P=0.038) and tumor node metastasis (TNM) stage (P=0.015). However, no significant associations were found between miR-193b expression and other clinical features, including age, gender, and tumor location.
Table 2.
Correlations between miR-193b expression and clinicopathologic features in 106 colorectal cancer patients
| Clinicopathological feature | Expression of miR-193b | |||
|---|---|---|---|---|
|
|
||||
| Total 106 | Low (n=53, 50%) | High (n=53, 50%) | P value (χ2 test) | |
| Age(years) | ||||
| <65 | 46 | 22 (47.83) | 24 (52.17) | 0.695 |
| ≥65 | 60 | 31 (51.67) | 29 (48.33) | |
| Gender | ||||
| Male | 55 | 29 (52.73) | 26 (47.27) | 0.560 |
| Female | 51 | 24 (47.06) | 27 (52.94) | |
| Tumor location | ||||
| Rectum | 58 | 27 (46.55) | 31 (53.45) | 0.435 |
| Colon | 48 | 26 (54.17) | 22 (45.83) | |
| Tumor size | ||||
| ≤5 cm | 54 | 21 (38.89) | 33 (61.11) | 0.020 |
| >5 cm | 52 | 32 (61.54) | 20 (38.46) | |
| CEA level | ||||
| ≤5 ng/ml | 62 | 25 (40.32) | 37 (59.68) | 0.006 |
| >5 ng/ml | 44 | 28 (66.67) | 16 (33.33) | |
| T classificattion | ||||
| T1-2 | 24 | 7 (29.17) | 17 (70.83) | 0.020 |
| T3-4 | 82 | 46 (56.10) | 36 (43.90) | |
| Lymph node metastasis | ||||
| Absent | 57 | 21 (36.84) | 36 (63.16) | 0.003 |
| Present | 49 | 32 (65.31) | 17 (34.69) | |
| Distant metastasis | ||||
| Absent | 88 | 40 (45.45) | 48 (54.55) | 0.038 |
| Present | 18 | 13 (72.72) | 5 (27.78) | |
| TNM stage(AJCC) | ||||
| Stage I | 17 | 5 (29.41) | 12 (70.59) | 0.015 |
| Stage II | 35 | 13 (37.14) | 22 (62.86) | |
| Stage III | 36 | 22 (61.11) | 14 (38.89) | |
| Stage IV | 18 | 13 (72.22) | 5 (27.78) | |
Values in parentheses indicate percentage values. The bold number represents the P-values with significant differences.
To determine the prognostic value of miR-193b for CRC, the relationship between miR-193b expression and the clinical follow-up data were analyzed using Kaplan-Meier survival curves and the log-rank test. The results revealed that high expression of miR-193b was inversely associated with overall survival (OS) (n=106, P=0.001, Figure 2), which indicates that OS is poorer in CRC patients with high miR-193b expression than in those with low miR-193b expression.
Figure 2.
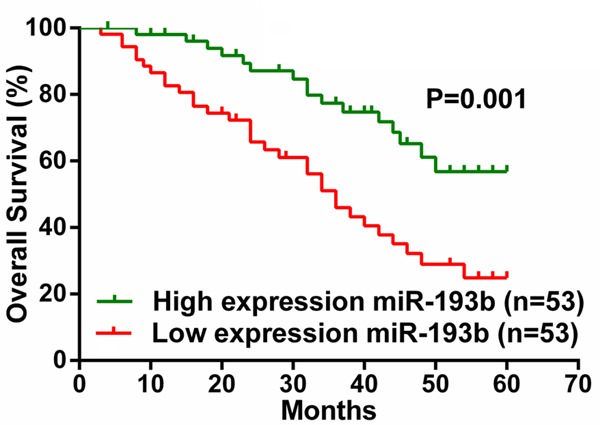
Comparison of overall survival in CRC patients with low or high miR-193b expression.
To directly identify the risk factors associated with OS in CRC patients, univariate and multivariate analyses were performed to confirm that miR-193b represents an independent risk factor for poor prognosis. Univariate Cox regression analysis showed that the miR-193b expression level, CEA level, and TNM stage were significantly associated with OS (Table 3). Furthermore, multivariate Cox regression analysis confirmed that the miR-193b expression level and TNM stage were independent predictors of OS in patients with CRC (Table 3). These data indicated that high expression of miR-193b may be a predictor for the diagnosis and prognosis of colorectal cancer patients.
Table 3.
Univariate and multivariate analyses of prognostic parameters for survival in 106 colorectal cancer patients
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Prognostic parameter | HR | 95% CI | P value | HR | 95% CI | P value |
| Expression of miR-193b (low vs. high) | 0.376 | 0.206-0.687 | 0.001 | 0.471 | 0.252-0.882 | 0.018 |
| Age (<65 vs. ≥65) | 1.583 | 0.868-2.887 | 0.134 | - | - | - |
| Gender (male vs. female) | 1.220 | 0.692-2.152 | 0.491 | - | - | - |
| Tumor Size (≤5 cm vs. >5 cm) | 1.504 | 0.841-2.691 | 0.169 | - | - | - |
| CEA level (≤5 ng/ml vs. >5 ng/ml) | 2.008 | 1.137-3.547 | 0.016 | 1.084 | 0.595-1.976 | 0.792 |
| Tumor location (rectum vs. colon) | 1.048 | 0.587-1.871 | 0.875 | - | - | - |
| TNM stage (I vs. II vs. III vs. IV) | 3.235 | 2.166-4.830 | 0.000 | 3.157 | 2.051-4.861 | 0.000 |
HR: Hazard ratio; CI: Confidence interval. The bold number represents the P-values with significant differences.
miR-193b inhibits CRC cell proliferation and invasion
To investigate the function of miR-193b in CRC, we transiently transfected the miR-193b mimics or miR-193b inhibitor into CRC cells and measured cellular functions (Figure 3A, 3E). We observed that the miR-193b mimic significantly inhibited the proliferation and invasion of HT29 and SW1116 cells compared with control (Figure 3B-D). However, in HCT116 and SW480, cells treated with the miR-193b inhibitor had a significantly higher proliferative and invasive capacity than the negative control (Figure 3F-H). These results showed that miR-193b promoted proliferation and invasion in colorectal cancer cells.
Figure 3.
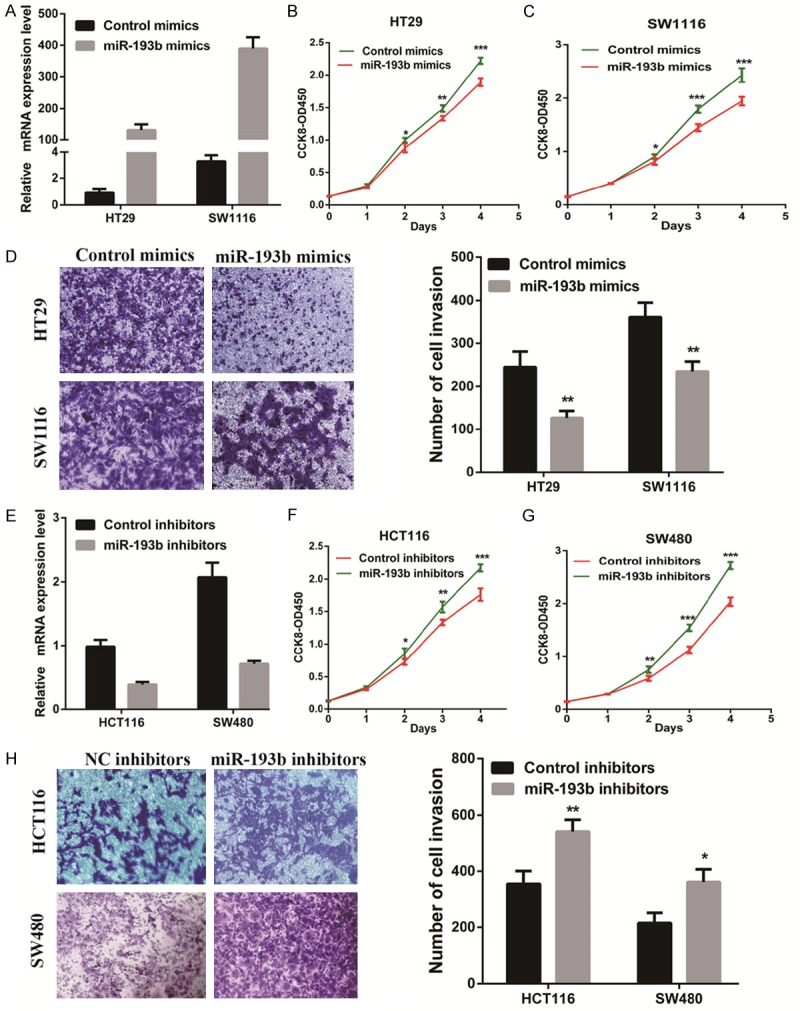
miR-193b inhibits CRC cell proliferation and invasion. (A) miR-193b knockdown efficiency was confirmed by RT-qPCR in CRC cells. (B, D) The effect of miR-193b knockdown on cell proliferation (B, C) or invasion (D) was evaluated by either the CCK-8 assay or Transwell assay, respectively. (E) miR-193b overexpression efficiency was confirmed by RT-qPCR in CRC cells. (F-H) The effect of miR-193b overexpression on cell proliferation (F, G) or invasion (H) was measured by either the CCK-8 assay or Transwell assay, respectively. The results are shown as the mean ± SEM (*P<0.05, **P<0.01, ***P<0.001) of triplicate determination from three independent experiments.
miR-193b inhibited tumor growth in vivo
To further confirm the role of miR-193b in tumor growth, we performed a subcutaneous tumor transplantation experiment. We found that the tumor xenograft volume and weight in nude mice treated with miR-193b were smaller than that in the mock-treated mice (Figure 4A, 4B). Furthermore, the tumor xenograft growth in the miR-193b-treated nude mice was slower than that in the mock group (Figure 4C).
Figure 4.
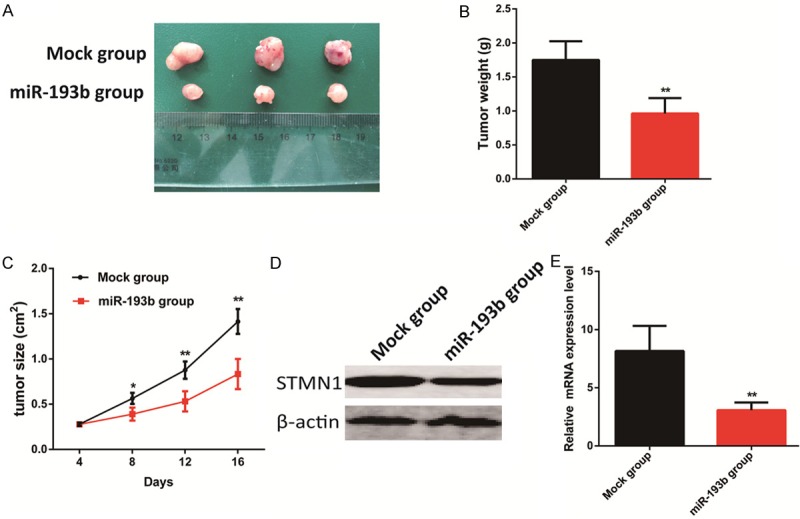
miR-193b inhibited tumor growth in vivo. (A, B) Tumor xenograft volume (A) and weight (B) in miR-193b-treated nude mice were smaller than that in the mock group. (C) Tumor xenograft growth in miR-193b-treated nude mice was slower than that in the mock group. (D, E) STMN1 expression in the tumor xenografts of miR-193b-treated nude mice was decreased compared with that in the mock-treated nude mice. The results are shown as the mean ± SEM (*P<0.05, **P<0.01).
STMN1 is a direct target of miR-193b
To explore the mechanism by which miR-193b affects the biological functions of CRC cells, we investigated the potential gene targets of miR-193b using target prediction programs including, Miranda (http://www.microrna.org/microrna/home.do) [16], TargetScan (http://www.targetscan.org/) [17], PicTar (http://pictar.mdc-berlin.de/) [18], and PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html) [19]. Our analyses revealed that STMN1, CADM1, and IGFBP5 are three potential targets of miR-193b. Then, we detected the mRNA expression of STMN1, CADM1, and IGFBP5 in the presence of the miR-193b mimic or miR-193b inhibitor. The results showed that STMN1 mRNA and protein expression were significantly reduced by treatment with the miR-193b mimic, but remarkably increased by treatment with the miR-193b inhibitor (Figure 5A-C). Furthermore, we found decreased STMN1 expression in the tumor xenografts of the miR-193b-treated nude mice compared with that in the mock-treated nude mice (Figure 4D, 4E).
Figure 5.
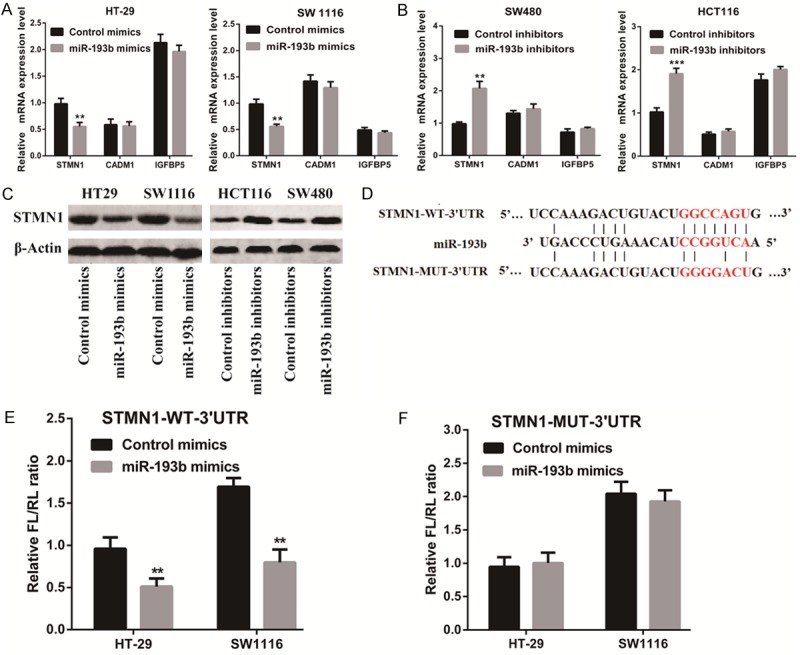
STMN1 is a direct target of miR-193b. (A, B) The mRNA levels of STMN1, CADM1, and IGFBP5 were detected in the presence of the miR-193b mimic (A) or miR-193b inhibitor (B). (C) The protein level of STMN1 was detected in the presence of the miR-193b mimic or miR-193b inhibitor by Western blotting. (D) The predicted miR-193b binding sites in the 3’-UTR of STMN1 were detected using bioinformatics prediction tools. The mutated site in the 3’-UTR of STMN1 is also shown. (E, F) CRC cells were co-transfected with either the miR-193b expression vector or empty vector, and either the STMN1 3’-UTR reporter plasmid (E) or its mutant form (F). The luciferase activity was detected 48 h after transfection. The results are shown as the mean ± SEM (**P<0.01, ***P<0.001) of triplicate determination from three independent experiments.
To verify whether STMN1 is a direct target of miR-193b, a human STMN1 3’ untranslated region (3’UTR) fragment containing the wild-type or mutant miR-193b-binding site was inserted downstream of the luciferase open reading frame (Figure 5D). The results showed that the relative luciferase activity of the reporter containing the wild-type STMN1 3’-UTR was markedly decreased upon miR-193b co-transfection, whereas the luciferase activity of the reporter containing the mutant binding site was unaffected (Figure 5E, 5F). These results strongly suggest that STMN1 is a direct target of miR-193b in CRC cells.
Silencing of STMN1 compromises the tumor progression of CRC in vitro
To reveal the role of STMN1 in CRC, we silenced STMN1 in HT29 and SW1116 cells with STMN1 siRNA and confirmed the transfection efficiency by Western blotting (Figure 6A). Consistent with the functions of STMN1 in endometrial carcinoma [20] and bladder carcinoma [21,22], silencing of STMN1 decreased CRC cell proliferation and invasion (Figure 6B-D).
Figure 6.
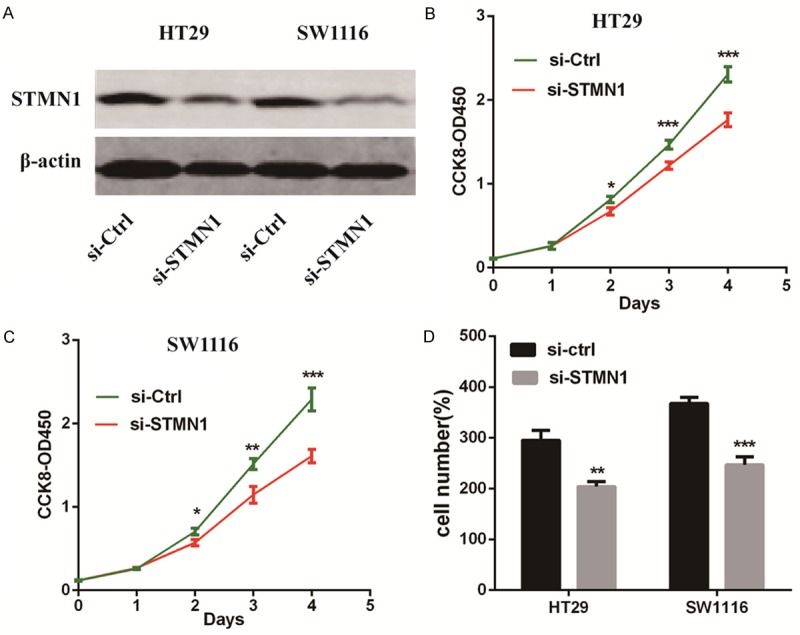
Silencing of STMN1 compromises CRC tumor progression in vitro. A. The efficacy of RNA interference in HT29 and SW1116 cells was verified by Western blotting. B and C. Silencing of STMN1 inhibited CRC cell proliferation as detected by CCK-8 assays. D. Silencing of STMN1 significantly decreased the invasive ability of CRC cells as indicated by Transwell assays. The results are shown as the mean ± SEM (*P<0.05, **P<0.01, ***P<0.001) of triplicate determination from three independent experiments.
Restoration of STMN1 abolishes the tumor suppressor role of miR-193b
To further confirm whether the tumor-suppressive role of miR-193b was mediated by STMN1, a gain-of-function study was performed. Firstly, we transfected HT29 and SW1116 cells with either control or miR-193b mimics along with either pcDNA3.1 or pcDNA3.1-STMN1 to detect the protein level of STMN1 (Figure 7A). Then, we performed CCK-8 assays and Transwell assays. We found that overexpression of STMN1 completely abolished the effects of miR-193b on cell proliferation and invasion in HT29 and SW1116 cells (Figure 7B, 7C).
Figure 7.

Restoration of STMN1 abolishes the tumor suppressor role of miR-193b. A. The protein level of STMN1 was measured in HT29 and SW1116 cells transfected with either the control or miR-193b mimics along with either pcDNA3.1 or pcDNA3.1-STMN1. B. The effect of the miR-193b mimic on the proliferative potential of HT29 and SW1116 cells was detected in the presence of pcDNA3.1-STMN1 treatment via CCK-8 assays. C. The effect of the miR-193b mimic on the invasive potential of HT29 and SW1116 cells was detected in the presence of pcDNA3.1-STMN1 treatment via Transwell assays. The results are shown as the mean ± SEM (**P<0.01, ***P<0.001) of triplicate determination from three independent experiments.
Discussion
Few advances have been made in the treatment for CRC over the past decade [2]. The discovery of miRNAs provides a novel insight into understanding the molecular mechanisms of and treatment for different cancers. The role of miRNAs in various cancers has been attracting more attention [23,24]. In this study, we demonstrated that the expression of miR-193b in CRC tissues was significantly downregulated compared with corresponding normal controls. Kaplan-Meier survival analysis showed that patients displaying a low miR-193b expression level exhibited a significantly shorter survival duration than those displaying a high miR-193b expression level.
Accumulating evidence has suggested that miR-193b acts as a tumor suppressor in various types of cancer, such as breast, gastric, cervical, and prostate cancers [25-28]. For instance, the expression level of miR-193b was significantly downregulated in endometrioid adenocarcinoma [29]. Chen et al. found that miR-193b was significantly downregulated in melanoma tissues, and overexpression of miR-193b in melanoma cell lines repressed cell proliferation [30]. Consistent with this observation in melanoma, we also demonstrated that miR-193b acts as a tumor suppressor in CRC cell proliferation and invasion. In addition, we found that the tumor sizes in the miR-193b-treated BALB/c mice were significantly smaller than that in the control group.
It is generally accepted that miRNAs exert their biological effects through regulating the expression of their downstream target genes [31]. In recent years, many studies have focused on the role of miRNAs in cellular growth, differentiation, and metastasis of cancer cells through interactions between the miRNAs and their target genes [9,14]. Studies show that STMN1 participates in the process of miR-193b-related regulation [32,33]. Consistent with studies in pancreatic cancer and melanoma, our research also identified STMN1 as a target of miR-193b in colorectal cancer. Four lines of evidence supported our finding. First, bioinformatics prediction tools identified a miR-193b binding site in the 3’-UTR of STMN1. Second, the miR-193b mimic reduced the protein and mRNA levels of STMN1, while the miR-193b inhibitor promoted the expression of STMN1 in CRC cells. Third, STMN1 3’-UTR-mediated luciferase activity was specifically responsive to transfection of the miR-193b mimic. Finally, restoration of STMN1 completely abrogated the tumor suppressor role of miR-193b. However, a specific miRNA has the potential to target multiple genes. miR-193b may perform its function through cooperative regulation of its other target genes, such as cyclin D1, MCL-1, and Rab22A [14,34,35]. In the future, other targets of miR-193b, besides STMN1, need to be identified in colorectal cancer.
STMN1, also known as oncoprotein 18 (Op 18), is a 19-kDa cytosolic protein that destabilizes microtubules in a phosphorylation-dependent manner [36,37]. STMN1 expression has been reported in various human cancers, such as breast and lung cancer [25,38]. In CRC, STMN1 promotes proliferation and migrationand is correlated with a more advanced pathological grade and poor overall survival [39-41]. These studies suggest that STMN1 may play a critical role in the progress of colorectal cancer, although the specific molecular mechanism is unclear. Schimmack et al. found that STMN1 could regulate phosphoinositide 3-kinase (PI3K) signaling in pancreatic cancer [42]. Therefore, the downstream signaling pathways of STMN1 in CRC require further investigation.
In summary, the present study describes the essential role of miR-193b in negatively regulating CRC progression and suggests a novel link between miR-193b and STMN1 in CRC. The miR-193b/STMN1 axis provides a novel insight into the pathogenesis of CRC, and might represent a potential therapeutic target for the treatment of CRC.
Acknowledgements
This work was supported by the grant from Municipal Commission of Health and Family Planning of Shanghai, China (no. 201540031 to Ming Zhong).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Eefsen RL, Vermeulen PB, Christensen IJ, Laerum OD, Mogensen MB, Rolff HC, Van den Eynden GG, Hoyer-Hansen G, Osterlind K, Vainer B, Illemann M. Growth pattern of colorectal liver metastasis as a marker of recurrence risk. Clin Exp Metastasis. 2015;32:369–381. doi: 10.1007/s10585-015-9715-4. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Huang J, Lyu H, Wang J, Liu B. MicroRNA regulation and therapeutic targeting of survivin in cancer. Am J Cancer Res. 2015;5:20–31. [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou S, Ye W, Ren J, Shao Q, Qi Y, Liang J, Zhang M. MicroRNA-381 increases radiosensitivity in esophageal squamous cell carcinoma. Am J Cancer Res. 2015;5:267–277. [PMC free article] [PubMed] [Google Scholar]
- 7.Annema W, von Eckardstein A. Dysfunctional high-density lipoproteins in coronary heart disease: implications for diagnostics and therapy. Transl Res. 2016;173:30–57. doi: 10.1016/j.trsl.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Rotllan N, Price N, Pati P, Goedeke L, Fernandez-Hernando C. microRNAs in lipoprotein metabolism and cardiometabolic disorders. Atherosclerosis. 2016;246:352–360. doi: 10.1016/j.atherosclerosis.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, Li M, Chen X, Zhang D, Wei L, Zhang Z, Wang S, Meng L, Zhu S, Li B. MicroRNA-373 promotes migration and invasion in human esophageal squamous cell carcinoma by inhibiting TIMP3 expression. Am J Cancer Res. 2016;6:1–14. [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, Yang Z, Wang H, Cao Z, Zhao Y, Gong C, Ma L, Wang X, Hu X, Chen S. MicroRNA-320a inhibits proliferation and invasion of breast cancer cells by targeting RAB11A. Am J Cancer Res. 2015;5:2719–2729. [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J. Considering Exosomal miR-21 as a Biomarker for Cancer. J Clin Med. 2016;5 doi: 10.3390/jcm5040042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitra AK, Chiang CY, Tiwari P, Tomar S, Watters KM, Peter ME, Lengyel E. Microenvironment-induced downregulation of miR-193b drives ovarian cancer metastasis. Oncogene. 2015;34:5923–5932. doi: 10.1038/onc.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Xu Y, Qiu W, Zhao D, Zhang Y. Tissue miR-193b as a Novel Biomarker for Patients with Ovarian Cancer. Med Sci Monit. 2015;21:3929–3934. doi: 10.12659/MSM.895407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaukoniemi KM, Rauhala HE, Scaravilli M, Latonen L, Annala M, Vessella RL, Nykter M, Tammela TL, Visakorpi T. Epigenetically altered miR-193b targets cyclin D1 in prostate cancer. Cancer Med. 2015;4:1417–1425. doi: 10.1002/cam4.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyhan MJ, O’Donovan TR, Boersma AW, Wiemer EA, McKenna SL. MiR-193b promotes autophagy and non-apoptotic cell death in oesophageal cancer cells. BMC Cancer. 2016;16:101. doi: 10.1186/s12885-016-2123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 18.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 19.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He X, Liao Y, Lu W, Xu G, Tong H, Ke J, Wan X. Elevated STMN1 promotes tumor growth and invasion in endometrial carcinoma. Tumour Biol. 2016;37:9951–8. doi: 10.1007/s13277-016-4869-5. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Cao J, Zhao X. miR-221 facilitates the TGFbeta1-induced epithelial-mesenchymal transition in human bladder cancer cells by targeting STMN1. BMC Urol. 2015;15:36. doi: 10.1186/s12894-015-0028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemdan T, Linden M, Lind SB, Namuduri AV, Sjostedt E, de Stahl TD, Asplund A, Malmstrom PU, Segersten U. The prognostic value and therapeutic target role of stathmin-1 in urinary bladder cancer. Br J Cancer. 2014;111:1180–1187. doi: 10.1038/bjc.2014.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoof CR, Botelho EL, Izzotti A, Vasques Ldos R. MicroRNAs in cancer treatment and prognosis. Am J Cancer Res. 2012;2:414–433. [PMC free article] [PubMed] [Google Scholar]
- 24.Hemmatzadeh M, Mohammadi H, Jadidi-Niaragh F, Asghari F, Yousefi M. The role of oncomirs in the pathogenesis and treatment of breast cancer. Biomed Pharmacother. 2016;78:129–139. doi: 10.1016/j.biopha.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 25.Kuang XY, Chen L, Zhang ZJ, Liu YR, Zheng YZ, Ling H, Qiao F, Li S, Hu X, Shao ZM. Stathmin and phospho-stathmin protein signature is associated with survival outcomes of breast cancer patients. Oncotarget. 2015;6:22227–22238. doi: 10.18632/oncotarget.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li SC, Ma R, Wu JZ, Xiao X, Wu W, Li G, Chen B, Sharma A, Bai S, Dun BY, She JX, Tang JH. Delineation of gastric cancer subtypes by co-regulated expression of receptor tyrosine kinases and chemosensitivity genes. Am J Transl Res. 2015;7:1429–1439. [PMC free article] [PubMed] [Google Scholar]
- 27.Howitt BE, Nucci MR, Drapkin R, Crum CP, Hirsch MS. Stathmin-1 expression as a complement to p16 helps identify high-grade cervical intraepithelial neoplasia with increased specificity. Am J Surg Pathol. 2013;37:89–97. doi: 10.1097/PAS.0b013e3182753f5a. [DOI] [PubMed] [Google Scholar]
- 28.Jeon TY, Han ME, Lee YW, Lee YS, Kim GH, Song GA, Hur GY, Kim JY, Kim HJ, Yoon S, Baek SY, Kim BS, Kim JB, Oh SO. Overexpression of stathmin1 in the diffuse type of gastric cancer and its roles in proliferation and migration of gastric cancer cells. Br J Cancer. 2010;102:710–718. doi: 10.1038/sj.bjc.6605537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu W, Lin Z, Zhuang Z, Liang X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur J Cancer Prev. 2009;18:50–55. doi: 10.1097/CEJ.0b013e328305a07a. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–2529. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Kong F, Wu K, Song K, He J, Sun W. miR-193b directly targets STMN1 and uPA genes and suppresses tumor growth and metastasis in pancreatic cancer. Mol Med Rep. 2014;10:2613–2620. doi: 10.3892/mmr.2014.2558. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Abi-Daoud M, Wang A, Yang X, Zhang X, Feilotter HE, Tron VA. Stathmin 1 is a potential novel oncogene in melanoma. Oncogene. 2013;32:1330–1337. doi: 10.1038/onc.2012.141. [DOI] [PubMed] [Google Scholar]
- 34.Long J, Ji Z, Jiang K, Wang Z, Meng G. miR-193b Modulates Resistance to Doxorubicin in Human Breast Cancer Cells by Downregulating MCL-1. Biomed Res Int. 2015;2015:373574. doi: 10.1155/2015/373574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, He M, Wang K, Sun G, Tang L, Xu Z. Tumor suppressive microRNA-193b promotes breast cancer progression via targeting DNAJC13 and RAB22A. Int J Clin Exp Pathol. 2014;7:7563–7570. [PMC free article] [PubMed] [Google Scholar]
- 36.Steinmetz MO. Structure and thermodynamics of the tubulin-stathmin interaction. J Struct Biol. 2007;158:137–147. doi: 10.1016/j.jsb.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Rubin CI, Atweh GF. The role of stathmin in the regulation of the cell cycle. J Cell Biochem. 2004;93:242–250. doi: 10.1002/jcb.20187. [DOI] [PubMed] [Google Scholar]
- 38.Nie W, Xu MD, Gan L, Huang H, Xiu Q, Li B. Overexpression of stathmin 1 is a poor prognostic biomarker in non-small cell lung cancer. Lab Invest. 2015;95:56–64. doi: 10.1038/labinvest.2014.124. [DOI] [PubMed] [Google Scholar]
- 39.Tan HT, Wu W, Ng YZ, Zhang X, Yan B, Ong CW, Tan S, Salto-Tellez M, Hooi SC, Chung MC. Proteomic analysis of colorectal cancer metastasis: stathmin-1 revealed as a player in cancer cell migration and prognostic marker. J Proteome Res. 2012;11:1433–1445. doi: 10.1021/pr2010956. [DOI] [PubMed] [Google Scholar]
- 40.Ogino S, Nosho K, Baba Y, Kure S, Shima K, Irahara N, Toyoda S, Chen L, Kirkner GJ, Wolpin BM, Chan AT, Giovannucci EL, Fuchs CS. A cohort study of STMN1 expression in colorectal cancer: body mass index and prognosis. Am J Gastroenterol. 2009;104:2047–2056. doi: 10.1038/ajg.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng P, Liu YX, Chen L, Liu XH, Xiao ZQ, Zhao L, Li GQ, Zhou J, Ding YQ, Li JM. Stathmin, a new target of PRL-3 identified by proteomic methods, plays a key role in progression and metastasis of colorectal cancer. J Proteome Res. 2010;9:4897–4905. doi: 10.1021/pr100712t. [DOI] [PubMed] [Google Scholar]
- 42.Schimmack S, Taylor A, Lawrence B, Schmitz-Winnenthal H, Fischer L, Buchler MW, Modlin IM, Kidd M, Tang LH. Stathmin in pancreatic neuroendocrine neoplasms: a marker of proliferation and PI3K signaling. Tumour Biol. 2015;36:399–408. doi: 10.1007/s13277-014-2629-y. [DOI] [PubMed] [Google Scholar]


