Abstract
Colorectal cancer (CRC) is a prevalent cancer with high mortality worldwide. This study was aimed to explore the functional effects of microRNA-195 (miR-195) on CRC cells and the underling mechanism involved. quantitative PCR (qPCR) was performed to monitor the expression of miR-195 in CRC tissues and cell lines. SW480 and SW620 cells were transfected with either miR-195 mimic or antisense oligonucleotides (ASO) of miR-195. Then cell viability, cell cycle and the expressions of CyclinB1, CyclinD1 and Cyclin-Dependent Kinase 2 (CDK2) were respectively detected by 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyltetrazolium bromide (MTT), flow cytometry, qPCR and Western blot. A target of miR-195 was predicted and verified in vitro by using TargetScan and microRNA database, Dual-Luciferase reporter assay, qPCR and Western blot. Further, the functions of the target on cell viability and cell cycle were detected by transfection with its expression vector. Moreover, the expressions of Wnt/β-catenin pathway proteins were detected by qPCR and Western blot. Results show that MiR-195 was decreased during CRC, and miR-195 overexpression inhibited cell viability, arrested cells in G2/M phase, and down-regulated CyclinB1, CyclinD1 and CDK2 (P < 0.05 or P < 0.01). Fibroblast growth factor 2 (FGF2) was a direct target of miR-195 and alleviated the inhibitive effects of miR-195 on cell viability and cell cycle progression (P < 0.05 or P < 0.01). Further, miR-195 specifically regulated Wnt/β-catenin pathway proteins (P < 0.01). All these findings suggest that miR-195 suppressed CRC cells proliferation via targeting FGF2 and blocking Wnt/β-catenin pathway.
Keywords: Colorectal cancer, miR-195, FGF2, proliferation, Wnt/β-catenin
Introduction
Colorectal cancer (CRC), also known as colon cancer, is the fourth most prevalent cancer and the leading cause of cancer mortality worldwide [1]. Main risk factors of CRC include increasing age, male gender, alcohol, red meat, obesity, insulin resistance, and metabolic syndrome [2]. In the past decade, remarkable advances have been made in the research for treating CRC. The development of new combinations of standard chemotherapy, as well as the introduction of new targeted therapies, largely improved the survival of CRC patients [3]. However, the 5-year relative survival for patients aged 65 years and older is remain unsatisfactory, as low as 60% [4]. Therefore, development of new therapy target for CRC treatment is still urgently needed.
MicroRNAs (miRNAs) are endogenous, evolutionarily conserved, and single chain noncoding small RNAs consisting of about 22 nucleotides, which inhibit mRNA transcription or induce mRNA degradation and shed new light on cancer research [5-7]. Nowadays, abnormal regulation of miRNAs expression has been found implicated in the development of CRC. Clinical investigations found that miR-92 was aberrantly in plasma of CRC patients and miR-92 was identified as a noninvasive molecular marker for CRC screening [8]. Preclinical studies showed that altered expressions of miR-21, miR-31, miR-143 and miR-145 were related to clinicopathologic features of CRC [9]. Furthermore, the anti-proliferative, chemosensitizer and pro-apoptotic roles for miR-143 have been found in CRC [10].
MiR-195, one of the miR-16/15/195/424/497 family members, has been shown to play a pivotal role in tumorigenesis, as a tumor suppressor [11,12]. In terms of CRC, several previous studies indicated that miR-195 was down-regulated in CRC tissues and cell lines [13,14]. Down-regulation of miR-195 has been shown to correlate with CRC cells proliferation, colony-formation and invasion as well as poor prognosis in this cancer [15]. However, the functional effects of miR-195 on CRC and its underling mechanism have not been exhaustively investigated. Therefore, in this study, human CRC cell lines SW480 and SW620 were employed and transfected with either miR-195 mimic or antisense oligonucleotides (ASO) of miR-195, then cell viability and cell cycle was assessed. Besides, a target of miR-195 was predicted and verified, and main factors in Wnt/β-catenin signaling pathway were detected to reveal the potential mechanisms. This study uncovered the important roles of miR-195 in CRC and might shed new light on the mechanism of miR-195 on this disease.
Materials and methods
Clinical samples
A total of 40 adults (25 males and 15 females; aged from 28-66 years) with CRC were enrolled in the current study, from November, 2012 to July, 2015. Tumor tissues and adjacent tissues (located < 3 cm away from the tumor) were obtained from these individuals who had undergone proctocolectomy with lymph node dissection for CRC, at the Affiliated Hospital of Qingdao University. All these samples were derived from initial surgery without either preoperative chemotherapy or radiotherapy. Tumor tissues and adjacent tissues were collected immediately after surgical removal and snap-frozen in liquid nitrogen until further use. This study was approved by our local Ethics Committee, and written informed consent was obtained from all subjects for the use of their tissues samples for research.
Cell culture and transfection
Human CRC cell lines SW480, SW620, HT29 and LOVO were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA), and the normal human intestinal epithelial cell line HIEC was purchased from Shanghai Institute of Cell Biology (Shanghai, China). All cells were cultured in RPMI-1640 (Gibco, Grand Island, NY) medium supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY), 100 U/mL penicillin, and 100 μg/mL streptomycin (Life Technologies, Cergy Pontoise, France), maintained at 37°C in a humidified air atmosphere containing 5% CO2 [16].
A Fibroblast growth factor 2 (FGF2) expression vector was constructed by sub-cloning the full-length FGF2 coding sequence into pcDNA3.1 (Sangon Biotech, Shanghai, China). The empty pcDNA-3.1 plasmid was used as its negative control. MiR-195 mimic, ASO of miR-195 and theirs corresponding controls (scrambled) were synthesized by RiboBio (Guangzhou, China). For transfection, SW480 and SW620 cells were plated onto 60-mm dishes at a density of 2 × 106 cells/well. At 24 h after the incubation, cells were transfected with miR-195 mimic, ASO of miR-195, or their corresponding controls, otherwise transfected with miR-195 mimic and/or FGF2 expression vector. All transfection were performed by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Cells were collected after 48 h of transfection for subsequent analyses.
Real-time quantitative PCR (qPCR)
Total RNA in cells and tissues were extracted by using Trizol reagent (Invitrogen, Carlsbad, CA). RNA (500 ng) from each samples were converted to cDNA by Transcriptor First Strand cDNA Synthesis Kit (Roche, USA). Real-time qPCR was conducted on the PCR System 7500 (ABI) by using FastSTART Universal SYBR Green Master (ROX; Roche, USA), according to the instructions of manufacture. Data were normalized to GAPDH or U6 snRNA expression, and were analyzed using the classic 2-ΔΔCt method [17]. All primers were synthesized by GenePharma (Shanghai, China).
Cell viability assay
Cell viability was measured by the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) assay. In brief, the transfected SW480 and SW620 cells were seeded in 96-well plates at a density of 5 × 103 cells/well. After 24 h of incubation, 20 μL MTT (0.5 mg/mL; Sigma-Aldrich, St. Louis, Mo, USA) was added into each well and incubated for another 3 h at 37°C. Subsequently 150 μL dimethyl sulfoxide (DMSO; Sigma-Aldrich) was added to stop the reaction. The optical density was detected with a Multiskan EX (Thermo, Finland) at a wavelength of 570 nm [18].
Cell cycle analysis
Cell cycle distribution was detected by using the Cell Cycle and Apoptosis Analysis Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. After transfection, cells were harvested and washed twice with phosphate-buffered saline (PBS), and then fixed in chilled 70% ethanol at 4°C overnight. Afterward, cells were re-suspended in 500 μL of PBS containing 0.2 mg/mL RNaseA and 50 μg/mL PI for 30 min at room temperature in the dark. Proportion of the cells in G0/G1, S and G2/M phase was analyzed by the ModFit software (Verity Software House, Topsham, ME, USA) [19].
Luciferase reporter assays
The 3’-untranslated region (3’-UTR) of FGF2 was amplified by PCR and placed in the pmiR-Report vector (Ambion). These vectors were co-transfected with miR-195 mimic or its control into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 48 h transfection, luciferase assay was carried out using the Dual-Luciferase reporter assay system (Promega) as previous described [20].
Western blot
After transfection, cellular proteins were extracted by lysis buffer (Beyotime, Shanghai, China), and the protein concentration was determined by Bicinchoninic Acid (BCA) Kit (Solarbio, Beijing, China). Protein samples were separated by the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDSPAGE) and transferred to nitrocellulose membranes (Maibio, Shanghai, China). After blocked with 5% nonfat milk for 1 h at room temperature, the membranes were incubated with specific primary antibodies against CyclinB1 (ab32053), CyclinD1 (ab134175), Cyclin-Dependent Kinase 2 (CDK2; ab6433), FGF2 (ab16828), β-catenin (ab32572), Ki-67 (ab15580), Glycogen Synthase Kinase 3β (GSK-3β; ab93926), phosphorylated GSK-3β (p-GSK-3β; ab131097), Transcription Factor 4 (TCF-4; ab60727), Lymphoid Enhancer Binding Factor 1 (LEF-1; ab53293) and GAPDH (ab8245) (Abcam, Cambridge, UK), at 4°C overnight. The membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 2 h at room temperature. Enhanced chemiluminescence (ECL) reagent (GE Healthcare, Little Chalfont, UK) was used to develop the signals, and the densitometric measurements were performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA) [21].
Statistical analysis
Data were represented as the mean ± standard deviation (SD) from three independent experiments. Data were analyzed by Student’s t test to calculate the different significance between two groups, using the GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA). P < 0.05 was considered statistically significant.
Results
MiR-195 expression was decreased during CRC
To explore the role of miR-195 in CRC, qPCR was performed to compare the expression of miR-195 between CRC tissues and paired adjacent tissues, as well as between CRC cell lines (SW480, SW620, HT29 and LOVO) and normal intestinal epithelial cell line (HIEC). Results in Figure 1A and 1B showed that, in CRC tissues and cell lines, the mRNA level expression of miR-195 was significantly lower than in theirs normal groups (P < 0.05 or P < 0.01). These data suggesting that, the expression of miR-195 might be implicated in the pathogenesis of CRC. In addition, SW480 and SW620 seemed to possess the lowest miR-195 expression; therefore, these two cell lines were selected for the following investigations.
Figure 1.
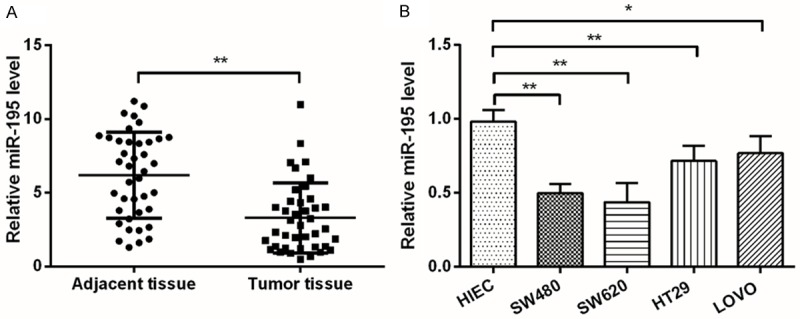
MiR-195 expression was decreased during CRC. A. The mRNA level expression of miR-195 in CRC tissues and paired adjacent tissues was measured by qPCR; B. The mRNA level of miR-195 in CRC cell lines, i.e., SW480, SW620, HT29 and LOVO, and in normal intestinal epithelial cell line HIEC, was measured by qPCR. MiR-195, microRNA-195; CRC, colorectal cancer; qPCR, quantitative PCR; *, P < 0.05; **, P < 0.01.
MiR-195 suppressed CRC cells viability and induced G2/M arrest
To observe the effects of miR-195 on CRC cells, miR-195 mimic, ASO of miR-195 or their corresponding controls were transfected into SW480 and SW620 cells. After the transfection, we sought to scan changes in cell viability and cell cycle. MTT assay (Figure 2A and 2B) showed that, cell viability was significantly inhibited by miR-195 overexpression, while was enhanced by miR-195 suppression (P < 0.05 or P < 0.01), in a time-dependent manner. Detection by flow cytometer (Figure 2C) showed that, G2/M phase cell proportion increased remarkably in miR-195 overexpressed cells when compared with its control. Further, qPCR and Western blot analyses (Figure 2D and 2E) displayed that, both the mRNA and protein levels of CyclinB1, CyclinD1 and CDK2 were down-regulated by miR-195 overexpression, while were up-regulated by miR-195 suppression (P < 0.05 or P < 0.01). Taken together, these data indicated the anti-proliferation role of miR-195 in CRC cells.
Figure 2.
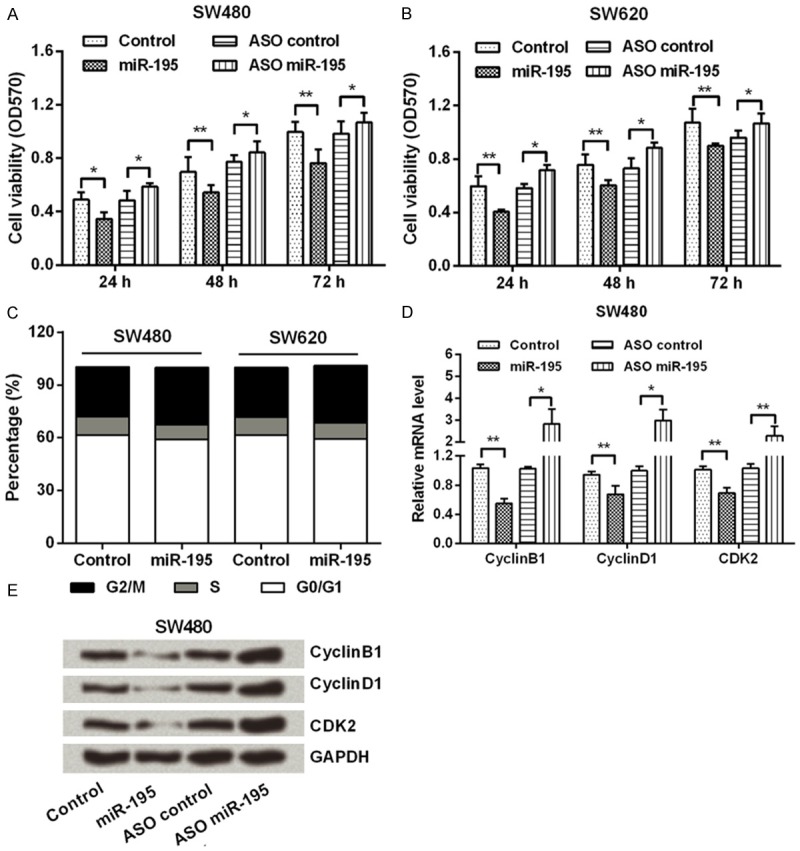
MiR-195 suppressed CRC cells viability and induced G2/M arrest. SW480 and SW620 cells were transfected with miR-195 mimic, ASO of miR-195 or their corresponding controls. Then, (A and B) cell viability was measured by MTT; (C) Cell cycle was detected by flow cytometer; and (D and E) the mRNA and protein level expressions of CyclinB1, CyclinD1 and CDK2 were determined by qPCR and Western blot. GAPDH acted as an internal control. MiR-195, microRNA-195; CRC, colorectal cancer; ASO, antisense oligonucleotides; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide; CDK2, Cyclin-Dependent Kinase 2; qPCR, quantitative PCR; *, P < 0.05; **, P < 0.01.
FGF2 was a direct target of miR-195
Here, TargetScan (www.targetscan.org) and microRNA database (www.microrna.org) were used to predict the target gene of miR-195. As the result showed in Figure 3A, FGF2 was predicted as a target of miR-195. Further, Dual-Luciferase reporter assay (Figure 3B and 3C) showed that miR-195 reduced the activity of the luciferase reporter fused to the 3’-UTR-WT of FGF2 (P < 0.01), but did not suppress that of the reporter fused to the Mut version (P > 0.05). These data suggested that miR-195 directly targeted FGF2. Additionally, qPCR and Western blot (Figure 3D and 3E) were performed to detect FGF2 expression in miR-195 overexpressed or suppressed cells. We found that both the mRNA and protein levels of FGF2 were negatively regulated by miR-195 (P < 0.05 or P < 0.01). In summary, FGF2 was a direct target of miR-195 and was negatively regulated by miR-195.
Figure 3.
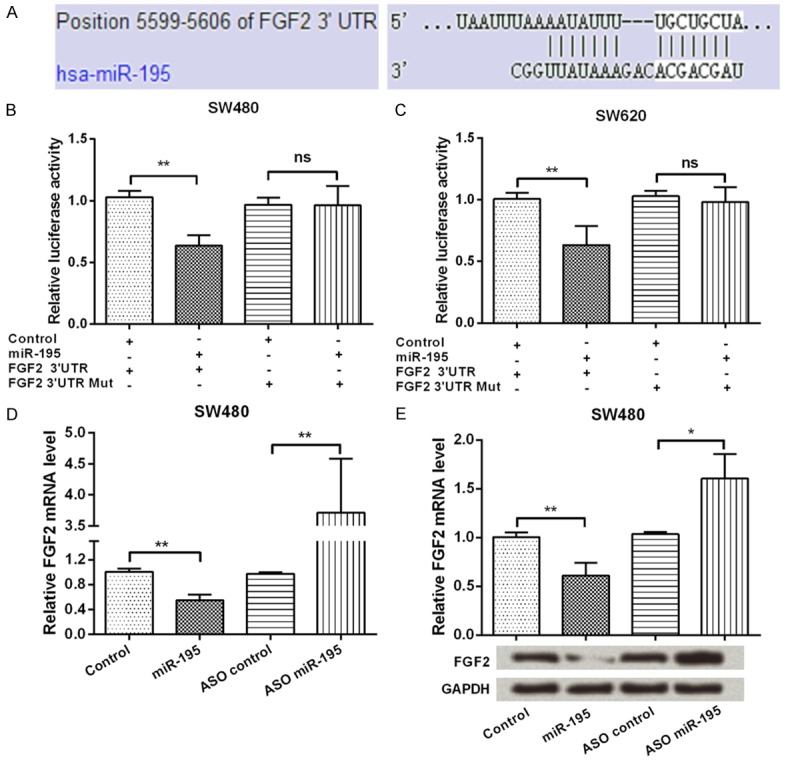
FGF2 was a direct target of miR-195. A. FGF2 was predicted as a target of miR-195 by using TargetScan and microRNA database; B and C. Dual-Luciferase reporter assay was performed to verify FGF2 was a direct target of miR-195; D and E. SW480 cells were transfected with miR-195 mimic, ASO of miR-195 or their corresponding controls, then the mRNA and protein level expressions of FGF2 were detected by qPCR and Western blotting. GAPDH acted as an internal control. FGF2, Fibroblast growth factor 2; miR-195, microRNA-195; ASO, antisense oligonucleotides; qPCR, quantitative PCR; ns, no significance; *, P < 0.05; **, P < 0.01.
FGF2 reduced the inhibitory effects of miR-195 on CRC cells proliferation
To explore whether miR-195 exerted its anti-proliferation role through FGF2, SW480 was transfected with miR-195 mimic, FGF2 expression vector, or both of them. Figure 4A and 4B showed that, FGF2 significantly enhanced cell viability and increased S-phase cells proportion (P < 0.05 or P < 0.01). Furthermore, FGF2 could notably reduce the inhibitory effects of miR-195 on cell viability (P < 0.05 or P < 0.01; Figure 4C). Additionally, FGF2 remarkably recovered the down-regulative effects of miR-195 on the expression of CyclinB1, CyclinD1 and CDK2 proteins (Figure 4D). These data indicated miR-195 modulated CRC cells proliferation might be via regulation of FGF2.
Figure 4.
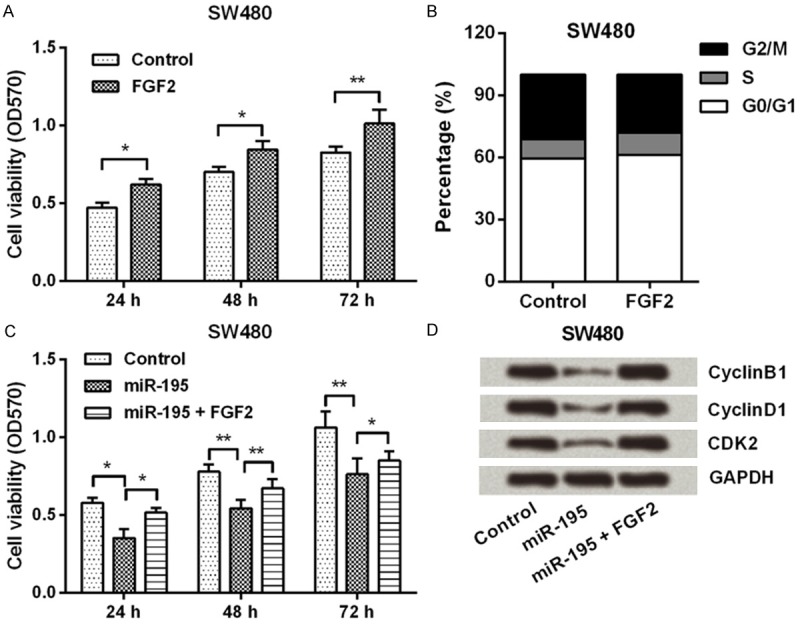
FGF2 reduced the inhibitory effects of miR-195 on CRC cells proliferation. A and B. SW480 cells were transfected with either FGF2 expression vector or its negative control, then cell viability and cell cycle were detected by MTT and flow cytometer; C and D. SW480 cells were transfected with miR-195 mimic or co-transfected with miR-195 mimic and FGF2 expression vector, then cell viability and the expression of CyclinB1, CyclinD1 and CDK2 were measured by MTT and Western blot. FGF2, Fibroblast growth factor 2; miR-195, microRNA-195; CRC, colorectal cancer; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide; CDK2, Cyclin-Dependent Kinase 2; *, P < 0.05; **, P < 0.01.
MiR-195 blocked Wnt/β-catenin pathway
To confirm the impact of miR-195 on cell proliferation, the expression of Ki-67, a proliferating marker, was measured. Western blotting data (Figure 5A) showed that, Ki-67 was down-regulated by miR-195 overexpression, while was up-regulated by miR-195 suppression. Further, to determine the signaling pathway by which miR-195 inhibited CRC cells proliferation, the expressions of Wnt/β-catenin pathway factors were determined. qPCR and Western blot assay (Figure 5A-D) displayed that, down-regulations of β-catenin, TCF-4 and LEF-1, as well as up-regulation of GSK-3β and p-GSK-3β, were found in miR-195 over-expressed cells. In contrast, these five factors were reversely regulated by miR-195 suppression. Thus, we inferred Wnt/β-catenin pathway was blocked by miR-195 in CRC cells.
Figure 5.
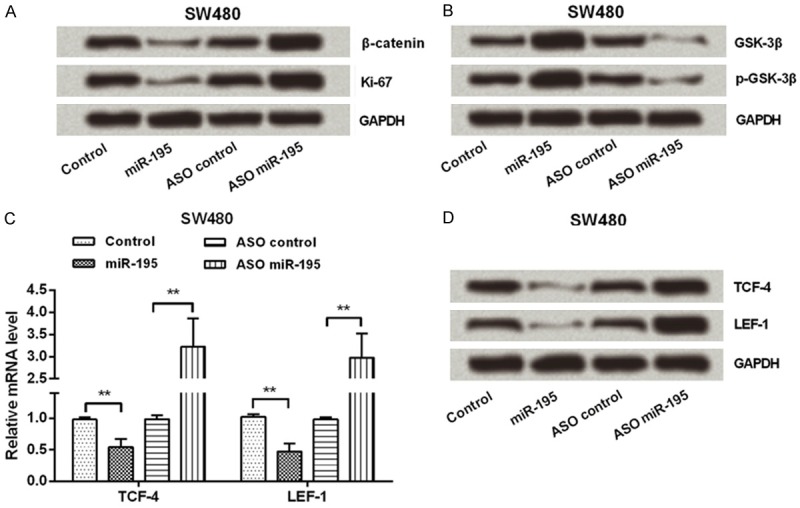
MiR-195 blocked Wnt/β-catenin pathway. SW480 cells were transfected with miR-195 mimic, ASO of miR-195 or their corresponding controls. Then, the protein expressions of (A) β-catenin and Ki-67s, as well as (B) GSK-3β and p-GSK-3β were measured by Western blot; (C and D) The mRNA and protein expressions of TCF-4 and LEF-1 were measured by qPCR and Western blot. GAPDH acted as an internal control. MiR-195, microRNA-195; ASO, antisense oligonucleotides; GSK-3β, Glycogen Synthase Kinase 3β; phosphorylated GSK-3β, p-GSK-3β; TCF-4, Transcription Factor 4; LEF-1, Lymphoid Enhancer Binding Factor 1; qPCR, quantitative PCR; **, P < 0.01.
Discussion
Accumulating evidence has revealed that miRNAs play pivotal roles in developmental and pathological processes of many human cancers, some of them being considered as tumor suppressor genes, while others recognized as oncogenes [22-24]. Currently, miR-195 has been identified as tumor suppressor genes in CRC. However, the underling mechanism in which miR-195 controls CRC has not been fully investigated. The data in this study showed that, miR-195 was remarkably decreased in CRC tissues and cell lines, and overexpression of miR-195 suppressed CRC cells viability and induced G2/M arrest. Besides, FGF2 was verified as a direct target of miR-195, and FGF2 could alleviate the inhibitory effects of miR-195 on CRC cells proliferation. Further in vitro investigations displayed that, miR-195 could block Wnt/β-catenin pathway in CRC cells.
Deregulated cell proliferation and unscheduled re-entry into the cell cycle are two main characteristics of most human tumor cells [25]. A vast body of literature has illustrated inhibition of tumor cell proliferation via arresting cells at G2/M or G0/G1 phase may provide therapeutic benefit against certain human neoplasias [25,26]. Functional studies have provided the evidences that miR-195 could suppress cell growth, and stagnate cell cycle progression at the G1/S transition in CRC cells [15,27]. In line with these previous studies, our data also suggested miR-195 could suppress CRC cells proliferation, while might be via arresting more cells at G2/M phase. CDKs are key mediators in cell cycle, and CDK activity requires binding of regulatory subunits known as Cyclins [25]. In primary CRC tissue, elevated CDK2 expression was found, and the impacts of CDK2 on cell proliferation were associated with CyclinB1 and CyclinD1 [28,29]. In the current study, the levels of CyclinB1, CyclinD1 and CDK2 were all down-regulated by miR-195 overexpression, further confirmed that miR-195 could control CRC cells proliferation via disrupting cell cycle.
FGFs have significant interaction with cell growth, differentiation, and functioning, and they play vital roles in maintaining tissue and repairing damage [30]. Currently, elevated expression of FGF2 has been reported in CRC cell lines, and FGF2 played a key role in promotion of cell proliferation [31,32]. In the present study, we revealed that FGF2 was a functional target of miR-195 in CRC cells. Further functional analyses showed that, FGF2 partly recovered the decreased cell viability and down-regulated CyclinB1, CyclinD1 and CDK2 expressions caused by miR-195 overexpression. Our data indicated that miR-195 exerted anti-proliferative functions in CRC at least in part via targeting FGF2.
The Ki-67 nuclear antigen is present throughout the cell cycle and its increases during mitosis. Immune-staining with Ki-67 provides a reliable means of rapidly evaluating the growth fraction of neoplastic human cell populations [33]. This study displayed that Ki-67 was down-regulated by miR-195 overexpression, further confirmed the anti-proliferation role of miR-195 in CRC cells. Wnt/β-catenin pathway is a crucial mechanism involved in the regulation of cell proliferation, and has been recognized as a key pathway during tumorigenesis [34,35]. In mammals, cytoplasmic β-catenin translocates to the nucleus and combines with the TCF/LEF family, as a result of the deactivation of GSK-3 by Wnt [36]. This event leads to the specific regulation of β-catenin target genes, including CyclinD1, and regulation of cell proliferation. In the current study, miR-195 overexpression notably down-regulated the levels of β-catenin, TCF-4 and LEF-1, while up-regulated GSK-3β and p-GSK-3β, suggesting that miR-195 affected cell proliferation via blocking Wnt/β-catenin pathway.
Taken together, we determined that miR-195 was frequently decreased in CRC tissues and cell lines. MiR-195 suppressed CRC cells proliferation via targeting FGF2 and blocking Wnt/β-catenin pathway. However, further study still needed to confirm these hypotheses.
Acknowledgements
There is no funding to support the work and all authors declare they have no conflict of interests.
Disclosure of conflict of interest
None.
References
- 1.Goncalves-Ribeiro S, Guillen Diaz-Maroto N, Berdiel-Acer M, Soriano A, Guardiola J, Martinez-Villacampa M, Salazar R, Capella G, Villanueva A, Martinez-Balibrea E, Mollevi DG. Carcinoma-associated fibroblasts affect sensitivity to oxaliplatin and 5FU in colorectal cancer cells. Oncotarget. 2016 doi: 10.18632/oncotarget.11121. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdelsatir AA, Husain NE, Hassan AT, Elmadhoun WM, Almobarak AO, Ahmed MH. Potential Benefit of Metformin as Treatment for Colon Cancer: the Evidence so Far. Asian Pac J Cancer Prev. 2015;16:8053–8058. doi: 10.7314/apjcp.2015.16.18.8053. [DOI] [PubMed] [Google Scholar]
- 3.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 4.Aan de Stegge WB, van Leeuwen BL, Elferink MA, de Bock GH. The Evaluation of More Lymph Nodes in Colon Cancer Is Associated with Improved Survival in Patients of All Ages. PLoS One. 2016;11:e0155608. doi: 10.1371/journal.pone.0155608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C, Xu N, Li YQ, Wang Y, Zhu ZT. Inhibition of SW620 human colon cancer cells by upregulating miRNA-145. World J Gastroenterol. 2016;22:2771–2778. doi: 10.3748/wjg.v22.i9.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 8.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 9.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 10.Borralho PM, Kren BT, Castro RE, da Silva IB, Steer CJ, Rodrigues CM. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276:6689–6700. doi: 10.1111/j.1742-4658.2009.07383.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Wang J, Ma H, Zhang J, Zhou X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med Oncol. 2012;29:919–927. doi: 10.1007/s12032-011-9880-5. [DOI] [PubMed] [Google Scholar]
- 12.Katoh M. Cardio-miRNAs and onco-miRNAs: circulating miRNA-based diagnostics for non-cancerous and cancerous diseases. Front Cell Dev Biol. 2014;2:61. doi: 10.3389/fcell.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Sheikh YA, Ghneim HK, Softa KI, Al-Jobran AA, Al-Obeed O, Mohamed MA, Abdulla M, Aboul-Soud MA. Expression profiling of selected microRNA signatures in plasma and tissues of Saudi colorectal cancer patients by qPCR. Oncol Lett. 2016;11:1406–1412. doi: 10.3892/ol.2015.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu J, Zhao L, Zhang P, Wang J, Xu N, Mi W, Jiang X, Zhang C, Qu J. MicroRNA-195 chemosensitizes colon cancer cells to the chemotherapeutic drug doxorubicin by targeting the first binding site of BCL2L2 mRNA. J Cell Physiol. 2015;230:535–545. doi: 10.1002/jcp.24366. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Qian L, Li X, Yan J. MicroRNA-195 inhibits colorectal cancer cell proliferation, colony-formation and invasion through targeting CARMA3. Mol Med Rep. 2014;10:473–478. doi: 10.3892/mmr.2014.2178. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Zhang Z, Sun L, Chai N, Tang S, Jin J, Hu H, Nie Y, Wang X, Wu K, Jin H, Fan D. MicroRNA-499-5p promotes cellular invasion and tumor metastasis in colorectal cancer by targeting FOXO4 and PDCD4. Carcinogenesis. 2011;32:1798–1805. doi: 10.1093/carcin/bgr213. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Lu L, Li C, Li D, Wang Y, Zhou C, Shao W, Peng J, You Y, Zhang X, Shen X. Cryptotanshinone inhibits human glioma cell proliferation by suppressing STAT3 signaling. Mol Cell Biochem. 2013;381:273–282. doi: 10.1007/s11010-013-1711-x. [DOI] [PubMed] [Google Scholar]
- 19.Pan XW, Zhao XH. In Vitro Proliferation and Anti-Apoptosis of the Papain-Generated Casein and Soy Protein Hydrolysates towards Osteoblastic Cells (hFOB1.19) Int J Mol Sci. 2015;16:13908–13920. doi: 10.3390/ijms160613908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi W, Bruce J, Lee M, Yue S, Rowe M, Pintilie M, Kogo R, Bissey PA, Fyles A, Yip KW, Liu FF. MiR-449a promotes breast cancer progression by targeting CRIP2. Oncotarget. 2016;7:18906–18. doi: 10.18632/oncotarget.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fantetti KN, Gray EL, Ganesan P, Kulkarni A, O'Donnell LA. Interferon gamma protects neonatal neural stem/progenitor cells during measles virus infection of the brain. J Neuroinflammation. 2016;13:107. doi: 10.1186/s12974-016-0571-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong H, Luo L, Hong S, Siu H, Xiao Y, Jin L, Chen R, Xiong M. Integrated analysis of mutations, miRNA and mRNA expression in glioblastoma. BMC Syst Biol. 2010;4:163. doi: 10.1186/1752-0509-4-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visone R, Croce C. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda T, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 26.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 27.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–121. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- 28.Hu B, Mitra J, van den Heuvel S, Enders GH. S and G2 phase roles for Cdk2 revealed by inducible expression of a dominant-negative mutant in human cells. Mol Cell Biol. 2001;21:2755–2766. doi: 10.1128/MCB.21.8.2755-2766.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JH, Kang MJ, Park CU, Kwak HJ, Hwang Y, Koh GY. Amplified CDK2 and cdc2 activities in primary colorectal carcinoma. Cancer. 1999;85:546–553. [PubMed] [Google Scholar]
- 30.Jibiki N, Saito N, Kameoka S, Kobayashi M. Clinical significance of fibroblast growth factor (FGF) expression in colorectal cancer. Int Surg. 2014;99:493–499. doi: 10.9738/INTSURG-D-14-00044.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galzie Z, Fernig DG, Smith JA, Poston GJ, Kinsella AR. Invasion of human colorectal carcinoma cells is promoted by endogenous basic fibroblast growth factor. Int J Cancer. 1997;71:390–395. doi: 10.1002/(sici)1097-0215(19970502)71:3<390::aid-ijc15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto K, Miyata Y, Nakayama T, Saito S, Suzuki R, Hayakawa F, Nishiwaki S, Mizuno H, Takeshita K, Kato H, Ueda R, Takami A, Naoe T. Fibroblast Growth Factor-2 facilitates the growth and chemo-resistance of leukemia cells in the bone marrow by modulating osteoblast functions. Sci Rep. 2016;6:30779. doi: 10.1038/srep30779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- 34.Moon RT, Brown JD, Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 35.Li K, Zhou ZY, Ji PP, Luo HS. Knockdown of beta-catenin by siRNA influences proliferation, apoptosis and invasion of the colon cancer cell line SW480. Oncol Lett. 2016;11:3896–3900. doi: 10.3892/ol.2016.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pecina-Slaus N. Wnt signal transduction pathway and apoptosis: a review. Cancer Cell Int. 2010;10:22. doi: 10.1186/1475-2867-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]


