Abstract
Human T-cell leukemia virus type 1 (HTLV-1) is an oncogenic retrovirus that is the etiological agent of adult T-cell leukemia (ATL). The HTLV-1 basic leucine zipper factor (HBZ), which is encoded by the minus strand of the provirus, is constitutively expressed in all ATL patient cells and likely contributes to the development and maintenance of ATL. Furthermore, the overexpression of the myeloid cell leukemia 1 (MCL1) protein is frequently observed in hematological cancers as well as several other types of cancers. Here, we found that the expression of HBZ in cells stabilized MCL1 protein expression and suppressed the MCL1-mediated release of cytochrome c from the mitochondria. This effect was mediated by inhibition of the ubiquitin-dependent degradation of MCL1. In a serial binding assay, HBZ interacted with cullin 1 (CUL1) through a head-to-tail interaction. The association between CUL1 and Skp1, which serves as the molecular scaffold for the components of SCF ubiquitin ligase complexes, was markedly repressed in the presence of HBZ. Mechanistic analysis indicated that HBZ abrogated the CUL1 association with Skp1, which in turn promoted the cellular expression of MCL1. This novel function of HBZ likely plays a role in the viral pathogenesis of HTLV-1 and provides important insights into our understanding of the development of ATL.
INTRODUCTION
Human T-cell leukemia virus type 1 (HTLV-1) infects at least 5 million to 10 million people worldwide and is the causative agent of adult T-cell leukemia (ATL) (1–4). In most cases, HTLV-1 infection is transmitted through breast milk, blood cells, and dendritic cells in vivo (5). The majority of HTLV-1 carriers do not develop any significant clinical symptoms throughout their lives; however, approximately 5% of HTLV-1-infected subjects progress to ATL (6). Although ATL was discovered 40 years ago, there is still no effective treatment for this disease, in part because the underlying mechanisms of HTLV-1-mediated oncogenesis have not been fully elucidated.
The HTLV-1 genome encodes three common retroviral structural and enzymatic proteins (gag, pol, and env proteins) and is flanked by long terminal repeats (LTR) at each end. There is a pX region located between the env gene and the 3′ LTR (7). The plus strand of the pX region encodes regulatory and accessory proteins, including p12I, p21I, p13II, p30II, Rex, and Tax (7). It is well established that the Tax protein is a potent oncoprotein that either strongly activates or inactivates the transcription of target genes as well as interacts with numerous cellular factors that promote the survival and immortalization of HTLV-1-infected T cells (8–13). Interestingly, the Tax protein was detected in only 40% of ATL cases (14, 15) due to nonsense mutations, insertions, deletions, and epigenetic alterations, such as DNA methylation and histone modifications of the 5′ LTR of the HTLV-1 provirus (16–19). These studies suggested that Tax is required for the virus to enhance viral spreading during the earliest stage of HTLV-1 infection but that it is not necessary for the development of ATL.
It has previously been reported that the HTLV-1 basic leucine zipper factor (HBZ) is transcribed from the 3′ LTR of the HTLV-1 provirus (20). The HBZ protein contains a transactivation domain in its N-terminal region and a basic leucine zipper (bZIP) domain in its C-terminal region (20, 21). Nuclear translocation of HBZ is directed by three potential nuclear localization signals (21). Also, HBZ contains a functional nuclear export signal motif within its N-terminal region and can shuttle between the cytoplasm and nucleus (22). Previous reports showed that HBZ interacts with various cellular factors and modulates their functional activities in both the cytoplasmic and nuclear compartments (22, 23). In contrast to Tax, HBZ is constitutively expressed in all ATL patient samples because its 3′ LTR is conserved and unmethylated in ATL patient cells (24, 25). Although these studies suggest that HBZ may play an important role in the pathogenesis of ATL, its function has not yet been identified (26).
A recent study reported that myeloid cell leukemia 1 (MCL1) enhances cell survival and antiapoptosis by suppressing cytochrome c release from mitochondria. However, the overexpression of the MCL1 protein was markedly increased in several types of human cancers, including breast cancer, lung cancer, and leukemia (27, 28). While these data suggest that the abnormal expression of MCL1 may play an important role in carcinogenesis, there are no reports of its role in the development of ATL. The purpose of this study was to investigate the role of HBZ in regulating MCL1 expression in HTLV-1-infected cells.
MATERIALS AND METHODS
Cell culture.
HEK293T, NIH 3T3, and HeLa cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Biowest), 100 U/ml penicillin, and 100 μg/ml streptomycin. Jurkat, MT-1, ATL43T, and SLB-1 cells were grown in RPMI 1640 medium (Sigma) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% FBS. All cells were maintained at 37°C in a 5% CO2 atmosphere.
Plasmid construction.
The coding region for CUL1 was isolated by reverse transcription-PCR (RT-PCR) from total RNA derived from human testicular tissue and was then cloned into the BamHI sites of pcDNA3 containing a 3× FLAG epitope tag, a 5× hemagglutinin (HA) epitope tag, or a 6× Myc epitope tag at the N terminus. Primer CUL1-full length (CUL1-full) was used, and the forward and reverse sequences were 5′-AAGGATCCATGTCGTCAACCCGGAGCC-3′ and 5′-AAGGATCCTTAAGCCAAGTAACTGTAGGTG-3′, respectively, where the underlined sequences here and in the primer sequences below denote restriction sites. The plasmids used for expression of CUL1 deletion mutants were generated by PCR using pcDNA3-6× Myc-CUL1-full as a template. The primers and their sequences were as follows: CUL1-A, 5′-AAGGATCCATGTCGTCAACCCGGAGCC-3′ (forward) and 5′-AAGGATCCCTATGTGTCAGCCAAAAATTGA-3′ (reverse); CUL1-B, 5′-AAGGATCCGAGAGATTTTATACCAGAGAGA-3′ (forward) and 5′-AAGGATCCTTATTGCTCGTTCAGATCTTTGCT-3′ (reverse); and CUL1-C, 5′-AAGGATCCTTCAAAAAGCACTTGACAAACT-3′ (forward) and 5′-AAGGATCCTTAAGCCAAGTAACTGTAGGTG-3′ (reverse). The HBZ-related plasmids and the expression plasmid for Tax have been described previously (22, 29). The coding region for MCL1 was obtained by RT-PCR from total RNA derived from human testicular tissue and was cloned into the EcoRI sites of pcDNA3 containing a 3× FLAG epitope tag or a 5× HA epitope tag at the N-terminal region. Primer MCL1 was used, and the forward and reverse sequences were as follows: 5′-AAGAATTCATGTTTGGCCTCAAAAGAAACGC-3′ and 5′-AAGAATTCCTATCTTATTAGATATGCCAAA-3′, respectively. The coding regions for Rbx1, Skp1, and Fbw7 were obtained by RT-PCR from total RNA derived from human testicular tissue and were cloned into the BamHI/XhoI (Rbx1 and Skp1) or EcoRI (Fbw7) sites of pcDNA3 containing a 3× FLAG epitope tag or a 6× Myc epitope tag at the N-terminal region. The primers and their sequences were as follows: Rbx1, 5′-AAGGATCCATGGCGGCAGCGATGGATGTGGAT-3′ (forward) and 5′-AACTCGAGCTAGTGCCCATACTTTTGGAATT-3′ (reverse); Skp1, 5′-AAGGATCCATGCCTTCAATTAAGTTGCAGA-3′ (forward) and 5′-AACTCGAGTCAAAGACAAAACTGTGTGCTA-3′ (reverse); and Fbw7, 5′-AAGAATTCATGAATCAGGAACTGCTCTCTGT-3′ (forward) and 5′-AAGAATTCTCACTTCATGTCCACATCAAAGT-3′ (reverse).
Immunoprecipitation and immunoblotting.
HEK293T or NIH 3T3 cells (1 × 106 cells per 6-cm-diameter dish) were transfected with expression plasmids using the Lipofectamine 2000 reagent (Thermo Scientific Lafayette) according to the manufacturer's instructions. After 36 h, the cells were lysed in 1 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.5% NP-40, 1 mM EDTA, 50 mM NaF, 2 mM Na3VO4, the Pefabloc SC Plus inhibitor [Roche Diagnostics], a cOmplete protease inhibitor cocktail tablet [Roche Diagnostics]), and the cell debris was removed by centrifugation for 15 min. Lysates were incubated with antibodies for 1 h at 4°C, and antibody complexes were captured using protein G-Sepharose beads (GE Healthcare Bio-Sciences AB) for 1 h at 4°C. The protein-bound beads were washed four times with lysis buffer, and the immunoprecipitated proteins were eluted in SDS sample buffer. The proteins were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane (Polyscreen; PerkinElmer), blocked with 7.5% skim milk–phosphate-buffered saline containing 0.05% Tween (PBST), probed with the appropriate antibodies, and developed by chemiluminescence. The antibodies used in this study were specific for FLAG (antibody M2; Sigma), Myc (antibody 9E10; Santa Cruz Biotechnology), HA (antibodies M132-3 [MBL] and 3F10 [Roche Diagnostics]), MCL1 (Abcam), α-tubulin (antibody Ab-1; Calbiochem), horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (True blot; eBioscience), HRP-conjugated goat anti-rat IgG (Jackson ImmunoResearch Laboratories), and HRP-conjugated goat anti-rabbit IgG (Dako). The anti-HBZ-specific polyclonal antibody and the anti-Tax-specific monoclonal antibody were previously described (22, 30).
Lentiviral vector and transfection.
The lentivirus-based transfection system was a generous gift from H. Miyoshi (Riken, Tsukuba, Japan). Short hairpin RNA (shRNA) targeting luciferase (Luc) was used as a negative control (pCS-RfA-CG-GFP-shLuc). HEK293T cells were transfected with pCS-RfA-CG-GFP-shHBZ or pCS-RfA-CG-GFP-shLuc, a vesicular stomatitis virus glycoprotein G (VSV-G)- and respiratory syncytial virus (RSV) Rev-expressing construct (pCMV-VSV-G-RSV-Rev), and a packaging construct (pMDLg-pRRE) in the presence of 25 mM chloroquine using the Lipofectamine 2000 reagent. At 10 h posttransfection, 10 mM sodium butyrate was added to the culture medium. After 16 h, the culture medium was changed and fresh medium containing 10 mM forskolin was added. Culture supernatants were collected 48 h after transfection and concentrated 100- to 200-fold by ultrafiltration. The titers of the concentrated viruses were measured on SLB-1 cells on the basis of their levels of green fluorescent protein (GFP) expression. Cells were infected at a multiplicity of infection of 20 in the presence of Polybrene (4 mg/ml).
In vivo ubiquitination assay.
HEK293T cells (1 × 106 cells per 6-cm-diameter dish) were transfected with expression plasmids using the Lipofectamine 2000 reagent. At 24 h posttransfection, 30 μM MG132 (Wako Pure Chemicals), a proteasome inhibitor, was added. After 15 h, transfected cells were lysed in buffer A (8 M urea, 30 mM imidazole, 0.5% Triton X-100, 100 mM HEPES-KOH [pH 7.5]) and incubated with 20 μM Ni-nitrilotriacetic acid (NTA) beads for 2 h at room temperature. The beads were washed with buffer B (8 M urea, 50 mM imidazole, 0.5% Triton X-100, 100 mM HEPES-KOH [pH 7.5], 0.5 M NaCl). The bound proteins were eluted with SDS sample buffer containing 5 mM EDTA and then subjected to SDS-PAGE, followed by immunoblot analysis (see Fig. 2A).
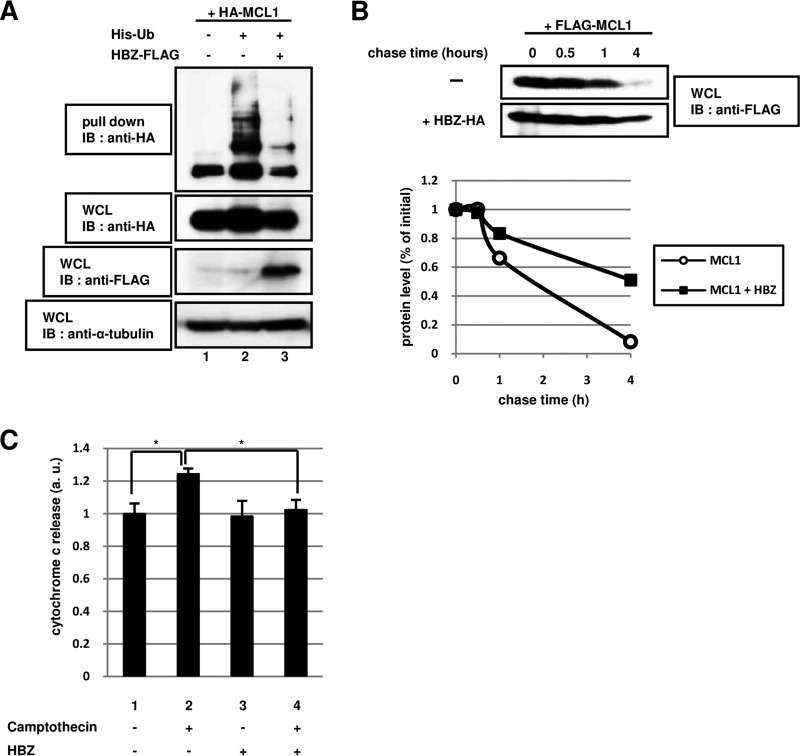
FIG 2.
The degradation of MCL1 by the ubiquitin-dependent pathway is controlled by HBZ. (A) NIH 3T3 cells were transfected (+) or not transfected (−) with 2 μg of pcDNA3-HA-MCL1 (HA-MCL1), pcDNA3-His-Ub (His-Ub), and pcDNA3-HBZ-FLAG (HBZ-FLAG), as indicated. After 24 h, the cells were treated with 20 μM MG132 (a proteasome inhibitor) for 15 h. Following purification with Ni-NTA beads, bound proteins were detected by immunoblotting with anti-HA antibody. Total protein levels in whole-cell lysates were analyzed by immunoblotting using anti-HA, anti-FLAG, and anti-α-tubulin antibodies. (B) HEK293T cells were transfected with 1 μg of a plasmid expressing FLAG-MCL1 with (+) or without (−) 3 μg of a plasmid expressing HBZ-HA. After 36 h, the cells were treated with 50 μM cycloheximide (a protein synthesis inhibitor) and then collected at the indicated times. Cell lysates were analyzed by immunoblotting using an anti-FLAG antibody. The intensity of each band was quantified and graphed. (C) HeLa cells were transfected (+) or not transfected (−) with 2 μg of pcDNA3-HBZ (HBZ). After 36 h, the cells were treated with 100 μM camptothecin for 6 h. The level of cytochrome c released was measured using a cytochrome c ELISA kit. Each value represents the mean from three independent experiments ± SE (*, P < 0.05). WCL, whole-cell lysates; IB, immunoblotting; a.u., arbitrary units.
HEK293T cells were transfected with expression plasmids using the Lipofectamine 2000 reagent. After treatment with 30 μM MG132 for 15 h, the cells were lysed in 1 ml of lysis buffer C (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate [Nakarai], Pefabloc SC Plus inhibitor, a cOmplete protease inhibitor cocktail tablet), and cell debris was removed by centrifugation for 15 min. The lysates were incubated with antibodies for 1 h at 4°C, and then antibody complexes were captured with protein G-Sepharose beads for 1 h. The beads were washed four times with lysis buffer C, and immunoprecipitants were eluted. Samples were subjected to SDS-PAGE followed by immunoblot analysis (see Fig. 6A).
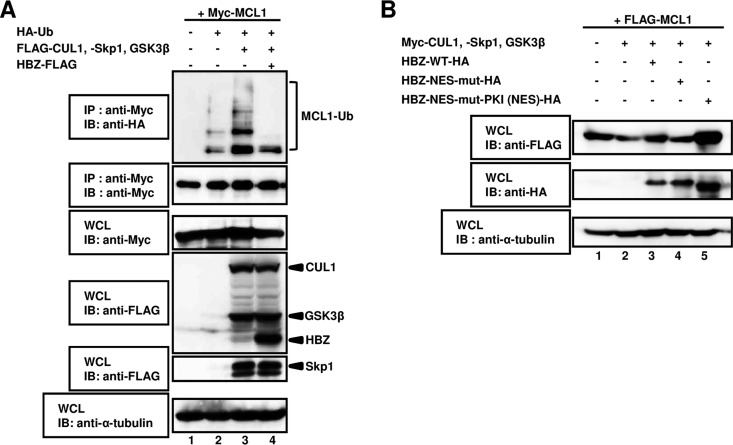
FIG 6.
HBZ controls the stabilization of MCL1 by inhibiting SCF E3 ligase activity. (A) HEK293T cells were cotransfected with 1 μg of a plasmid expressing Myc-MCL1 with (+) or without (−) 0.5 μg of a plasmid expressing HA-Ub, 2 μg of a plasmid expressing FLAG-CUL1, FLAG-Skp1, and FLAG-tagged glycogen synthase kinase 3β (FLAG-GSK3β), and 5 μg of a plasmid expressing HBZ-FLAG. After 24 h, cells were treated with MG132 for 15 h. The cell extracts were prepared and subjected to immunoprecipitation using an anti-Myc antibody, followed by immunoblot analysis with anti-HA and anti-Myc antibodies. Total protein levels in whole-cell lysates were analyzed by immunoblotting using anti-Myc, anti-FLAG, and anti-α-tubulin antibodies. (B) HEK293T cells were cotransfected with 0.5 μg of a plasmid expressing FLAG-MCL1 with (+) or without (−) 1 μg of a plasmid expressing Myc-CUL1, Myc-Skp1, or Myc-GSK3β and 2.5 μg of a plasmid expressing HBZ-WT-HA, HBZ-NES-mut-HA, or HBZ-NES-mut-PKI (NES)-HA. After 36 h, the cell lysates were then subjected to SDS-PAGE, followed by immunoblotting with anti-FLAG, anti-HA, and anti-α-tubulin antibodies. WCL, whole-cell lysates; IB, immunoblotting; IP, immunoprecipitation.
Pulse-chase experiment.
HEK293T cells were seeded on 6-well plates (3 × 105 cells/well) and transfected with the appropriate expression plasmids. After 36 h, the cells were treated with 50 μM cycloheximide to inhibit protein synthesis and were chased for the intervals indicated below. Harvested cells were resolved by SDS-PAGE and analyzed by Western blotting. Band intensities were measured using a LAS 3000 image analyzer (Fuji film).
Quantification of cytochrome c release.
Detection of cytochrome c release was performed using a cytochrome c enzyme-linked immunosorbent assay (ELISA) kit (R&D Systems). HeLa cells were cultured and treated with 100 μM camptothecin, which induces the release of cytochrome c, or control diluent for 6 h. The cell extracts were processed according to the manufacturer's protocol.
Semiquantitative RT-PCR analysis.
Total RNA was extracted with the Isogen reagent (Nippon Gene) following the manufacturer's instructions. Then, total RNA was reverse transcribed using SuperScript IV reverse transcriptase (Invitrogen) following the manufacturer's directions. Samples were analyzed by semiquantitative PCR of MCL1 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and the resulting PCR products were separated by electrophoresis on a 1.5% agarose gel. The primer sequences used were as follows: for MCL1, 5′-AAGGATCCATGTTTGGCCTCAAAAGAAACGCGG-3′ (forward) and 5′-AAGAAAAGCAGCCTCGCGGGGGT-3′ (reverse), and for GAPDH, 5′-ATGGGGAAGGTGAAGGTCGG-3′ (forward) and 5′-TGGAGGGATCTCGCTCCTGG-3′ (reverse).
RESULTS
HBZ regulates the stability of the MCL1 protein.
Previous reports described an association between abnormal levels of the myeloid cell leukemia 1 (MCL1) protein and the development of some cancers (27, 28, 31). However, scientific studies do not suggest a link between the level of MCL1 protein expression in HTLV-1-infected cells and the pathogenesis of adult T-cell leukemia (ATL) has not been studied. HTLV-1 bZIP factor (HBZ) is thought to be necessary for the development of ATL. Therefore, we examined whether the steady-state level of the MCL1 protein in cells is controlled by HBZ. HBZ is expressed as an unspliced isoform (HBZ-US) and as a spliced isoform (HBZ-SI) in ATL patient cells (32). Moreover, the amino acid sequences of HBZ-US and HBZ-SI are highly conserved, with the only difference being 6 amino acids in the N terminus (32). This experiment used the HBZ-US (HBZ) expression vector. HEK293T cells were transfected with a plasmid expressing FLAG-tagged MCL1 and an HA-tagged HBZ expression vector. Overexpression of HBZ resulted in increased MCL1 protein levels (Fig. 1A, lane 2).
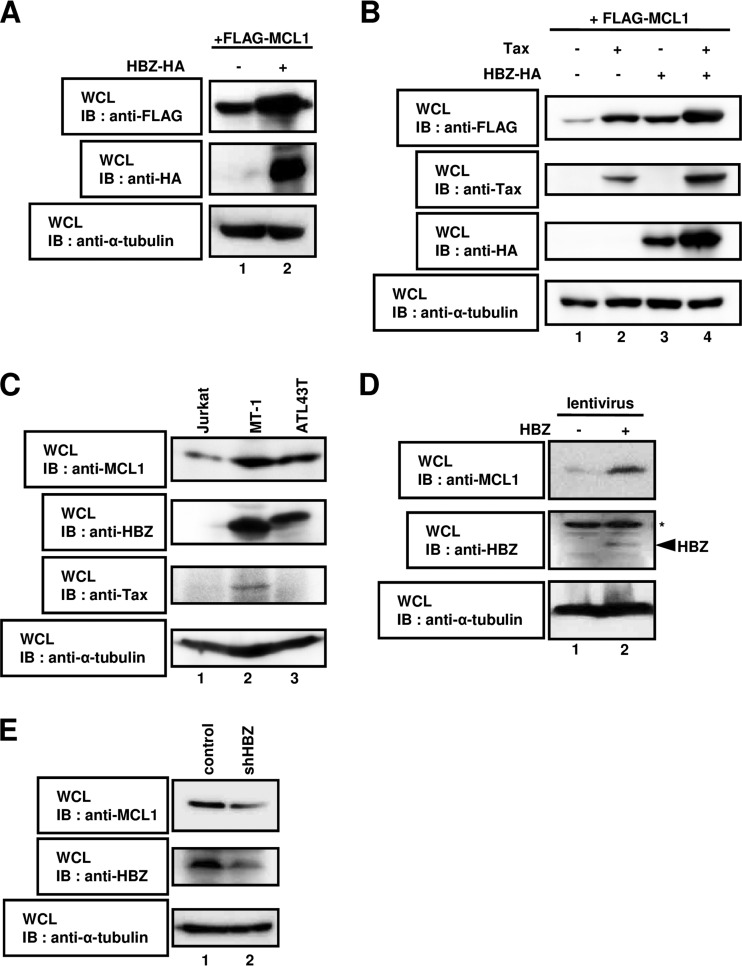
FIG 1.
HBZ regulates the stability of the MCL1 protein. (A) HEK293T cells were cotransfected with 1 μg of a plasmid expressing FLAG-MCL1 with (lane +) or without (lane −) 3 μg of a plasmid expressing HBZ-HA. After 36 h, cell lysates were subjected to SDS-PAGE, followed by immunoblotting with anti-FLAG, anti-HA, or anti-α-tubulin antibodies. (B) HEK293T cells were cotransfected with 0.5 μg of a plasmid expressing FLAG-MCL1 with (lanes +) or without (lanes −) 2.5 μg of a plasmid expressing Tax and HBZ-HA. After 36 h, cell lysates were subjected to SDS-PAGE, followed by immunoblotting with anti-FLAG, anti-Tax, anti-HA, and anti-α-tubulin antibodies. (C) Lysates prepared from Jurkat, MT-1, and ATL43T cells were subjected to immunoblot analysis with anti-MCL1, anti-HBZ, anti-Tax, and anti-α-tubulin antibodies. (D) The lentivirus-based expression system was used to create Jurkat cell lines that carried the empty vector (lane 1) or that stably expressed HBZ (lane 2). Whole-cell lysates were prepared and subjected to immunoblot analysis with anti-MCL1, anti-HBZ, and anti-α-tubulin antibodies. *, nonspecific bands. (E) SLB-1 cells were transduced with lentiviral particles containing shRNA targeting luciferase (control; lane 1) or HBZ (shHBZ; lane 2). Cell lysates were analyzed by immunoblotting using anti-MCL1, anti-HBZ, and anti-α-tubulin antibodies. WCL, whole-cell lysates; IB, immunoblotting.
It was recently reported that MCL1 production is also modulated by Tax (33). In this regard, we postulated that the level MCL1 may be increased by its coexpression with Tax and HBZ. We performed Western blot analysis of MCL1 in HEK293T cells expressing FLAG-MCL1 with or without exogenous Tax and/or HBZ expression (Fig. 1B). Similar to the findings presented in a previous report, the overexpression of Tax resulted in the stabilization of MCL1 (Fig. 1B, lane 1 versus lane 2) (33). As expected, coexpression of Tax and HBZ dramatically increased the level of MCL1 protein (Fig. 1B, lane 1 versus lane 4).
Next, we examined the expression level of endogenous MCL1 in an HTLV-1-uninfected cell line (Jurkat cells) and HTLV-1-infected cell lines (MT-1 and ATL43T cells). The cells were collected and subjected to immunoblotting with the antibodies indicated below. As shown in Fig. 1C, the expression level of endogenous MCL1 was increased in HTLV-1-infected cells (lanes 2 and 3) compared to HTLV-1-uninfected cells (lane 1).
In our previous report, we established Jurkat cell lines (human T lymphocytes) that constitutively expressed HBZ or the empty control vector using a lentiviral expression system (22) and examined the endogenous levels of the MCL1 protein using immunoblot analysis. The MCL1 protein levels were significantly increased in cells expressing HBZ (Fig. 1D, lane 2) compared to cells expressing the empty control vector (Fig. 1D, lane 1).
To determine whether the expression of short hairpin RNA (shRNA) targeting HBZ in cells reduces endogenous MCL1 protein levels, shRNA against the HBZ plasmid construct was introduced into the HTLV-1-infected cell line SLB-1. As shown in Fig. 1E, specific knockdown of endogenous HBZ resulted in reduced expression of MCL1 (lane 2). Collectively, our findings revealed that the stabilization of MCL1 is enhanced by expression of HBZ. It is possible that the increased expression of MCL1 by HBZ occurs at the transcriptional level. To confirm this point, we performed a semiquantitative RT-PCR assay. When HEK293T cells were expressed with HBZ, the amount of MCL1 was decreased compared with that in empty vector-transfected cells (see Fig. S1 in the supplemental material). These data indicate that the MCL1 mRNA level was significantly downregulated in the cells, in spite of the prolonged stability of the MCL1 protein by the expression of HBZ.
HBZ stabilizes the MCL1 protein by suppressing its ubiquitination.
It is generally accepted that most intracellular proteins are degraded by the ubiquitin (Ub)-proteasome system, and the majority of proteasomal substrates are targeted for degradation by conjugation to polyubiquitin chains. We therefore investigated whether expression of HBZ modulates the ubiquitination of MCL1 in cells. Histidine (His)-tagged Ub, HA-MCL1, and HBZ-FLAG were coexpressed in HEK293T cells, as indicated in Fig. 2A. After the cells were allowed to rest for 24 h posttransfection, the cells were treated with the proteasome inhibitor MG132 for 16 h. Total cell lysates from transfected cells were purified using Ni-NTA beads and subjected to immunoblot analysis to detect polyubiquitinated MCL1. The expression of HBZ dramatically reduced the level of ubiquitination of MCL1 compared to that in cells that did not express HBZ (Fig. 2A, lane 2 versus lane 3).
In addition to determining whether HBZ regulated the steady-state levels of MCL1 via ubiquitination, we performed a pulse-chase analysis of MCL1 in HEK293T cells expressing MCL1 with or without exogenous HBZ expression. The levels of the MCL1 protein declined more slowly in cells that expressed MCL1 and HBZ (Fig. 2B, bottom) than in cells that expressed MCL1 alone (Fig. 2B, top). These data indicate that HBZ enhanced the stability of the MCL1 protein by inhibiting its ubiquitination and degradation.
The antiapoptotic BCL-2 family member MCL1 has been shown to play a key role in protecting against apoptosis by inhibiting cytochrome c release from the mitochondria (34, 35). Therefore, we postulated that the release of cytochrome c would be suppressed through MCL1 protein stabilization by HBZ. Cytochrome c levels were measured in transiently transfected HeLa cells that were treated or not treated camptothecin for 6 h (Fig. 2C). As a positive control in these experiments, camptothecin increased the level of cytochrome c release in untransfected cells (Fig. 2C, lane 1 versus lane 2). On the other hand, camptothecin treatment attenuated cytochrome c release in HBZ-overexpressing cells (Fig. 2C, lane 2 versus lane 4), thus suggesting that HBZ may be involved in the inhibition of cytochrome c release by stabilizing the MCL1 protein.
Identification of CUL1 as a cellular factor that interacts with HBZ.
In order to clarify the mechanism by which the MCL1 protein is stabilized by HBZ expression in cells, we focused on the Skp-cullin-F box protein (SCF) complex, which is the well-recognized cellular E3 ubiquitin ligase for MCL1. Cullin 1 (CUL1) is an essential component of the SCF E3 ubiquitin ligase complex (36). We identified CUL1 to be an HBZ-interacting protein. It is known that the regulation of MCL1 stability is controlled by SCF-mediated ubiquitination (37). Thus, the interaction between HBZ and CUL1 is expected to affect the stability of MCL1 via inhibition of SCF-mediated degradation. To analyze the physiological interaction of HBZ with CUL1 in mammalian cells, we performed a coimmunoprecipitation (co-IP) experiment. In this study, HEK293T cells were transfected with plasmids expressing HA-tagged HBZ-US or HBZ-SI plus a FLAG-tagged CUL1 expression vector. At 36 h after transfection, total cell lysates were harvested and subjected to immunoprecipitation with an anti-HA antibody, followed by SDS-PAGE and Western blotting using an anti-FLAG antibody. HBZ-US and HBZ-SI were identified to be CUL1 binding partners (Fig. 3A, lanes 2 and 3).
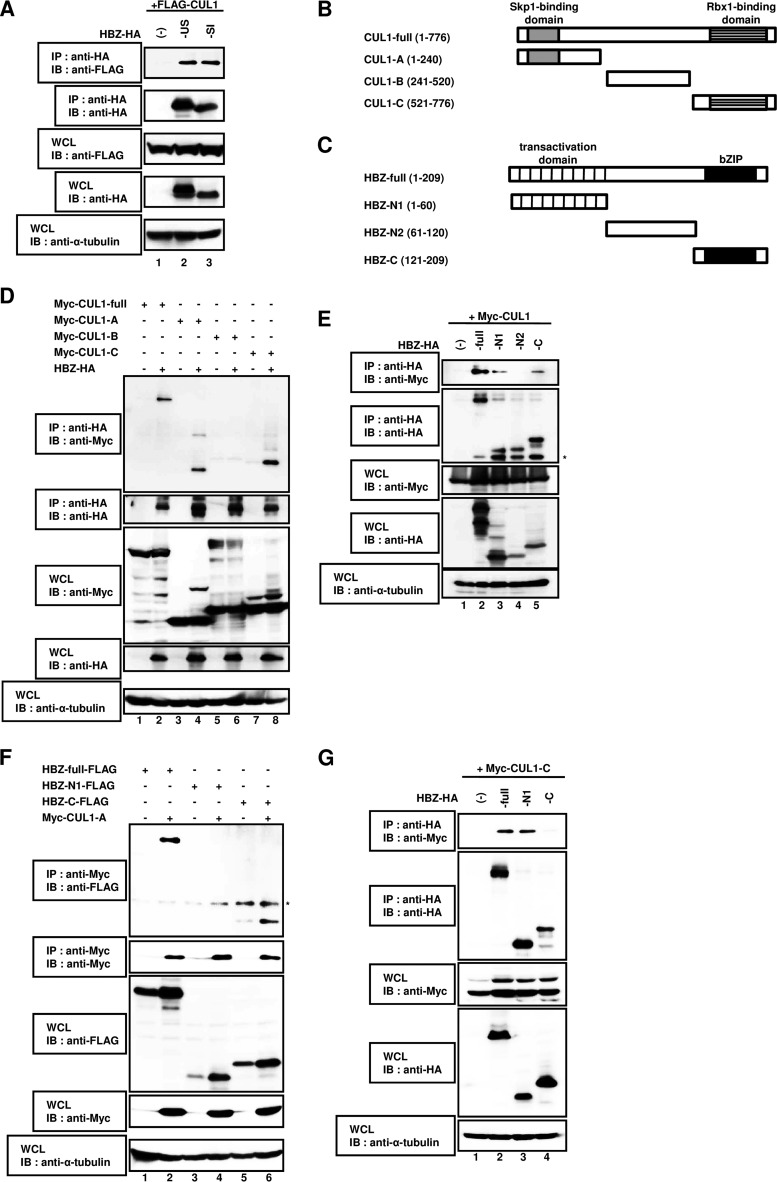
FIG 3.
The association between HBZ and CUL1 occurs by a head-to-tail interaction. (A) HEK293T cells were cotransfected with 3 μg of a plasmid expressing FLAG-CUL1 with (+) or without (−) 3 μg of a plasmid expressing the HBZ unspliced form-HA (-US) or the HBZ spliced isoform-HA (-SI). After 36 h, cell extracts were prepared and subjected to immunoprecipitation using an anti-HA antibody, followed by immunoblot analysis with anti-FLAG and anti-HA antibodies. Total protein levels in whole-cell lysates were analyzed by immunoblotting using anti-FLAG, anti-HA, and anti-α-tubulin antibodies. (B) Schematic diagram of full-length CUL1 (CUL1-full) and the CUL1 deletion mutants (CUL1-A, CUL1-B, and CUL1-C) used in this study. Characteristic domains of CUL1 are indicated. (C) Schematic diagram of full-length HBZ (HBZ-full) and the deletion mutants (HBZ-N1, HBZ-N2, and HBZ-C) used in this study. Characteristic domains of HBZ are indicated. (D) HEK293T cells were cotransfected with 3 μg of a plasmid expressing HBZ-HA with or without 3 μg of a plasmid expressing Myc-full-length CUL1, CUL1-A, CUL1-B, or CUL1-C. After 36 h, cell extracts were prepared and subjected to immunoprecipitation using an anti-HA antibody, followed by immunoblot analysis with anti-Myc and anti-HA antibodies. Total protein levels in whole-cell lysates were analyzed by immunoblotting using anti-Myc, anti-HA, and anti-α-tubulin antibodies. (E) HEK293T cells were cotransfected with 3 μg of a plasmid expressing Myc-CUL1 with or without 3 μg of a plasmid expressing full-length HBZ-HA, HBZ-N1-HA, HBZ-N2-HA, or HBZ-C-HA. After 36 h, cell extracts were prepared and subjected to immunoprecipitation using the anti-HA antibody, followed by immunoblot analysis with anti-Myc and anti-HA antibodies. Total protein levels in whole-cell lysates were analyzed by immunoblotting using anti-Myc, anti-HA, and anti-α-tubulin antibodies. (F) HEK293T cells were cotransfected with 3 μg of a plasmid expressing Myc-CUL1-A with or without 3 μg of a plasmid expressing HBZ-full-FLAG, HBZ-N1-FLAG, or HBZ-C-FLAG. After 36 h, cell extracts were prepared and subjected to immunoprecipitation using an anti-Myc antibody, followed by immunoblot analysis with anti-FLAG and anti-Myc antibodies. Total protein levels in whole-cell lysates were analyzed by immunoblotting using anti-FLAG, anti-Myc, and anti-α-tubulin antibodies. (G) HEK293T cells were cotransfected with 3 μg of a plasmid expressing Myc-CUL1-C with or without 3 μg of a plasmid expressing HBZ-full-HA, HBZ-N1-HA, or HBZ-C-HA. After 36 h, cell extracts were prepared and subjected to immunoprecipitation using the anti-HA antibody, followed by immunoblot analysis with anti-Myc and anti-HA antibodies. Total protein levels in whole-cell lysates were analyzed by immunoblotting using anti-Myc, anti-HA, and anti-α-tubulin antibodies. *, nonspecific bands; WCL, whole-cell lysates; IB, immunoblotting; IP, immunoprecipitation.
To define the HBZ and CUL1 sequences responsible for the interaction, we constructed deletion mutants (Fig. 3B and C) and performed a co-IP assay. The following experiments used the HBZ-US (HBZ) expression vector. As shown in Fig. 3D, HBZ was detected in the immunocomplexes isolated from cells expressing full-length CUL1 (CUL1-full), CUL1-A, and CUL1-C (lanes 2, 4, and 8, respectively) but not CUL1-B (lane 6). Likewise, full-length HBZ (HBZ-full), HBZ-N1, and HBZ-C interacted with CUL1 (Fig. 3E, lanes 2, 3, and 5, respectively) but not HBZ-N2 (Fig. 3E, lane 4). In order to identify more precisely which domains of HBZ are necessary for the association with CUL1, we performed co-IP assays using different combinations of HBZ and CUL1 expression vectors. The C-terminal region of HBZ interacted with CUL1-A (Fig. 3F, lane 6), while the N-terminal region of HBZ interacted with CUL1-C (Fig. 3G, lane 3). Taken together, these findings indicate that the head of HBZ interacts with the tail of CUL1 through a head-to-tail association.
Interaction between HBZ and CUL1 in the cytoplasm.
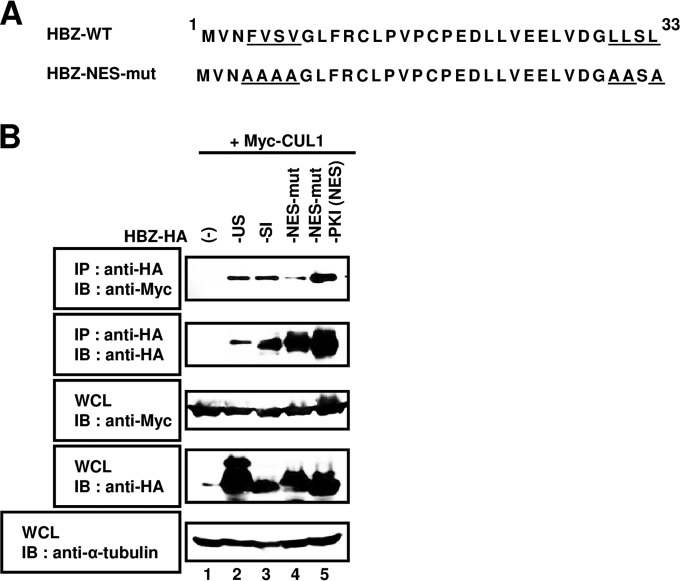
HBZ is known to have both a nuclear localization signal (NLS) and a nuclear export signal (NES), and it constantly shuttles between the nucleus and cytoplasm (21, 22). CUL1 is localized predominantly to the cytoplasm, thus suggesting that it would not interact with the NES mutant of HBZ that is retained in the nucleus. To determine whether an interaction between CUL1 and HBZ occurred in the cytoplasm, HBZ-related plasmids were generated as previously described and co-IP assays were performed (Fig. 4) (22). Lysates were immunoprecipitated with an anti-HA antibody and then immunoblotted using an anti-Myc antibody. The HA-HBZ-NES mutant (mut) that was not exported to the cytoplasm demonstrated a reduced interaction with CUL1 (Fig. 4B, lanes 2 and 3 versus lane 4). In contrast, we observed an association between CUL1 and HBZ-NES-mut-PKI (NES), which was exported to the cytoplasm (Fig. 4B, lane 5). Collectively, our results reveal that the nuclear export of HBZ is essential for its interaction with CUL1.
FIG 4.
HBZ and CUL1 interact in the cytoplasm. (A) Schematic diagram of the HBZ-NES mutant (HBZ-NES-mut) used in this study. Amino acids altered to alanine are underlined. (B) HEK293T cells were cotransfected with 3 μg of pcDNA3-Myc-CUL1 with (+) or without (−) 3 μg of pcDNA3-HBZ-unspliced form-HA (-US), pcDNA3-HBZ-spliced isoform-HA (-SI), pcDNA3-HBZ-NES-mut-HA (-NES-mut), or pcDNA3-HBZ-NES-mut-PKI-HA [-PKI (NES)]. After 36 h, cell extracts were prepared and subjected to immunoprecipitation (IP) using anti-HA antibody, followed by immunoblotting (IB) with anti-Myc and anti-HA antibodies. Total protein levels in whole-cell lysates (WCL) were analyzed by immunoblotting using anti-Myc, anti-HA, and anti-α-tubulin antibodies.
HBZ abrogates the interaction of CUL1 with Skp1.
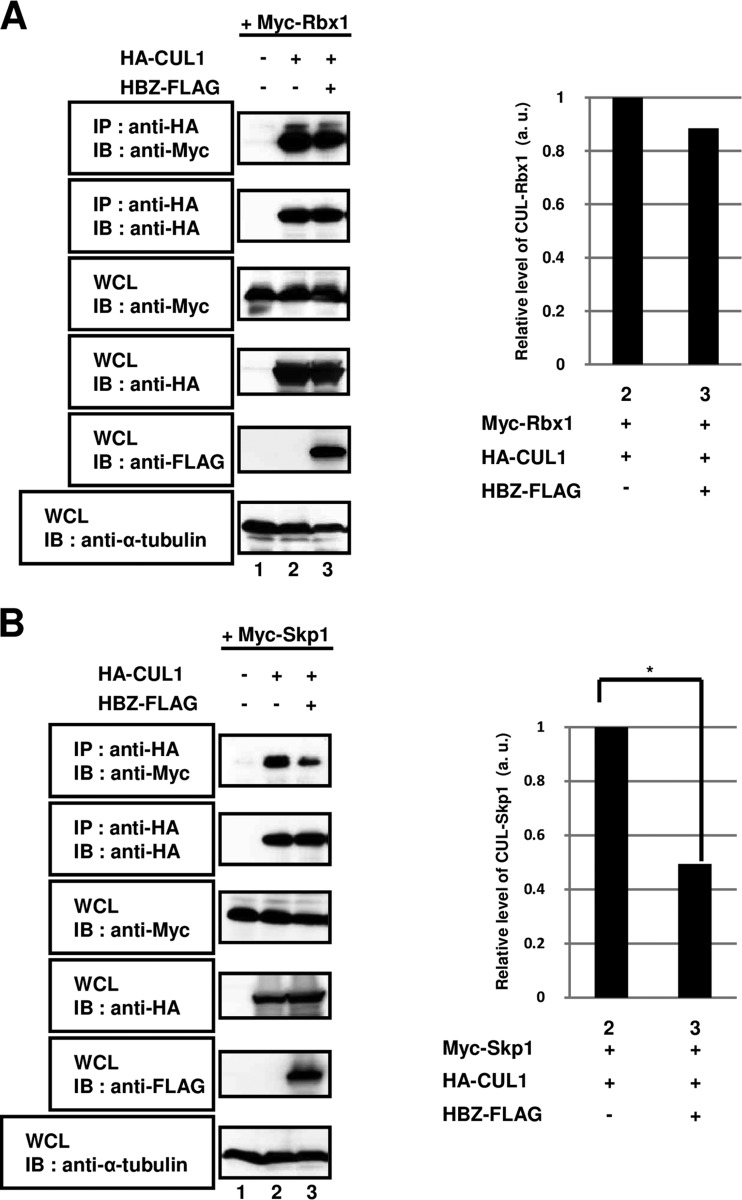
The CUL1 protein serves as a scaffold for the SCF complex, with its N-terminal and C-terminal regions binding to an adaptor protein (Skp1) and interacting with a RING protein (Rbx1), respectively (38). We postulated that the association of CUL1 with Rbx1 and/or Skp1 would be impacted by its interaction with HBZ in HEK293T cells. First, we tested whether HBZ affected the heterodimerization of CUL1 and Rbx1 in a co-IP assay. As shown in Fig. 5A, the formation of heterodimers between CUL1 and Rbx1 was not affected in cells overexpressing HBZ (lane 2 versus lane 3). We next investigated whether the interaction between CUL1 and Skp1 was altered by the overexpression of HBZ. As shown in Fig. 5B, when CUL1 and Skp1 together with HBZ were coexpressed in cells, the amount of the CUL1-Skp1 heterodimers was significantly decreased compared with that in control cells (lane 2 versus lane 3). These results suggest that HBZ strongly blocked the binding of Skp1 to CUL1 and thus prevented the formation of CUL1-Skp1 complexes.
FIG 5.
HBZ abrogates the interaction of CUL1 with Skp1 but not with Rbx1. HEK293T cells were cotransfected with 2 μg of a plasmid expressing Myc-Rbx1 (A) or Myc-Skp1 (B) with (+) or without (−) 1 μg of a plasmid expressing HA-CUL1 and 5 μg of a plasmid expressing HBZ-FLAG, as indicated. After 36 h, cell extracts were prepared and subjected to immunoprecipitation using anti-HA antibodies, followed by immunoblot analysis with anti-Myc and anti-HA antibodies. Total protein levels in whole-cell lysates were analyzed by immunoblotting using anti-Myc, anti-HA, anti-FLAG, and anti-α-tubulin antibodies. The intensity of each band was quantified and graphed (*, P < 0.05). WCL, whole-cell lysates; IB, immunoblotting; IP, immunoprecipitation.
HBZ inhibits SCF ubiquitin ligase activity.
We investigated whether the expression of HBZ leads to decreased polyubiquitination of MCL1 via inhibition of SCF E3 ubiquitin ligase activity. The expression vector FLAG-MCL1 was transfected into HEK293T cells together with vectors containing Myc-tagged components of the SCF complex. Lysates of the transfected cells were immunoprecipitated with an anti-Myc antibody, and the immunoprecipitates were resolved by SDS-PAGE followed by immunoblotting with an anti-HA antibody. Similar to previous reports, MCL1 polyubiquitination levels were directly related to SCF ubiquitin ligase activity (Fig. 6A, lane 2 versus lane 3). On the other hand, expression of MCL1 and SCF-related complexes together with HBZ dramatically decreased the level of ubiquitinated MCL1 (Fig. 6A, lane 3 versus lane 4). These results reveal that SCF-mediated ubiquitination of MCL1 was attenuated by overexpression of HBZ.
In Fig. 4B, we showed that the nuclear export ability of HBZ is essential for the association with CUL1. Therefore, to determine whether the nuclear export of HBZ to the cytoplasm is important for the stabilization of the MCL1 protein, HEK293T cells were transfected with an MCL expression vector with or without wild-type (WT) HBZ (HBZ-WT), HBZ-NES-mut, HBZ-NES-mut-PKI (NES), and SCF expression vectors (Fig. 6B). As previously reported (37), the amount of MCL1 protein was significantly decreased via SCF E3 ligase-mediated degradation (Fig. 6B, lane 1 versus lane 2). On the other hand, expression of the SCF complex together with HBZ-WT or HBZ-NES-mut-PKI (NES) (Fig. 6B, lane 2 versus lanes 3 and 5) but not HBZ-NES-mut (Fig. 6B, lane 2 versus lane 4) restored MCL1 levels. These findings suggest that the cytoplasmic translocation of HBZ is required for the inhibition of MCL1 protein degradation by suppressing SCF-dependent ubiquitination.
DISCUSSION
In this study, we revealed that HBZ promotes the stabilization of MCL1 by inhibiting SCF E3 ubiquitin ligase activity (Fig. 1 and 2) through its interaction with CUL1, which is a member of the SCF complex. An interaction between HBZ and CUL1 in the cytoplasm of mammalian cells was confirmed (Fig. 3 and 4). Furthermore, it was reported that CUL1 forms the structural scaffold of the SCF complex and binds to Skp1 and Rbx1. On the basis of our findings, it is possible that HBZ blocks the interaction of CUL1 with Skp1 but not that with Rbx1 (Fig. 5).
It is well established that several viral oncoproteins modulate the ubiquitin-proteasome pathway in order to avoid host cell defenses. For example, the high-risk human papillomavirus (HPV) E6 oncoprotein forms a complex with the host cell protein E6AP ubiquitin ligase, leading to the degradation of substrate proteins, such as p53 and PDZ domain-containing proteins (39, 40). In addition, the human hepatitis B virus (HBV) X protein (HBx) is a potent transcriptional activator that binds to the CUL4A-based E3 ubiquitin ligase that contains DDB1 (41). Similarly, both the Vpr and Vpx proteins encoded by the human immunodeficiency virus (HIV) also interact with DDB1 (42, 43). Therefore, these proteins are involved in the induction of cell cycle arrest at the G2 phase and the promotion of abnormal cell behavior. We have reported in this study that the SCF complex, which acts as an E3 ubiquitin ligase, was repressed by the overexpression of HBZ. Hence, like many other viral proteins, HBZ hijacks the host ubiquitin ligase-mediated pathway to control the degradation of subcellular factors. In addition, many intracellular proteins are degraded by the SCF ubiquitin-proteasome pathway. One of the cell cycle regulators, p21, is well-known as a substrate of the SCF complex. Indeed, enhanced stabilization of p21 was detected in cells transiently expressing HBZ (see Fig. S2 in the supplemental material). It was previously reported that p21 is overexpressed at high levels in HTLV-1-infected cell lines as well as in ATL and HTLV-1-associated myelopathy/topical spastic paraparesis patient samples (44). The stability of p21 may be regulated by disruption of the function of HBZ on the SCF complex. In addition, the c-Jun protein, which is involved in the regulation of malignant transformation and tumor progression, is one such protein (45). Interestingly, in a previous report, the expression level of c-Jun protein in HTLV-1-infected cells was significantly higher than that in uninfected cells (46). The dramatic downregulation of SCF activity by HBZ may indirectly promote c-Jun stability.
A previous study showed that transient expression of MCL1 inhibits the autophagy induced by nutrient deprivation in cells, whereas the stable knockdown of endogenous MCL1 by shRNA markedly promotes autophagy under starvation conditions (47). Autophagy is an intracellular bulk degradation system and has emerged as a critical player in the control of host defense against viral infection. Recently, we reported that starvation-induced autophagy in cells constitutively expressing HBZ was significantly suppressed by inhibition of the mTOR signaling pathway through an interaction between HBZ and GADD34 (22). Our results presented in this report suggest that HBZ strongly inhibits autophagy not only by interacting with GADD34 but also by suppressing MCL1 degradation. Moreover, HBZ may play a key role in determining the cell fate.
There is a lot of evidence to suggest that abnormal expression of the MCL1 protein contributes to the progression of human cancers, including prostate, breast, ovarian, and cervical cancers as well as melanoma and leukemia (27, 28, 31). Moreover, knockdown of MCL1 expression using an shRNA system in several cancer cell lines resulted in a remarkable inhibition of tumor growth (47). Therefore, it has been suggested that the MCL1 protein may lead to the malignant progression of many human tumors. This study showed that the association between HBZ and CUL1 led to an increase in the level of the MCL1 protein in cells by interfering with the SCF-mediated polyubiquitination/degradation pathway. Thus, HBZ may contribute to the development of ATL by stabilizing MCL. Furthermore, a recent study reported increased MCL1 stability by the HTLV-1 Tax protein during cell transformation (33). Tax and HBZ are known to have opposing functions on the regulation of signaling pathways (48, 49). In HTLV-1-infected cells, there are data to suggest cooperation between HBZ and Tax proteins to promote MCL1 stabilization (33). However, the expression of Tax during the late stage of HTLV-1 infection is undetectable because the 5′ LTR of HTLV-1 undergoes changes, such as mutations and deletions (16–19). It is possible that the dysfunction of CUL1 through its interaction with HBZ plays a crucial role in the stabilization of MCL1 in ATL cells that no longer express the Tax protein.
Future studies will be required to more clearly elucidate the role of HBZ in HTLV-1-infected and ATL patient cells. The classification of ATL into the acute, lymphoma, chronic, and smoldering subtypes has been proposed. Although the chronic and smoldering types of ATL have an indolent course, approximately 70% of cases of indolent-type ATL progress to the aggressive type of ATL (50). The elucidation of the molecular mechanisms by which HBZ contributes to leukemogenesis may facilitate the discovery of a therapeutic target in ATL as well as the establishment of a strategy for preventing blast crisis in the chronic and smoldering types ATL.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Miyoshi for providing the lentivirus-based transfection system and Y. Tan for technical assistance.
This work was supported by JSPS KAKENHI grant numbers JP15H06790 (to R.M.) and JP26430128 (to T.O.). In addition, this work was supported in part by research funds from the Takeda Science Foundation (to T.O.), the Suzuken Memorial Foundation (to T.O.), the Japanese Leukemia Research Foundation (to T.O. and R.M.), a Sasakawa scientific research grant (to R.M.), and the L'Oréal-UNESCO for Women in Science Fellowship Program (to R.M.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00450-16.
REFERENCES
- 1.Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita KI, Shirakawa S, Miyoshi I. 1981. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci U S A 78:6476–6480. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida M, Seiki M, Yamaguchi K, Takatsuki K. 1984. Monoclonal integration of human T-cell leukemia provirus in all primary tumors of adult T-cell leukemia suggests causative role of human T-cell leukemia virus in the disease. Proc Natl Acad Sci U S A 81:2534–2537. doi: 10.1073/pnas.81.8.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verdonck K, González E, Van Dooren S, Vandamme AM, Vanham G, Gotuzzo E. 2007. Human T-lymphotropic virus 1: recent knowledge about an ancient infection. Lancet Infect Dis 7:266–281. doi: 10.1016/S1473-3099(07)70081-6. [DOI] [PubMed] [Google Scholar]
- 4.Gessain A, Cassar O. 2012. Epidemiological aspects and world distribution of HTLV-1 infection. Front Microbiol 3:388. doi: 10.3389/fmicb.2012.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutartre H, Clavière M, Journo C, Mahieux R. 2016. Cell-free versus cell-to-cell infection by human immunodeficiency virus type 1 and human T-lymphotropic virus type 1: exploring the link among viral source, viral trafficking, and viral replication. J Virol 90:7607–7617. doi: 10.1128/JVI.00407-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuoka M, Jeang KT. 2007. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer 7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- 7.Bai XT, Nicot C. 2012. Overview on HTLV-1 p12, p8, p30, p13: accomplices in persistent infection and viral pathogenesis. Front Microbiol 3:400. doi: 10.3389/fmicb.2012.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida M. 2001. Multiple viral strategies of HTLV-1 for dysregulation of cell growth control. Annu Rev Immunol 19:475–496. doi: 10.1146/annurev.immunol.19.1.475. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida M. 2005. Discovery of HTLV-1, the first human retrovirus, its unique regulatory mechanisms, and insights into pathogenesis. Oncogene 24:5931–5937. doi: 10.1038/sj.onc.1208981. [DOI] [PubMed] [Google Scholar]
- 10.Hasegawa H, Sawa H, Lewis MJ, Orba Y, Sheehy N, Yamamoto Y, Ichinohe T, Tsunetsugu-Yokota Y, Katano H, Takahashi H, Matsuda J, Sata T, Kurata T, Nagashima K, Hall WW. 2006. Thymus-derived leukemia-lymphoma in mice transgenic for the Tax gene of human T-lymphotropic virus type I. Nat Med 12:466–472. doi: 10.1038/nm1389. [DOI] [PubMed] [Google Scholar]
- 11.Simonis N, Rual JF, Lemmens I, Boxus M, Hirozane-Kishikawa T, Gatot JS, Dricot A, Hao T, Vertommen D, Legros S, Daakour S, Klitgord N, Martin M, Willaert JF, Dequiedt F, Navratil V, Cusick ME, Burny A, Van Lint C, Hill DE, Tavernier J, Kettmann R, Vidal M, Twizere JC. 2012. Host-pathogen interactome mapping for HTLV-1 and -2 retroviruses. Retrovirology 9:26. doi: 10.1186/1742-4690-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohsugi T, Wakamiya M, Morikawa S, Matsuura K, Kumar JM, Kumasaka T, Yamaguchi K. 2013. Invasion of histiocytic sarcoma into the spinal cord of HTLV-1 tax transgenic mice with HTLV-1-associated myelopathy/tropical spastic paraparesis-like disease. Oncol Res 20:403–410. doi: 10.3727/096504013X13657689383058. [DOI] [PubMed] [Google Scholar]
- 13.Romanelli MG, Diani E, Bergamo E, Casoli C, Ciminale V, Bex F, Bertazzoni U. 2013. Highlights on distinctive structural and functional properties of HTLV Tax proteins. Front Microbiol 4:271. doi: 10.3389/fmicb.2013.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamiya S, Matsuoka M, Etoh K, Watanabe T, Kamihira S, Yamaguchi K, Takatsuki K. 1996. Two types of defective human T-lymphotropic virus type I provirus in adult T-cell leukemia. Blood 88:3065–3073. [PubMed] [Google Scholar]
- 15.Miyazaki M, Yasunaga J, Taniguchi Y, Tamiya S, Nakahata T, Matsuoka M. 2007. Preferential selection of human T-cell leukemia virus type 1 provirus lacking the 5′ long terminal repeat during oncogenesis. J Virol 81:5714–5723. doi: 10.1128/JVI.02511-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furukawa Y, Kubota R, Tara M, Izumo S, Osame M. 2001. Existence of escape mutant in HTLV-I tax during the development of adult T-cell leukemia. Blood 97:987–993. doi: 10.1182/blood.V97.4.987. [DOI] [PubMed] [Google Scholar]
- 17.Koiwa T, Hamano-Usami A, Ishida T, Okayama A, Yamaguchi K, Kamihira S, Watanabe T. 2002. 5′-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J Virol 76:9389–9397. doi: 10.1128/JVI.76.18.9389-9397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeda S, Maeda M, Morikawa S, Taniguchi Y, Yasunaga J, Nosaka K, Tanaka Y, Matsuoka M. 2004. Genetic and epigenetic inactivation of tax gene in adult T-cell leukemia cells. Int J Cancer 109:559–567. doi: 10.1002/ijc.20007. [DOI] [PubMed] [Google Scholar]
- 19.Taniguchi Y, Nosaka K, Yasunaga J, Maeda M, Mueller N, Okayama A, Matsuoka M. 2005. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology 2:64. doi: 10.1186/1742-4690-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard JM. 2002. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J Virol 76:12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hivin P, Frédéric M, Arpin-André C, Basbous J, Gay B, Thébault S, Mesnard JM. 2005. Nuclear localization of HTLV-I bZIP factor (HBZ) is mediated by three distinct motifs. J Cell Sci 118:1355–1362. doi: 10.1242/jcs.01727. [DOI] [PubMed] [Google Scholar]
- 22.Mukai R, Ohshima T. 2014. HTLV-1 HBZ positively regulates the mTOR signaling pathway via inhibition of GADD34 activity in the cytoplasm. Oncogene 33:2317–2328. doi: 10.1038/onc.2013.181. [DOI] [PubMed] [Google Scholar]
- 23.Ma G, Yasunaga J, Matsuoka M. 2016. Multifaceted functions and roles of HBZ in HTLV-1 pathogenesis. Retrovirology 13:16. doi: 10.1186/s12977-016-0249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan J, Ma G, Nosaka K, Tanabe J, Satou Y, Koito A, Wain-Hobson S, Vartanian JP, Matsuoka M. 2010. APOBEC3G generates nonsense mutations in human T-cell leukemia virus type 1 proviral genomes in vivo. J Virol 84:7278–7287. doi: 10.1128/JVI.02239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, Totoki Y, Chiba K, Sato-Otsubo A, Nagae G, Ishii R, Muto S, Kotani S, Watatani Y, Takeda J, Sanada M, Tanaka H, Suzuki H, Sato Y, Shiozawa Y, Yoshizato T, Yoshida K, Makishima H, Iwanaga M, Ma G, Nosaka K, Hishizawa M, Itonaga H, Imaizumi Y, Munakata W, Ogasawara H, Sato T, Sasai K, Muramoto K, Penova M, Kawaguchi T, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Nakamaki T, Ishiyama K, Miyawaki S, Yoon SS, Tobinai K, Miyazaki Y, Takaori-Kondo A, Matsuda F, et al. 2015. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet 47:1304–1315. doi: 10.1038/ng.3415. [DOI] [PubMed] [Google Scholar]
- 26.Satou Y, Yasunaga J, Zhao T, Yoshida M, Miyazato P, Takai K, Shimizu K, Ohshima K, Green PL, Ohkura N, Yamaguchi T, Ono M, Sakaguchi S, Matsuoka M. 2011. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog 7:e1001274. doi: 10.1371/journal.ppat.1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song L, Coppola D, Livingston S, Cress D, Haura EB. 2005. Mcl-1 regulates survival and sensitivity to diverse apoptotic stimuli in human non-small cell lung cancer cells. Cancer Biol Ther 4:267–276. doi: 10.4161/cbt.4.3.1496. [DOI] [PubMed] [Google Scholar]
- 28.Ding Q, He X, Xia W, Hsu JM, Chen CT, Li LY, Lee DF, Yang JY, Xie X, Liu JC, Hung MC. 2007. Myeloid cell leukemia-1 inversely correlates with glycogen synthase kinase-3 beta activity and associates with poor prognosis in human breast cancer. Cancer Res 67:4564–4571. doi: 10.1158/0008-5472.CAN-06-1788. [DOI] [PubMed] [Google Scholar]
- 29.Akita K, Kawata S, Shimotohno K. 2005. P21WAF1 modulates NF-kappaB signaling and induces anti-apoptotic protein Bcl-2 in Tax-expressing rat fibroblast. Virology 332:249–257. doi: 10.1016/j.virol.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 30.Akagi T, Ono H, Nyunoya H, Shimotohno K. 1997. Characterization of peripheral blood T-lymphocytes transduced with HTLV-1 Tax mutants with different trans-activating phenotypes. Oncogene 14:2071–2078. doi: 10.1038/sj.onc.1201045. [DOI] [PubMed] [Google Scholar]
- 31.Uddin S, Bhat AA, Krishnankutty R, Mir F, Kulinski M, Mohammad RM. 2016. Involvement of F-BOX proteins in progression and development of human malignancies. Semin Cancer Biol 36:18–32. doi: 10.1016/j.semcancer.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Murata K, Hayashibara T, Sugahara K, Uemura A, Yamaguchi T, Harasawa H, Tsuruda K, Okazaki T, Koji T, Miyanishi T, Yamada Y, Kamihira S. 2006. A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization. J Virol 80:2495–2505. doi: 10.1128/JVI.80.5.2495-2505.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi YB, Harhaj EW. 2014. HTLV-1 Tax stabilizes MCL-1 via TRAF6-dependent K63-linked polyubiquitination to promote cell survival and transformation. PLoS Pathog 10:e1004458. doi: 10.1371/journal.ppat.1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuconati A, Mukherjee C, Perez D, White E. 2003. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev 17:2922–2932. doi: 10.1101/gad.1156903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. 2003. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426:671–676. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 36.Deshaies RJ. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol 15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 37.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, Xiao Y, Christie AL, Aster J, Settleman J, Gygi SP, Kung AL, Look T, Nakayama KI, DePinho RA, Wei W. 2011. SCF (FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 39.Handa K, Yugawa T, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. 2007. E6AP-dependent degradation of DLG4/PSD95 by high-risk human papillomavirus type 18 E6 protein. J Virol 81:1379–1389. doi: 10.1128/JVI.01712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Zapien D, Ruiz FX, Poirson J, Mitschler A, Ramirez J, Forster A, Cousido-Siah A, Masson M, Vande Pol S, Podjarny A, Travé G, Zanier K. 2016. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 529:541–545. doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee TH, Elledge SJ, Butel JS. 1995. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol 69:1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Rouzic E, Belaïdouni N, Estrabaud E, Morel M, Rain JC, Transy C, Margottin-Goguet F. 2007. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6:182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava S, Swanson SK, Manel N, Florens L, Washburn MP, Skowronski J. 2008. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog 4:e1000059. doi: 10.1371/journal.ppat.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de La Fuente C, Santiago F, Chong SY, Deng L, Mayhood T, Fu P, Stein D, Denny T, Coffman F, Azimi N, Mahieux R, Kashanchi F. 2000. Overexpression of p21(waf1) in human T-cell lymphotropic virus type 1-infected cells and its association with cyclin A/cdk2. J Virol 74:7270–7283. doi: 10.1128/JVI.74.16.7270-7283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. 2004. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 46.Hooper WC, Rudolph DL, Lairmore MD, Lal RB. 1991. Constitutive expression of c-jun and jun-B in cell lines infected with human T-lymphotropic virus types I and II. Biochem Biophys Res Commun 181:976–980. doi: 10.1016/0006-291X(91)92032-F. [DOI] [PubMed] [Google Scholar]
- 47.Germain M, Nguyen AP, Le Grand JN, Arbour N, Vanderluit JL, Park DS, Opferman JT, Slack RS. 2011. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J 30:395–407. doi: 10.1038/emboj.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuoka M. 2010. HTLV-1 bZIP factor gene: its roles in HTLV-1 pathogenesis. Mol Aspects Med 31:359–366. doi: 10.1016/j.mam.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Mesnard JM, Barbeau B, Césaire R, Péloponèse JM. 2015. Roles of HTLV-1 basic zip factor (HBZ) in viral chronicity and leukemic transformation. Potential new therapeutic approaches to prevent and treat HTLV-1-related diseases. Viruses 7:6490–6505. doi: 10.3390/v7122952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takasaki Y, Iwanaga M, Imaizumi Y, Tawara M, Joh T, Kohno T, Yamada Y, Kamihira S, Ikeda S, Miyazaki Y, Tomonaga M, Tsukasaki K. 2010. Long-term study of indolent adult T-cell leukemia-lymphoma. Blood 115:4337–4343. doi: 10.1182/blood-2009-09-242347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.