Abstract
Chromosome transmission fidelity during mitosis is of critical importance for the fitness of an organism, as mistakes will lead to aneuploidy, which has a causative role in numerous severe diseases. Proper segregation of chromosomes depends on interdependent processes at the microtubule-kinetochore interface and the spindle assembly checkpoint. Here we report the discovery of a new element essential for chromosome transmission fidelity that implicates inositol pyrophosphates (IPPs) as playing a key role in this process. The protein is Asp1, the Schizosaccharomyces pombe member of the highly conserved Vip1 family. Vip1 enzymes are bifunctional: they consist of an IPP-generating kinase domain and a pyrophosphatase domain that uses such IPPs as substrates. We show that Asp1 kinase function is required for bipolar spindle formation. The absence of Asp1-generated IPPs resulted in errors in sister chromatid biorientation, a prolonged checkpoint-controlled delay of anaphase onset, and chromosome missegregation. Remarkably, expression of Asp1 variants that generated higher-than-wild-type levels of IPPs led to a faster-than-wild-type entry into anaphase A without an increase in chromosome missegregation. In fact, the chromosome transmission fidelity of a nonessential chromosome was enhanced with increased cellular IPPs. Thus, we identified an element that optimized the wild-type chromosome transmission process.
INTRODUCTION
In eukaryotes, the faithful transmission of genetic information from one generation to the next is tightly controlled. Genome instability generally has a strong negative impact on the fitness of an organism. Decreased chromosome segregation fidelity during cell division will give rise to cell progeny which have lost or gained chromosomes. Aneuploidy is a distinguishing feature of many types of cancer and neurological diseases and has been implicated as an important factor in the aging process (1, 2). It is a poorly understood paradox that genome plasticity can also be advantageous for the propagation of cells or an organism. Aneuploid tumor cells have a proliferative advantage, and large-scale chromosome changes are a common feature of human-pathogenic fungi such as Candida. In the latter case, these alterations facilitate an adaption to the environment resulting in increased survival (3).
Chromosome segregation is a precise process: in model yeasts such as the fission yeast Schizosaccharomyces pombe where in vivo chromosome loss rates can be determined easily, it is approximately 0.01% (reviewed in reference 4). Chromosome transmission fidelity depends on the formation and function of the bipolar spindle, the multicomponent kinetochore complex assembled on centromeric chromatin, and the correct connections between these two structures. Biorientation of sister chromatids requires the binding of plus ends of spindle microtubules (MTs) to attachment sites on the kinetochore in such a way that sisters are bound to MTs polymerized from opposing spindle poles. Incorrect attachments are common and are corrected by the Aurora B kinase family, whose phosphorylation of specific kinetochore proteins weakens incorrect MT-kinetochore binding (reviewed in reference 5). The process of MT-kinetochore attachment is monitored by the spindle assembly checkpoint (SAC) (reviewed in reference 6), which also seems to play an active role in achieving chromosome biorientation (7).
Spindle assembly in S. pombe is similar to that in other eukaryotes and can be divided into three phases corresponding to prometaphase (I), metaphase/anaphase A (II), and anaphase B (III). Phase I spindles rapidly elongate to approximately 1.5 μm, while phase II spindles are much less dynamic, with a length of up to 3 μm. Phase III spindles rapidly elongate to 10 to 12 μm (8, 9). In phase II, sister chromosomes need to become bioriented, and the cell will stay in this phase until biorientation has been achieved.
In this work, we describe a new modulator of all 3 spindle phases: inositol pyrophosphates (IPPs) generated by the Asp1 kinase. Asp1 is a member of the highly conserved Vip1 protein family. Vip1 proteins have a dual-domain structure that consists of an N-terminal “RimK”/ATP-grasp superfamily kinase domain and a C-terminal domain with homology to histidine-acid phosphatases (10–13). Both domains are enzymatically active. The function of the N-terminal kinase domain has been studied extensively, and it has been demonstrated that the enzymatic activity is highly specific: generation of diphospho-myo-inositol polyphosphates (here called inositol pyrophosphates [IPPs]) by adding a phosphate at position 1 on the fully phosphorylated inositol ring (11, 12, 14, 15). We showed recently that in vitro the C-terminal Asp1 domain reduces the IPP output of the kinase domain, indicating that it has phosphatase activity (16). Again, this enzymatic activity is highly substrate specific, as in vitro analysis showed that the C-terminal Asp1 domain hydrolyzes the diphosphate exclusively at position 1 of the inositol ring (17). Vip1 members regulate a wide variety of eukaryotic processes ranging from the human antiviral response to plant defense mechanisms against insects (10, 11, 18–20). In fission yeast, the Vip1 member Asp1 regulates the dimorphic switch, which is a dramatic morphological alteration in response to changing environmental conditions (21). How the Asp1 kinase controlled dimorphic switch functions at the molecular level is not fully understood. However, we have recently shown that fungal Vip1 family members regulate morphogenesis by modulating the interphase MT cytoskeleton (16).
Our present analysis now extends the number of cellular processes controlled by Vip1 kinases, as we find that mitotic fidelity is dependent on Asp1 kinase function.
MATERIALS AND METHODS
Strains and media.
All S. pombe strains used are listed in Table 1. New strains were obtained by crossing the initial strains, followed by random spore analysis and phenotype determination. Strains were grown in full medium (YE5S) or minimal medium (MM) with supplements (22). For nmt81::gfp-atb2+ expression, cells were grown in MM without thiamine for 24 h at 30°C before analysis.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source (alternative strain name) |
|---|---|---|
| UFY2156 | h− SV40::GFP-Atb2+::LEU2 his3-D1 | Yeast Genetic Resource Center, Osaka, Japan |
| UFY1511 | h+ asp1D333A::Kanr his3-D1 ade6-M210 leu1-32 ura4-D18 | U. Fleig |
| UFY1579 | h+ asp1H397A::Kanr his3-D1 ade6-M210 leu1-32 ura4-D18 | U. Fleig |
| UFY2279 | h− asp1H397A::Kanr SV40::GFP-Atb2+::LEU2 | This study |
| UFY2280 | h− asp1D333A::Kanr SV40::GFP-Atb2+::LEU2 his3-D1 | This study |
| UFY605 | h− his3-D1 ade6-M210 leu1-32 ura4-D18 | K. Gould (425) |
| UFY857 | h− Kanr::nmt81::GFP-Atb2+ leu1-32 | T. Toda |
| UFY1763 | h+ asp1H397A::Kanr Kanr::nmt81::GFP-Atb2+ leu1-32 ura4-D18 | U. Fleig |
| UFY1529 | h+ asp1D333A::Kanr Kanr::nmt81::GFP-Atb2+ leu1-32 ura4-D18 | U. Fleig |
| UFY2249 | h− Sad1-mCherry::Kanr LacI-GFP::his7+ LacO-repeat::lys1+ ura4-D18 leu1-32 | This study |
| UFY2242 | h− asp1D333A::Kanr Sad1-mCherry::Kanr LacI-GFP::his7+ LacO-repeat::lys1+ | This study |
| UFY2357 | h+ ark1+-GFP::Kanr sid4+-mCherry::Natr | Silke Hauf (SK402) |
| UFY2363 | h− ark1+-GFP::Kanr Sid4+-mCherry::Natr asp1D333A::Kanr | This study |
| UFY1543 | h+ asp1D333A::Kanr mph1Δ::ura4+ his3-D1 ade6-M216 leu1-32 ura4-D18 | This study |
| UFY2337 | h− asp1D333A::Kanr mph1Δ::ura4+ SV40::GFP-Atb2+::LEU2 ura4-D18 | This study |
| UFY2367 | h− asp1H397A::Kanr Sad1-mCherry::Kanr LacI-GFP::his7+, LacO-repeat::lys1+ ura4-D18 leu1-32 | This study |
| UFY2646 | h− asp1D333A::Kanr mph1Δ::ura4+ LacI-GFP::his7+ LacO-repeat::lys1+ Sad1-mCherry::Kanr his− lys1-131 ura4-D18 leu1-32 | This study |
| UFY2627 | h− mph1Δ::ura4+ LacI-GFP::his7+ LacO-repeat::lys1+ Sad1-mCherry::Kanr leu1-32 ura4-D18 his− lys1-131 | This study |
| UFY138 | h− mph1Δ::ura4+ leu1-32 ura4-D18 ade6-M216 | Shalley Sazer |
| UFY103 | h− ade6-M210 leu1-32 ura4-D6 Ch16[ade6-M216] | Mitsuhiro Yanagida (10.2.2) |
| UFY2336 | leu1-32::SV40::Atb2+-GFP[LEU2] mad2Δ::ura4+ ura4-D18 | This study |
| UFY2335 | leu1-32::SV40::Atb2+-GFP[LEU2] asp1D333A::Kanr mad2Δ::ura4+ ura4-D18 | This study |
| UFY1790 | h− asp1D333A::Kanr ade6-M210 leu1-32 | This study |
| UFY139 | h− mad2Δ::ura4+ leu1-32 ura4-D18 ade6-M210 | Shelley Sazer |
| UFY2215 | h− asp1D333A::Kanr mad2Δ::ura4+ ura4-D18 leu1-32 | This study |
| UFY229 | h− cut7-466 leu1-32 | Iain Hagan (138) |
| UFY2422 | h+ klp6Δ::Kanr leu1 ura4 his2/7 | Takashi Toda (AR018) |
| UFY302 | h− pkl1Δ::his3+ leu1-32 ura4-D18 ade6-210 his3-D1 | Robin Allshire and Alison Pidoux (ALP192) |
For live-cell imaging, cells were preincubated overnight at 30°C or 25°C, diluted to 105 cells/ml, and grown for additional 24 h in MM. Microscopy slides were prepared by patching cells on agarose pads as described previously (23).
Microscopy.
For live-cell imaging, a Zeiss spinning-disk confocal microscope equipped with an AxioCam MRm camera (photomicrographs in Fig. 2 and in Fig. S3 in the supplemental material) or a Rolera EM-C2 (QImaging) camera (all other photomicrographs) and AxioVision and Zen2011/2012 software was used. Image processing was done with Adobe Photoshop CS2 (Adobe Systems Software Ireland Limited, Dublin, Ireland). A z-stack of 15 z slices with a distance of 0.5 μm was recorded and a maximum-intensity projection (MIP) created. The time interval between images was 10 to 30 s. For technical reasons, microscopic analysis was done at 20°C. For spindle midzone measurements, images were taken every 10 s. The MIP image of the spindle with the thinnest midzone was used for measurement. The signal intensity of green fluorescent protein (GFP) fluorescence in live-cell images was determined with ImageJ 1.44 (NIH).
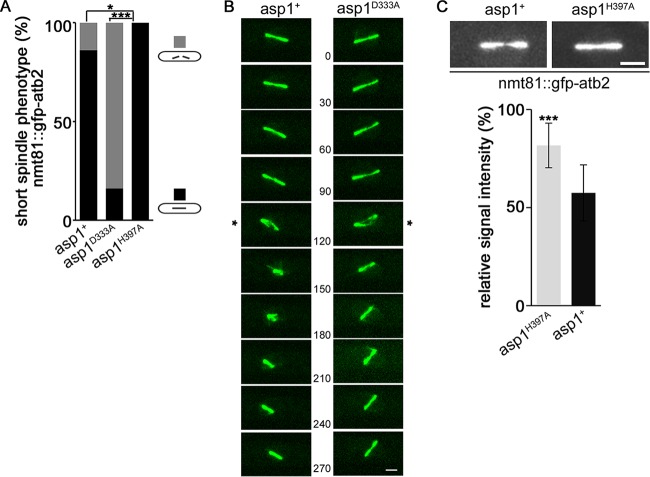
FIG 2.
Aberrant spindle phenotype of an nmt81::gfp-atb2+ strain rescued by expression of Asp1H397A. (A) Quantification of spindle phenotypes (gray and black bars show broken and nonbroken spindles, respectively) for the indicated strains expressing nmt81::gfp-atb2+ (asp1+ strain, n = 37; asp1H397A strain, n = 31; asp1D333A strain, n = 21; *, P < 0.05; ***, P < 0.001 [as determined by χ2 test]). (B) Example of collapsing (*) asp1+ and asp1D333A spindles in the nmt81::gfp-atb2+strain background. Time between images, 30 s. Bar, 2 μm. (C) Top, photomicrographs of asp1+ and asp1H397A short spindles. Bar, 2 μm. Bottom, relative signal intensity of spindle midzone in asp1+ (57.5 ± 14.3) and asp1H397A (81.6 ± 11.4) cells expressing nmt81::gfp-atb2+. Quantification of the fluorescence signal was as described for Fig. 1B (n = 9/strain; ***, P < 0.0005 using two-sample t test).
Western blot analysis.
To determine expression of the plasmid-borne variant encoding positions 1 to 364 of Asp1 fused to GFP (asp11–364-gfp) via the nmt1+ promoter, transformants were grown in MM with or without thiamine for 24 h at 30°C before protein extraction. Preparation of protein extracts, determination of protein concentrations, and Western blot analysis were carried out as described previously (16) using anti-GFP antibody (monoclonal mouse; Roche) and anti-γ-tubulin antibody (monoclonal mouse; Sigma).
Ch16 chromosome loss assay.
The Ch16-containing strain was pregrown on MM without adenine to select for the presence of the Ch16 chromosome in cells. Such cells were transformed with a control vector or a plasmid expressing asp11–364 via the thiamine-repressible nmt1+ promoter. Transformants were grown under plasmid and Ch16 selective conditions in the absence or presence of 5 μg/ml thiamine for 20 h at 25°C. Up to 30,000 cells per transformant were plated on plasmid-selective plates with a limiting amount of adenine and with or without thiamine for 10 days at 25°C. Ch16 chromosome loss results in adenine prototrophy and red cells. Thus, chromosome loss in a growing colony was identified as red sectors within the white colony.
RESULTS
Asp1-generated IPPs are required for spindle assembly.
To determine whether Asp1-generated IPPs had a role in spindle assembly, we looked at spindle structures using GFP–α-tubulin strains that expressed endogenously one of the three Asp1 variants Asp1, Asp1D333A, or Asp1H397A (21). Asp1 is encoded by the wild-type asp1+ gene, while asp1D333A generates an Asp1 variant that has a single amino acid change at position 333, which is the key catalytic residue for IPP kinase activity (10). Thus, Asp1D333A cannot generate IPPs (10, 16). The Asp1H397A variant has a mutation in a highly conserved histidine residue, which abolishes the C-terminal pyrophosphatase activity (16). Thus, the IPP output of Asp1H397A is higher than that of the wild-type Asp1 protein (up to twice as much in an in vitro assay [16]). Therefore, the asp1D333A strain cannot generate Asp1-made IPPs, the asp1+ strain generates wild-type IPP amounts, and the asp1H397A strain generates amounts of IPP that are increased compared to those of the wild-type strain.
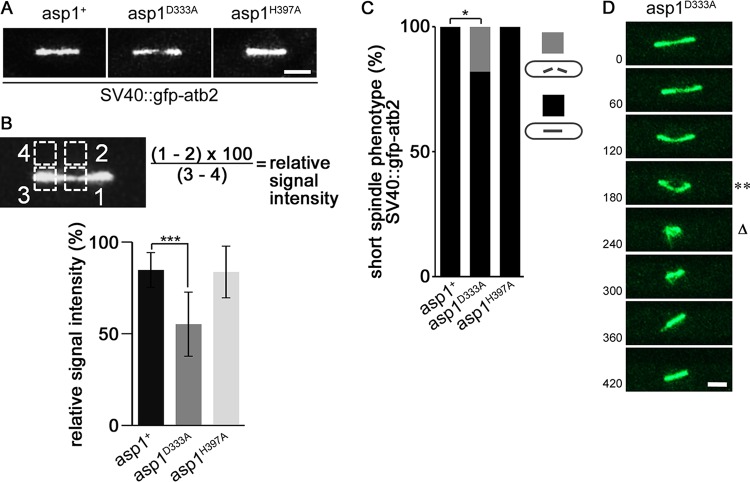
The presence of GFP–α-tubulin (SV40::GFP-Atb2) (24) in the different asp1 strains did not affect their growth on rich YE5S medium but slightly increased their sensitivity to the MT-destabilizing drug thiabendazole (TBZ) (see Fig. S1A in the supplemental material). Live-cell imaging of the spindles in the various mutant strains revealed that in contrast to asp1+ spindles, asp1D333A spindles were often aberrant, as they displayed an abnormally thin spindle midzone (Fig. 1A; Movies S1 and S2 in the supplemental material show an asp1+ spindle and an asp1D333A spindle, respectively). The spindle midzone consists of interpolar MTs coming from the two separated spindle pole bodies (SPBs). The plus ends of such MTs interdigitate via specific MT-associated proteins, thus giving stability to the spindle midzone (reviewed in reference 25). The asp1D333A mutant spindles were between 3 and 5 μm in length, and the relative fluorescence signal intensity of the spindle midzone was significantly reduced compared to that of the midzones of asp1+ spindles (84.7 ± 9.5 and 55.1 ± 17.5 for asp1+ and asp1D333A, respectively) (Fig. 1B). The thin spindle midzone phenotype was not observed in the asp1+ and asp1H397A strains (Fig. 1A and B). Interestingly, 18% of asp1D333A mitotic cells showed spindle breakage (Fig. 1C). A typical example of such a phenotype in which the spindle collapsed and then reformed is shown in Fig. 1D.
FIG 1.
Asp1-generated IPPs control spindle midzone stability. (A) Photomicrographs of asp1+, asp1D333A, and asp1H397A cells with a short (3- to 5-μm) spindle expressing SV40::GFP-Atb2+. Bar, 2 μm. (B) Quantification of the fluorescence signal of the spindle midzone. The thinnest part of a spindle midzone was measured (square 1), and the fluorescent background was subtracted (square 2). For relative signal intensity this value was set in relation to the fluorescence signal of a spindle end (square 3) minus fluorescent background (square 4). Relative signal intensities of spindle midzones: asp1+, 84.7 ± 9.5; asp1D333A, 55.1 ± 17.5; and asp1H397A, 83.6 ± 14.1 (n = 9/strain; ***, P < 0.0005 as determined by two-sample t test). (C) Quantification of spindle phenotypes (gray and black bars show broken and nonbroken spindles, respectively) determined for the indicated strains expressing SV40::GFP-Atb2+ (asp1+ strain, n = 24; asp1H397A strain, n = 22; asp1D333A strain, n = 17; *, P < 0.05 as determined using χ2 test). (D) Representative example of an asp1D333A SV40::GFP-Atb2+ cell. Time between images, 60 s. Bar, 2 μm. **, collapsing spindle; Δ, broken spindle.
Although expression of a GFP-tubulin construct is necessary for live-cell imaging of the MT cytoskeleton, it has been noted that expression of such constructs can affect MT behavior and dynamics (26). For example, it has been reported that strains expressing nmt81::GFP-Atb2 at the atb2+ locus have altered MT properties (24, 27–29). The reason for this is unclear. However, it has been shown that the ratio of tagged GFP–α-tubulin to untagged α-tubulin is higher in the nmt81::GFP-Atb2 strain than in the SV40::GFP-Atb2 strain (26). We found that under our assay conditions, the presence of nmt81::GFP-Atb2 in the asp1+ strain gave rise to short spindles that broke in 16% of cases (Fig. 2A and B, left panel). We therefore asked whether in the nmt81::GFP-Atb2 background the frequency of the asp1D333A spindle phenotype was increased relative to that in wild-type cells. We found that short spindles collapsed in 86% of nmt81::GFP-Atb2 asp1D333A cells (Fig. 2A and B, right panel). Movie S3 in the supplemental material shows an example of a cell in which the thin spindle midzone collapsed, generating two spindle halves which reformed into a bipolar spindle. In some instances, these spindles collapsed again (see Movie S3). In general, broken spindles reformed and were able to undergo spindle elongation in spindle phase III (an example is shown in Movie S3). The asp1D333A strain fitness was not reduced severely by the presence of nmt81::GFP-Atb2 (see Fig. S1B in the supplemental material).
Intriguingly, the collapsed spindle phenotype of the nmt81::GFP-Atb2 strain was not observed for the asp1H397A nmt81::GFP-Atb2 strain (Fig. 2A). We thus compared the relative fluorescence signal intensities of the spindle midzones of asp1+ and asp1H397A spindles in the nmt81::GFP-Atb2 background. The relative fluorescence signal intensity of the spindle midzone was significantly increased for asp1H397A spindles compared to asp1+ spindles (Fig. 2C) (signal intensities of 81.6 ± 11.4 and 57.5 ± 14.3 for asp1H397A and asp1+, respectively). We conclude that Asp1-generated IPPs regulate spindle midzone stability: asp1D333A cells, which have no Asp1 kinase activity, often show abnormally thin spindle midzones and a high number of collapsing spindles, while asp1H397A cells have more stable spindle midzones than wild-type cells expressing nmt81::GFP-Atb2.
Spindle collapse occurs prior to chromosome segregation.
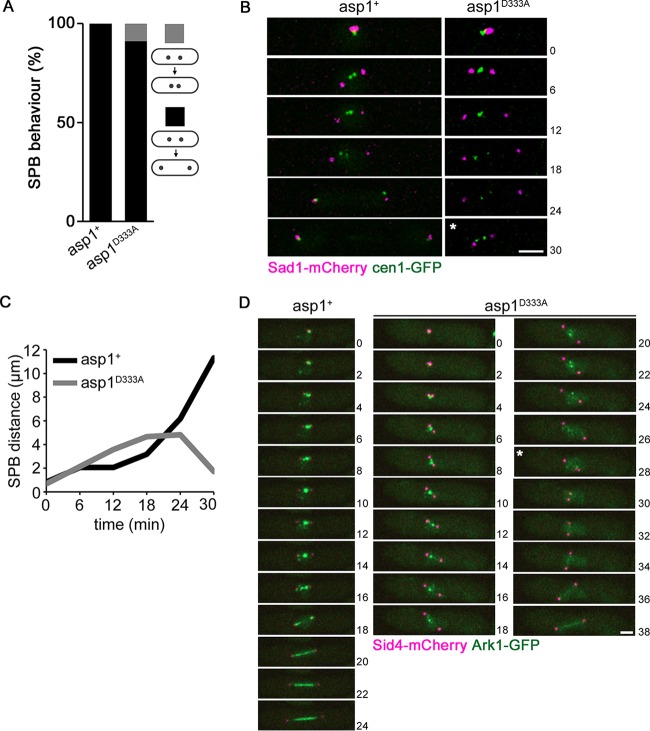
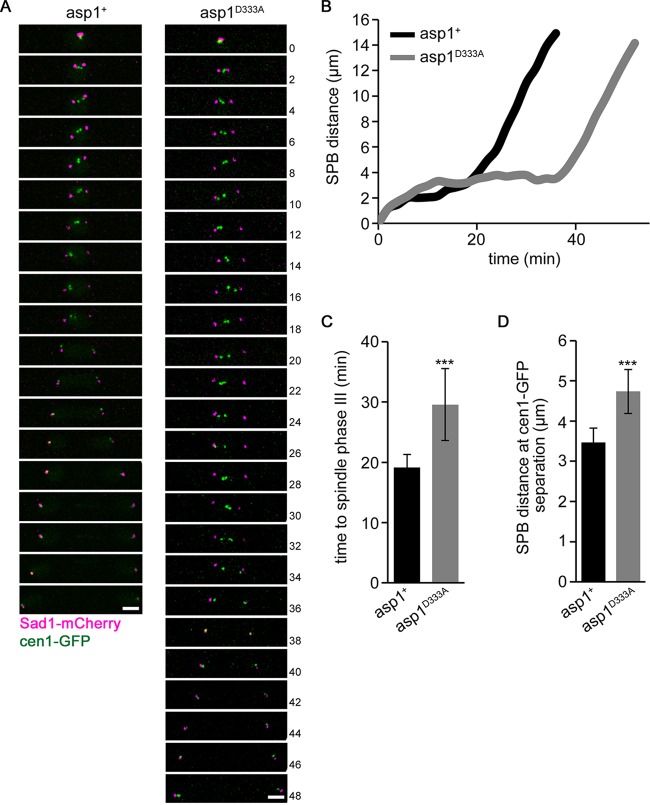
In order to determine at which mitotic stage spindle collapse occurred, we used a strain where the segregation behavior of chromosome I sisters and the SPBs could be monitored. The collapsing spindles observed in asp1D333A strains expressing either SV40::GFP-Atb2 or nmt81::GFP-Atb2 were spindles of maximally 5 μm in length. Metaphase spindles are usually shorter, but abnormally long metaphase spindles can occur due to altered force-generating mechanisms (reviewed in references 30 and 31). To analyze the segregation behavior of the SPBs and chromosome I sisters in asp1D333A mitotic cells, we used the SPB component Sad1-mCherry and cen1-GFP (centromere I-linked GFP) (8, 32, 33). The presence of Sad1-mCherry cen1-GFP did not negatively affect growth of the asp1D333A strain (see Fig. S2A in the supplemental material). However, live-cell imaging of cells undergoing mitosis revealed that in 9% of mitotic cells, the distance between the SPBs first increased and then drastically decreased, indicating spindle collapse (Fig. 3A and B). In the asp1D333A cell shown in Fig. 3B and C, the distance between the SPBs increased to 4.8 μm, followed by spindle collapse and a reduced distance between the SPBs of just 1.7 μm. In all cases analyzed, the cen1-GFP signals had not been segregated to the SPBs prior to spindle collapse.
FIG 3.
Spindle breakage occurs prior to sister chromatid segregation. (A) Quantification of SPB segregation phenotype determined for the indicated strains expressing cen1-gfp Sad1-mCherry (asp1+ strain, n = 31; asp1D333A strain, n = 45) (gray and black bars show collapsing and noncollapsing spindles, respectively). (B) Time-lapse images of mitotic asp1+ and asp1D333A cells expressing cen1-GFP Sad1-mCherry. Time between images, 6 min. Bar, 2 μm. *, time of spindle collapse. (C) Diagrammatic representation of the Sad1-mCherry images shown in panel B. (D) Localization of Ark1-GFP and Sid4-mCherry (= SPB position) in mitotic asp1+ (left) und asp1D333A (right) cells. Spindle collapse (*) was observed in the asp1D333A cell. Time between images, 2 min. Bar, 2 μm.
The subcellular localization of the highly conserved Aurora kinase Ark1 is an indicator of the mitotic stage (34). Ark1 localizes in close proximity to centromeres/kinetochores in early mitosis and relocalizes to the spindle at the onset of anaphase B (34, 35). We thus analyzed Ark1-GFP localization in strains expressing SPB-localized Sid4-mCherry (36) and either asp1+ or asp1D333A (growth of such strains is shown in Fig. S2B in the supplemental material). As has been described previously for wild-type (asp1+) cells, upon entry into mitosis, a strong Ark1-GFP signal between the two separated SPBs is observed, demonstrating proximal centromere/kinetochore localization (Fig. 3D, left panel). Entry in anaphase B results in loss of Ark1-GFP at the centromere/kinetochore and relocalization to the spindle (Fig. 3D, left panel). A similar Ark1-GFP localization was seen for asp1D333A cells in early mitosis (Fig. 3D, right panels). However, the Ark1-GFP centromere/kinetochore signal persisted approximately twice as long as that of wild-type cells and separated into several distinct spots that moved between the SPBs (Fig. 3D, right panels). These spots represent kinetochores not aligned in a bipolar manner. Upon spindle collapse (time point 28 min; the distance between SPBs changed from 4.5 to 1.1 μm), faint spot-like Ark1-GFP signals were observed, suggesting that spindle breakage occurs prior to anaphase B. Following spindle collapse, the spindle regenerated and the cell was able to finish mitosis, as indicated by the spindle localization of Ark1-GFP (Fig. 3D, right panels). Six of 33 analyzed asp1D333A cells expressing Ark1-GFP Sid4-mCherry showed spindle collapse, and in these cases Ark1-GFP localization was comparable to that in the example shown. Interestingly in five of six cases with breaking spindles, the Ark1-GFP signal was greatly reduced just before spindle breakage. In addition, the Ark1-GFP signal intensity of asp1D333A mitotic cells was reduced compared to that of asp1+ cells. Measurement of the Ark1-GFP signal in prometaphase cells revealed a significant reduction in relative fluorescence signal intensity (asp1+, 16.3 ± 3.4; asp1D333A, 10.8 ± 3.3) (see Fig. S3A to C in the supplemental material).
Thus, our analysis of the cen1-GFP or Ark1-GFP subcellular localization in asp1D333A cells shows that spindle collapse occurs prior to anaphase A completion.
Spindle phase I and III dynamics are regulated by Asp1 kinase activity.
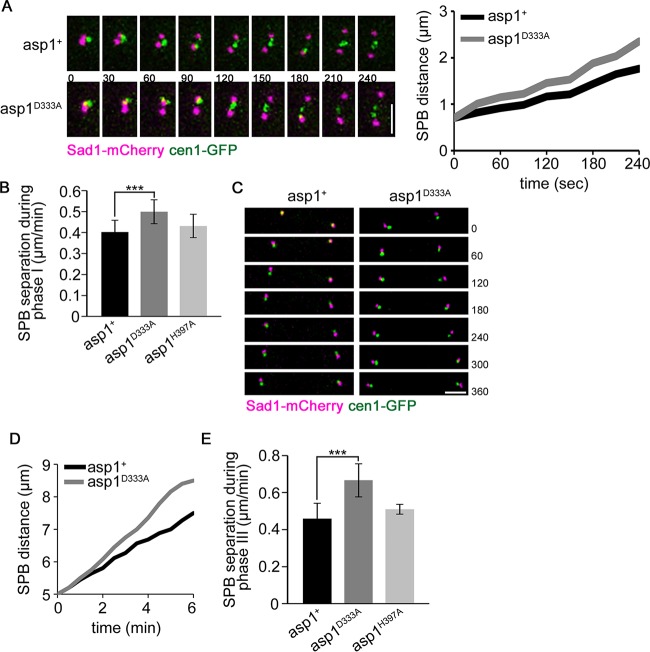
To determine if spindle formation (spindle phase I) and spindle elongation in anaphase B (phase III) were modulated by Asp1 kinase function, we determined the speed of SPB separation in the various asp1 variant strains using Sad1-mCherry and cen1-GFP. For this analysis, we chose spindles that did not collapse. For spindle phase I, the measurements were done as follows: the first appearance of two Sad1-mCherry signals was defined as the starting point for the measurements, followed by determining the distance between the two separated SPB Sad1-mCherry signals for the successive 4 min (examples for asp1+ and asp1D333A cells are shown in Fig. 4A). At the end of the measurement, the average distance between SPBs of asp1+ cells was 1.76 μm, while that between SPBs of asp1D333A cells was 2.35 μm. SPBs in asp1+ cells separated on average with a speed of 0.40 ± 0.06 μm/min, in accordance with previous measurements of spindle elongation at 20°C (8) (Fig. 4B). However, SPB separation in asp1D333A cells was significantly faster; the average speed of separation was 0.50 ± 0.06 μm/min (Fig. 4B). Spindle phase I was not changed significantly in asp1H397A cells compared to wild-type asp1+ cells (Fig. 4B) (average speed of SPB separation in asp1H397A cells, 0.43 ± 0.06 μm/min).
FIG 4.
Spindle phases I and III are modulated by Asp1 kinase activity. (A) Left, live-cell imaging of Sad1-mCherry cen1-GFP-expressing wild-type (asp1+) and asp1D333A cells during prometaphase (t = 0 is first separation of SPBs). Time between images, 30 s. Bar, 2 μm. Right, distance between SPBs shown diagrammatically. (B) Diagrammatic representation of SPB separation speed during spindle phase I (n = 20/strain; ***, P < 0.0005 for the asp1D333A strain compared to the asp1+ strain [two-sample t test]). (C) Merged images of a wild-type (asp1+) and an asp1D333A anaphase B cell expressing Sad1-mCherry cen1-GFP. Time between images, 60 s. Bar, 2 μm. Cells with centromere signals with 5-μm distance (t = 0) were monitored by time-lapse microscopy to determine the speed of SPB separation. The time between images was 1 min, and cells were photographed for 6 min. At this time the average SPB distances were 7.5 and 8.5 μm for asp1+ and asp1D333A cells, respectively. (D) Diagrammatic representation of SPB separation of the cells shown in panel C. (E) Quantification of SPB separation during spindle phase III of the indicated strains: asp1+ strain, 0.46 ± 0.08 μm/min; asp1D333A strain, 0.67 ± 0.09 μm/min; asp1H397A strain, 0.51 ± 0.03 μm/min (n = 13/strain; ***, P < 0.0005 for the asp1D333A strain compared to the asp1+ strain [two-sample t test]). Start of measurement, separated cen1-GFP signal and a spindle length of 5 μm. Endpoint of measurement, 6 min after start of measurement.
Similarly, spindle phase III, which encompasses spindle elongation by sliding apart of the polymerizing MT plus ends (9), was also modulated by IPPs. The speed of SPB separation in the various asp1 strains was determined by analyzing mitotic cells with separated, SPB-proximal cen1-GFP signals. In the examples shown in Fig. 4C and D, the asp1+ cell elongated the anaphase B spindle at a rate of 0.43 μm/min, while the asp1D333A cell had a rate of 0.62 μm/min. In general we found that in asp1D333A cells, spindle phase III elongation was significantly faster than in asp1+ cells (Fig. 4E). In the majority of cases, the altered phase III dynamics observed for asp1D333A phase III spindles did not result in spindle collapse. However, in some cells, phase III spindle collapse occurred (see Movie S4 in the supplemental material).
Analysis of SPB separation speed in asp1H397A cells revealed that spindle phase III was comparable to that of wild-type cells (Fig. 4E).
Entry into anaphase A is prolonged significantly in asp1D333A cells.
To avoid aneuploidy, sister chromosomes need to establish bioriented attachments to spindle MTs before entry into anaphase A can proceed. Live-cell imaging of Sad1-mCherry cen1-GFP-expressing asp1+ and asp1D333A cells revealed that the mutant cell type took significantly longer to enter anaphase A (examples are shown in Fig. 5A and B). In the cases shown, the asp1+ cell needed 18 min from the time of SPB separation to segregation of cen1-GFP signals to the SPBs, while the asp1D333A cell needed 34 min for this process. Analysis of further asp1+ and asp1D333A mitotic cells revealed that the time needed for asp1D333A cells to proceed to anaphase A was on average 54% longer than that for asp1+ cells (Fig. 5C).
FIG 5.
Asp1 kinase activity modulates entry into anaphase A. (A) Merged images of a mitotic cen1-GFP Sad1-mCherry wild-type (asp1+) or asp1D333A cell. Time between images, 2 min. Bars, 2 μm. (B) Diagrammatic representation of SPB distance versus time for the cells shown in panel A. (C) Diagrammatic representation of the time needed from prometaphase (separation of the SPBs) to anaphase A (segregation of cen1-GFP signals to SPB) in asp1+ and asp1D333A cells. (asp1+ cells, 19.2 ± 2.1 min; asp1D333A cells, 29.6 ± 5.9 min; n = 19/strain; ***, P < 0.0005 for asp1D333A cells compared to asp1+ cells [Welch test]). (D) Quantification of SPB distance at the time of cen1-GFP segregation in cen1-GFP Sad1-mCherry-expressing strains (asp1+ strain, 3.5 ± 0.4 μm; asp1D333A strain, 4.7 ± 0.5 μm; n = 19/strain; ***, P < 0.0005 for the asp1D333A strain compared to the asp1+ strain [two-sample t test]).
Prolonged delay before entry into anaphase leads to elongation of the S. pombe metaphase-like spindle. Indeed, we found that asp1D333A anaphase A spindles were significantly longer than those of wild-type cells: asp1+ cells segregated their cen1-GFP signals at an average spindle length of 3.5 μm, while asp1D333A spindles were 4.7 μm at the onset of anaphase (Fig. 5D). Therefore, we tested whether the SAC was activated in the mutant cells, causing the observed delay.
SAC components Mph1 and Mad2 are required for the mitotic delay of asp1D333A cells.
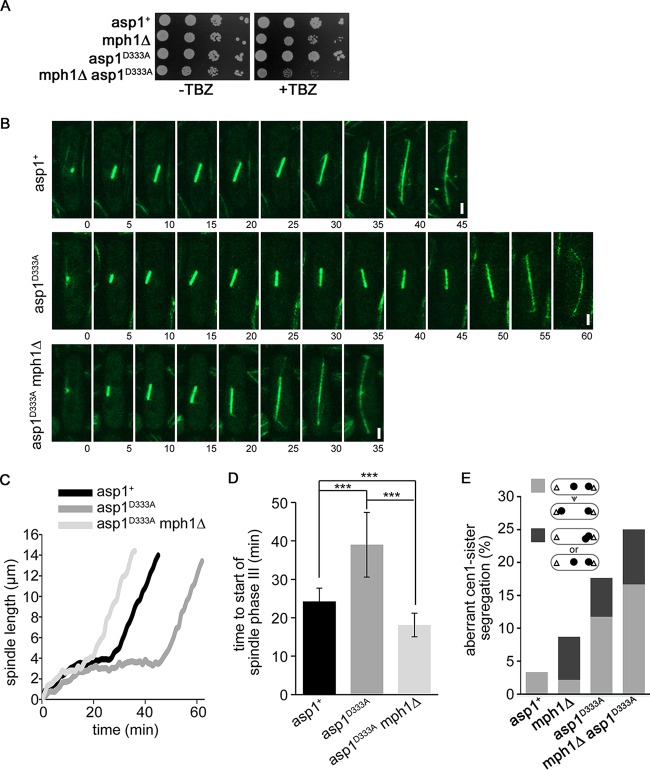
Chromosome biorientation is a prerequisite for entry into anaphase A and is monitored by the SAC (reviewed in references 6 and 37). To determine if the mitotic delay of asp1D333A cells was caused by the SAC, we analyzed mitosis in SV40::GFP-Atb2+ asp1D333A mph1Δ (deletion of the mph1+ open reading frame [ORF]) cells (growth of strains is shown in Fig. S2C in the supplemental material). The S. pombe Mph1 protein is part of the SAC Mps1 kinase family and is at the top of the SAC signaling cascade (36, 38–42). asp1D333A mph1Δ cells had a reduced fitness when grown on medium containing TBZ, implying that a functional Mph1 protein was necessary for the fitness of the asp1D333A strain (Fig. 6A). Importantly, we found that the prolonged mitotic delay seen for asp1D333A cells was abolished in asp1D333A mph1Δ mitotic cells (examples are shown in Fig. 6B and C). Entry into spindle phase III (rapid spindle elongation) was on average twice as fast in asp1D333A mph1Δ cells than in asp1D333A cells (Fig. 6D).
FIG 6.
asp1D333A cells require Mph1 for the prolonged delay into anaphase. (A) Serial dilution patch tests (104 to 101 cells) of the viability of the indicated strains on YE5S plates with (+TBZ) or without (−TBZ) 4 μg/ml TBZ. The plates were incubated for 4 days at 25°C. (B) Confocal live-cell images of the indicated strains expressing SV40::GFP-Atb2+. Time between images, 5 min. Bars, 2 μm. (C) Diagrammatic illustration of mitotic spindle length over time for the cells shown in panel B. (D) Quantification of time required for transition from prometaphase to anaphase B due to spindle elongation of the indicated strains (asp1+ strain, n = 16; asp1D333A strain, n = 16; asp1D333A mph1Δ strain, n = 13; ***, P < 0.0005 for the asp1D333A mph1Δ [two-sample t test] and asp1D333A [Welch test] strains compared to the asp1+ strain and for the asp1D333A mph1Δ [Welch test] strain compared to the asp1D333A strain). (E) Phenotypes of aberrant cen1-GFP segregation behavior, i.e., lagging cen1-GFP signal that moved to the correct SPB during the time of measurement (light gray) or cen1-GFP signals both present at one SPB or a cen1-GFP signal that did not move to either SPB (dark gray) (asp1+, n = 62; mph1Δ, n = 46; asp1D333A, n = 34; asp1D333A mph1Δ, n = 36).
Our analysis of the segregation behavior of cen1-GFP signals revealed that asp1D333A cells showed chromosome missegregation. Eighteen percent of the asp1D333A mitotic cells either displayed a lagging chromosome phenotype which was resolved during our measurements (Fig. 6E, light gray bars) or showed chromosome nondisjunction or chromosomes that were not moved to either SPB (Fig. 6E; these two phenotypes were scored together and are shown as dark gray box). Loss of Mph1 led to a further increase of the aberrant phenotype as determined for asp1D333A mph1Δ cells (Fig. 6E).
Next we analyzed whether the prolonged mitotic delay of asp1D333A cells was also dependent on a functional Mad2 protein (43, 44). asp1D333A mad2Δ cells showed reduced growth on medium containing the MT-destabilizing drug TBZ (Fig. 7A). Importantly, the prolonged delay seen for asp1D333A cells was abolished in asp1D333A mad2Δ mitotic cells (representative examples are shown in Fig. 7B). Entry into spindle phase III (rapid spindle elongation) was significantly faster in asp1D333A mad2Δ cells than in asp1D333A cells (Fig. 7C).
FIG 7.

asp1D333A cells require Mad2 for the prolonged delay into anaphase. (A) Serial dilution patch tests (104 to 101 cells) of the indicated strains grown on YE5S plates without and with 8 μg/ml TBZ. The plates were incubated for 5 days at 25°C. (B) Diagrammatic illustration of the spindle length over time. A representative example was chosen for each of the indicated strains expressing SV40::GFP-Atb2+. (C) Quantification of the time required for the transition from prometaphase to anaphase B for the indicated strains (asp1D333A mad2Δ strain, 19.3 ± 4.3 min; asp1D333A strain, 36.2 ± 13.3 min; mad2Δ strain, 13.9 ± 2.0 min; n for all strains = 20. ***, P < 0.005 (two-sample t test).
We conclude that asp1D333A cells, which cannot produce Asp1-generated IPPs, have defects in kinetochore-MT association and chromosome biorientation leading to chromosome missegregation. The SAC is active in such cells and can partially “rescue” the aberrant phenotype.
asp1H397A-expressing cells proceed to anaphase A faster than wild-type cells.
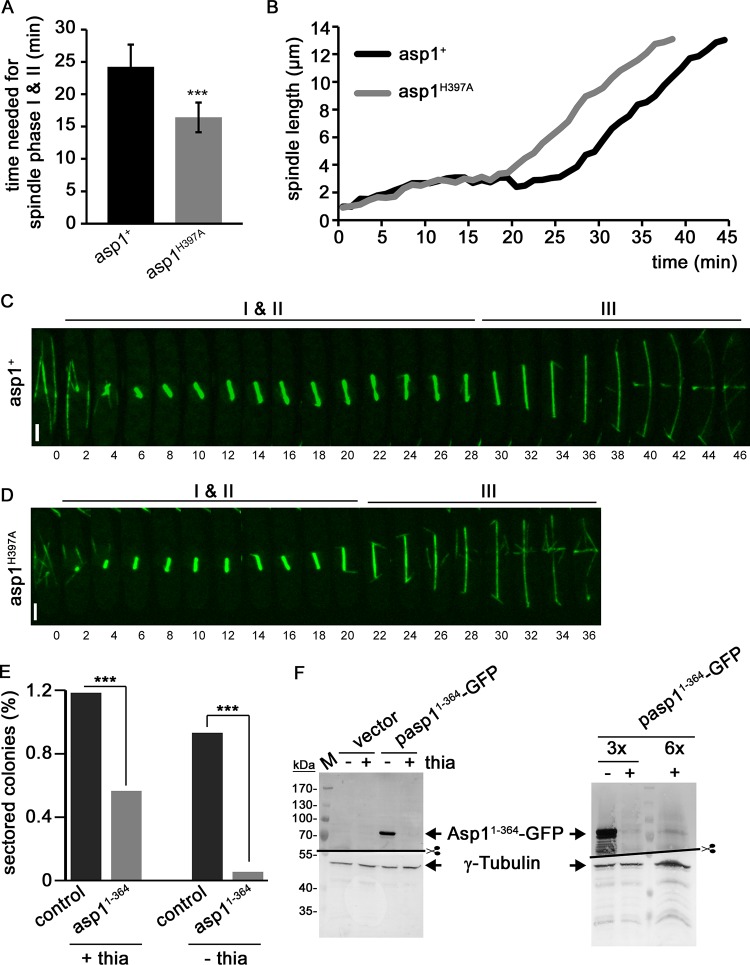
Our finding that the absence of Asp1-generated IPPs led to a prolonged SAC-controlled delay of the entry into anaphase A raised the question of whether excess cellular IPPs might have the opposite effect. Therefore, we determined the length of spindle phases I and II in an asp1H397A strain expressing SV40::GFP-Atb2. asp1+ wild-type cells took an average of 24.3 min before entry into phase III, while this time was reduced significantly to an average of 16.4 min in asp1H397A-expressing cells (Fig. 8A). An example for each strain is shown in Fig. 8B to D. In this example, the times to spindle phase III were 28 and 20 min for asp1+ and asp1H397A cells, respectively. Measurement of the time required to enter anaphase A was also assessed in asp1+ and asp1H397A cells expressing Sad1-mCherry cen1-GFP. In general, we found that cells of these strains needed less time for entry into anaphase A than the SV40::GFP-Atb2-expressing strains, possibly because the presence of SV40::GFP-Atb2 influenced spindle formation and chromosome biorientation. Nevertheless, on average asp1H397A cells showed a significantly earlier segregation of the cen1-GFP signals than asp1+ cells (see Fig. S4 in the supplemental material).
FIG 8.
asp1H397A cells reach spindle phase III faster than wild-type cells and show increased transmission fidelity of the Ch16 chromosome. (A) Diagrammatic representation of the times needed for spindle phases I and II as determined for asp1+ and asp1H397A cells expressing SV40::GFP-Atb2+ n = 16/strain; asp1+ cells, 24.3 ± 3.4 min; asp1H397A cells, 16.4 ± 2.3 min; ***, P < 0.0005 for asp1H397A compared to asp1+ cells [two-sample t test]). (B) Diagrammatic illustration of mitotic spindle length over time for the cells shown in panels C and D. (C and D) Confocal live-cell images of cells of the indicated strains expressing SV40::GFP-Atb2+. Time between images, 2 min. Bars, 2 μm. (E) Diagrammatic representation of Ch16 loss (percent sectored colonies) in cells transformed with a control vector (control) or a plasmid expressing asp11–364 via the nmt1+ promoter. +thia and −thia, presence or absence of thiamine, respectively, resulting in low or high Asp11–364 expression. For +thia: control, n = 19,411; Asp11–364, n = 10,075. For −thia: control, n = 21,582; Asp11–364, n = 21,488. (F) Western blot analysis of Asp11–364-GFP expressed in the presence or absence of thiamine. The blot on the right was overloaded (3 or 6 times the amount of protein used for the left blot) to visualize the Asp11–364 signal when thiamine was in the growth medium. γ-Tubulin protein was used as an internal control.
We next determined whether the faster entry into anaphase A led to increased chromosome missegregation. This was not the case. Missegregation of cen1-GFP signals in asp1+ Sad1-mCherry cen1-GFP or asp1H397A Sad1-mCherry cen1-GFP cells was not observed (asp1+, n = 62; asp1H397A, n = 58). Furthermore, the absence of mph1+ in these strains resulted in missegregation in 3 of 46 cells in the asp1+ background and 1 of 44 cells in the asp1H397A background. Thus, the observed reduction of the time required for anaphase A entry did not affect the fidelity of chromosome segregation.
We next determined the chromosome transmission fidelity of the nonessential Ch16 minichromosome (45) in cells that generated more IPPs than wild-type cells. The 550-kb Ch16 minichromosome can be maintained stably in a haploid S. pombe strain but is transmitted less faithfully than an endogenous chromosome, thus representing a sensitized system for the analysis of chromosome transmission fidelity (46). Loss of the minichromosome can be scored by the appearance of red sectors in a white colony (46). A Ch16-containing strain was transformed with a plasmid expressing the asp1 variant asp11–364 via the thiamine-repressible promoter nmt1+ (47). Asp11–364 (the Asp1 kinase domain) generates up to twice as much IPPs as full-length Asp1 in vitro (16). In addition, the plasmid-borne expression via the nmt1+ promoter further ensured excess intracellular IPP production.
We found that approximately 1% of the colonies of the Ch16-containing strain transformed with a control vector were sectored, demonstrating loss of the minichromosome (Fig. 8E). This number was reduced 2-fold in transformants that expressed low levels of asp11–364 (i.e., basal expression of the nmt1+ promoter in the presence of thiamine in the medium [+thia in Fig. 8E and F]). Furthermore, Ch16 minichromosome loss was further reduced when asp11–364 was expressed at a high level (i.e., derepression of the nmt1+ promoter in the absence of thiamine in the medium [−thia in Fig. 8E and F]). In this case, loss of the minichromosome was reduced from 0.93% for the vector control transformants to 0.053% for cells expressing asp11–364 (Fig. 8E). This is a 17-fold reduction in Ch16 loss for asp11–364-expressing cells compared to cells transformed with the control vector.
Whether transmission fidelity of endogenous chromosomes was also increased in asp1H397A cells compared to wild-type cells could not be determined conclusively due to the low number of segregation errors occurring for endogenous chromosomes. However, we found that in the presence of a small amount of TBZ (5 μg/ml), 76% of mph1Δ cells segregated the cen1-GFP signal correctly, while this number was increased to 83% in the mph1Δ asp1H397A strain (n = 51 and 59, respectively).
Expression of Asp1365–920 influences growth of strains with mutant motor proteins.
To determine which process in spindle formation might be affected by Asp1 kinase function, we expressed the truncated Asp1 versions Asp11–364 and Asp1365–920 (Asp1 kinase and phosphatase domains, respectively) via the nmt1+ promoter on a plasmid in various strains with defective components of the mitotic spindle. We have shown previously that in vitro and presumably also in vivo, the presence of the Asp1 pyrophosphatase domain results in lower IPP output, while expression of the Asp1 kinase domain has the opposite effect (16). Interestingly, we found that the growth of pkl1Δ, klp6Δ, and cut7-466ts mutant strains was affected in opposite manners by expression of Asp1365–920. Klp6 is part of the heterodimer kinesin-8 Klp5/Klp6 which acts as an MT plus-end depolymerase, kinesin-14 Pkl1 is needed for anchoring of microtubule ends to the SPB, and the essential, spindle midzone-localized kinesin-5 Cut7 organizes bipolar MT interdigitation at the midzone (48–54). Klp5/6- and Pkl1-generated forces at the spindle act antagonistically to those of Cut7. Under the conditions tested, expression of Asp11–364 had little effect on the growth of the strains (Fig. 9), but expression of Asp1365–920 rescued the temperature-sensitive nongrowth phenotype of the cut7-466ts strain at 28°C (Fig. 9A) by increasing the number of surviving cells (Fig. 9B). In contrast, expression of Asp1365–920 led to reduced growth of the klp6Δ strain and in particular of the pkl1Δ strain (Fig. 9C and D).
FIG 9.
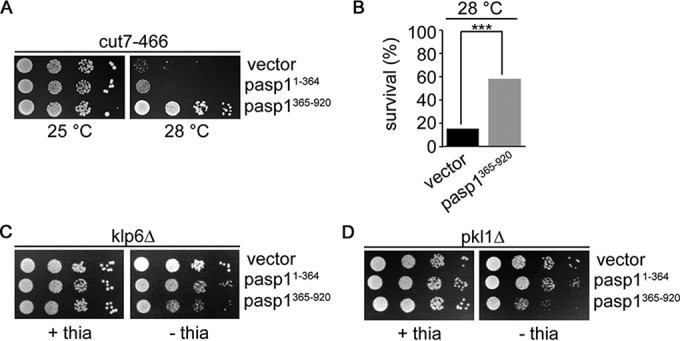
Plasmid-borne Asp1365–920 expression rescues the temperature-sensitive nongrowth phenotype of the cut7-466 strain. (A) Serial dilution patch tests (104 to 101 cells) of the cut7-466 strain transformed with vector (control), asp11–364, or asp1365–920 plasmid. Transformants were grown in thiamine-free medium under plasmid selective conditions at the indicated temperatures for 6 days. (B) Diagrammatic illustration of the survival of transformed cut7-466 cells grown at the restrictive temperature of 28°C. Transformants were pregrown at 25°C under plasmid selective conditions that allowed expression of the asp1 variants before plating cells at 28°C. Colony formation was assayed for 1,000 cells/transformant. ***, P < 0.005 (χ2 test). (C and D) Serial dilution patch tests (104 to 101 cells) of the indicated strains transformed with the same plasmids as for panel A. The absence of thiamine results in derepression of the nmt1+ promoter, which controls asp11–364 and asp1365–920 expression. Plates were incubated at 25°C for 5 days.
DISCUSSION
In this work we have discovered a mitotic function of the S. pombe member of the highly conserved Vip1 family. Asp1 kinase function is needed for the execution of all three stages of mitosis and controls bipolar spindle stability, chromosome biorientation, and the fidelity of chromosome transmission. This is the first report of a role for the high-energy IPP molecules in the regulation of fission yeast mitosis. Our previous analysis of Vip1 kinase interphase function in S. pombe and two distantly related filamentous fungi revealed that this enzyme family controls conserved biological processes (16). Thus, it is likely that our present findings will also be relevant for other mitotic systems. Indeed, a Saccharomyces cerevisiae synthetic genetic array analysis identified several negative interactions between vip1Δ (deletion of the Vip1-encoding ORF) and genes encoding mitotic components (55).
Spindle elongation dynamics are controlled by Asp1 kinase function.
The bipolar spindle consists of kinetochore-MTs and interpolar MTs polymerized from both SPBs. Interpolar MTs from one SPB interdigitate with interpolar MTs from the other SPB at the spindle midzone to form a stable bipolar spindle cytoskeleton (reviewed in reference 25). Asp1 kinase function was required for all three spindle phases. Formation of the metaphase spindle (spindle phase I) requires the concerted action of MT-associated proteins and MT motor proteins such as the kinesin-5 class, which are responsible for sliding the antiparallel MTs apart. asp1D333A cells showed a 46% faster separation of the duplicated SPBs than wild-type cells during the prometaphase stage. Furthermore, such cells often showed aberrant thin spindle midzones and frequent spindle collapse. Remarkably, we made the observation that in asp1D333A cells, spindle collapse was often followed by immediate chromosome segregation. A similar behavior has been described for the cut7.24 klp6Δ double mutant (56). Cut7 and Klp6 belong to the kinesin-5 and kinesin-8 families, respectively (48, 57), and are antagonistic spindle motors. The authors suggested that the “transient spindle shrinkage” might bring MT plus ends closer to kinetochores, thus facilitating attachment (56). This might also be the case in asp1D333A cells, as such cells also have defects in chromosome-MT attachment.
How might Asp1-generated IPPs regulate spindle formation? IPPs are known to modulate cellular processes by two means: they regulate the function of a protein/protein complex either via reversible binding or by pyrophosphorylation (58, 59). Thus, direct binding of Asp1 to the IPP target protein might not be necessary. As Asp1 is a nuclear and cytoplasmic protein (reference 60 and our unpublished observations), it is feasible that IPPs generated in the nucleus modulate a component(s) of the mitotic spindle. Our previous analysis of the interphase microtubule cytoskeleton in asp1D333A cells revealed that most microtubule parameters analyzed were altered, resulting in more dynamic microtubules (16). The EB1 family member Mal3, associated with spindle midzones, is not required for Asp1-mediated microtubule modulation (16, 61). Furthermore, localization of the midzone bundler Ase1/Prc1 was seemingly normal in asp1D333A cells (references 62 and 63 and our unpublished observation). However, lowering intracellular IPP levels by plasmid-borne expression of Asp1365–920 rescued the temperature-sensitive nongrowth phenotype of the cut7-466 mutant strain, while negatively affecting growth of kinesin-14 pkl1Δ and kinesin-8 klp6Δ deletion strains. As Klp5/6 and Pkl1 act antagonistically to Cut7 and vice versa (53, 56), our results propose a function for IPPs in controlling the processes that balance the Cut7-generated outward force at the spindle.
Asp1-generated IPPs modulate chromosome transmission fidelity and control chromosome biorientation.
Chromosome segregation errors in cells unable to produce Asp1-made IPPs were of the following two types: (i) chromosome nondisjunction, or the inability of chromosomes to be moved to either SPB, or (ii) chromosomes that lagged behind during anaphase. The first phenotype is a consequence of defective binding of spindle MTs to the kinetochore and can be detected by the SAC. Indeed, we found that chromosome biorientation was significantly delayed in asp1D333A cells compared to wild-type cells. This delay was SAC dependent. Thus, Asp1-generated IPPs are required for biorientation of chromosomes.
Lagging chromosomes are not detected by the SAC. This behavior is caused by merotelic attachments where a single kinetochore is bound by MTs from either SPB. In mammalian cells the majority of aneuploidy events appear to arise by merotelic kinetochore-MT attachments (64). One of the consequences of merotelic attachments is the induction of the DNA damage response in human cell lines, leading to chromosomal aberrations in the progeny. This observation links aberrant mitosis with chromosome rearrangements (65).
Merotelic attachments are caused by hyperstabilized kinetochore-MT associations that need to be resolved by the Aurora B kinase-containing chromosome passenger complex present at the centromere/kinetochore (reviewed in references 66 and 67). Interestingly we found that the fluorescence intensity of the S. pombe Aurora kinetochore Ark1-GFP signal was reduced significantly in prometaphase asp1D333A cells compared to wild-type cells. Abnormally low Ark1 concentrations might explain why erroneous kinetochore-MT attachments cannot be converted into correctly bioriented attachments, leading to an increased number of lagging chromosomes in asp1D333A cells. Thus, Asp1-generated IPPs might be required for Ark1 kinetochore recruitment. How this might be achieved is at present unclear. However, it has been shown recently that spindle MTs via the conserved EB1 family play a role in the centromere targeting of human Aurora B (68). Thus, as IPPs modulate the MT cytoskeleton, the possibility exists that they affect Ark1 kinetochore targeting via this route.
Intriguingly, we found that cells that express the Asp1H397A variant are able to enter anaphase A faster than wild-type cells, possibly because they were able to establish the correct chromosome biorientation faster. Furthermore, the transmission fidelity of the nonessential 550-kb Ch16 chromosome was increased in Asp11–364-expressing cells. Thus, higher-than-wild-type IPP levels increased the fidelity of Ch16 chromosome transmission beyond the frequency seen for the wild-type situation. Our data propose a direct correlation between intracellular IPP levels and genome integrity. How IPP levels modulate kinetochore-MT attachments is not clear. However, we previously found a direct correlation between IPP levels and the residence time of interphase MT plus ends at the cell cortex (16). Thus, IPPs might also control the residence time of spindle MT plus ends at the kinetochore interface, thus facilitating correct bipolar connections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jennifer Pöhlmann for help with Fig. S1 in the supplemental material and Robin Allshire, Kathleen Gould, Keith Gull, Iain Hagan, Silke Hauf, Alison Pidoux, Shelley Sazer, Takashi Toda, Mitsuhiro Yanagida, and the Yeast Genetic Resource Centre for the gifts of reagents used in this work. We are grateful to Shelley Sazer for critical input and careful reading of the manuscript and to the Center for Advanced Imaging (CAi) at the Heinrich-Heine-University, Düsseldorf, for help with microscopic analysis.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sector.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sector.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00330-16.
REFERENCES
- 1.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet 36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 2.Lott IT, Head E. 2005. Alzheimer disease and Down syndrome: factors in pathogenesis. Neurobiol Aging 26:383–389. doi: 10.1016/j.neurobiolaging.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Harrison BD, Hashemi J, Bibi M, Pulver R, Bavli D, Nahmias Y, Wellington M, Sapiro G, Berman J. 2014. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol 12:e1001815. doi: 10.1371/journal.pbio.1001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santaguida S, Amon A. 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- 5.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. 2012. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol 13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacristan C, Kops GJ. 2015. Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol 25:21–28. doi: 10.1016/j.tcb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Akera T, Goto Y, Sato M, Yamamoto M, Watanabe Y. 2015. Mad1 promotes chromosome congression by anchoring a kinesin motor to the kinetochore. Nat Cell Biol 17:1124–1133. doi: 10.1038/ncb3219. [DOI] [PubMed] [Google Scholar]
- 8.Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M. 1998. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell 9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallavarapu A, Sawin K, Mitchison T. 1999. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol 9:1423–1426. doi: 10.1016/S0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- 10.Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, Haystead TA, Ribeiro AA, York JD. 2007. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- 11.Choi JH, Williams J, Cho J, Falck JR, Shears SB. 2007. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem 282:30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fridy PC, Otto JC, Dollins DE, York JD. 2007. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J Biol Chem 282:30754–30762. doi: 10.1074/jbc.M704656200. [DOI] [PubMed] [Google Scholar]
- 13.Gokhale NA, Zaremba A, Shears SB. 2011. Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem J 434:415–426. doi: 10.1042/BJ20101437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin H, Fridy PC, Ribeiro AA, Choi JH, Barma DK, Vogel G, Falck JR, Shears SB, York JD, Mayr GW. 2009. Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J Biol Chem 284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Falck JR, Hall TM, Shears SB. 2011. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat Chem Biol 8:111–116. doi: 10.1038/nchembio.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pöhlmann J, Risse C, Seidel C, Pohlmann T, Jakopec V, Walla E, Ramrath P, Takeshita N, Baumann S, Feldbrugge M, Fischer R, Fleig U. 2014. The Vip1 inositol polyphosphate kinase family regulates polarized growth and modulates the microtubule cytoskeleton in fungi. PLoS Genet 10:e1004586. doi: 10.1371/journal.pgen.1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Nair VS, Holland AA, Capolicchio S, Jessen HJ, Johnson MK, Shears SB. 2015. Asp1 from Schizosaccharomyces pombe binds a [2Fe-2S](2+) cluster which inhibits inositol pyrophosphate 1-phosphatase activity. Biochemistry 54:6462–6474. doi: 10.1021/acs.biochem.5b00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pulloor NK, Nair S, Kostic AD, Bist P, Weaver JD, Riley AM, Tyagi R, Uchil PD, York JD, Snyder SH, Garcia-Sastre A, Potter BV, Lin R, Shears SB, Xavier RJ, Krishnan MN. 2014. Human genome-wide RNAi screen identifies an essential role for inositol pyrophosphates in type-I interferon response. PLoS Pathog 10:e1003981. doi: 10.1371/journal.ppat.1003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laha D, Johnen P, Azevedo C, Dynowski M, Weiss M, Capolicchio S, Mao H, Iven T, Steenbergen M, Freyer M, Gaugler P, de Campos MK, Zheng N, Feussner I, Jessen HJ, Van Wees SC, Saiardi A, Schaaf G. 2015. VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in Arabidopsis. Plant Cell 27:1082–1097. doi: 10.1105/tpc.114.135160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feoktistova A, McCollum D, Ohi R, Gould KL. 1999. Identification and characterization of Schizosaccharomyces pombe asp1(+), a gene that interacts with mutations in the Arp2/3 complex and actin. Genetics 152:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pöhlmann J, Fleig U. 2010. Asp1, a conserved 1/3 inositol polyphosphate kinase, regulates the dimorphic switch in Schizosaccharomyces pombe. Mol Cell Biol 30:4535–4547. doi: 10.1128/MCB.00472-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194:795–823. doi: 10.1016/0076-6879(91)94059-L. [DOI] [PubMed] [Google Scholar]
- 23.Tran PT, Paoletti A, Chang F. 2004. Imaging green fluorescent protein fusions in living fission yeast cells. Methods 33:220–225. doi: 10.1016/j.ymeth.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 24.Bratman SV, Chang F. 2007. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell 13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winey M, Bloom K. 2012. Mitotic spindle form and function. Genetics 190:1197–1224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snaith HA, Anders A, Samejima I, Sawin KE. 2010. New and old reagents for fluorescent protein tagging of microtubules in fission yeast; experimental and critical evaluation. Methods Cell Biol 97:147–172. doi: 10.1016/S0091-679X(10)97009-X. [DOI] [PubMed] [Google Scholar]
- 27.Kerres A, Jakopec V, Fleig U. 2007. The conserved Spc7 protein is required for spindle integrity and links kinetochore complexes in fission yeast. Mol Biol Cell 18:2441–2454. doi: 10.1091/mbc.E06-08-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia MA, Vardy L, Koonrugsa N, Toda T. 2001. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J 20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakopec V, Topolski B, Fleig U. 2012. Sos7, an essential component of the conserved S. pombe Ndc80-MIND-Spc7 complex, identifies a new family of fungal kinetochore proteins. Mol Cell Biol 32:3308–3312. doi: 10.1128/MCB.00212-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goshima G, Scholey JM. 2010. Control of mitotic spindle length. Annu Rev Cell Dev Biol 26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- 31.Costa J, Fu C, Khare VM, Tran PT. 2014. csi2p modulates microtubule dynamics and organizes the bipolar spindle for chromosome segregation. Mol Biol Cell 25:3900–3908. doi: 10.1091/mbc.E14-09-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagan I, Yanagida M. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol 129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakazawa N, Nakamura T, Kokubu A, Ebe M, Nagao K, Yanagida M. 2008. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J Cell Biol 180:1115–1131. doi: 10.1083/jcb.200708170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen J, Paris J, Willer M, Philippe M, Hagan IM. 2001. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J Cell Sci 114:4371–4384. [DOI] [PubMed] [Google Scholar]
- 35.Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y. 2007. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev 21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinrich S, Windecker H, Hustedt N, Hauf S. 2012. Mph1 kinetochore localization is crucial and upstream in the hierarchy of spindle assembly checkpoint protein recruitment to kinetochores. J Cell Sci 125:4720–4727. doi: 10.1242/jcs.110387. [DOI] [PubMed] [Google Scholar]
- 37.Overlack K, Krenn V, Musacchio A. 2014. When Mad met Bub. EMBO Rep 15:326–328. doi: 10.1002/embr.201438574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millband DN, Hardwick KG. 2002. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol Cell Biol 22:2728–2742. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Windecker H, Langegger M, Heinrich S, Hauf S. 2009. Bub1 and Bub3 promote the conversion from monopolar to bipolar chromosome attachment independently of shugoshin. EMBO Rep 10:1022–1028. doi: 10.1038/embor.2009.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito D, Saito Y, Matsumoto T. 2012. Centromere-tethered Mps1 pombe homolog (Mph1) kinase is a sufficient marker for recruitment of the spindle checkpoint protein Bub1, but not Mad1. Proc Natl Acad Sci U S A 109:209–214. doi: 10.1073/pnas.1114647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol 14:746–752. doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- 42.Lara-Gonzalez P, Westhorpe FG, Taylor SS. 2012. The spindle assembly checkpoint. Curr Biol 22:R966–980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Saitoh S, Ishii K, Kobayashi Y, Takahashi K. 2005. Spindle checkpoint signaling requires the mis6 kinetochore subcomplex, which interacts with mad2 and mitotic spindles. Mol Biol Cell 16:3666–3677. doi: 10.1091/mbc.E05-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinrich S, Geissen EM, Kamenz J, Trautmann S, Widmer C, Drewe P, Knop M, Radde N, Hasenauer J, Hauf S. 2013. Determinants of robustness in spindle assembly checkpoint signalling. Nat Cell Biol 15:1328–1339. doi: 10.1038/ncb2864. [DOI] [PubMed] [Google Scholar]
- 45.Niwa O, Matsumoto T, Yanagida M. 1986. Construction of a mini-chromosome by deletion and itsmitotic and meiotic behaviour in fission yeast. Mol Gen Genet 203:397–405. doi: 10.1007/BF00422063. [DOI] [Google Scholar]
- 46.Fleig U, Sen-Gupta M, Hegemann JH. 1996. Fission yeast mal2+ is required for chromosome segregation. Mol Cell Biol 16:6169–6177. doi: 10.1128/MCB.16.11.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maundrell K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127–130. doi: 10.1016/0378-1119(93)90551-D. [DOI] [PubMed] [Google Scholar]
- 48.Hagan I, Yanagida M. 1992. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature 356:74–76. doi: 10.1038/356074a0. [DOI] [PubMed] [Google Scholar]
- 49.Garcia MA, Koonrugsa N, Toda T. 2002. Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J 21:6015–6024. doi: 10.1093/emboj/cdf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia MA, Koonrugsa N, Toda T. 2002. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol 12:610–621. doi: 10.1016/S0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- 51.Erent M, Drummond DR, Cross RA. 2012. S. pombe kinesins-8 promote both nucleation and catastrophe of microtubules. PLoS One 7:e30738. doi: 10.1371/journal.pone.0030738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR. 2009. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol Biol Cell 20:963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yukawa M, Ikebe C, Toda T. 2015. The Msd1-Wdr8-Pkl1 complex anchors microtubule minus ends to fission yeast spindle pole bodies. J Cell Biol 209:549–562. doi: 10.1083/jcb.201412111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Syrovatkina V, Tran PT. 2015. Loss of kinesin-14 results in aneuploidy via kinesin-5-dependent microtubule protrusions leading to chromosome cut. Nat Commun 6:7322. doi: 10.1038/ncomms8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniel JA, Keyes BE, Ng YP, Freeman CO, Burke DJ. 2006. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics 172:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Syrovatkina V, Fu C, Tran PT. 2013. Antagonistic spindle motors and MAPs regulate metaphase spindle length and chromosome segregation. Curr Biol 23:2423–2429. doi: 10.1016/j.cub.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.West RR, Malmstrom T, McIntosh JR. 2002. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci 115:931–940. [DOI] [PubMed] [Google Scholar]
- 58.Lee YS, Mulugu S, York JD, O'Shea EK. 2007. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. 2004. Phosphorylation of proteins by inositol pyrophosphates. Science 306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- 60.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, Horinouchi S, Yoshida M. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 61.Beinhauer JD, Hagan IM, Hegemann JH, Fleig U. 1997. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol 139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loiodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. 2005. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell 16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. 2005. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell 16:1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol 153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. 2011. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 66.Ruchaud S, Carmena M, Earnshaw WC. 2007. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 67.Bakhoum SF, Thompson SL, Manning AL, Compton DA. 2009. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol 11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banerjee B, Kestner CA, Stukenberg PT. 2014. EB1 enables spindle microtubules to regulate centromeric recruitment of Aurora B. J Cell Biol 204:947–963. doi: 10.1083/jcb.201307119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.