Abstract
K-Ras must localize to the plasma membrane and be arrayed in nanoclusters for biological activity. We show here that K-Ras is a substrate for cyclic GMP-dependent protein kinases (PKGs). In intact cells, activated PKG2 selectively colocalizes with K-Ras on the plasma membrane and phosphorylates K-Ras at Ser181 in the C-terminal polybasic domain. K-Ras phosphorylation by PKG2 is triggered by activation of AMP-activated protein kinase (AMPK) and requires endothelial nitric oxide synthase and soluble guanylyl cyclase. Phosphorylated K-Ras reorganizes into distinct nanoclusters that retune the signal output. Phosphorylation acutely enhances K-Ras plasma membrane affinity, but phosphorylated K-Ras is progressively lost from the plasma membrane via endocytic recycling. Concordantly, chronic pharmacological activation of AMPK → PKG2 signaling with mitochondrial inhibitors, nitric oxide, or sildenafil inhibits proliferation of K-Ras-positive non-small cell lung cancer cells. The study shows that K-Ras is a target of a metabolic stress-signaling pathway that can be leveraged to inhibit oncogenic K-Ras function.
INTRODUCTION
Ras proteins are small GTPases that regulate important cellular signaling cascades to control cell growth, proliferation, and differentiation (1). The three Ras isoforms, H-, N-, and K-Ras4B (here, K-Ras), are ubiquitously expressed in mammalian cells. Ras proteins must be localized to the inner leaflet of the plasma membrane (PM) by a C-terminal membrane anchor for biological activity. In the case of K-Ras, the anchor comprises a posttranslationally attached C-terminal cysteine farnesyl-methyl ester operating in concert with a polybasic motif of 6 lysine residues (2, 3). Electrostatic interactions between the K-Ras C-terminal polybasic domain and the negatively charged inner leaflet of the PM provide membrane affinity (2, 4–9). Maintenance of K-Ras on the PM also requires the chaperone protein PDEδ (10). Cytosolic PDEδ binds K-Ras released from the PM as a result of endocytosis and unloads K-Ras in the perinuclear region in response to Arl2/3 binding, whence K-Ras translocates to the recycling endosome (RE) for redelivery to the PM by vesicular transport (11). Ras proteins on the PM are spatially organized into nanodomains, called nanoclusters, that are required for high-fidelity signal transduction by the Ras/mitogen-activated protein kinase (MAPK) pathway (12–14). Ras GTP nanoclusters contain ∼6 to 7 Ras proteins, are <20 nm in diameter, and are exclusive platforms for Raf recruitment and MEK/extracellular signal-regulated kinase (ERK) activation. Perturbation of the spatiotemporal dynamics of Ras nanoclustering disrupts cellular signaling (15, 16).
We have used a high-content cell-based screen (HCS) to identify multiple chemical compounds that mislocalize K-Ras from the PM and abrogate K-Ras signal transmission (17, 18). One group of compounds disrupts the cellular phosphatidylserine (PtdSer) distribution or PtdSer levels with a consequent reduction in the PtdSer content of the inner leaflet of the PM (18–21). Since PtdSer is an anionic phospholipid that provides much of the electrostatic surface potential to the inner PM, a reduced PtdSer PM content leads to K-Ras dissociation. A second group of compounds includes a diverse set of mitochondrial inhibitors (22–24) that all triggered activation of the master metabolic kinase, AMP-activated protein kinase (AMPK). In dissecting the molecular mechanism, we identified cyclic GMP (cGMP)-dependent protein kinases (PKGs) regulated by nitric oxide (NO) signaling to be novel K-Ras kinases. We show that PKGs directly phosphorylate K-Ras at Ser181 to regulate multiple aspects of K-Ras–PM interactions. Our data further show that pharmacological activators of this signaling pathway can inhibit proliferation of K-Ras-positive non-small cell lung cancer (NSCLC) cells.
MATERIALS AND METHODS
Cell culture and reagents.
Madin-Darby canine kidney (MDCK) and baby hamster kidney (BHK) cells were maintained in Dulbecco's modified Eagle medium (Gibco) supplemented with 2 mM l-glutamine and 10% fetal bovine serum (FBS) or 10% donor calf serum, respectively. A549 cells were maintained in Ham's F-12K medium (ATCC) supplemented with 10% FBS with 2 mM l-glutamine. H358, H441, and H2122 cells were maintained in RPMI 1640 (ATCC) supplemented with 10% FBS and 2 mM l-glutamine. All cell lines were grown at 37°C in 5% CO2. Antibodies against phospho-p42/44 mitogen-activated protein kinase (MAPK; ERK1/2; T202/T204; catalog number 4370), phospho-Akt (Ser473; catalog number 4060), phospho-endothelial nitric oxide synthase (phospho-eNOS; S1177; catalog number 9570), phospho-AMPKα (T172; catalog number 2535), phospho-vasodilator-stimulated phosphoprotein (phospho-VASP; S239; catalog number 3114), total VASP (catalog number 3132), phospho-CRaf (S338; catalog number 9427), and phospho-MARCKS (S152/156; catalog number 2741) were from Cell Signaling Technology. Antiactin (catalog number A2228) and antiphosphoserine (catalog number ab9332) antibodies were obtained from Sigma-Aldrich and Abcam, respectively. Neoantimycin and oligomycin A were obtained from BioAustralis (Australia). 8-Br-cGMP (catalog number BML-CN205), l-NG-nitroarginine methyl ester (l-NAME; catalog number ALX-105-003), and diethylamine nitric oxide (DEA-NO; catalog number ALX-430-034) were from Enzo Life Sciences. Metformin (catalog number D150959) was from Sigma-Aldrich, and aminoimidazole-4-carboxamide riboside (AICAR; catalog number 9944) was from Cell Signaling Technology. Sildenafil citrate (catalog number RSC01) was obtained from Biotang Inc.
In vitro phosphorylation.
All glutathione S-transferase (GST)-tagged K-Ras proteins and GST-tagged RhoA proteins were produced in Escherichia coli BL21 cells. Overnight cultures were diluted 1:10 and grown for 1 h at 37°C, protein expression was induced by adding 1 mM isopropyl-1-thio-β-d-galactopyranoside, and cultures were grown for an additional 3 h at 37°C. Bacteria were harvested by centrifugation, the pellet was resuspended in bacterial lysis buffer (50 mM Tris-HCl [pH 7.4], 200 mM NaCl, 5 mM MgCl2), and the bacteria were lysed by sonication. Lysates were cleared by centrifugation at 16,000 × g for 10 min, and proteins were isolated by incubation with glutathione-Sepharose beads (GE Healthcare) for 1 h at 4°C. The beads were washed in bacterial lysis buffer, and the proteins bound to the beads were quantified by the Bradford assay. For in vitro kinase reactions, beads containing 3 μg wild-type or mutant K-Ras or RhoA were incubated with 100 ng PKG in kinase reaction buffer (30 mM HEPES [pH 7.0], 10 mM MgCl2, 10 mM, β-glycerol phosphate, 1 mM dithiothreitol, 5 μM 8-chlorophenylthio-cGMP, 10 μCi [γ-32PO4]ATP) for 5 min at 30°C. After 5 min, cold ATP was added to 5 μM and the reaction mixture was incubated at 30°C for an additional 5 min. Reactions were stopped by washing the beads in phosphate-buffered saline (PBS) containing 0.1% NP-40. The beads were boiled in SDS sample buffer, and phosphorylation was analyzed by SDS-PAGE/autoradiography. Equal loading of the GST-tagged proteins was confirmed by Western blotting with anti-GST antibody (Santa Cruz Biotechnology).
Detecting K-Ras phosphorylation in vivo.
MDCK cells stably expressing monomeric enhanced green fluorescent protein (mGFP)-tagged K-Ras (mGFP–K-Ras) or phosphosite mutants were treated with PKG activators when they were at a confluence of ∼70% on a 10-cm dish. Cells were harvested with 1 ml of binding buffer (50 mM Tris [pH 7.5], 1% Triton X-100, 5 mM MgCl2, 25 mM NaF, 75 mM NaCl, 5 mM Na-pyrophosphate, 20 mM β-glycerophosphate, 100 μM Na3VO4, 1 mM dithiothreitol, leupeptin [3.33 μg/ml], aprotinin [0.33 μg/ml]). Cell debris was cleared by centrifugation, and the supernatant was incubated with 10 ml binding buffer plus 15 μl GFP-Trap beads (50% slurry; catalog number GFA-0050; Chromotek) overnight at 4°C. The beads were washed 3 times with 1 ml binding buffer and then incubated in 80 μl 2× sample buffer for 5 min at 95°C. Protein samples were resolved on 8% SDS-polyacrylamide gels and transferred to a polyvinylidene difluoride (PDVF) membrane using a semidry transfer apparatus. The PVDF membrane was then incubated with 4% paraformaldehyde and 0.01% glutaraldehyde for 30 min at room temperature (RT) to increase the immunogenicity of the phosphoproteins (25), washed 3 times with Tris-buffered saline with 0.1% Tween 20 (TBS-T), and blocked with 10% bovine serum albumin in TBS-T for 2 h at RT. The membrane was incubated with antiphosphoserine antibody (1:1,000) overnight at 4°C. The signal was detected by enhanced chemiluminescence as described below. The extra fixation of the PVDF membrane used in this protocol, including the ratio of paraformaldehyde to glutaraldehyde, and the blocking time were optimized to maximize antiphosphoserine immunodetection.
Electron microscopy (EM) and spatial mapping.
Plasma membrane sheets were prepared and fixed as previously described (18, 26, 27). For univariate analysis, plasma membrane sheets were labeled with anti-green fluorescent protein (anti-GFP) antibody conjugated to 4.5-nm gold particles. For bivariate analysis, plasma membrane sheets were labeled with anti-monomeric enhanced red fluorescent protein (anti-mRFP) antibody conjugated to 2-nm gold particles and anti-GFP antibody conjugated to 6-nm gold particles. Digital images of the immunogold-labeled plasma membrane sheets were taken in a transmission electron microscope. Intact 1-μm2 areas of the plasma membrane sheet were identified using ImageJ software, and the (x, y) coordinates of the gold particles were determined (26, 27). K-functions (28) were calculated and standardized on the 99% or 95% confidence interval (CI) for univariate or bivariate functions, respectively (26, 27, 29). In the case of univariate functions, a value of L(r) − r greater than the CI indicates significant clustering, and the maximum value of the function (Lmax) estimates the extent of clustering. Differences between replicated point patterns were analyzed by constructing bootstrap tests as described previously (14, 29), and the statistical significance against the results obtained with 1,000 bootstrap samples was evaluated. In the case of bivariate functions, under the null hypothesis that there is no spatial interaction between the two populations of gold particles, Lbiv(r) − r has an expected value of 0 for all values of r. As for L(r) − r, values of Lbiv(r) − r greater than the CI indicate significant coclustering of the two populations of gold. For a simple summary statistic, the Lbiv(r) − r function is standardized (Std) on the 95% confidence interval and integrated over a fixed range to derive the LBI parameter, as follows:
LBI has an expected value of 100 for proteins that do not cocluster. Differences between LBI values are evaluated in bootstrap tests on the actual Lbiv(r) − r function.
Western blotting.
Cells were washed in cold PBS and lysed in buffer containing 50 mM Tris HCl (pH 7.5), 75 mM NaCl, 25 mM NaF, 5 mM MgCl2, 5 mM EGTA, 1 mM dithiothreitol, 100 μM Na3VO4, and 1% NP-40 plus protease inhibitors. SDS-PAGE and immunoblotting with the antibody specified below were performed using 20 μg of each lysate. The signal was detected by enhanced chemiluminescence (SuperSignal; Pierce, Thermo Fisher Scientific, Rockford, IL) and imaged using a FluorChemQ imager (Alpha Inotech, San Leandro, CA). Quantification of intensities was performed using FluorChemQ software.
Cell proliferation assay.
Cancer cells (5,000 per well) were plated on a 96-well plate (catalog number 137101; Thermo Scientific). Twenty-four hours later, the cells were incubated with the compounds indicated below for 3 days. Fresh complete growth medium with the compounds was replaced every 24 h. Cell proliferation was assayed using a CyQuant proliferation assay kit (catalog number C7027; Invitrogen) according to the manufacturer's instructions. Plates were read using a Tecan Infinite 200 plate reader (excitation λ = 480 nm, emission λ = 520 nm).
siRNA-mediated PKG2 knockdown.
MDCK cells (1.0 × 105 cells) stably expressing mGFP-tagged K-Ras with a G12V substitution (mGFP–K-RasG12V) were mixed with 80 nM PKG2-targeting small interfering RNA (siRNA) [sense sequence, 5′-CUGCUUGGAAGUGGAAUACUA(dT)(dT)-3′; antisense sequence, 5′-UAGUAUUCCACUUCCAAGCAG(dT)(dT)-3′] (30) or a nontargeting scramble siRNA control (catalog number D-001810-10-01; Dharmacon) and a transfection reagent (DharmaFECT I; Dharmacon), and the mixture was plated in triplicate in a 6-well plate. After 48 h, the plated cells were retransfected with 80 nM siRNA. After a further 48 h, the cells were either treated with compounds or harvested for mRNA extraction. To verify knockdown, exon-exon boundary-spanning primers specific for canine PKG2 exons 14 and 15 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) exons 2 and 3 were designed: for PKG2 exons 14 and 15, 5′-GACATTCTGTGGGACTCCTGAGTA-3′ and 5′-CCTGCAAAGCCTCCGAATCAAATC-3′, respectively, and for GAPDH exons 2 and 3, 5′-ATTTGGCCGTATTGGGCGC-3′ and 5′-TCCTGGAAGATGGAGATGGAC-3′, respectively.
cDNA was generated from MDCK cells stably expressing mGFP–K-RasG12V after transfection with PKG2 or scrambled siRNA and amplified by PCR, and the products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining.
Confocal microscopy.
Cells were fixed with 4% paraformaldehyde, followed by 50 mM NH4Cl treatment to quench aldehyde groups. Cells were imaged in a Nikon A1R confocal microscope using 60× objectives.
Statistical analysis.
Prism software (version 5.0c; GraphPad Software) was used for one-way analysis of variance (ANOVA) testing and two-tailed t tests.
MD.
Atomistic molecular dynamics (MD) simulations were performed on wild-type K-Ras and the phospho-membrane-targeting motif of K-Ras (phospho-tK) in a negatively charged bilayer following a previously described protocol (31). Briefly, a symmetric bilayer of 104 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine and 1-hexadecanoyl-2–(9Z-octadecenoyl)-sn-glycero-3-phospho-l-serine lipids and two peptides per leaflet was solvated by TIP3P water molecules using a simulation of a box of 110 by 65 by 95 Å3, and charge-neutralizing sodium ions were added. The resulting 650,000-atom system was energy minimized for 2,000 steps, with lipid and protein heavy atoms being fixed. It was then equilibrated, using a time step of 1 fs, for 200 ps, with the lipid phosphate atoms and protein heavy atoms being harmonically restrained with a force constant (k) of 4 kcal/mol/Å2. This was followed by four steps of 100-ps runs with k scaled by 0.75, 0.50, 0.25, and 0. The resulting system was simulated for 1 μs with a 2-fs time step, with all bonds involving hydrogens being restrained by SHAKE, and by the use of particle mesh Ewald electrostatics with 12-Å and 14-Å cutoffs for nonbonded interactions and pair-list updates, respectively. Constant pressure (105 Pa) and temperature (310 K) were maintained using the Nose-Hoover Langevin piston and the Langevin thermostat methods, respectively. The force field was CHARMM27 (32) for proteins and CHARMM36 for lipids (33). Simulations were conducted with the NAMD2.9 program (34) for 1 μs.
RESULTS
AMPK activation triggers dissociation of K-Ras from the PM.
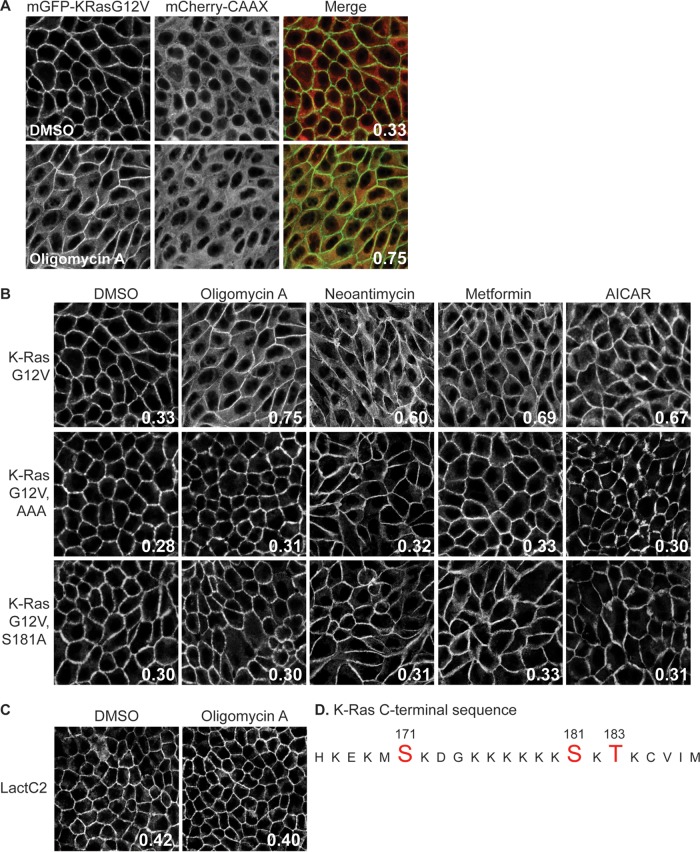
Among a set of compounds that displace K-RasG12V but not H-RasG12V from the PM in an HCS (18), we found an oligomycin family and a rare neoantimycin family of Streptomyces-derived molecules and the antidiabetic drug metformin (22–24, 35) (Fig. 1A and B). All of these compounds have been reported to inhibit various aspects of mitochondrial function (22–24, 35). Since inhibition of mitochondrial activity depletes cellular ATP levels and activates AMPK (36, 37), we tested whether direct AMPK activation would also mislocalize K-RasG12V. To this end, we used a confocal imaging assay that measures the colocalization of mGFP–K-RasG12V with a generic endomembrane marker, mCherry-CAAX (18). In this assay, the extent of colocalization of mGFP–K-RasG12V and mCherry-CAAX was quantified by use of a Manders coefficient, which provides an estimate of the fraction of K-Ras displaced from the PM (Fig. 1A). Figure 1 shows that the direct AMPK activator aminoimidazole-4-carboxamide riboside (AICAR) very effectively displaces K-RasG12V from the PM. To explore the molecular mechanism, we first examined the cellular distribution of PtdSer, because the PM PtdSer content is critical for K-Ras PM binding (18–21). We analyzed the localization of the PtdSer probe LactC2 by confocal imaging (4, 38). Oligomycin A had no effect on the distribution of LactC2 (Fig. 1C), indicating that AMPK-mediated K-RasG12V PM mislocalization is not a consequence of the loss of PtdSer from the inner PM. In contrast, K-RasG12V rendered insensitive to phosphorylation by three alanine substitutions at Ser171, Ser181, and Thr183 in the polybasic domain (the K-RasG12V AAA mutant; Fig. 1C) (39) was not mislocalized by any of the mitochondrial inhibitors (Fig. 1B). Similarly, a K-RasG12V mutant with a single alanine mutation at Ser181 (K-RasG12V S181A) was insensitive to oligomycin A, metformin, neoantimycin, or AICAR treatment (Fig. 1B). Taken together, these results suggest that AMPK-induced K-RasG12V dissociation from the PM is mediated through K-Ras phosphorylation at Ser181.
FIG 1.
AMPK activation mislocalizes K-RasG12V from the PM as a result of C-terminal phosphorylation. (A and B) MDCK cells stably coexpressing mCherry-CAAX, a general endomembrane marker (67), and mGFP–K-RasG12V, mGFP–K-RasG12V T171A, S181A, or S183A (AAA), or mGFP–K-RasG12V S181A were treated with 2.6 nM oligomycin A, 5.2 nM neoantimycin, 1 mM metformin, or 1 mM AICAR for 48 h, and cells were fixed and imaged using a confocal microscope. K-Ras mislocalization from the PM was quantified as the colocalization of mGFP–K-RasG12V and mCherry-CAAX using Manders coefficients. (A) A representative image showing the green (mGFP) and red (mCherry) channels used to calculate the Manders coefficients. (B) The Manders coefficient is shown on each representative image of drug-treated cells. (C) MDCK cells stably coexpressing mCherry-CAAX and mGFP-LactC2 were treated with 2.6 nM oligomycin A for 48 h and then fixed and imaged using a confocal microscope. K-Ras or LactC2 mislocalization from the PM was quantified using Manders coefficients, as described in the legend to panel A, with the value being shown on each image. (D) K-Ras C-terminal sequence. Amino acids in red are putative phosphorylation sites. DMSO, dimethyl sulfoxide.
K-Ras is a direct substrate of PKGs.
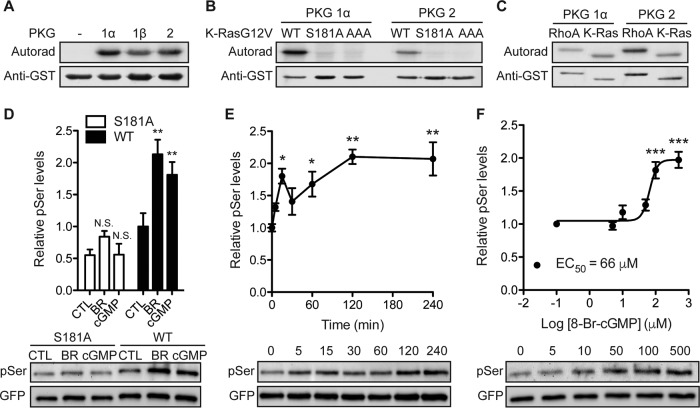
Protein kinase C (PKC) can phosphorylate K-Ras at S181 and to a lesser extent at S171 and T183 (39) (Fig. 1D). There is, however, no clear signaling pathway from AMPK to PKC, whereas there is such a pathway to cGMP-dependent protein kinase (PKG). Moreover, the consensus phosphorylation site for PKG, -Arg-Lys-Lys-X-Ser-, where X can be any amino acid (40), resembles the K-Ras polybasic motif (Fig. 1D). We first examined if K-Ras is a PKG substrate by performing in vitro kinase assays with recombinant PKG1α, PKG1β, and PKG2 and using RhoA, a known PKG substrate, as a positive control (41). All three PKG isoforms, when activated by cGMP, phosphorylated K-RasG12V (Fig. 2A) but did not phosphorylate the K-RasG12V AAA or K-RasG12V S181A mutant, indicating that K-Ras is a PKG substrate with Ser181 as the target phosphorylation site (Fig. 2B). We next developed an in vivo assay to measure K-Ras phosphorylation at Ser181 by endogenous protein kinases. MDCK cells stably expressing mGFP–K-RasG12V or mGFP–K-RasG12V S181A were treated with cell-permeant 8-Br-cGMP, and mGFP immunoprecipitates were immunoblotted with an antiphosphoserine antibody after fixation of the PVDF transfer membranes in paraformaldehyde and glutaraldehyde (25). In this assay, 8-Br-cGMP significantly increased the K-RasG12V but not the K-RasG12V S181A phosphoserine content (Fig. 2D), suggesting that PKG activation in vivo phosphorylates K-Ras at Ser181. Treatment with 8-Br-cGMP dose dependently and time dependently stimulated K-Ras phosphorylation (Fig. 2E and F). In the same assay, bryostatin-1, a potent PKC activator (39), elevated the K-RasG12V phosphoserine content to the same extent as 8-Br-cGMP but had a minimal effect on K-RasG12V S181A (Fig. 2D).
FIG 2.
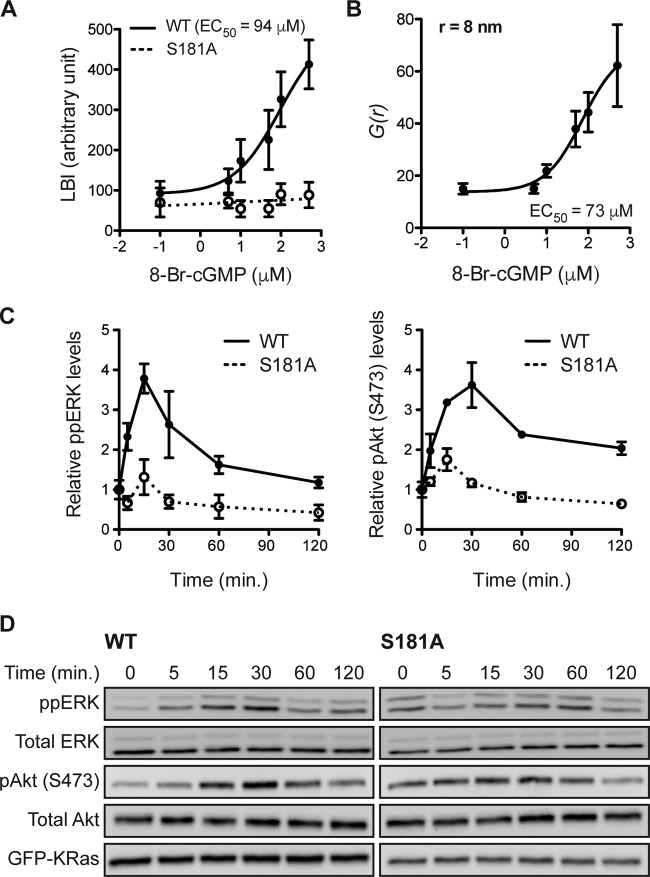
K-Ras is a substrate for cGMP-activated protein kinases. (A) Recombinant GST–K-Ras was incubated in vitro with purified PKG1α, PKG1β, or PKG2 and [γ-32PO4]ATP. Phosphate incorporation was analyzed by autoradiography (Autorad) (top), and equal loading was determined by anti-GST immunoblotting (bottom). (B and C) In vitro kinase experiments as described in the legend to panel A but including recombinant GST–K-RasG12V with the S181A and AAA (AAA = S171A, S181A, T183A) mutations (B) or recombinant GST-tagged RhoA (C). (D) MDCK cells stably expressing mGFP–K-RasG12V (wild type [WT]) or mGFP–K-RasG12V S181A (S181A) were treated with 1 μM bryostatin-1 (BR) for 5 min or 500 μM 8-Br-cGMP (cGMP) for 15 min. mGFP–K-Ras proteins were immunopurified and immunoblotted with an antiphosphoserine (pSer) antibody after fixation in 4% paraformaldehyde and 0.01% glutaraldehyde. The membranes were stripped and reblotted with an anti-GFP antibody. The graph shows the mean phospho-K-Ras levels ± SEM from three independent experiments, with representative blots being shown. Significant differences between control (CTL; vehicle-treated) and drug-treated cells were assessed using one-way ANOVA tests (**, P < 0.01; N.S., not significant). (E and F) MDCK cells stably expressing mGFP–K-RasG12V were treated with 500 μM 8-Br-cGMP for the indicated times (E) or the indicated concentrations for 15 min (F). Phosphorylated K-Ras was detected as described in the legend to panel D. The graph shows the mean phospho-K-Ras levels ± SEM from three independent experiments, with representative blots being shown. EC50, 50% effective concentration. Significant differences between cGMP-treated and control (PBS-treated) cells were assessed using one-way ANOVA tests (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
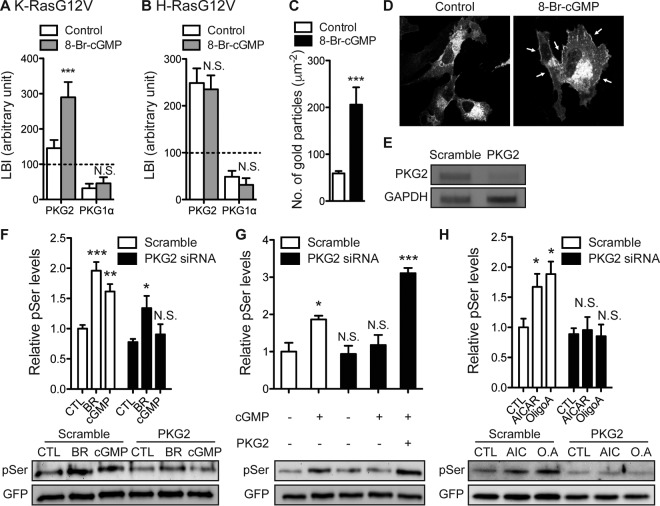
To identify the most likely candidate K-Ras kinase among the PKG isoforms, we used electron microscopy (EM) to spatially map the nanoscale PM distribution of different PKGs with respect to K-RasG12V. Intact basal PM sheets were prepared from MDCK cells coexpressing mGFP-PKG1α or mGFP-PKG2 and mCherry–K-RasG12V or mCherry-H-RasG12V. The PM sheets were colabeled with anti-GFP and anti-RFP antibodies conjugated directly to 6-nm or 2-nm gold particles, respectively, and the immunogold particle point patterns revealed by EM were analyzed using spatial statistics. The integrated bivariate K-function (LBI), which quantifies the extent of coclustering of the 6-nm or 2-nm gold particles, showed that under nonstimulated conditions, PKG2 but not PKG1α coclustered with K-RasG12V and H-RasG12V on the PM (Fig. 3A and B). After treatment with 8-Br-cGMP, the PKG2 levels on the inner PM increased significantly (Fig. 3C and D), and this newly recruited PKG2 selectively coclustered with K-RasG12V (Fig. 3A and B). Concordantly, siRNA knockdown of PKG2 totally abolished 8-Br-cGMP-stimulated K-Ras phosphorylation, which was rescued by ectopic PKG2 expression (Fig. 3E to G). Knockdown of PKG2 expression also abolished the K-Ras phosphorylation induced by AICAR and oligomycin (Fig. 3H). Interestingly, K-Ras phosphorylation in response to bryostatin-stimulated PKC activation was also partially abrogated in PKG2-knockdown cells (Fig. 3F), suggesting that PKC, in addition to directly phosphorylating K-Ras, may also do so indirectly by causing activation of PKG2 (39). Taking these results together, we conclude that PKG2 selectively interacts with K-Ras on the PM and is the major PKG isoform that phosphorylates K-Ras in response to AMPK activation.
FIG 3.
PKG2 phosphorylates K-Ras. (A and B) Intact basal PM sheets from MDCK cells coexpressing mGFP-PKG1α or mGFP-PKG2 and mCherry–K-RasG12V (A) or mCherry-H-RasG12V (B) were treated with 100 μM 8-Br-cGMP for 15 min, labeled with anti-GFP–6-nm gold and anti-RFP–2-nm gold, and imaged by EM. Spatial analyses performed using bivariate K-functions [Lbiv(r) − r] that determine whether one gold population is clustered with respect to the other are shown as LBI values (mean ± SEM), where LBI is a defined integral of the cognate Lbiv(r) − r curve. Significant differences between bivariate K-functions for 8-Br-cGMP-treated and control (PBS-treated) cells were evaluated in bootstrap tests (***, P < 0.001; N.S., not significant). (C) Intact PM sheets from MDCK cells expressing PKG2-mGFP and treated with 100 μM 8-Br-cGMP for 6 h were labeled with anti-GFP–4.5-nm gold. PM recruitment was assayed as the mean number of gold particles per square micrometer ± SEM (n = 20). Significant differences between 8-Br-cGMP-treated and control (PBS-treated) cells were evaluated in two-tailed t tests (***, P < 0.001). (D) BHK cells expressing PKG2-mGFP treated with 500 μM 8-Br-cGMP for 15 min were fixed with 4% paraformaldehyde and imaged using a confocal microscope. Arrows, enhanced PKG2 recruitment to the PM. (E) cDNA from MDCK cells stably expressing mGFP–K-RasG12V after transfection with PKG2-targeting siRNA or scrambled siRNA was amplified with primers specific for PKG2 exons 14 and 15 or GAPDH exons 2 and 3. PCR products were resolved by electrophoresis and visualized by ethidium bromide staining. Images representative of those from three independent experiments are shown. (F) MDCK cells stably expressing mGFP–K-RasG12V and transfected with scrambled or PKG2-targeting siRNA were treated with 500 μM 8-Br-cGMP for 15 min or 1 μM bryostatin-1 (BR) for 5 min. Phosphorylated K-Ras was detected as described in the legend to Fig. 2D. The graph shows the mean phospho-K-Ras levels ± SEM from three independent experiments, with representative blots being shown. Significant differences between control and drug-treated cells were assessed using one-way ANOVA tests (*, P < 0.05; **, P < 0.01; ***, P < 0.001; N.S., not significant). (G) MDCK cells stably expressing mGFP–K-RasG12V and transfected with scrambled or PKG2-targeting siRNA were treated with 500 μM 8-Br-cGMP for 15 min or left untreated. PKG2 was ectopically expressed where indicated. Phosphorylated K-Ras was detected as described in the legend to Fig. 2D. The graph shows the mean phospho-K-Ras levels ± SEM from three independent experiments, with representative blots being shown. Significant differences from the control (untreated cells without PKG2 knockdown) were assessed using one-way ANOVA tests (*, P < 0.05; ***, P < 0.001; N.S., not significant). (H) MDCK cells stably expressing mGFP–K-RasG12V and transfected with scrambled or PKG2-targeting siRNA were treated with 5 mM AICAR (AIC) or 0.5 μM oligomycin A (O.A) for 6 h. Phosphorylated K-Ras was detected as described in the legend to Fig. 2D. The graph shows the mean phospho-K-Ras levels ± SEM from three independent experiments, with representative blots being shown. Significant differences between control (vehicle-treated) cells and drug-treated cells were assessed using one-way ANOVA tests (*, P < 0.05; N.S., not significant).
AMPK and eNOS signaling regulates K-Ras interaction with the PM.
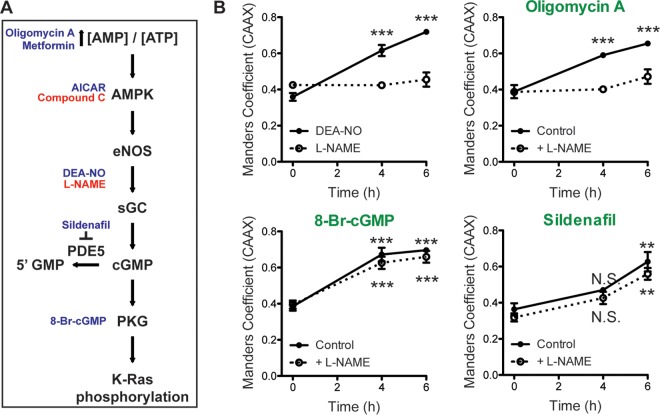
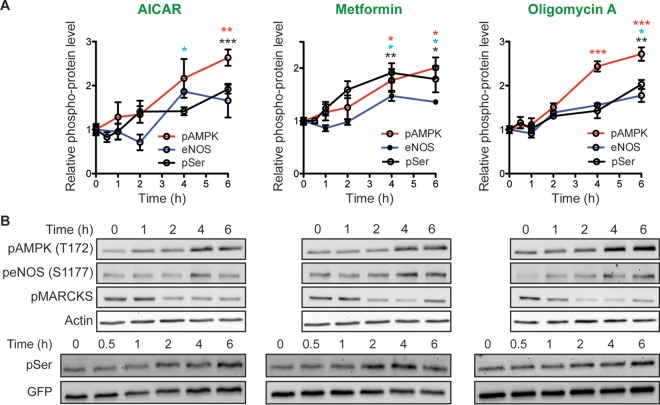
AMPK is sensitive to the cellular AMP concentration/ATP concentration ratio and is activated by metabolic stresses that inhibit ATP production or increase ATP consumption (36). Active AMPK regulates metabolism, cell signaling, transcription, and ion transport (36). One of the effectors involved in cell signaling is endothelial nitric oxide synthase (eNOS). We therefore tested whether AMPK-mediated K-Ras dissociation from the PM is a consequence of enhanced NO production (Fig. 4A). Both DEA-NO, which is an NO donor, and oligomycin A triggered K-Ras PM dissociation, which was blocked if cells were coincubated with l-NAME, an NOS inhibitor (Fig. 4B). NO activates soluble guanylyl cyclase (sGC) to produce cGMP. An alternative mechanism to increase cGMP levels is to inhibit PDE5, which hydrolyzes cGMP. Concordantly, sildenafil, a PDE5 inhibitor, triggered K-Ras PM dissociation. The effects of sildenafil (and 8-Br-cGMP) were not blocked by l-NAME, which inhibits proximally in the signaling pathway (Fig. 4B). Activation of AMPK and eNOS by oligomycin A, AICAR, and metformin followed the same time course as K-Ras PM dissociation (Fig. 5A and B). However, none of these compounds stimulated phosphorylation of MARCKS, a PKC substrate, suggesting that AMPK-mediated K-Ras PM dissociation is PKC independent (Fig. 5A and B). Together these pharmacological data are fully consistent with the AMPK → eNOS → PKG signaling pathway regulating K-Ras PM interactions. One caveat here, however, is that while we have shown that the endogenous PKG2 signaling pathway efficiently phosphorylates ectopically expressed GFP–K-Ras, we cannot formally evaluate the phosphorylation of endogenous K-Ras due to the limitation of available reagents.
FIG 4.
Activation of AMPK signaling induces K-Ras displacement from the PM. (A) The AMPK → PKG signaling pathway leading to K-Ras phosphorylation, with activators indicated in blue and inhibitors indicated in red. (B) MDCK cells stably coexpressing mGFP–K-RasG12V and mCherry-CAAX were treated with 0.5 μM oligomycin A, 100 μM 8-Br-cGMP, or 100 nM sildenafil in the presence (dotted line) or absence (solid line) of 1 mM l-NAME for 4 and 6 h. K-RasG12V PM dissociation was quantified using Manders coefficients from three independent experiments. Significant difference between drug-treated and control (vehicle-treated) cells were assessed using one-way ANOVA tests (N.S., not significant; **, P < 0.01; ***, P < 0.001).
FIG 5.
Activation of AMPK signaling phosphorylates K-Ras. MDCK cells stably expressing mGFP–K-RasG12V were treated with 5 mM AICAR, 5 mM metformin, or 0.5 μM oligomycin A for the indicated times, and cell lysates were immunoblotted for phospho-AMPK (pAMPK; T172), phospho-eNOS (peNOS; S1177), phospho-MARKCS (pMARCKS), and phosphorylated K-Ras as described in the legend to Fig. 1D. (A) The graphs show the mean phosphorylated AMPK, eNOS, and K-Ras levels ± SEM from three independent experiments. Significant differences between control (vehicle-treated) cells and drug-treated cells were assessed using a one-way ANOVA test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (B) Representative blots are shown, with actin and GFP blots being used as loading controls (top and bottom, respectively).
K-Ras phosphorylation regulates spatial organization and PM interaction.
K-Ras phosphorylated on S181 in response to PKC activation laterally segregates on the PM away from nonphosphorylated K-Ras (42). We observed a similar lateral reorganization in response to PKG activation. Intact basal PM sheets from 8-Br-cGMP-treated MDCK cells coexpressing mGFP–K-RasG12V S181D, a phosphomimetic mutant (39, 42), and mCherry–K-RasG12V were analyzed using EM and integrated bivariate K-functions (LBI values.) These data show that 8-Br-cGMP drives coclustering of K-RasG12V with K-RasG12V S181D (Fig. 6A), with the dose-response curve being similar to that for K-Ras phosphorylation measured by Western blotting (Fig. 2F). In control experiments, 8-Br-cGMP treatment did not stimulate coclustering of mRFP–K-RasG12V S181A with mGFP–K-RasG12V S181D (Fig. 6A). PKG-mediated phosphorylation also increased the pair correlation function [g(r)] of the K-Ras spatial distribution, indicating that phosphorylated K-Ras is more tightly packed in nanoclusters than nonphosphorylated K-Ras (Fig. 6B). To determine the consequences of acute PKG-mediated K-Ras phosphorylation on signal output, MDCK cells stably expressing K-RasG12V or K-RasG12V S181A were treated with 8-Br-cGMP for various time intervals, and the levels of diphosphorylated ERK (ppERK) and phosphorylated Akt (pAkt; S473) were measured using immunoblotting. The results show that PKG activation acutely but transiently enhanced ERK and Akt activation in K-RasG12V-expressing cells but had a minimal effect on cells expressing K-RasG12V S181A (Fig. 6C and D). We conclude that this signaling profile represents an integration of enhanced signal output from phospho–K-Ras nanoclusters at early time points, which is mitigated at later time points as phosphorylated K-Ras is progressively lost from the PM.
FIG 6.
K-Ras phosphorylation differentially regulates PM interactions. (A) Intact PM sheets prepared from MDCK cells coexpressing mGFP–K-RasG12V S181D and mCherry–K-RasG12V (solid line) or mRFP–K-RasG12V S181A (dotted line) after 8-Br-cGMP treatment for 6 h were labeled with anti-GFP–6-nm gold and anti-RFP–2-nm gold, respectively. The extent of coclustering of the two proteins was analyzed using bivariate K-functions, shown as the summary statistic LBI. The graph shows the mean LBI values ± SEM from ≥20 sheets for each data point. The LBI value for coclustering of mGFP–K-RasG12V with mCherry–K-RasG12V was measured to be 355, which implies that 500 μM 8-Br-cGMP induces the nearly complete overlap of the mGFP–K-RasG12V S181D and mCherry–K-RasG12V gold populations, consistent with ∼100% mCherry–K-RasG12V phosphorylation. (B) Intact PM sheets from MDCK cells expressing mGFP–K-RasG12V were treated for 6 h with 8-Br-cGMP, labeled with anti-GFP–4.5-nm gold, and visualized by EM. Spatial organization was analyzed at each 8-Br-cGMP concentration using the pair correlation function g(r) (r = 8 nm; data are means ± SEMs for 20 PM sheets). (C and D) MDCK cells stably expressing mGFP–K-RasG12V (wild type [WT]) or mGFP–K-RasG12V S181A (S181A) were serum starved overnight and treated with 100 μM 8-Br-cGMP for various time intervals in the absence of serum. Cell lysates were quantitatively immunoblotted for ppERK or pAkt (S473). The graphs shows the means ± SEMs from three independent experiments (C), with representative blots being shown (D). Total ERK, total Akt, and GFP blots were included as loading controls.
Phosphorylation does not immediately dissociate K-Ras from the PM.
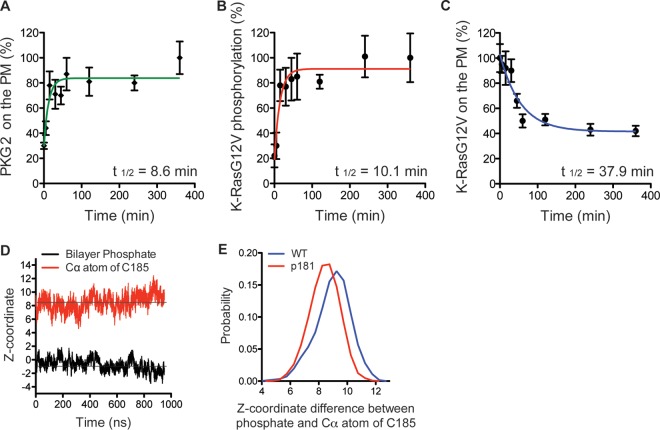
We next examined in more detail the kinetics of K-RasG12V phosphorylation in situ on the PM after PKG activation in BHK cells. We first monitored mGFP-PKG2 recruitment to the PM using EM of intact PM sheets. PKG2 recruitment to the PM after 8-Br-cGMP treatment was rapid with a calculated half-life (t1/2) of 8.6 min (Fig. 7A). Next, intact PM sheets from cells expressing mGFP–K-RasG12V S181D and mCherry–K-RasG12V were prepared after treatment with 8-Br-cGMP, and colocalization of K-RasG12V S181D and K-RasG12V was analyzed using bivariate EM. These data show that within 15 min of PKG activation, K-RasG12V is maximally phosphorylated, as indicated by an LBI value equal to that for the positive control; the calculated t1/2 for the LBI increase—and, hence, the inferred rate of K-Ras phosphorylation—was 10.1 min (Fig. 7B). This matches the time course of PKG2 recruitment and the time scale of K-Ras phosphorylation measured directly by immunoblotting. In contrast, the actual loss of K-RasG12V from the PM, again measured by EM, was much slower with a calculated t1/2 of 37.9 min (Fig. 7C). These data suggest that whereas activated PKG2 is rapidly recruited to the PM and quantitatively phosphorylates K-RasG12V, phosphorylation does not directly trigger the dissociation of K-Ras from the PM.
FIG 7.
Phosphorylation at Ser181 does not directly dissociate K-Ras from the PM. (A and C) Intact PM sheets from BHK cells expressing PKG2-mGFP (A) or mGFP–K-RasG12V (C) and treated with 500 μM 8-Br-cGMP over a time course were labeled with anti-GFP–4.5-nm gold. The PM levels of each protein were assayed as the mean number of gold particles per square micrometer ± SEM (n = 20 PM sheets). Values were normalized to the values at 0 min and 360 min for K-RasG12V and PKG2, respectively. (B) PM sheets from BHK cells coexpressing mGFP–K-RasG12V S181D and mCherry–K-RasG12V and treated with 500 μM 8-Br-cGMP over a time course were labeled with anti-GFP–6-nm gold and anti-RFP–2-nm gold, respectively. The extent of coclustering was analyzed using bivariate K-functions, summarized as LBI values, and used as an estimate of the extent of phosphorylation by calibration with a positive-control LBI value as described in the legend to Fig. 5A. Each data point is the mean for ≥20 PM sheets ± SEM. (D) Time evolution of the average z-position of the C-α atom of the farnesylated cysteine (red) of phospho-tK and that of the phosphate group of the lipids (black), which is centered to zero. The average for the four peptides in the system was calculated. (E) Distribution of the difference between the z-position of the phosphate group of the bilayer and that of the C-α atom of farnesylated cysteine in wild-type (WT) tK (blue) and phospho-tK (p181; red). The left-shifted distribution of phospho-tK indicates deeper bilayer insertion.
This somewhat surprising result prompted us to directly examine the PM interactions of phosphorylated K-Ras. To this end, we used a computational approach that provides atomistic information on the structure and dynamics of K-Ras–membrane complexes (31, 43–46). We simulated the minimal membrane-targeting motif of K-Ras (tK) with and without phosphorylation at Ser181 bound to a phospholipid bilayer containing 20% PtdSer. To directly check if phosphorylation induces spontaneous membrane dissociation, we added the phosphate on Ser181 after the peptide was already inserted into and equilibrated in the bilayer. For better statistics, we included four peptides per system and ran each simulation for 1 μs. The starting structures for the simulations were from a previous wild-type tK simulation (43), and the simulation details are described in reference 31. Analysis of the trajectories showed that neither phospho-tK nor tK dissociated from the bilayer during the entire simulation length, suggesting that phosphorylation does not dislodge K-Ras from the membrane. For example, Fig. 7D shows the time evolution of the average z-position of the C-α atom of the farnesylated cysteine of phospho-tK as well as the z-position of the phosphate moiety of the lipids that is centered to zero (i.e., a negative value indicates insertion). It is clear from Fig. 7D that phospho-tK remains deeply inserted in the bilayer during the entire simulation. In fact, the C terminus of phospho-tK inserts slightly deeper into the bilayer core than wild-type tK (Fig. 7D and E).
PKG signaling inhibits the growth of K-Ras-positive non-small cell lung cancer cells.
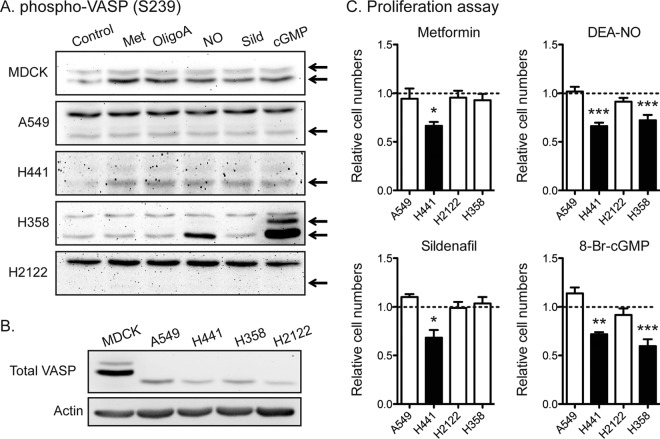
In K-Ras-positive cancer cells, prolonged activation of the AMPK → eNOS → PKG pathway phosphorylates oncogenic K-Ras, displaces it from the PM, and hence, reduces oncogenic K-Ras signaling and K-Ras-dependent cell growth. Since elements of this pathway are variably expressed, we monitored the Ser238 phosphorylation of vasodilator-stimulated phosphoprotein (VASP), a ubiquitously expressed PKG substrate (47), as an assay of PKG activity (48–50). A set of K-Ras-positive NSCLC cell lines was treated with metformin, oligomycin A, DEA-NO, sildenafil, or 8-Br-cGMP, and phospho-VASP levels were measured using immunoblotting. MDCK cells were used as a control cell line. In the MDCK and H441 cell lines, all the PKG activators increased phospho-VASP levels, whereas only DEA-NO and 8-Br-cGMP stimulated VASP phosphorylation in the H358 cell line (Fig. 8A and B). There were no detectable responses in the A549 and H2122 cell lines. These data suggest that all elements of the AMPK → eNOS → sGC → PKG pathway are intact in the MDCK and H441 cell lines, whereas only the lower segment of sGC → PKG is present in the H358 cell line and the pathway is not present at all in the A549 and H2122 cell lines. We next performed cell proliferation assays. Consistent with the integrity of the PKG signaling pathway data, all PKG activators inhibited the proliferation of H441 cells, only DEA-NO and 8-Br-cGMP inhibited the proliferation of H358 cells, and the A549 and H2122 cell lines were not sensitive to any PKG activator.
FIG 8.
Activators of AMPK → eNOS → PKG signaling inhibit the proliferation of NSCLC cells expressing the target pathway. (A and B) NSCLC cell lines were treated with 5 mM metformin (Met), 0.5 μM oligomycin A (OligoA), 0.1 mM DEA-NO (NO), 100 nM sildenafil (Sild), or 500 μM 8-Br-cGMP (cGMP). Cell lysates were blotted for phospho-VASP (S239) and total VASP. Representative blots from three independent experiments are shown. Actin is shown as a loading control. MDCK cells stably expressing K-RasG12V were used as a positive-control cell line. (C) NSCLC cells were plated on a 96-well plate and treated with the indicated activators for 4 days. Complete growth medium with activators was replaced every 24 h. Cell proliferation was analyzed using a CyQuant proliferation assay kit. The graph shows the mean cell proliferation ± SEM (n = 4 experiments) relative to that for the control cells (PBS-treated cells for metformin, DEA-NO, and 8-Br-cGMP and dimethyl sulfoxide-treated cells for sildenafil). Closed bars, pVASP levels increased after drug treatment, as described in the legend to panels A and B. Significant differences between activator-treated and control cells were assessed using one-way ANOVA tests (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
In this study, we identified PKG2 to be a novel K-Ras kinase that phosphorylates K-Ras at Ser181 in response to activation of AMPK. PKG is activated by the AMPK-mediated activation of eNOS, which generates cGMP through NO activation of soluble guanylyl cyclase. Using multiple activators of this pathway to drive K-Ras phosphorylation, we identified two separate consequences of K-Ras phosphorylation. Phospho-K-RasG12V laterally segregates from nonphospho-K-RasG12V to form nanoclusters that acutely retune the K-Ras signal output to enhance both phosphatidylinositol 3-kinase–Akt and Raf-MAPK activation. This is mitigated, however, by a progressive loss of phosphorylated K-Ras from the PM, which subsequently abrogates K-Ras signaling. Thus, AMPK activation can both retune K-Ras signaling and silence it.
The identification of PKG2 among the three expressed isoforms of PKG as the K-Ras kinase is based on several observations. First, knockdown of PKG2 expression was sufficient to completely abolish AMPK-mediated K-Ras phosphorylation. Second, EM experiments showed that acute activation of PKG2 but no other PKG isoform resulted in stable recruitment to K-Ras PM nanoclusters. Third, the time course of PKG2 recruitment to the PM closely matched that of K-Ras phosphorylation, with both processes occurring at rates with t1/2s of 8 to 10 min. Finally, alone among the PKG isoforms, PKG2 is N-myristoylated and has an adjacent positively charged sequence that, like K-Ras, may provide additional PM affinity; cGMP activation of PKG2 appears to increase PM affinity, presumably by enhancing the display of the N-terminal lipid anchor. Little is known about the mechanism of PKG2 PM recruitment. A recent study showed that PKG2 interacts with Rab11 (51), which would potentially target PKG2 to the recycling endosome, a distribution consistent with the confocal imaging shown in Fig. 3D. In the context of the current study, it is therefore possible that PKG recruitment is mediated by forward vesicular trafficking from the recycling endosome, with cGMP-activated PKG2 then being trapped on the PM as a result of increased PM affinity.
The identification of PKG2 as a K-Ras kinase with its attendant agonists allowed us to investigate the mechanism of K-Ras loss from the PM. Surprisingly, displacement of K-Ras from the PM did not occur immediately after phosphorylation; the process had a much longer t1/2 of 37.9 min. Thus, adding one phosphate group conferring a negative charge to the K-Ras polybasic domain at Ser181 was not sufficient to disrupt the electrostatic interaction between K-Ras and the PM, or else PM loss would have been concordant with phosphorylation. This was validated by MD simulations, which showed that phosphorylation of the K-Ras polybasic domain, if K-Ras is already bound to a PtdSer-containing membrane bilayer, does not trigger displacement. Indeed, phosphorylation results in deeper insertion of the phosphorylated anchor into the membrane bilayer. Therefore, we need to consider an alternative scenario whereby phosphorylated K-Ras is removed from the PM.
In this context, it has recently been shown that K-Ras is actively maintained on the PM by an elegant spatial organizing system (11). In brief, K-Ras removed from the PM by endocytosis is delivered to the cytosol because endosomes rapidly lose PtdSer asymmetry after internalization (52). Soluble K-Ras is captured by PDEδ, which unloads K-Ras in the perinuclear region in response to Arl2/3 binding, whence K-Ras binds to the recycling endosome (RE) for redelivery to the PM by vesicular transport (10, 11) (Fig. 9A). The rate of PM loss of K-RasG12V after phosphorylation (t1/2 = 37.9 min) is consistent with the time scale of endosomal recycling (53). We therefore propose that when PKG2 is activated, it is recruited to the PM and phosphorylates K-Ras. Phosphorylated K-Ras is delivered to the cytosol by the same process by which nonphosphorylated K-Ras is delivered and interacts normally with PDEδ, since binding is solely mediated through the farnesyl group (54); it is then released by Arl2/3 at the recycling endosome. At this point, phosphorylated K-Ras in solution must rebind to the negatively charged RE membrane; a recent MD simulation study showed that the prenyl anchor of K-Ras phosphorylated on Ser181 is unable to efficiently insert into an ionic phospholipid bilayer because of electrostatic repulsion (55). Thus, although K-Ras, when phosphorylated while bound to a PtdSer-containing membrane, does not dissociate, soluble phosphorylated K-Ras is unable to rebind to such a membrane and thus accumulates in the cytosol. In the context of the intact cell, compromised rebinding to the recycling endosome abrogates phosphorylated K-Ras forward transport to the PM, leading to progressive redistribution to the cytosol and endomembrane (Fig. 9B).
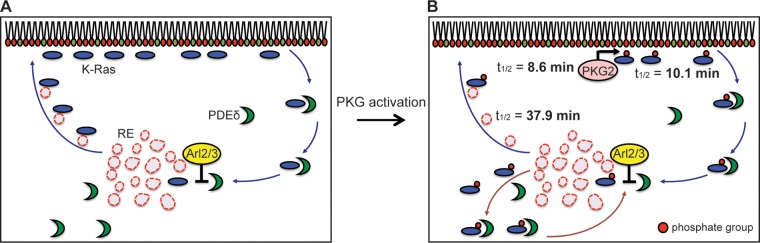
FIG 9.
A model for PKG-mediated K-Ras PM mislocalization. (A) K-Ras removed from the PM on endocytic vesicles is released to the cytosol when PtdSer asymmetry is lost, and cytosolic PDEδ binds to soluble K-Ras and unloads it in the perinuclear region in response to Arl2/3 binding, whence K-Ras is translocated to the recycling endosome (RE) for redelivery to the PM by vesicular transport (11). (B) On activation, PKG2 is recruited to the PM (t1/2 = 8.6 min) and colocalizes with K-Ras, where PKG2 phosphorylates K-Ras at Ser181 (t1/2 = 10.1 min). Endocytosed phospho-K-Ras translocates to the RE by the same process as nonphosphorylated K-Ras involving PDEδ and Arl2/3. The phosphorylated PBD significantly reduces the affinity of K-Ras for the anionic RE membrane, resulting in inefficient forward trafficking of K-Ras to the PM and the consequent accumulation in the cytosol and on endomembranes (t1/2 = 37.9 min).
Previous work has shown that the use of ionomycin in combination with bryostatin to robustly activate PKC also mislocalizes K-Ras from the PM (39). Under these conditions, K-Ras is lost from the PM much more rapidly than it is after PKG2 activation. However, since PKC, unlike PKG2, is able to efficiently phosphorylate K-Ras on both Ser171 and Ser181, the presence of multiple phosphate groups may cause the loss of K-Ras directly from the PM. It is also possible that activated PKC has more extensive access to K-Ras in multiple cellular compartments than just the K-Ras on the PM. Calcium fluxes induced by ionomycin also acutely reduce the electrostatic potential of the PM and separately cause K-Ras displacement (4). Interestingly, we observed that PKC-mediated phosphorylation was partially abrogated after PKG2 knockdown. Taken together with previous observations that PKCα can directly activate eNOS (56), it is possible that, in addition to directly phosphorylating K-Ras, PKC also indirectly activates another K-Ras kinase, PKG2.
Two studies have shown that PKC activators can suppress the growth of K-Ras tumors in nude mice by stimulating K-Ras phosphorylation (39, 57). Similarly and consistent with the loss of phosphorylated K-Ras from the PM as a result of prolonged activation of PKG, we observed inhibition of cell proliferation in K-Ras-positive NSCLC cells upon chronic activation of the AMPK → eNOS → sGC → PKG signaling pathway. Only NSCLC cells with an intact PKG pathway were sensitive to activators of the pathway, and the response to specific activators correlated with which components were expressed, identifying important biomarkers for predicting NSCLC cell responsiveness. LKB1/STK11 is a critical AMPK-activating kinase that was originally identified to be the product of a tumor suppressor gene (58). Germ line-inactivating mutations of LKB1 cause Peutz-Jeghers syndrome, which results in the development of benign polyps in the gastrointestinal tract, mucocutaneous pigmentation (59), and a predisposition to the development of various cancers of the lung, pancreas, colon, breast, small intestine, and ovary (60). Intriguingly, oncogenic mutant K-Ras is predominantly found in these cancers, suggesting a correlation of the oncogenic activity of these two proteins. Correspondingly, LKB1 nonsense mutations occur in some 30% of K-Ras-positive NSCLC cells (61, 62). Metabolic stress in these tumors would consequently fail to activate AMPK and inter alia also therefore fail to remove K-Ras from the PM, thus sustaining the K-Ras signal output. Thus, the loss of the LKB1 function in K-Ras-positive cancers would potentially prevent the downregulation of K-Ras signaling under conditions of ATP depletion.
Conversely, Counter and colleagues reported that elevated eNOS activity contributes to the development of K-Ras-driven pancreatic ductal adenocarcinomas (PDACs) (63). Advanced PDACs with an induced genetic deficiency of eNOS or treatment of mice with a NOS inhibitor limited the development of preinvasive pancreatic lesions and somewhat extended the mouse life span (63). It is therefore tempting to speculate that in different cellular contexts the different outcomes from K-Ras phosphorylation assume different levels of importance, such that in certain tissues, retuning of the K-Ras signal output, albeit with the attendant loss of total K-Ras from the PM, may be desirable. In this context, acute PKG activation in vascular smooth muscle cells enhanced the MAPK signal output (64), whereas PKG activation in small cell lung carcinoma cells inhibited epidermal growth factor-mediated MAPK stimulation (65). The inference from the current study that the chronic loss of PKG2 signaling allows enhanced K-Ras signaling is supported by observations in PKG2-null mice that develop marked crypt hyperplasia in the colonic epithelium, while ectopic PKG2 expression in colorectal cancer cell lines inhibits proliferation (66).
In sum we have discovered that PKG2 is a novel K-Ras kinase that regulates K-Ras PM interactions in response to the activation of AMPK, rendering K-Ras signaling sensitive to metabolic stress and other signal cascades that generate NO, although to what extent PKG2-mediated suppression of K-Ras signaling contributes to a metabolic stress response remains to be elucidated. We also identified components of the AMPK, eNOS, and PKG pathway to be a set of biomarkers and tractable drug targets for K-Ras-positive cancers. Thus, K-Ras tumors that express all or some components of the AMPK → PKG signaling pathway are predictably sensitive to specific defined activators of the pathway, which opens up interesting therapeutic options. For example, FDA-approved metformin, used for the treatment of type II diabetes mellitus, and sildenafil, used for the treatment of erectile dysfunction, are likely to have efficacy in K-Ras-positive cancer cells, where these agents activate PKG.
ACKNOWLEDGMENTS
This work was supported by a grant to J.F.H. from the Cancer Research and Prevention Institute of Texas (CPRIT; RP130059), a National Institutes of Health (NIH) Pathway to Independence Award (K99CA188593) to K.-J.C., a grant to A.A.G. from NIH (R01GM100078), Extreme Science and Engineering Discovery Environment (XSEDE; project MCB150054) grants to R.J.C. from the Australian Research Council (DP120100183 and LP120100088) and The University of Queensland, Institute for Molecular Bioscience, and grants to D.E.C. from the University of California, San Diego, Department of Medicine.
REFERENCES
- 1.Hancock JF. 2003. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol 4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 2.Hancock JF, Paterson H, Marshall CJ. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63:133–139. doi: 10.1016/0092-8674(90)90294-O. [DOI] [PubMed] [Google Scholar]
- 3.Hancock JF, Magee AI, Childs JE, Marshall CJ. 1989. All Ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 4.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. 2008. Membrane phosphatidylserine regulates surface charge and protein localization. Science 319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 5.Okeley NM, Gelb MH. 2004. A designed probe for acidic phospholipids reveals the unique enriched anionic character of the cytosolic face of the mammalian plasma membrane. J Biol Chem 279:21833–21840. doi: 10.1074/jbc.M313469200. [DOI] [PubMed] [Google Scholar]
- 6.Roy MO, Leventis R, Silvius JR. 2000. Mutational and biochemical analysis of plasma membrane targeting mediated by the farnesylated, polybasic carboxy terminus of K-ras4B. Biochemistry 39:8298–8307. doi: 10.1021/bi000512q. [DOI] [PubMed] [Google Scholar]
- 7.Plowman SJ, Ariotti N, Goodall A, Parton RG, Hancock JF. 2008. Electrostatic interactions positively regulate K-Ras nanocluster formation and function. Mol Cell Biol 28:4377–4385. doi: 10.1128/MCB.00050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutierrez L, Magee AI, Marshall CJ, Hancock JF. 1989. Post-translational processing of p21ras is two-step and involves carboxyl-methylation and carboxy-terminal proteolysis. EMBO J 8:1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock JF, Cadwallader K, Paterson H, Marshall CJ. 1991. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of Ras proteins. EMBO J 10:4033–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, Wittinghofer A, Bastiaens PI. 2012. The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol 14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 11.Schmick M, Vartak N, Papke B, Kovacevic M, Truxius DC, Rossmannek L, Bastiaens PI. 2014. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell 157:459–471. doi: 10.1016/j.cell.2014.02.051. [DOI] [PubMed] [Google Scholar]
- 12.Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. 2007. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol 9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 13.Kholodenko BN, Hancock JF, Kolch W. 2010. Signalling ballet in space and time. Nat Rev Mol Cell Biol 11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plowman SJ, Muncke C, Parton RG, Hancock JF. 2005. H-Ras, K-Ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci U S A 102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohnke M, Schmitt S, Ariotti N, Piggott AM, Parton RG, Lacey E, Capon RJ, Alexandrov K, Abankwa D. 2012. Design and application of in vivo FRET biosensors to identify protein prenylation and Nanoclustering inhibitors. Chem Biol 19:866–874. doi: 10.1016/j.chembiol.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, Cho KJ, Plowman SJ, Hancock JF. 2012. Nonsteroidal anti-inflammatory drugs alter the spatiotemporal organization of Ras proteins on the plasma membrane. J Biol Chem 287:16586–16595. doi: 10.1074/jbc.M112.348490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Hoeven D, Cho KJ, Ma X, Chigurupati S, Parton RG, Hancock JF. 2013. Fendiline inhibits K-Ras plasma membrane localization and blocks K-Ras signal transmission. Mol Cell Biol 33:237–251. doi: 10.1128/MCB.00884-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho KJ, Park JH, Piggott AM, Salim AA, Gorfe AA, Parton RG, Capon RJ, Lacey E, Hancock JF. 2012. Staurosporines disrupt phosphatidylserine trafficking and mislocalize Ras proteins. J Biol Chem 287:43573–43584. doi: 10.1074/jbc.M112.424457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho KJ, Park JH, Hancock JF. 2013. Staurosporine: a new tool for studying phosphatidylserine trafficking. Commun Integr Biol 6:e24746. doi: 10.4161/cib.24746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salim AA, Xiao X, Cho KJ, Piggott AM, Lacey E, Hancock JF, Capon RJ. 2014. Rare Streptomyces sp. polyketides as modulators of K-Ras localisation. Org Biomol Chem 12:4872–4878. doi: 10.1039/c4ob00745j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho KJ, van der Hoeven D, Zhou Y, Maekawa M, Ma X, Chen W, Fairn GD, Hancock JF. 2015. Inhibition of acid sphingomyelinase depletes cellular phosphatidylserine and mislocalizes K-Ras from the plasma membrane. Mol Cell Biol 36:363–374. doi: 10.1128/MCB.00719-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salim AA, Tan L, Huang XC, Cho KJ, Lacey E, Hancock JF, Capon RJ. 2016. Oligomycins as inhibitors of K-Ras plasma membrane localisation. Org Biomol Chem 14:711–715. doi: 10.1039/C5OB02020D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salim AA, Cho KJ, Tan L, Quezada M, Lacey E, Hancock JF, Capon RJ. 2014. Rare Streptomyces N-formyl amino-salicylamides inhibit oncogenic K-Ras. Org Lett 16:5036–5039. doi: 10.1021/ol502376e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iglesias DA, Yates MS, van der Hoeven D, Rodkey TL, Zhang Q, Co NN, Burzawa J, Chigurupati S, Celestino J, Bowser J, Broaddus R, Hancock JF, Schmandt R, Lu KH. 2013. Another surprise from metformin: novel mechanism of action via K-Ras influences endometrial cancer response to therapy. Mol Cancer Ther 12:2847–2856. doi: 10.1158/1535-7163.MCT-13-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki A, Arawaka S, Sato H, Kato T. 2015. Sensitive Western blotting for detection of endogenous Ser129-phosphorylated alpha-synuclein in intracellular and extracellular spaces. Sci Rep 5:14211. doi: 10.1038/srep14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancock JF, Prior IA. 2005. Electron microscopic imaging of Ras signaling domains. Methods 37:165–172. doi: 10.1016/j.ymeth.2005.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prior IA, Muncke C, Parton RG, Hancock JF. 2003. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol 160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ripley BD. 1977. Modelling spatial patterns. J R Stat Soc Series B Stat Methodol 39:172–192. [Google Scholar]
- 29.Diggle PJ, Mateu J, Clough HE. 2000. A comparison between parametric and non-parametric approaches to the analysis of replicated spatial point patterns. Adv Appl Probab 32:331–343. [Google Scholar]
- 30.Rangaswami H, Marathe N, Zhuang S, Chen Y, Yeh JC, Frangos JA, Boss GR, Pilz RB. 2009. Type II cGMP-dependent protein kinase mediates osteoblast mechanotransduction. J Biol Chem 284:14796–14808. doi: 10.1074/jbc.M806486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakash P, Zhou Y, Liang H, Hancock JF, Gorfe AA. 2016. Oncogenic K-Ras binds to an anionic membrane in two distinct orientations: a molecular dynamics analysis. Biophys J 110:1125–1138. doi: 10.1016/j.bpj.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FT, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 33.Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD Jr, Pastor RW. 2010. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B 114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. 2005. Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toogood PL. 2008. Mitochondrial drugs. Curr Opin Chem Biol 12:457–463. doi: 10.1016/j.cbpa.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Hardie DG. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 37.Hardie DG. 2006. Neither LKB1 nor AMPK are the direct targets of metformin. Gastroenterology 131:973. doi: 10.1053/j.gastro.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 38.Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. 2011. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol 194:257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, Perez de Castro I, Li C, Thompson CB, Cox AD, Philips MR. 2006. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell 21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Tegge W, Frank R, Hofmann F, Dostmann WR. 1995. Determination of cyclic nucleotide-dependent protein kinase substrate specificity by the use of peptide libraries on cellulose paper. Biochemistry 34:10569–10577. [DOI] [PubMed] [Google Scholar]
- 41.Rolli-Derkinderen M, Sauzeau V, Boyer L, Lemichez E, Baron C, Henrion D, Loirand G, Pacaud P. 2005. Phosphorylation of serine 188 protects RhoA from ubiquitin/proteasome-mediated degradation in vascular smooth muscle cells. Circ Res 96:1152–1160. doi: 10.1161/01.RES.0000170084.88780.ea. [DOI] [PubMed] [Google Scholar]
- 42.Barcelo C, Paco N, Beckett AJ, Alvarez-Moya B, Garrido E, Gelabert M, Tebar F, Jaumot M, Prior I, Agell N. 2013. Oncogenic K-ras segregates at spatially distinct plasma membrane signaling platforms according to its phosphorylation status. J Cell Sci 126:4553–4559. doi: 10.1242/jcs.123737. [DOI] [PubMed] [Google Scholar]
- 43.Janosi L, Gorfe AA. 2010. Segregation of negatively charged phospholipids by the polycationic and farnesylated membrane anchor of Kras. Biophys J 99:3666–3674. doi: 10.1016/j.bpj.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorfe AA, Babakhani A, McCammon JA. 2007. H-Ras protein in a bilayer: interaction and structure perturbation. J Am Chem Soc 129:12280–12286. doi: 10.1021/ja073949v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. 2007. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J Med Chem 50:674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 46.Gorfe AA, McCammon JA. 2008. Similar membrane affinity of mono- and di-S-acylated Ras membrane anchors: a new twist in the role of protein lipidation. J Am Chem Soc 130:12624–12625. doi: 10.1021/ja805110q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. 1992. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J 11:2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deguchi A, Soh JW, Li H, Pamukcu R, Thompson WJ, Weinstein IB. 2002. Vasodilator-stimulated phosphoprotein (VASP) phosphorylation provides a biomarker for the action of exisulind and related agents that activate protein kinase G. Mol Cancer Ther 1:803–809. [PubMed] [Google Scholar]
- 49.Smolenski A, Bachmann C, Reinhard K, Honig-Liedl P, Jarchau T, Hoschuetzky H, Walter U. 1998. Analysis and regulation of vasodilator-stimulated phosphoprotein serine 239 phosphorylation in vitro and in intact cells using a phosphospecific monoclonal antibody. J Biol Chem 273:20029–20035. doi: 10.1074/jbc.273.32.20029. [DOI] [PubMed] [Google Scholar]
- 50.Butt E, Abel K, Krieger M, Palm D, Hoppe V, Hoppe J, Walter U. 1994. cAMP- and cGMP-dependent protein kinase phosphorylation sites of the focal adhesion vasodilator-stimulated phosphoprotein (VASP) in vitro and in intact human platelets. J Biol Chem 269:14509–14517. [PubMed] [Google Scholar]
- 51.Reger AS, Yang MP, Koide-Yoshida S, Guo E, Mehta S, Yuasa K, Liu A, Casteel DE, Kim C. 2014. Crystal structure of the cGMP-dependent protein kinase II leucine zipper and Rab11b protein complex reveals molecular details of G-kinase-specific interactions. J Biol Chem 289:25393–25403. doi: 10.1074/jbc.M114.575894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeung T, Heit B, Dubuisson JF, Fairn GD, Chiu B, Inman R, Kapus A, Swanson M, Grinstein S. 2009. Contribution of phosphatidylserine to membrane surface charge and protein targeting during phagosome maturation. J Cell Biol 185:917–928. doi: 10.1083/jcb.200903020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grant BD, Donaldson JG. 2009. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol 10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. 2002. The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J 21:2095–2106. doi: 10.1093/emboj/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jang H, Abraham SJ, Chavan TS, Hitchinson B, Khavrutskii L, Tarasova NI, Nussinov R, Gaponenko V. 2015. Mechanisms of membrane binding of small GTPase K-Ras4B farnesylated hypervariable region. J Biol Chem 290:9465–9477. doi: 10.1074/jbc.M114.620724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fukumura D, Kashiwagi S, Jain RK. 2006. The role of nitric oxide in tumour progression. Nat Rev Cancer 6:521–534. doi: 10.1038/nrc1910. [DOI] [PubMed] [Google Scholar]
- 57.Wang MT, Holderfield M, Galeas J, Delrosario R, To MD, Balmain A, McCormick F. 2015. K-Ras promotes tumorigenicity through suppression of non-canonical Wnt signaling. Cell 163:1237–1251. doi: 10.1016/j.cell.2015.10.041. [DOI] [PubMed] [Google Scholar]
- 58.Shackelford DB, Shaw RJ. 2009. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer 9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, Jarvinen H, Kristo P, Pelin K, Ridanpaa M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA. 1998. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 60.Giardiello FM, Welsh SB, Hamilton SR, Offerhaus GJ, Gittelsohn AM, Booker SV, Krush AJ, Yardley JH, Luk GD. 1987. Increased risk of cancer in the Peutz-Jeghers syndrome. N Engl J Med 316:1511–1514. doi: 10.1056/NEJM198706113162404. [DOI] [PubMed] [Google Scholar]
- 61.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, et al. . 2008. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsumoto S, Iwakawa R, Takahashi K, Kohno T, Nakanishi Y, Matsuno Y, Suzuki K, Nakamoto M, Shimizu E, Minna JD, Yokota J. 2007. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene 26:5911–5918. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lampson BL, Kendall SD, Ancrile BB, Morrison MM, Shealy MJ, Barrientos KS, Crowe MS, Kashatus DF, White RR, Gurley SB, Cardona DM, Counter CM. 2012. Targeting eNOS in pancreatic cancer. Cancer Res 72:4472–4482. doi: 10.1158/0008-5472.CAN-12-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Komalavilas P, Shah PK, Jo H, Lincoln TM. 1999. Activation of mitogen-activated protein kinase pathways by cyclic GMP and cyclic GMP-dependent protein kinase in contractile vascular smooth muscle cells. J Biol Chem 274:34301–34309. doi: 10.1074/jbc.274.48.34301. [DOI] [PubMed] [Google Scholar]
- 65.Tao Y, Gu YJ, Cao ZH, Bian XJ, Lan T, Sang JR, Jiang L, Wang Y, Qian H, Chen YC. 2012. Endogenous cGMP-dependent protein kinase reverses EGF-induced MAPK/ERK signal transduction through phosphorylation of VASP at Ser239. Oncol Lett 4:1104–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang R, Kwon IK, Thangaraju M, Singh N, Liu K, Jay P, Hofmann F, Ganapathy V, Browning DD. 2012. Type 2 cGMP-dependent protein kinase regulates proliferation and differentiation in the colonic mucosa. Am J Physiol Gastrointest Liver Physiol 303:G209–G219. doi: 10.1152/ajpgi.00500.2011. [DOI] [PubMed] [Google Scholar]
- 67.Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. 1999. Endomembrane trafficking of Ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]