Abstract
RasGAP SH3-domain-Binding Protein 1 (G3BP1) has been implicated in cell growth, migration, and metastasis of some cancers, yet its function in hepatocellular carcinoma (HCC) remains to be explored. In the present study, we reported that G3BP1 was upregulated in HCC tissues compared with adjacent non-cancerous liver tissues both in mRNA and protein levels, and its high expression was significantly correlated with poor prognosis of HCC patients. Functional analyses demonstrated that forced expression of G3BP1 in HCC cells promoted cell migration, and silenced expression of G3BP1 by RNA interference caused opposite effects. Moreover, G3BP1 knockdown attenuated the distant metastasis capacity of HCC cells through tail vein injection approach in nude mice model. At molecular mechanism, we found G3BP1 knockdown decreased Slug expression, and increased the expression of the epithelial cell marker E-cadherin. Overexpression of Slug could restore the phenotype of G3BP1 silencing induced cell migration inhibition. Together, our data establish G3BP1 as an oncogenic factor involved in the metastasis of HCC and suggest that G3BP1 might serve as a novel predictor for patients’ outcome.
Keywords: G3BP1, hepatocellular carcinoma, Slug, tumor metastasis
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer in the world and has the second highest mortality, with 520,000 people per year for males and 224,000 cases per year for females according to the International Agency for Research on Cancer. Treatment of HCC is limited to surgical removal and there is no effective systemic chemotherapy available for HCC patients because of the liver’s natural resistance [1]. For early stage patients, surgery is the best option, the five-year survival rate ranges from 41% to 74% [2]. If patients were diagnosed in the late stages when surgery is not an option, the chemotherapy have little effect. Therefore, it is important to find early diagnose marker and new therapies for this disease. Additionally, HCC also shows a propensity to metastasize and infiltrate adjacent and more distant tissues. Metastasis is thought to be the major cause for poor prognosis and low survival rate of HCC. To date, the underlying molecular mechanisms in HCC metastasis are still not fully understood.
Ras GTPase-activating protein SH3 domain-binding proteins (G3BPs) are a class of RNA-binding proteins. The name of G3BP was designated due to the early report of G3BP1 interacting with the SH3 domain of RasGAP [3]. Many reports have shown that G3BP1 is involved in multiple cell functions, including stress granule formation [4], RNA metabolism [5], cell growth [6], motility [7], and apoptosis [8]. G3BP1 is known to contain a RNA recognition motif (RRM) by which exerts its mRNA-stabilizing or mRNA-degrading effects. The dysregulation of G3BP1 interactions with its target mRNAs may result in cancer development and progression. Some cancer related gene, such as c-MYC [9], CTNNB1 [10], and PMP22 [11], have been implicated in the regulation of G3BP1 by this way. In human sarcomas [12], breast cancer [13], lung cancer [14], gastric cancer [15] and colon cancer [16], G3BP1 has been found to be highly expressed. High expression of G3BP1 could promote proliferation and motility of gastric cancer or breast cancer cells. Downregulation of G3BP1 in sarcoma xenografts prevented tumor invasion and completely blocked lung metastasis in mouse models. Through binding with p53, G3BP1 led to the redistribution of p53 from the nucleus to the cytoplasm and affected cell apoptosis [17]. These reports suggest the important roles of G3BP1 in tumorigenesis, however, the connection between G3BP1 and HCC still remains unknown.
In the current work, we investigated the expression and functional role of G3BP1 in HCC. We found that G3BP1 was frequently upregulated in HCC samples and upregulated G3BP1 expression was correlated with poor survival. Further cell biological results indicated that G3BP1 promoted HCC cell migration and Slug protein was identified as one of main effectors of G3BP1. Thus, our study provides new insights into the role of G3BP1 in tumor and suggested that G3BP1 may be a potential target for HCC treatment.
Materials and methods
Human liver cancer specimens
The use of the tissue material for research was approved by the committee on ethics of biomedicine research at Shanghai East Hospital, Tongji University School of Medicine. 37 paired of human HCC and adjacent liver specimens were collected from patients suffered from HCC with informed consent who underwent resection of the primary tumor at the department of Shanghai East hospital. Before RNA extraction, all the samples were snap-frozen in liquid nitrogen and stored at -80°C. Human liver cancer tissue microarray (HLiv-HCC180Sur-05) was obtained from Shanghai Outdo Bioteck, China. All the patients had well-documented clinical histories and follow-up information. G3BP1 was immunohistochemical stained using an anti-G3BP1 antibody (13057-2-AP, Proteintech, Wuhan, China) at a dilution of 1:500. The staining results were scored to four stages according to the percentage of positive cells and staining intensity: negative (-), slight positive (+), moderate positive (++), or strong positive (+++). Classifying of stages was conducted in a blinded manner to prevent bias from knowledge of clinical data.
Cell lines and culture conditions
Human HCC cell lines (Huh7, Sk-Hep1, and Hep3B) were obtained from Shanghai Cell Bank of Chinese Academy of Sciences. YY8013 and WRL68 cells were taken from our laboratory stocks. Cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Corning, US) with 10% fetal bovine serum and 100 µg/mL penicillin/streptomycin at 37°C in a humidified 5% CO2 incubator.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from the tissue samples or the cultured cell lines using RNAiso Plus reagent (TaKaRa, Japan) and was reverse transcribed into cDNA by using PrimeScriptTM RT reagent kit (TaKaRa, Japan) according to manufacturers’ protocols. G3BP1 and SLUG expression were examined on an ABI 7500 Real Time System with SYBR green reagent (TaKaRa, Japan). The 2-∆∆Ct method was used for quantification as previously described [18], and b-actin was used as an endogenous control. Each experiment was conducted for at least three times. The following primer pairs were used in this study. G3BP1-qF: 5’-ATGCAGTCTACGGACAGAAAGA-3’, G3BP1-qR: 5’-GAGCATCAACATGGCGAATCT-3’; Slug-qF: 5’-GATGCCGCGCTCCTTCCTGG-3’, Slug-qR: 5’-GGGGGACTCACTCGCCCCAA-3’; β-actin-qF: 5-CCTGGCACCCAGCACAATG-3, β-actin-qR: 5-GGGCCGGACTCGTCATACT-3.
RNA interference and cell transfection
Transfection of siRNA or plasmids was carried out using Lipofectamine 3000 (Initrogen, CA, USA) according to the manufacturer’s recommendation. A specific siRNA against G3BP1, non-specifc control (NC), were synthesized by GenePharma (Shanghai, China). The sequences were designed as follow: siG3BP1, sense 5-GAGUGCGAGAACAACGAAU-3 and siNC sense 5-UUCUCCGAACGUGUCACGUdTdT-3).
Plasmid and lentivirus construction
To overexpress G3BP1 and SLUG, the expression plasmids, pEnter-G3BP1 (GeneBank Accession Number: NM_005754) and pEnter-SLUG (GeneBank Accession Number: NM_003068) were purchased from Vigene Biosciences, Shandong, China. RNA interference lentivirus LV-shG3BP1 and LV-shNC was packaged and purchased from GenePharma, Shanghai using corresponding sequences of siG3BP1 and siNC, respectively. The G3BP1 overexpression lentivirus was purchased from Shanghai Tuzhu Biotech. Stably infected cell lines with lentivirus were isolated by puromycin selection.
Cell migration
Transwell assays were used to measure cell migration capability using a modified 24-well Boyden chamber with a membrane as previously described [19]. 24 h after transfection, cells were resuspended and counted. About 3~5×104 cells in 400 μl DMEM without FBS were placed into the upper chambers. The lower chambers were filled with 800 μl complete medium with 5% FBS as a chemo attractant stimulus. After incubation for 30 h at 37°C, non-invading cells were removed from the top of the chamber with a cotton swab. Migrated cells on the bottom surface of the filter were fixed, stained with 0.5% crystal violet, and counted in five random fields under a microscope and the average number of five fields was calculated. All assays were performed in triplicate and repeated three times.
Wound healing assay
Briefly, HCC cells were cultured to about 90% confluence in a 6-well plate at 37°C and 5% CO2. A wound about 2 mm width was created by scratching cells with a sterile 100 μL plastic tip. Cells were washed with PBS to remove floating cells, then 2 mL 0.5% FBS-containing DMEM was added. The light microscope (Nikon, Japan) was used to determine wound healing at 0 h, 24 h and 48 h, with a microscope at 200× magnification. The experiments were performed in triplicate.
Western blot analysis
Total proteins from cultured cells were extracted in RIPA buffer, and the protein concentration was determined using the BCA protein assay kit (TaKaRa, Japan). Cell lysates were electrophoresed on SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride membrane (EMD Millipore, USA). Membranes were blocked with 5% non-fat milk in Phosphate Buffered Saline (PBST) buffer (0.05% Tween-20) for 1 h and probed with primary antibodies, followed by fluorescent secondary antibody and bounded proteins were visualized by using the Odyssey Infared Imaging System (Li-COR, USA) according to the manufacturer’s instructions. The antibodies used in this study included: G3BP1 (1:2000, BD Biosciences, #611126), β-actin (1:500, Santa Cruz Biotechnology, sc-58673), Phospho-Erk1/2 (Thr202/Tyr204) (1:1000, Cell Signaling Technology, #4370), Erk1/2 (Thr202/Tyr204) (1:1000, Cell Signaling Technology, #4695), Phospho-Akt (Ser473) (1:1000, Cell Signaling Technology, #4051), Akt (1:1000, Cell Signaling Technology, #9272), E-Cadherin (1:1000, Cell Signaling Technology, #3195), Vimentin (1:500, Cell Signaling Technology, #5741), Slug (1:500, Cell Signaling Technology, #9585).
Animal studies
Male athymic nude mice were obtained from Animal Center of the Chinese Academy of Science (Shanghai, China) and maintained in sterile laminar flow cabinets. 2×106 of cells (WRL68-LV-shNC, WRL68-LV-shADAR and YY8103-LV-Vec, YY8103-LV-G3BP1) were tail vein-injected into these mice (n=5 per group, 5-6 weeks of age), respectively. 50 days after injection, mice were sacrificed and lung metastasis was calculated by the obvious lung metastatic tumors on the surface. The lung tissues were fixed in 4% paraformaldehyde and suffered with H&E staining and histopathological examination. All experimental procedures involving animals were in accordance with the Guide for the Care and Use of Laboratory Animals and approved by the Ethics Committee of Shanghai East Hospital.
Statistical analysis
The χ2 test was used to analyze the relationships between G3BP1 expression and various clinicopathological parameters. Survival curves were calculated using the Kaplan-Meier method and compared by the log-rank test. The significance of in vitro and in vivo data analysis was performed using a two-tailed Student t-test and the data were presented as mean ± SD. P<0.05 was considered statistically significant. “*” indicates P<0.05; “**” indicates P<0.01.
Results
G3BP1 was frequently upregulated in HCC and predicted poor prognosis
We firstly examined G3BP1 expression in 37 paired of HCC tissues by quantitative RT-PCR. The results showed that G3BP1 was significantly higher in cancer tissues compared with corresponding non-cancerous liver tissues. Among them, 18/37 (48.6%) were upregulated at least 1.5-fold as shown in Figure 1A. Meanwhile, a HCC tissue microarray with available clinical information was immunohistochemical stained with a specific antibody against G3BP1. Consistent with the qRT-PCR results, G3BP1 was significantly overexpressed in 40/76 (52.6%) of HCC samples relative to their corresponding non-cancerous tissues based on G3BP1 staining intensity. Two representative cases of the HCC samples handled with immunohistochemical staining were shown in Figure 1B. Most importantly, the overall survival of these patients was markedly decreased in G3BP1 strong staining group (Figure 1C), indicating that high expression of G3BP1 was closely associated with poor outcomes of HCC patients. However, G3BP1 expression was not statistically correlated with any other clinicopathological characteristic, such as age, gender, tumor size, differentiation and JACC stage. These collective findings suggested that G3BP1 is overexpressed in HCC and its expression could predict prognosis of patients.
Figure 1.
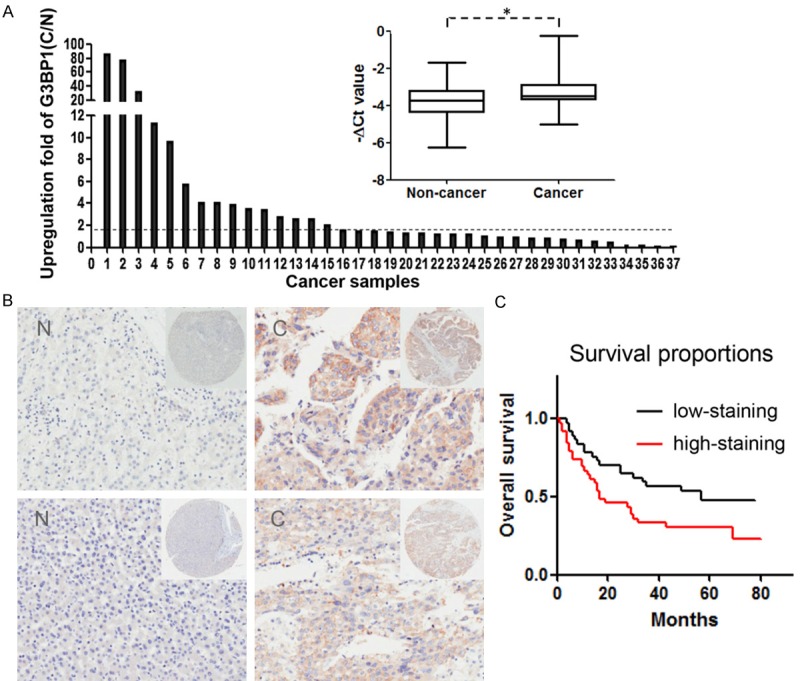
G3BP1 expression is upregulated in human HCC samples. A. The expression of G3BP1 in 37 matched pairs of HCC and non-cancerous tissues was detected by quantitative real-time PCR where β-actin was used as internal control (right), P value as indicated was calculated by paired t test. *P<0.05. The change fold (C/N) of G3BP1 expression in each sample pair was respectively indicated as a column (left). B. The protein expression of G3BP1 was performed with immunohistochemical staining on a HCC tissue microarray. Two representative clinical cases of immunohistochemical staining are shown. Magnification: ×40 (upper right) and ×200 (bottom). C. The result of Kaplan-Meier survival analysis exhibited that patients with high expression of G3BP1 (red line; n=39) had shorter overall survival time than those with low G3BP1 expression (black line; n=37; P=0.02, Log-rank test).
G3BP1 enhanced cell migration in vitro
To determine whether G3BP1 plays a role on cell migration, the G3BP1 overexpression plasmid was transiently transfected into Hep3B and YY8103 cells. Transwell assay data demonstrated that overexpressed G3BP1 significantly promoted cell migration in the two HCC cells (Figure 2A). Conversely, knocking down G3BP1 with specific small interference RNA in Sk-Hep-1 and WRL68 cells strongly reduced the migrated cell number when compared with the controls (Figure 2B). Furthermore, wound healing assay was also employed to detect the effect of G3BP1 on cell migration. As shown in Figure 2C, stable silencing of G3BP1 restrained cell motility in Sk-Hep-1 and WRL68 cells. These results indicated that G3BP1 plays an important role on HCC cell migration in vitro.
Figure 2.
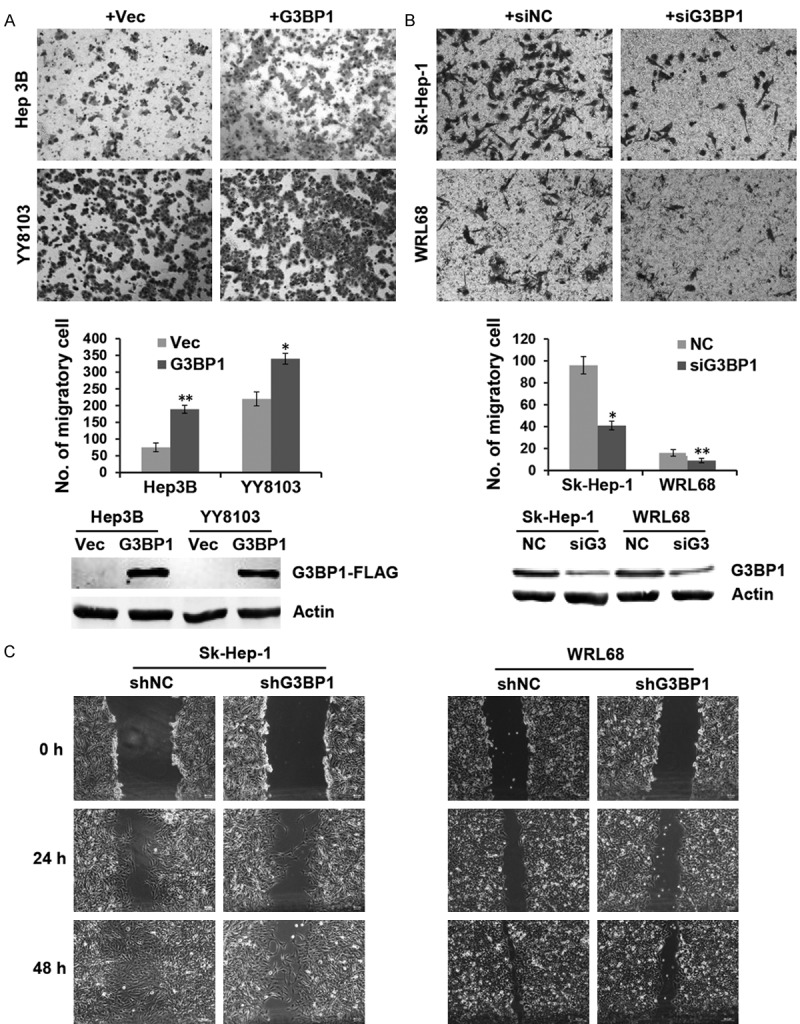
G3BP1 promotes HCC cell motility. A. Representative fields of YY8103 and Hep3B cells transfected with G3BP1 overexpression plasmid or empty vector in the transwell assay and average number of migration cells per field was evaluated. *P<0.05, **P<0.01. B. Representative fields of WRL68 and Sk-Hep-1 cells were evaluated by transwell assay after transfected with siG3BP1, where siNC was used as a control. Cell numbers were counted in five randomly selected microscopic fields. *P<0.05, **P<0.01. The overexpression and knockdown efficiency of G3BP1 was determined by western blotting. C. Wound healing experiment was used to analyze the motility ability of WRL68 and Sk-Hep-1 cells in which G3BP1 was silenced, photographs were taken at 0, 24, and 48 h.
Silencing of G3BP1 suppressed HCC cell metastasis in vivo
To investigate the in vivo role of G3BP1 on metastasis of HCC cells, we injected 2×106 of HCC cells into tail veins of nude mice (n=5, per group) for the observation of lung metastasis. The final results showed that silencing of G3BP1 strongly reduced the nodule number formed on the lung surface from mice receiving WRL68-LV-shG3BP1 cells than that formed from mice receiving WRL68-LV-shNC cells (Figure 3A). In contrast, overexpression of G3BP1 in YY8103 cells increased the formation of tumor nodule on the lung surface of nude mice (Figure 3B). Histological analysis confirmed the presence of metastatic tumors in the lungs of these mice (Figure 3C). These data suggested that G3BP1 may be involved in the positive regulation of metastasis of HCC cells in vivo.
Figure 3.
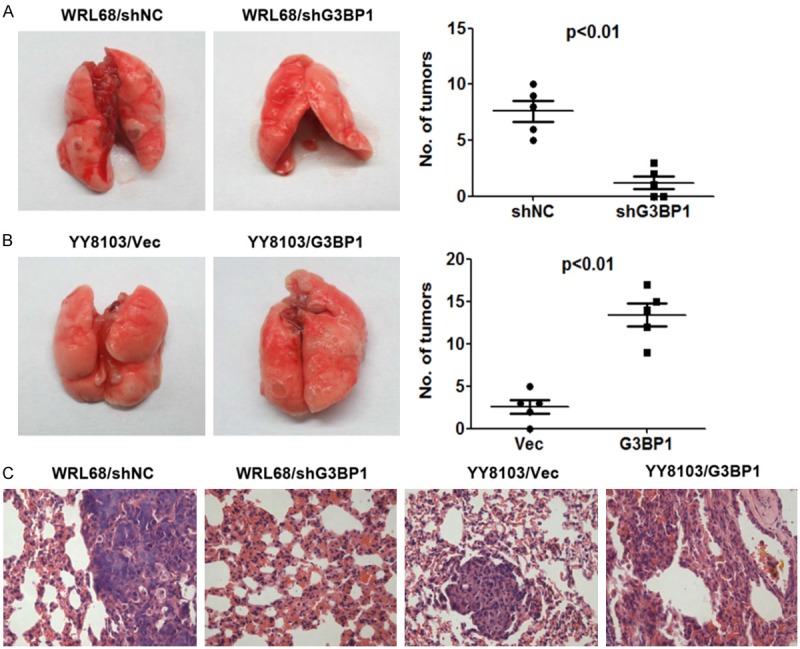
G3BP1 knockdown suppresses the metastasis of HCC cells in nude mice. A. G3BP1 knockdown inhibited lung metastasis of WRL68 cells. B. G3BP1 overexpression increased the number of lung metastasis of YY8103 cells. Lung metastatic tumor nodules were observed in lung surface after 50 days receiving tail vein injection with indicated cells. The mean number of tumor nodules from each group (n=5) were counted. C. The hematoxylin and eosin (HE) stained lung sections are shown. Magnification: ×400.
G3BP1 promoted Slug expression
To explore the molecular mechanisms by which G3BP1 promoted HCC cell migration, we examined the expression of some known tumor metastasis-related molecules, including E-cadherin, FAK, Slug, and Vimentin through western blotting analysis. In addition, the Ras downstream signaling pathway factors such as Akt and Erk were also examined. As shown in Figure 4A, only the protein level of Slug was obviously downregulated when G3BP1 was knocked down in Sk-Hep-1 and WRL68 cells, indicating that G3BP1 promoted Slug expression. Although we did not found the protein expression of E-cadherin in the two cell lines, qRT-PCR data revealed that the mRNA level of E-cadherin was upregulated in G3BP1 silenced cells (Figure 4B). Importantly, the regulation of Slug and E-cadherin by G3BP1 was confirmed in Huh7 cells (Figure 4C). Furthermore, overexpression of G3BP1 increased and knockdown of G3BP1 decreased the mRNA level of Slug (Figure 4D, 4E), suggesting that G3BP1 affected Slug expression in mRNA level.
Figure 4.
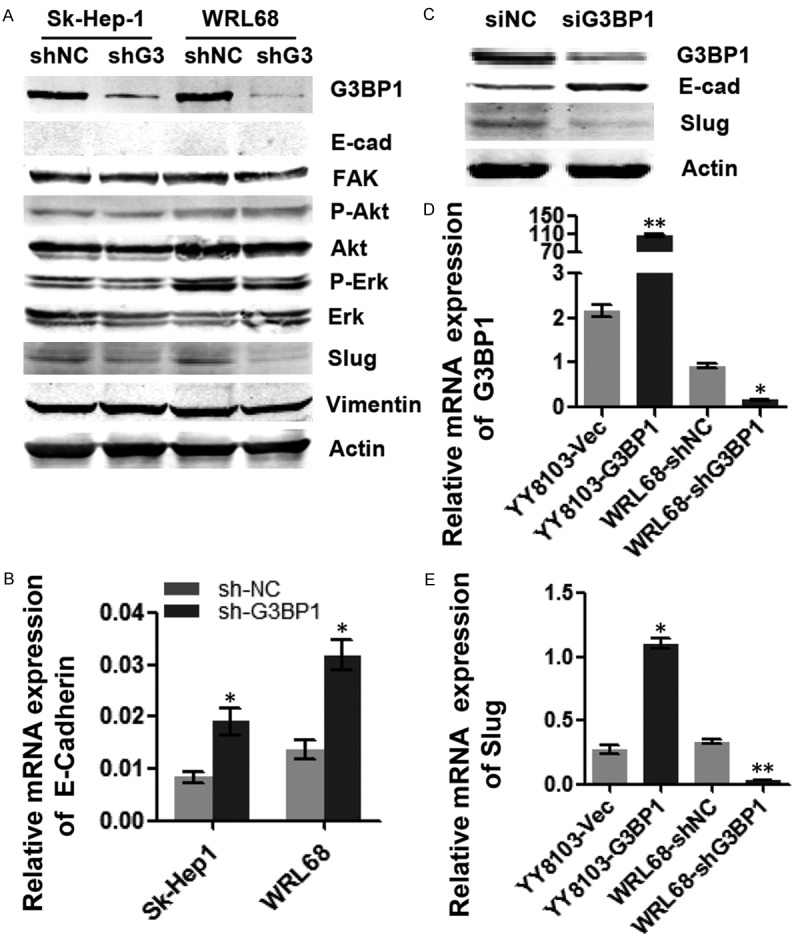
G3BP1 upregulates Slug expression. A. The expression levels of tumor-related factors were detected by western blotting in Sk-Hep1/LV-shNC, Sk-Hep1-1/LV-shG3BP1 and WRL68/LV-shNC, WRL68/LV-shG3BP1 cells. G3BP1 silencing decreased the expression of Slug on protein level. B. Knocking down G3BP1 increased the expression of E-cadherin in mRNA level in above cells. C. G3BP1 knockdown suppressed the expression of Slug and upregulated the expression of E-cadherin in Huh7 cells by western blotting. D and E. The expression of G3BP1 and Slug in G3BP1 stably silenced HCC cells as indicated by qRT-PCR analysis. *P<0.05, **P<0.01.
Slug overexpression restored cell migration defect induced by G3BP1 knockdown
To further address the relationship between G3BP1 and Slug, we attempted to overexpress Slug in G3BP1 knockdown cells and observe whether Slug could rescue the cell migration defect. As expected, silencing G3BP1 led to significant inhibition of cell migration in Sk-Hep-1 and WRL68 cells, while overexpressing Slug in these cells restored the migratory inhibition induced by G3BP1 knockdown (Figure 5), suggesting that Slug may function as the downstream effector of G3BP1 and both the two factors play crucial roles in tumor cell migration.
Figure 5.
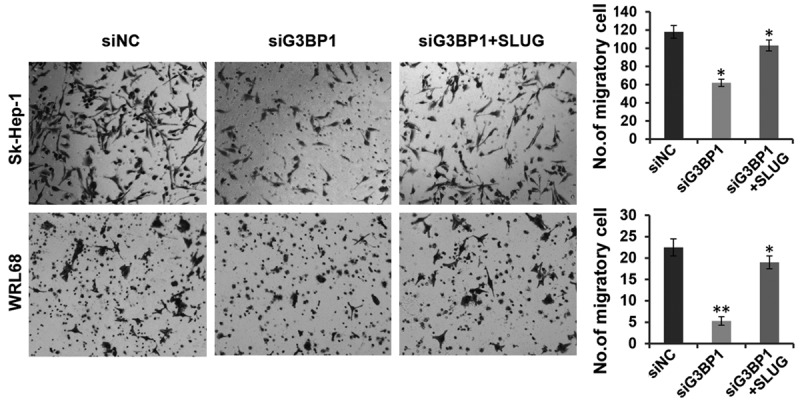
Overexpression of Slug restores the inhibition induced by G3BP1 knockdown on cell migration. The migratory ability of WRL68 and Sk-Hep-1 cells with indicated transfections were evaluated by transwell assays. The data is shown as the means ± SD, *P<0.05, **P<0.01.
Discussion
G3BP1 is an mRNA-binding protein that regulates mRNA stability and stress granule formation. Recent studies have implicated its involvement in the regulation of tumorigenesis. Overexpression of G3BP1 has been found in several cancers, however, little is known about its expression and roles in HCC. In this study, we investigated the expression of G3BP1 in HCC tissues and corresponding non-cancerous liver tissues and analyzed the association between G3BP1 expression and clinical parameters. We further evaluated the effects of G3BP1 on HCC cell migration in vitro and metastasis in vivo. Finally, we also explored the underlying mechanism by which G3BP1 regulated cell migration.
Our results showed that G3BP1 was frequently upregulated in HCC samples whether in mRNA level or protein level, in accordance with G3BP1 expression in other cancers. Statistical analysis revealed that high expression of G3BP1 was correlated with a shorter periods of postoperative survival of HCC patients. These results highlighted that G3BP1 could play important roles in the promotion of HCC progression and act as a poor prognosis marker. Subsequently, we provided evidence that G3BP1 enhanced HCC cell migration in vitro and promoted metastasis in nude mice model. However unexpected, we found that G3BP1 had no significant effect on HCC cell growth when CCK-8 method and colony formation assay were performed (data not shown). This difference with other cancer might reflect the tissue-specific or cancer type-specific. Further analysis for this is deserved to do in future.
As known, RasGAPs are main suppressors of Ras activity, G3BP1 has been reported to modulate Ras signaling pathway via interacting with RasGAP [20]. In our case, G3BP1 silencing had no markedly influence on the phosphorylation level of Akt and Erk [14,15], the key downstream kinase of Ras signaling. On the other hand, we found that silencing of G3BP1 significantly downregulated Slug expression and vice versa. Slug is one of the most highly studied members of the SNAIL family of zinc finger transcription factors. It contributes to tumor progression through induction of an Epithelial-Mesenchymal Transition (EMT) [21-23], which has been shown to occur in the initiation of metastasis for cancer progression [24,25]. Overexpression of G3BP1 has been shown to induce the EMT in breast cancer [13]. In this study, the obvious EMT was not observed in G3BP1 overexpressed HCC cells, but the epithelial cell marker E-cadherin was indeed upregulated in G3BP1 knockdown cells, implying Slug and E-cadherin mediates G3BP1 induced cell migration, although the EMT appeared not occur in the HCC cells examined. Furthermore, the restore experiments supported the notion that Slug functions as downstream effector of G3BP1 during cell migration.
In conclusion, we present evidence that G3BP1 is upregulated in HCC and plays an important role on metastasis via promoting Slug expression for the first time. Our findings suggest that G3BP1 may serve as a potential target for therapy of HCC and a valuable predictor for poor prognosis.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No. 81472576), the Outstanding Young Foundation (No. PWRq2015-09) and Science and Technology Development Foundation (No. PKJ2015-Y56) of Pudong New District, Shanghai, and Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (No. PWZxq2014-04).
Disclosure of conflict of interest
None.
References
- 1.Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, Hu T, Jiang L, Li J. Mammalian drug effl ux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 2016;370:153–164. doi: 10.1016/j.canlet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Wong R, Frenette C. Updates in the management of hepatocellular carcinoma. Gastroenterol Hepatol (N Y) 2011;7:16–24. [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs SD, Bryant SS, Jove R, Sanderson SD, Smithgall TE. The Ras GTPase-activating protein (GAP) is an SH3 domain-binding protein and substrate for the Src-related tyrosine kinase, Hck. J Biol Chem. 1995;270:14718–14724. doi: 10.1074/jbc.270.24.14718. [DOI] [PubMed] [Google Scholar]
- 4.Tsai WC, Gayatri S, Reineke LC, Sbardella G, Bedford MT, Lloyd RE. Arginine demethylation of G3BP1 promotes stress granule assembly. J Biol Chem. 2016;291:22671–22685. doi: 10.1074/jbc.M116.739573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irvine K, Stirling R, Hume D, Kennedy D. Rasputin, more promiscuous than ever: a review of G3BP. Int J Dev Biol. 2004;48:1065–1077. doi: 10.1387/ijdb.041893ki. [DOI] [PubMed] [Google Scholar]
- 6.Kociok N, Esser P, Unfried K, Parker F, Schraermeyer U, Grisanti S, Toque B, Heimann K. Upregulation of the RAS-GTPase activating protein (GAP)-binding protein (G3BP) in proliferating RPE cells. J Cell Biochem. 1999;74:194–201. [PubMed] [Google Scholar]
- 7.Taniuchi K, Nishimori I, Hollingsworth MA. The N-terminal domain of G3BP enhances cell motility and invasion by posttranscriptional regulation of BART. Mol Cancer Res. 2011;9:856–866. doi: 10.1158/1541-7786.MCR-10-0574. [DOI] [PubMed] [Google Scholar]
- 8.Oi N, Yuan J, Malakhova M, Luo K, Li Y, Ryu J, Zhang L, Bode AM, Xu Z, Li Y, Lou Z, Dong Z. Resveratrol induces apoptosis by directly targeting Ras-GTPase-activating protein SH3 domain-binding protein 1. Oncogene. 2015;34:2660–2671. doi: 10.1038/onc.2014.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallouzi IE, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, Capony JP, Tocque B, Tazi J. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol Cell Biol. 1998;18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bikkavilli RK, Malbon CC. Arginine methylation of G3BP1 in response to Wnt3a regulates beta-catenin mRNA. J Cell Sci. 2011;124:2310–2320. doi: 10.1242/jcs.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winslow S, Leandersson K, Larsson C. Regulation of PMP22 mRNA by G3BP1 affects cell proliferation in breast cancer cells. Mol Cancer. 2013;12:156. doi: 10.1186/1476-4598-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somasekharan SP, El-Naggar A, Leprivier G, Cheng H, Hajee S, Grunewald TG, Zhang F, Ng T, Delattre O, Evdokimova V, Wang Y, Gleave M, Sorensen PH. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol. 2015;208:913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Ma Y, Zhang S, Liu H, He H, Li N, Gong Y, Zhao S, Jiang JD, Shao RG. Involvement of Ras GTPase-activating protein SH3 domain-binding protein 1 in the epithelial-to-mesenchymal transition-induced metastasis of breast cancer cells via the Smad signaling pathway. Oncotarget. 2015;6:17039–17053. doi: 10.18632/oncotarget.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shim JH, Su ZY, Chae JI, Kim DJ, Zhu F, Ma WY, Bode AM, Yang CS, Dong Z. Epigallocatechin gallate suppresses lung cancer cell growth through Ras-GTPase-activating protein SH3 domain-binding protein 1. Cancer Prev Res (Phila) 2010;3:670–679. doi: 10.1158/1940-6207.CAPR-09-0185. [DOI] [PubMed] [Google Scholar]
- 15.Min L, Ruan Y, Shen Z, Jia D, Wang X, Zhao J, Sun Y, Gu J. Overexpression of Ras-GTPase-activating protein SH3 domain-binding protein 1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2015;67:677–688. doi: 10.1111/his.12695. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Zhang S, He H, Zhao W, Chen J, Shao RG. GAP161 targets and downregulates G3BP to suppress cell growth and potentiate cisplaitin-mediated cytotoxicity to colon carcinoma HCT116 cells. Cancer Sci. 2012;103:1848–1856. doi: 10.1111/j.1349-7006.2012.02361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MM, Wiederschain D, Kennedy D, Hansen E, Yuan ZM. Modulation of p53 and MDM2 activity by novel interaction with Ras-GAP binding proteins (G3BP) Oncogene. 2007;26:4209–4215. doi: 10.1038/sj.onc.1210212. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Han P, Fu Y, Liu J, Wang Y, He J, Gong J, Li M, Tan Q, Li D, Luo Y, Han J, Liu J, Tu W, Wang Y, Tian D, Yan W. Netrin-1 promotes cell migration and invasion by down-regulation of BVES expression in human hepatocellular carcinoma. Am J Cancer Res. 2015;5:1396–1409. [PMC free article] [PubMed] [Google Scholar]
- 20.Annibaldi A, Dousse A, Martin S, Tazi J, Widmann C. Revisiting G3BP1 as a RasGAP binding protein: sensitization of tumor cells to chemotherapy by the RasGAP 317-326 sequence does not involve G3BP1. PLoS One. 2011;6:e29024. doi: 10.1371/journal.pone.0029024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao X, Sun B, Sun D, Liu T, Che N, Gu Q, Dong X, Li R, Liu Y, Li J. Slug promotes hepatocellular cancer cell progression by increasing sox2 and nanog expression. Oncol Rep. 2015;33:149–156. doi: 10.3892/or.2014.3562. [DOI] [PubMed] [Google Scholar]
- 22.Guo Q, Ning F, Fang R, Wang HS, Zhang G, Quan MY, Cai SH, Du J. Endogenous Nodal promotes melanoma undergoing epithelial-mesenchymal transition via Snail and Slug in vitro and in vivo. Am J Cancer Res. 2015;5:2098–2112. [PMC free article] [PubMed] [Google Scholar]
- 23.de Herreros AG, Peiro S, Nassour M, Savagner P. Snail family regulation and epithelial mesenchymal transitions in breast cancer progression. J Mammary Gland Biol Neoplasia. 2010;15:135–147. doi: 10.1007/s10911-010-9179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith BN, Bhowmick NA. Role of EMT in Metastasis and Therapy Resistance. J Clin Med. 2016;5 doi: 10.3390/jcm5020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alves CC, Carneiro F, Hoefl er H, Becker KF. Role of the epithelial-mesenchymal transition regulator Slug in primary human cancers. Front Biosci (Landmark Ed) 2009;14:3035–3050. doi: 10.2741/3433. [DOI] [PubMed] [Google Scholar]


