Abstract
Dysregulation of long non-coding RNAs (lncRNAs) play important roles in tumor development and progression. The long non-coding RNA CCAT2 has been identified to be up-regulated in gastric cancer (GC). However, the detailed molecular mechanism of CCAT2 involved in GC progression is still unknown. The aim of this study was to explore the expression and role of CCAT2 in GC progression. In the study, the expression levels of CCAT2 were significantly up-regulated in 108 cases GC tissues compared with adjacent non-tumor tissues by qRT-PCR analysis. Higher CCAT2 expression was correlated with tumor size, lymph node metastasis and Tumor Node Metastasis (TNM) stage in GC patients. Multivariate analysis showed that lymph node metastasis, TNM stage and the expression of CCAT2 were independent prognostic indicator for disease-free survival (DFS) and the over survival time (OS) for GC patients. Further function analysis demonstrated that knockdown of CCAT2 inhibited the cell migration and invasion, whereas, the overexpression of CCAT2 showed the opposite results in GC cells. Our results also demonstrated that CCAT2 promoted the GC cells epithelial-mesenchymal transition (EMT) by downregulated the E-cadherin expression and upregulated the ZEB2, Vimentin and N-cadherin expression. Moreover, RNA immunoprecipitation (RIP) and Chromatin immunoprecipitation (ChIP) revealed that CCAT2 interacted with EZH2 and regulated the E-cadherin and LATS2 expression. Thus, our results demonstrated that CCAT2 functioned as an oncogene in GC and was involved in gastric cancer progression. Targeting CCAT2 might be a potential therapeutic target for GC.
Keywords: Gastric cancer, CCAT2, epithelial-mesenchymal transition, E-cadherin, LATS2
Introduction
Gastric cancer (GC) is one of the most common digestive malignant tumors and the second leading cause of cancer deaths worldwide [1]. Over the past decades, although the incidence of gastric cancer has decreased due to improved living standards, a reduction in chronic H. pylori infection and remarkable advances in diagnosis and treatment, unfortunately, the 5-year survival rate for advanced-stage patients remains low [2,3]. Therefore, it is essential to elucidate the underlying molecular mechanisms of initiation and metastasis and to develop novel therapeutic approaches for GC.
In the recent years, some studies have revealed that long non-coding RNAs, consisting of more than 200 nucleotides (nt) in length, are involved in molecular mechanism to regulate cell structure, function, and physiological development [4,5]. Long non-coding RNAs have been confirmed to play key roles in cancer development and progression [6]. For example, lncRNA H19 expression was higher in gastric cancer (GC) tissues and over-expression of H19 promoted the features of GC including proliferation, migration, invasion and metastasis [7]. LncRNA HOTAIR was also significantly up-regulated in cancerous tissues than that in corresponding normal mucosa, and higher expression of HOTAIR significantly correlated with peritoneal metastasis in GC patients [8]. Long non-coding RNA Linc00152 acted as an oncogene, which was involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer [9].
CCAT2 is a novel non-coding RNA located at 8q24. Ling et al found CCAT2 was highly over-expressed in microsatellite-stable colorectal cancer and promoted tumor growth, metastasis, and chromosomal instability [10]. Another study reported that CCAT2 was over-expressed in breast cancer tissues and up-regulated cell migration and down-regulated chemosensitivity to 5-FU in an rs6983267-independent manner [11]. Wang et al confirmed that up-regulation of lncRNA CCAT2 was correlated with gastric cancer progression and might be a potential molecular biomarker for predicting the prognosis in GC patients [12]. These investigation suggested that CCAT2 functioned as an oncogene in cancer development and progression, however, the detailed function of CCAT2 in gastric cancer was still unknown.
In present study, we showed that CCAT2 was significantly up-regulated in GC tissues and higher CCAT2 expression was correlated with poor survival outcome. Furthermore, knockdown of CCAT2 inhibited the cell migration, invasion and promoted the GC cells epithelial-mesenchymal transition (EMT) by downregulating the E-cadherin expression and upregulated the ZEB2, Vimentin and N-cadherin expression. Moreover, we also revealed that CCAT2 interacted with EZH2, LSD1, and H3k27me3, which regulated the E-cadherin and LATS2 expression. Therefore, these results indicated that CCAT2 functioned as an oncogene and was involved in gastric cancer progression.
Methods
Patient tissue samples
The gastric cancer tissue and adjacent normal tissue were obtained from 108 patients who had undergone surgical resection between March 2008 and February 2013 at the Hebei Chest Hospital and Hebei general Hospital. All of the patients were diagnosed by two experienced pathologists. The fresh tissue samples were obtained and then immediately frozen in liquid nitrogen, and stored at -80°C for further analysis. TNM staging classification was based on criteria of American Joint Committee on Cancer (AJCC, 6th edition). The written informed consent had been obtained from all the patients, and this study was approved by the Ethical Committee of Hebei Chest Hospital.
Cell lines and culture
The human gastric epithelial cell line GES-1 and the four gastric cancer cell lines (SGC7901, MKN45, BGC-823 and MKN-28) were purchased from Shanghai Institute of Cell Biology (Shanghai, China). All of the cell lines were maintained routinely in RPMI Media 1640 (Gibco) supplemented with 100 U/mL penicillin, and 100 μg/mL streptomycin. All cells were maintained in a humidified incubator containing 5% carbon dioxide at 37°C.
Cell transfection
Two siRNA oligos specifically targeting CCAT2 were constructed by GenePharma Co. Ltd (Shanghai, China) and two silencing sequences were si-CCAT2-1, 5’-GUGCAACUCUGCAAUUUAAUU-3’, si-CCAT2-2, 5’-UUAAAUUGCAGAGUUGCACUU-3’. The siRNA against EZH2 was 5’-GACUCUGAAUGCAGUUGCUTT-3’. GC Cells were transfected with siRNAs oligos by using Lipofectamine 3000 reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol. The CCAT2 full length sequence was subcloned into the pcDNA3.1 vector (Invitrogen). Ectopic CCAT2 expression was achieved through pcDNA3.1-CCAT2 transfection using lipofectamine 3000 (Invitrogen) and the empty pcDNA3.1 vector was used as a control.
RNA extraction and qRT-PCR
Total RNA was extracted from tissues or cells by using the Trizol reagent (TAKALA, Dalian, China) according to the manufacturer’s instructions. The quantitative real-time polymerase chain reaction (PCR) was performed by using SYBR-green PCR Master Mix in a Fast Real-time PCR 7500 System (Applied Biosystems). The gene-specific primers were as follows: CCAT2, forward: 5’-AGACAGTGCCAGCCAACC-3’, reverse: 5’-TGCCAAACCCTTCCCTTA-3’; E-cadherin, forward: 5’-TCT TCCAGGAACCTCTGTGATG-3’; reverse: 5’-CAATGCCGCCATCGCTTACACC-3’, LATS2, forward: 5’-ACCCCAAAGTTCGGACCTTAT-3’, 5’-reverse: CATTTGCCGGTTCACTTCTGC-3’. the GAPDH was used as an internal control, forward: 5’-AATGGACAACTGGTCGTGGAC-3’, and reverse: 5’-CCCTCCAGGGGATCTGTTTG-3’. The reactions were carried out at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. Fold change of mRNA was calculated by the equation 2-ΔΔCt.
Cell proliferation, migration and invasion assays
Cell proliferation was measured in MKN45 and BGC-823 cells using the Cell Counting Kit-8 assays (Dojindo) according to the manufacturer’s instructions. 3000 cells per well was seeded in 96-well plate. The 100 μl of cell suspension was placed in 96-well plate and pre-incubated for 12 h. Then, 10 μl of CCK-8 solution was added to each well. The cell number was evaluated at 0, 24 h, 48 h, 72 h, 96 h and the absorbance was 450 nm using Epoch Microplate Spectrophotometer (Bio Tek). The cell migration and invasion assays were measured by transwell invasion assay. Briefly, GC cells in different groups cultivated with DMEM medium without fetal bovine serum were put on the upper chamber coated (the invasion assays using BD Matrigel) in the 24-well pates. DMEM medium with 10% fetal bovine serum was put to the lower chamber. After 48 h incubation, cotton swabs were used to wipe off the GC cells from the upper chamber. The number of cells migrating or invasion to the lower chamber was calculated by inverted microscopy after Crystal violet staining. And the number of and the mean of number of cells in each field represented the invasive ability of the cells.
Western blotting analysis
The cells were lysed using RIPA Lysis buffer (Beyotime, China) supplemented with a Protease and Phosphatase Inhibitor Cocktail (Sigma). Cell lysates were centrifuged and then the protein concentration was calculated using the Pierce BCA protein assay kit (Beyotime, China). Proteins were separated by a 10% polyacrylamide gel and transferred to a PVDF membrane (Millipore, USA), then detected with anti-E-cadherin (1:1000, CST, USA), Vimentin (1:1000, CST, USA), N-cadherin (1:1000, CST, USA), ZEB2 (1:1000, CST, USA), LATS2 (1:1000, CST, USA) and The GAPDH (1:1000, CST, USA) was as the internal control. The protein bands were analyzed using Image J software.
RNA immunoprecipitation (RIP)
RNA immunoprecipitation (RIP) experiments were performed according to previous describe [13], and was used by a Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) according to the manufacturer’s instructions. EZH2 (CST, USA), LSD1 (CST, USA) and H3 trimethyl Lys 27(CST, USA) antibodies was used for RIP assays.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as described previously [14], and an EZ-CHIP KIT was used according to the manufacturer’s instruction (Millipore, USA). EZH2, LSD1 and H3 trimethyl Lys 27 antibody was used in the experiments. Quantification of immunoprecipitated DNA was detected by qRT-PCR with SYBR Green Mix (Takara). The ChIP data was calculated as a percentage relative to the input DNA.
Statistical analysis
The SPSS 18.0 was used for the statistical analysis. All of the values were analyzed as the mean ± SD and performed at least three independent experiments. Data were evaluated by using two-tailed Student’s t-test and one-way ANOVA. The P < 0.05 was considered to be statistically significant.
Results
CCAT2 is up-regulated in GC tissues and cells
QRT-PCR was used to analyze CCAT2 expression levels in 108 cases GC tissues and adjacent normal tissues. The results showed that CCAT2 was markedly up-regulated in GC tissues, compared with the adjacent normal tissues (Figure 1A, P < 0.05). We further detected the association between CCAT2 expression and clinicopathological features in GC patients. The GC patients were divided into two groups according to the CCAT2 median expression levels (2.92) in the GC tissues: lower CCAT2 expression group (n=52) (the change of relative expression < median CCAT2 fold) and higher CCAT2 expression group (n=56) (the change of relative expression > median CCAT2 fold). The results indicated that CCAT2 expression levels were significantly correction with tumor size, lymph node metastasis and TNM stage (P < 0.05, Table 1), but did not correlate with age, gender, Histological grade and so on in GC patients (P > 0.05, Table 1). Moreover, Multivariate analysis showed that lymph node metastasis, TNM stage and the expression of CCAT2 were independent prognostic indicator for disease-free survival (DFS) and the over survival (OS) time for GC patients. Patients with higher CCAT2 expression had a significantly poor disease-free survival (DFS) and the over survival (OS) time than those with lower expression (Figure 1B, 1C; Tables 2, 3). These results indicated that CCAT2 could act as a oncogene in GC patients.
Figure 1.
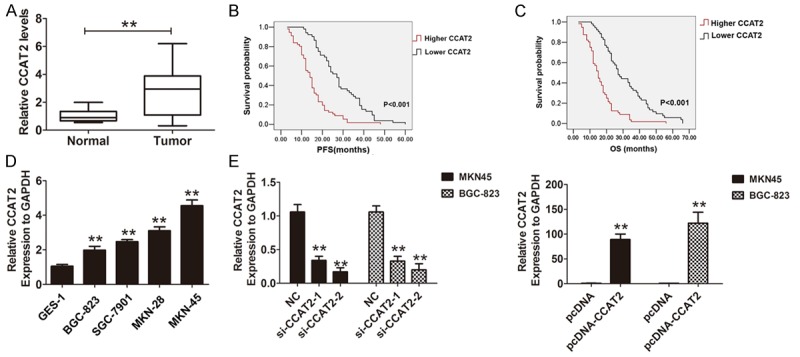
CCAT2 was up-regulated in GC tissues and cells. (A) The Relative expression of CCAT2 in 108 cases gastric tissues and adjacent normal tissues was analyzed by using qRT-PCR assays and normalized to GAPDH expression. Data are presented as mean ± S.D. (B, C) The expression of CCAT2 was classified into two groups according to the median value expression level in GC tissues sample. Kaplan-Meier method and the log-rank test were used to analyze the progression-free survival curves and the overall survival curves of GC patients in higher and lower CCAT2 expression groups. (D) The Relative expression of CCAT2 in four gastric cells and human gastric epithelial cell line GES-1 was analyzed by using qRT-PCR assays and normalized to GAPDH expression. Data are presented as mean ± S.D. (E) The Relative expression of CCAT2 was analyzed after silencing CCAT2 in MKN45 and BCG-823 cells and (E) was evaluated by transfecting pcDNA3.1-CCAT2 into MKN45 and BCG-823 cells, the normal control was GAPDH expression. The mean values and S.D were calculated from triplicates of a representative experiment. **P < 0.05.
Table 1.
Correlation between CCAT2 expression levels and clinicopathologic parameters in 108 GC patients was analyzed
| CCAT2 expression level | ||||
|---|---|---|---|---|
|
|
||||
| Clinicopathologic feathers | GC patients | Lower | Higher | χ2 test p-value |
| Gender | 0.749 | |||
| Female | 44 | 22 | 22 | |
| Male | 64 | 30 | 34 | |
| Age (years) | 0.106 | |||
| ≤ 60 | 62 | 34 | 28 | |
| > 60 | 46 | 18 | 28 | |
| Tumor size | 0.036** | |||
| < 5 cm | 68 | 38 | 30 | |
| > 5 cm | 40 | 24 | 26 | |
| Histological grade | 0.175 | |||
| High, middle differentiation | 55 | 30 | 25 | |
| Low differentiation | 53 | 22 | 31 | |
| Lymph node metastasis | ||||
| No | 50 | 35 | 15 | 0.001** |
| Yes | 58 | 17 | 41 | |
| Local invasion | 0.076 | |||
| T1, T2 | 59 | 33 | 26 | |
| T3, T4 | 49 | 19 | 30 | |
| TNM stage | 0.001** | |||
| I-II | 54 | 38 | 16 | |
| III-IV | 54 | 14 | 40 | |
p-value < 0.05 was considered statistically significant.
Table 2.
Univariate and multivariate analysis of disease-free survival in GC patients (n=108)
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| Age | 1.335 | 0.919-1.979 | 0.151 | |||
| Gender | 0.839 | 0.553-1.272 | 0.408 | |||
| Tumor size | 1.213 | 0.786-1.645 | 0.118 | |||
| Histological grade | 1.129 | 0.712-1.446 | 0.457 | |||
| Lymph node metastasis | 2.146 | 1.138-3.569 | 0.008** | 1.778 | 1.210-2.612 | 0.003** |
| Local invasion | 1.313 | 0.812-1.844 | 0.123 | |||
| TNM stage | 3.604 | 2.353-5.552 | 0.001** | 2.508 | 1.687-3.728 | 0.001** |
| CCAT2 | 2.526 | 1.700-3.755 | 0.001** | 2.305 | 1.554-3.418 | 0.001** |
p-value < 0.05 was considered statistically significant.
Table 3.
Univariate and multivariate analysis of overall survival in GC patients (n=108)
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| Age | 1.378 | 0.901-1.923 | 0.189 | |||
| Gender | 0.805 | 0.521-1.282 | 0.469 | |||
| Tumor size | 1.385 | 0.568-2.166 | 0.286 | |||
| Histological grade | 1.169 | 0.668-1.421 | 0.385 | |||
| Lymph node metastasis | 2.053 | 1.391-3.303 | 0.001** | 1.913 | 1.301-2.813 | 0.001** |
| Local invasion | 1.332 | 0.823-1.855 | 0.154 | |||
| TNM stage | 3.545 | 2.221-5.345 | 0.001** | 2.442 | 1.553-3.556 | 0.001** |
| CCAT2 | 2.346 | 1.537-3.556 | 0.001** | 2.108 | 1.442-3.202 | 0.001** |
p-value < 0.05 was considered statistically significant.
CCAT2 promotes cell proliferation, migration and invasion in vitro
We further investigated the role of CCAT2 in human GC cancer cells, the results suggested that CCAT2 was higher expression in four GC cells compared with human gastric epithelial cell line GES-1 (Figure 1D). Two si-CCAT2 oligos targeting CCAT2 were designed and transfected into MKN45 and BGC-823 cells, the results demonstrated that CCAT2 was effectively knocked down and especially was used by the si-CCAT2-2. Thus, the si-CCAT2-2 was used to perform the knockdown of CCAT2 in following experiments. The pcDNA3.1-CCAT2 was used to over-expression of CCAT2 in MKN 45 and BGC-823 cells (Figure 1E). Furthermore, we explored the effects of CCAT2 knock-down on cells proliferation abilities by CCK8 assays and the results showed that CCAT2 knockdown significantly inhibited the cells growth abilities in MKN45 and BGC-823 cells (Figure 2A, 2B). Moreover, we found that CCAT2 knockdown also significantly inhibited cell migration and invasion in MKN45 cells, and promoted the cell migration and invasion by introducing pcDNA3.1 into BGC-823 cells (Figure 2C-F). Therefore, our results demonstrated that CCAT2 could promote GC cell proliferation, migration and invasion in vitro.
Figure 2.
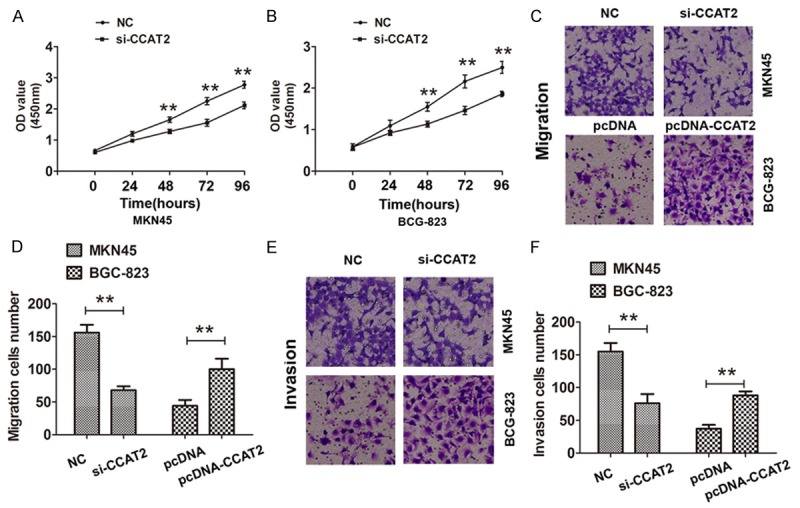
The effects of CCAT2 on GC cell proliferation migration, and invasion in vitro. A, B. CCK8 cell proliferation assays were used to determine the cell proliferation abilities in si-CCAT2-transfected MKN45 cells or pcDNA-CCAT2-transfected BCG-823 cells. C, D. Transwell migration assays were used to determine the cell migration abilities in si-CCAT2-transfected MKN45 cells or pcDNA-CCAT2-transfected BCG-823 cells. E, F. Transwell invasion assays were used to determine the cell invasion abilities in si-CCAT2-transfected MKN45 cells or pcDNA-CCAT2-transfected BCG-823 cells. The mean values and S.D were calculated from triplicates of a representative experiment. **P < 0.05.
CCAT2 promotes the epithelial-mesenchymal transition (EMT) in GC cells
To further study the role of CCAT2 in GC development, we investigated whether CCAT2 promoted the epithelial-mesenchymal transition (EMT) in MKN45 and BGC-823 cells. The results showed that knockdown of CCAT2 in MKN45 cells decreased the transcription factor ZEB2 and EMT mesenchymal markers Vimentin and N-cadherin expression, but up-regulating the epithelial marker E-cadherin expression (Figure 3A). Over-expression of CCAT2 in BGC-823 cells markedly inhibited the protein expression of E-cadherin, but up-regulated that of Vimentin and N-cadherin and transcription factor ZEB2, which contributed to promote GC cells EMT (Figure 3B). Previous study demonstrated that ZEB2 was a vital EMT inducer through suppressing E-cadherin expression or inducing Vimentin expression in human cancer [15]. Taken together, the present data indicated that CCAT2 could directly promote ZEB2 expression and regulate EMT process in GC cells.
Figure 3.
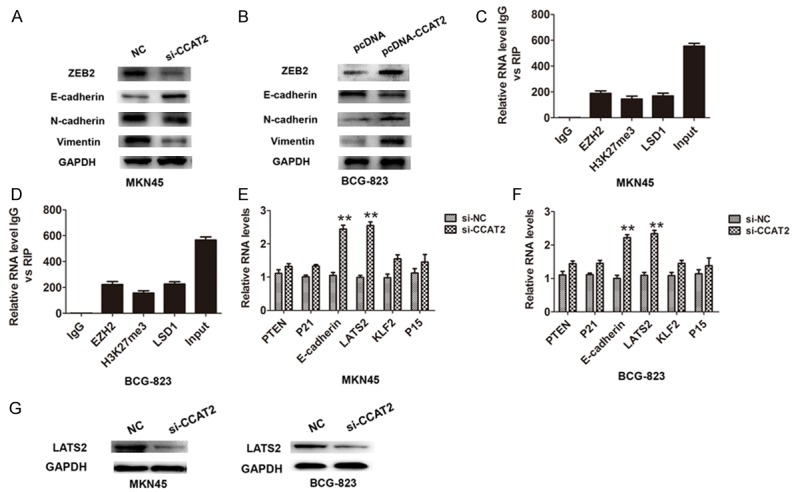
CCAT2 promoted GC cells epithelial-to-mesenchymal-transition and interacted with EZH2, H3k27me3 and LSD1. A, B. Western-blotting analysis was used to detect the protein expression of Transcription factor ZEB2 and EMT markers (E-cadherin, N-cadherin and Vimentin) in si-CCAT2-transfected MKN45 cells or pcDNA-CCAT2-transfected BCG-823 cells. C, D. RNA levels in immunoprecipitates with EZH2, LSD1, and H3K27me3 were determined by qRT-PCR analysis in MKN45 and BCG-823 cells. Expression levels of CCAT2 were presented as fold enrichment relative to IgG immunoprecipitate. E, F. The levels of PTEN,P21, E-cadherin, LAST2,KLF2 and P15 mRNA were detected by qRT-PCR after knockdown of CCAT2 in MKN45 and BCG-823 cells. The mean values and S.D were calculated from triplicates of a representative experiment. **P < 0.05. G. The protein expression of LAST2 were detected by western-blotting analysis after knockdown of CCAT2 in MKN45 and BCG-823 cells.
CCAT2 interacts with EZH2, H3K27me3 and LSD1 in GC cells
Recent evidences have reported that lncRNAs recruited polycomb-group proteins to specific loci and repressed gene expression, and some lncRNAs have been shown to physically associate with Polycomb Repressive Complex 2 (PRC2 complex) [16]. Furthermore, the RNA immunoprecipitation was performed and the results showed that the endogenous CCAT2 was enriched in the anti-EZH2, anti-H3K27me3 and anti-LSD1 immunoprecipitation (RIP) fraction relative to the input compared to the IgG fraction in MKN45 and BGC-823 cells (Figure 3C, 3D). Together, our results demonstrated a specific correlation between EZH2, H3K27me3 or LSD1 and CCAT2.
CCAT2 interacts with EZH2, H3k27me3, and LSD1 occupancy and epigenetically regulates the expression of E-cadherin and LATS2
Based on our findings that CCAT2 bound to EZH2, H3K27me3 and LSD1, we detected some targets of EZH2 and found that E-cadherin and LATS2 expression were significantly up-regulated when CCAT2 was knockdown in MKN45 and BGC-823 cells (Figure 3E, 3F). The protein expression analysis confirmed that the expression of LATS2 were also up-regulated in both cells (Figure 3G). Furthermore, the E-cadherin and LATS2 mRNA expression levels were also increased after EZH2 silencing in MKN45 and BGC-823 cells (Figure 4A, 4B). Further experiments to evaluate whether CCAT2 functioned as a transcriptional repression through EZH2 and targeted the E-cadherin and LATS2 promoters, we performed the ChIP assays and demonstrated that knockdown of CCAT2 significantly decreased the binding of EZH2, H3K27me3 and LSD1 levels across the E-cadherin and LATS2 promoters (Figure 4C, 4D). Moreover, functional experiments also demonstrated that knockdown of CCAT2 inhibited the cell proliferation and the effects were reversed by co-transfection of si-LATS2 and si-CCAT2 into MKN45 and BCG-823 cells. These results indicated that CCAT2 interacted with EZH2, H3k27me3, and LSD1 occupancy and epigenetically modulated the expression of E-cadherin and LATS2.
Figure 4.
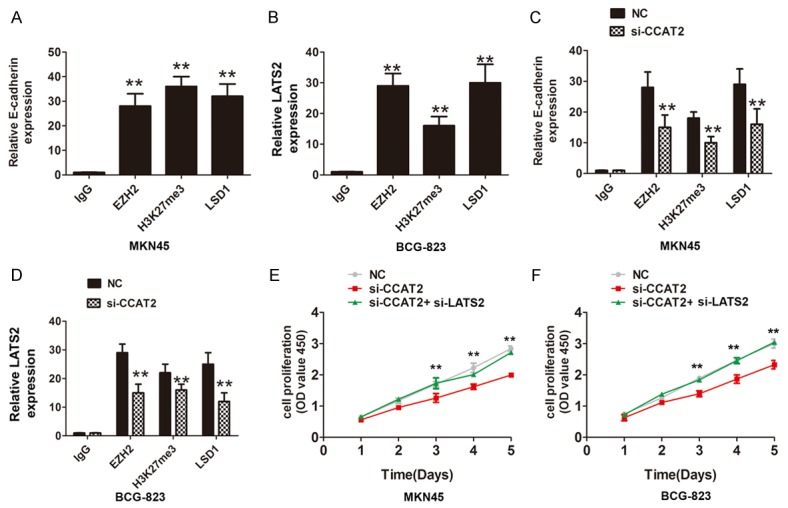
CCAT2 interacted with EZH2, H3k27me3 and LSD1 and repressed the LATS2 expression. A, B. Chromatin immunoprecipitation-qPCR was used to analyze EZH2 and LSD1 occupancy, H3K27me3 binding to E-cadherin and LATS2 promoter regions in MKN45 and BCG-823 cells, C, D. Chromatin immunoprecipitation-qPCR was used to analyze EZH2 and LSD1 occupancy, H3K27me3 binding to the LATS2 promoter regions after knockdown of CCAT2 in MKN45 and BCG-823 cells, the IgG was acted as a negative control. The mean values and S.D were calculated from triplicates of a representative experiment. **P < 0.05. E, F. CCK8 cell proliferation assays were performed to evaluate the cell growth when transfected si-NC, si-CCAT2 or si-CCAT2+si-LATS2 in MKN45 cells or BCG-823 cells.
CCAT2 inhibits the tumor growth and up-regulated the expression of E-cadherin and LATS2 in vivo
To explore the effect of lncRNA CCAT2 on tumor growth in vivo, we constructed stable cell lines by using the lentivirus vector to knockdown CCAT2 in MKN45 cells, the results indicated that the tumor volume in CCAT2 knockdown group was significantly smaller than that in the control group and the tumor growth increased slower in CCAT2 knockdown group than that in the control group (Figure 5A, 5B). Furthermore, we also demonstrated that E-cadherin and LATS2 expression were up-regulated in CCAT2 knockdown group than that in the control group (Figure 5C, 5D). Thus, our results indicated that CCAT2 inhibited the tumor growth and up-regulated the expression of E-cadherin and LATS2 in vivo.
Figure 5.
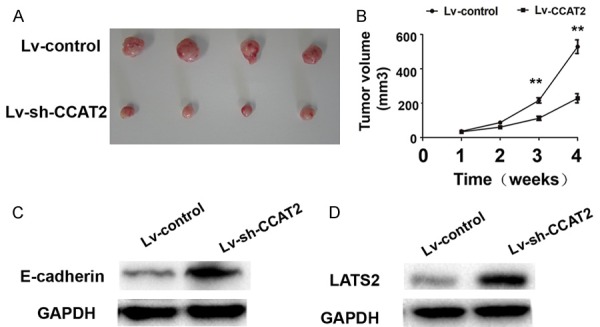
CCAT2 inhibited the tumor growth and up-regulated the expression of E-cadherin and LATS2 in vivo. (A) The stable CCAT2 knockdown MKN45 cells were used for the in vivo study. The nude mice carrying tumors from lv-shRNA-CCAT2 group and lv-control group were shown and (B) tumor growth curves were measured per week after the injection of MKN45 cells in lv-shRNA-CCAT2 group and lv-control group were shown. (C, D) Western-blotting analysis was performed to detect the expression of E-cadherin and LATS2 in tumor tissues in lv-shRNA-CCAT2 knockdown and control group. The mean values and S.D were calculated from triplicates of a representative experiment. **P < 0.05.
Discussion
LncRNAs play a critical role in cancer development and progression, previously some study reported that abnormally expressed cancer-related lncRNAs were identified involving in GC and affected the development and progression of GC [17,18]. For example, H19 was up-regulated in GC and promoted cell proliferation by partly inactivating p53, decreasing its activity, and suppressing the expression of the p53 target protein Bax [19]. Xu et al reported that high expression level of HOTAIR was a predictor of poor over-all survival in GC patients and suppression of HOTAIR could reverse GC cells EMT process [20]. Other lncRNA such as, SPRY4-IT1 [21], LEIGC [22], TUG1 [23], NEAT1 [24] and so on, were also involving in GC carcinogenesis. In our study, we proved that the expression levels of CCAT2 was up-regulated in GC tissues and higher CCAT2 expression had a significantly poor disease-free survival (DFS) and the over survival (OS) time than those with lower expression in GC patients. Over-expression of CCAT2 promoted the GC cell proliferation, migration and invasion, which indicated the oncogene role of CCAT2 in GC.
Further we found that over-expression of CCAT2 promoted the epithelial-mesenchymal transition (EMT) by up-regulating the ZEB2 expression and inhibiting the E-cadherin expression. Zheng et al demonstrated that up-regulation of long non-coding RNA CCAT2 indicated a poor prognosis for prostate cancer and promoted metastasis by affecting epithelial-mesenchymal transition [25], which was consistent with our findings. Moreover, our results revealed the potential Molecular Mechanisms that CCAT2 interacted with EZH2, H3k27me3 and LSD1 occupancy and epigenetically regulated the expression of E-cadherin and LATS2. Recent studies had identified many promoter-associated RNAs that altered gene transcription through interaction with protein complexes. Several studies have suggested that the LncRNAs were related to interact with EZH2 and inhibited EZH2 related target genes. Kong et al reported that Long non-coding RNA PVT1 indicated a poor prognosis of gastric cancer and promoted cell proliferation through epigenetically regulating p15 and p16 [26]. Liu et al reported that HOTAIR epigenetically silenced miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer [27]. Long non-coding RNA HOXA-AS2 also been indicated to promote gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expression [28]. Our results demonstrated that CCAT2 also bound to EZH2 and LSD1 and promoted GC progression through epigenetically regulating E-cadeherin and LAST2.
In conclusion, our results revealed that the lncRNA CCAT2 was over-expression in GC and higher CCAT2 predicted a poor survival outcome in GC patients. Further analysis suggested that the lncRNA CCAT2 recruited EZH2 to repress E-cadherin and LATS2, which promoted gastric cancer progression. To sum up, our study revealed CCAT2 served as an oncogene in GC and provided a therapeutic target for GC patients.
Acknowledgements
The authors would like to thank Cardiovascular Department, Hebei Chest Hospital, for its general help.
Disclosure of conflict of interest
None.
Authors’ contribution
Yong-Jun Wang and Pei Lv contributed in study concept and design, execution of experiments and writing of the manuscript. Jian-Zhen Liu, Yi Dang and Jiang-Yan Gao performed statistical analysis and interpretation of data. Yong Wang, Yong-Jun Wang and Pei Lv collected tissue samples and clinical data and performed the experiments.
References
- 1.Kanda M, Kodera Y. Recent advances in the molecular diagnostics of gastric cancer. World J Gastroenterol. 2015;21:9838–9852. doi: 10.3748/wjg.v21.i34.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 3.Wöhrer SS, Raderer M, Hejna M. Palliative chemotherapy for advanced gastric cancer. Ann Oncol. 2004;15:1585–1595. doi: 10.1093/annonc/mdh422. [DOI] [PubMed] [Google Scholar]
- 4.Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okugawa Y, Toiyama Y, Hur K. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui P, Zhang Y, Huang G. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, Buiga R, Pop V, Irimie A, Fodde R, Bedrosian I, Martens JW, Foekens JA, Berindan-Neagoe I, Calin GA. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4:1748–1762. doi: 10.18632/oncotarget.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ, Hu JH. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2015;8:779–785. [PMC free article] [PubMed] [Google Scholar]
- 13.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, De W, Wang KM, Wang ZX. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, De W, Shu YQ, Wang ZX. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Wang Y, Shan B, Han J, Zhu H, Lv Y, Fan X, Sang M, Liu XD, Liu W. The downregulation of miR-200c/141 promotes ZEB1/2 expression and gastric cancer progression. Med Oncol. 2015;32:428. doi: 10.1007/s12032-014-0428-3. [DOI] [PubMed] [Google Scholar]
- 16.Benetatos L, Voulgaris E, Vartholomatos G, Hatzimichael E. Non-coding RNAs and EZH2 interactions in cancer: long and short tales from the transcriptome. Int J Cancer. 2013;133:267–274. doi: 10.1002/ijc.27859. [DOI] [PubMed] [Google Scholar]
- 17.Zhang F, Zhang L, Zhang C. Long noncoding RNAs and tumorigenesis: genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol. 2016;37:163–175. doi: 10.1007/s13277-015-4445-4. [DOI] [PubMed] [Google Scholar]
- 18.Deng K, Wang H, Guo X, Xia J. The cross talk between long, non-coding RNAs and microRNAs in gastric cancer. Acta Biochim Biophys Sin (Shanghai) 2016;48:111–116. doi: 10.1093/abbs/gmv120. [DOI] [PubMed] [Google Scholar]
- 19.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 20.Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, Yu PF, Cheng XD. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng W, Wu G, Fan H, Wu J, Feng J. Long noncoding RNA SPRY4-IT1 predicts poor patient prognosis and promotes tumorigenesis in gastric cancer. Tumour Biol. 2015;36:6751–6758. doi: 10.1007/s13277-015-3376-4. [DOI] [PubMed] [Google Scholar]
- 22.Han Y, Ye J, Wu D, Wu P, Chen Z, Chen J, Gao S, Huang J. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. BMC Cancer. 2014;14:932. doi: 10.1186/1471-2407-14-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang E, He X, Yin D, Han L, Qiu M, Xu T, Xia R, Xu L, Yin R, De W. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109. doi: 10.1038/cddis.2015.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571–1579. doi: 10.1007/s00432-016-2152-1. [DOI] [PubMed] [Google Scholar]
- 25.Zheng J, Zhao S, He X, Zheng Z, Bai W, Duan Y, Cheng S, Wang J, Liu X, Zhang G. The up-regulation of long non-coding RNA CCAT2 indicates a poor prognosis for prostate cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2016;480:508–514. doi: 10.1016/j.bbrc.2016.08.120. [DOI] [PubMed] [Google Scholar]
- 26.Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM, Xia R, Wan L, Sun M, Wang ZX, De W, Zhang ZH. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH, Xu TP, De W, Liu BR, Wang ZX. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. doi: 10.1038/cddis.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie M, Sun M, Zhu YN, Xia R, Liu YW, Ding J, Ma HW, He XZ, Zhang ZH, Liu ZJ, Liu XH, De W. Long noncoding RNA HOXA-AS2 promotes gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expression. Oncotarget. 2015;6:33587–33601. doi: 10.18632/oncotarget.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]


