Abstract
microRNAs (miRNAs) have been demonstrated to contribute to tumor progression and metastasis, and have been proposed to be key regulators of diverse biological processes. In this study, we report that miR-4295 is deregulated in bladder cancer tissues and cell lines. To characterize the role of miR-4295 in bladder cancer cells, we performed functional assays. The overexpression of miR-4295 significantly promoted bladder cancer cell proliferation, colony formation, and migration. Moreover, its downregulation induced cell cycle arrest and apoptosis of bladder cancer cells. Furthermore, a luciferase reporter assay and rescue experiment indicated that miR-4295 directly targets BTG1 by binding its 3’UTR. In conclusion, these results demonstrate that miR-4295 acts as an oncogene and may be a potential biomarker for bladder cancer diagnosis and treatment.
Keywords: MiR-4295, BTG1, bladder cancer, cell growth
Introduction
Bladder cancer (BC), a malignant tumor in the urogenital tract, has become the fifth most common cancer and is one of the major causes of death in cancer patients worldwide. In middle-aged and elderly men, bladder cancer is the second most prevalent malignancy after prostate cancer [1,2]. Many patients who are diagnosed with bladder cancers are found to have advanced stage disease with distal metastases, which leads to poor prognosis. This feature of BCs largely results from the nature of BC, as it is often asymptomatic or non-specifically symptomatic in the early stages [3]. Additionally, there are limited therapy options for enhancing patient survival rate because the molecular mechanisms underlying BC are not well understood. Accordingly, it is crucial to clarify the key factors mediating bladder cancer growth and metastasis and to understand their molecular mechanism of action in order to design effective therapies.
Increasing evidence has suggested that there may be multiple molecular factors involved in bladder carcinogenesis and tumor progression. MicroRNAs (miRNAs) are endogenous RNA molecules of approximately 18-25 nucleotides that regulate the protein translation of target genes by base-pairing with the 3’-untranslated CDS region (3’-UTR) of the mRNAs [4,5]. A number of miRNAs have been shown to act as key regulators in bladder cancer, including miRNA (miR)29c, miR-124, miR-186, miR-24, miR-485, and miR-21 [6-10]. MiR-4295 was found to be a novel oncogenic miRNA that acts by targeting USP28 in non-small cell lung cancer [11]. In addition, RUNX3 is downregulated in glioma by the Myc-regulated miR-4295. However, there has not been evidence indicating the role of miR-4295 in bladder cancer.
In the present study, we investigated the expression of miR-4295 in bladder carcinomas and adjacent normal tissues, and found that the levels of miR-4295 were significantly decreased in the normal tissue. In addition, functional studies were conducted to assess the roles of miR-4295 in bladder cancer cell lines. Furthermore, bioinformatics analyses showed that miR-4295 targeted the 3’-UTR of BTG1 mRNA to inhibit its translation, and this was confirmed by a luciferase reporter assay.
Methods and materials
Human tissue samples
The tissue specimens and paired noncancerous bladder tissues used in this study were obtained from the Department of Urology of the China-Japan Union Hospital of Jilin University (Changchun, China) after surgical resection. All samples were immediately snapped frozen in liquid nitrogen and stored at -80°C for the subsequent experiments. Informed consent approving the use of their tissues for research purposes was obtained from each patient. The study protocol was approved by the Institute Research Ethics Committee at the China-Japan Union Hospital of Jilin University.
Cell culture and transfection
Hbc was established by the Cancer Research Institute of Kunming Medical College, 1986. The human bladder cancer cell lines, Hbc and T24, were cultured in RPMI-1640 (Invitrogen, CA, USA), and UM-UC3, Hcv29 and BIU-87 cells were cultured in MEM (Invitrogen, CA, USA) medium supplemented with 10% fetal bovine serum (Gibco, Australia), penicillin and streptomycin (100 IU/ml). The cells were maintained at 37°C under 5% CO2 atmosphere in a humidified incubator.
The CDS sequence of BTG1 was synthesized by Genewiz (Beijing, China) and cloned into pcDNA3.1(+) vector (Promega, WI, USA) between the BamHI and HindIII sites. Bladder cancer cells (T24 and Hbc) were cultured at 3×104 cells/well in 96-well plates or 5×105 cells/well in 6-well plates until they reached 80% confluence. Subsequently, the cells were transiently transfected with miRNA mimics, miRNA inhibitors (genepharma, Shanghai, China) or the pcDNA3.1(+)/BTG1 vectors using Lipofectamine 2000 (Invitrogen, CA, USA) according to the manufacturer’s instructions. To assess the transfection efficiency, for all experiments, qRT-PCR was performed 24 hours after transfection.
Real-time-PCR analysis
Total RNA was extracted using Trizol reagent (Invitrogen, CA, USA) according to the instructions. cDNA was synthesized with specific stem-loop primers and the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, CA, USA). The levels of gene expression were quantified on an iQ5 real-time PCR detection system (Bio-Rad, MA, USA) with SYBR Premix EX Taq (TaKaRa, Shiga, Japan). Gene expression was defined based on the threshold cycle (Ct) normalized to that of the housekeeping gene, GAPDH, or small nuclear RNA, U6, as internal controls. The fold change of gene expression was calculated with the 2-ΔΔCt method. PCR amplification was performed using the following thermal conditions: 95°C for 30 s followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. The PCR conditions for amplification of miR-4295 were: 95°C for 20 s, followed by 40 cycles of 95°C for 10 s and 60°C for 20 s, with a final incubation at 70°C for 5 s. All the primers were synthesized by Genewiz (Beijing, China) and the primer sequences used are as follows.
Cell viability assay
T24 and Hbc cells were cultured at 3×104 cells/well in 96-well plates until they reached 80% confluence. Cell proliferation was measured at 24, 48, 72 and 96 hours after transfection using WST-8 staining with Cell Counting Kit-8 (Byotime, Haimen, China).
Western blot assay
Protein was extracted from the transfected T24 and Hbc cells. The protein lysate was separated on a 10% SDS-PAGE gel and transferred to PVDF membranes (Bio-Rad, CA, USA). Nonspecific binding was blocked by incubating the PVDF membranes with 5% nonfat milk containing 0.1% Tween-20 for 2 hours at room temperature. The membrane was incubated with the primary antibodies, including BTG1 (Abcam, Cambridge, England) and GAPDH (Abcam, Cambridge, England), in TBST at 4°C overnight. After washing with TBST thrice, the PVDF membranes were incubated with horseradish peroxidase-conjugated sheep anti-mouse IgG for 2 h at 37°C. Finally, the proteins were visualized using ECL detection kits (Millipore, MA, USA).
Luciferase assay
The oligonucleotides with wild-type or mutant type 3’-UTR region of BTG1 (containing binding site with miR-4295) were synthesized by Genewiz (Beijing, China). The pmiRGLO vectors (Promega, WI, USA) were constructed with wild type BTG1 sequences or mutant BTG1 sequences. T24 and Hbc cells were seeded in 12-well plates and co-transfected with miR-control/miR-4295 mimics and WT-BTG1 3’-UTR vector/MUT-BTG1 3’-UTR vector using Lipofectamine 2000 (Invitrogen, CA, USA). The activities of Renilla and firefly luciferases in the cell lysates were measured using the Dual-Luciferase assay kit (Promega, WI, USA), and the quotient of Renilla/firefly luciferase activities (Rluc/Luc) was taken as the normalized data.
Colony formation assay
At 24 h after transfection, 500 cells were seeded into 6-well plates and cultured for 14 days. Afterwards, the cells were fixed with absolute methanol for 15 min and stained with crystal violet for 20 min. The number of colonies formed was counted in 10 different fields of vision, and the mean value was calculated. The experiment was performed independently three times for each cell line.
Cell migration assay
At 48 hours after transfection, the cells were trypsinized and seeded in 8 μm pore size transwell chambers (Corning, Cambridge, USA) at a density of 8×104 per well. The cells were cultured in RPMI 1640 medium with 2% FBS, and 600 μl of RPMI 1640 supplemented with 10% FBS was added to the lower chamber. Then, 24 hours later, the migrated cells were fixed with absolute methanol for 30 min, while the cells that had not migrated were removed using cotton swabs. Then, the cells on the bottom surface of the membrane were stained with 0.1% crystal violet. Five visual fields of ×200 magnification of each insert were randomly selected, and the number of cells was counted under a light microscope.
Cell cycle analysis
At 24 h after transfection, the cells (1×106) were harvested and fixed in 75% ice-cold ethanol. Before cell cycle analysis, the cells were treated with bovine pancreatic 2 μg/ml RNase (Sigma, USA) for 30 min, and subsequently incubated in 20 μg/ml propidium iodide (Sigma, USA) for 20 min. Cell cycle analysis was performed on a FACS-Calibur System (BD Biosciences, NJ, USA). The data were analyzed with the ModFit LT software package and the cell cycle distribution is shown as the percentage of cells in the G0/G1, S, and G2/M populations. Each experiment was conducted in triplicate.
Statistical analysis
All statistical analyses and graphing were performed using SPSS version 17.0. The data were expressed as the mean ± standard deviation (SD) for three independent experiments. Student’s t-test was used to evaluate the significant difference of two groups of data in all the pertinent experiments. A p value <0.05 was regarded as statistically significant.
Results
MiR-4295 was upregulated, while BTG1 was downregulated in bladder carcinoma and cell lines
The detailed sequence of miR-4295 obtained from miRBase database is shown in Figure 1A. Real-time RT-PCR was used to evaluate the expression of miR-4295 in bladder carcinomas and adjacent normal tissues. MiR-4295 was found to be markedly upregulated in bladder carcinomas compared with the adjacent normal tissue (Figure 1B). To further verify the role of miR-4295 in bladder cancer, the expression of miR-4295 in human bladder cancer cell lines (Hcv29, UM-UC-3, BIu87, T24 and Hbc) was evaluated. Consistently, the expression of miR-4295 was elevated in all the four cell lines, to different degrees, compared with that in Hcv29 cells (Figure 1E). Next, real-time RT-PCR was used to evaluate the expression of BTG1 in bladder carcinomas tissues and cell lines. BTG1 expression was notably decreased in bladder carcinomas tissues and bladder cancer cell lines, in comparison with that in adjacent normal tissues and Hcv cells (Figure 1C, 1F). To explore whether a relationship between BTG1 and miR-4295 exists, correlation analysis was performed (Figure 1D).
Figure 1.
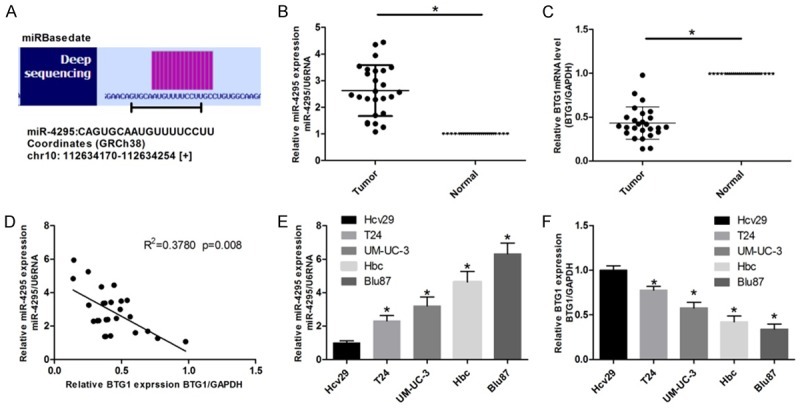
The aberrant expression of miR-4295 and BTG1 in bladder carcinoma and cell lines. A. The deep sequencing of miR-4295. B, C. Real-time PCR was used to assess the expression of miR-4295 and BTG1 in bladder carcinomas and paired adjacent normal tissues. D. A correlation analysis was performed to evaluate the correlation between miR-4295 and BTG1 expression. E, F. Real-time PCR was used to assess the expression of miR-4295 and BTG1 in bladder cancer cell lines and in the normal Hcv29 cells. The bars represent the mean ± SD of three independent experiments. **P <0.01, *P <0.05.
miR-4295 overexpression promotes bladder cancer cells growth and cell cycle progression
T24 and Hbc cells were transfected with miR-4295 mimics. CCK-8 assay was performed every 24 hours, as described in the methods section, and the proliferation curve is shown. The results showed that miR-4295 overexpression resulted in significantly increased cell viability at 3 days after transfection in both T24 and Hbc cells (Figure 2A, 2B). Similar trends were also observed for colony formation rate. MiR-4295 overexpression led to a significantly higher colony formation rate (Figure 2C) in T24 and Hbc cells. In addition, the cell migration assay was performed to further investigate the role of miR-4295 in bladder cancer cells. As expected, for both T24 and Hbc, the transfection of miR-4295 significantly increased the number of cells that had migrated (Figure 2D). The cell cycle analysis of T24 and Hbc cells by flow cytometry showed that there was a significant decrease in the percentage of cells in the G1/G0 phase and an increase in the percentage of cells in S phase (Figure 2E). These results indicate that the overexpression of miR-4295 promoted the proliferation, migration and cell cycle progression of bladder cancer cells in vitro.
Figure 2.
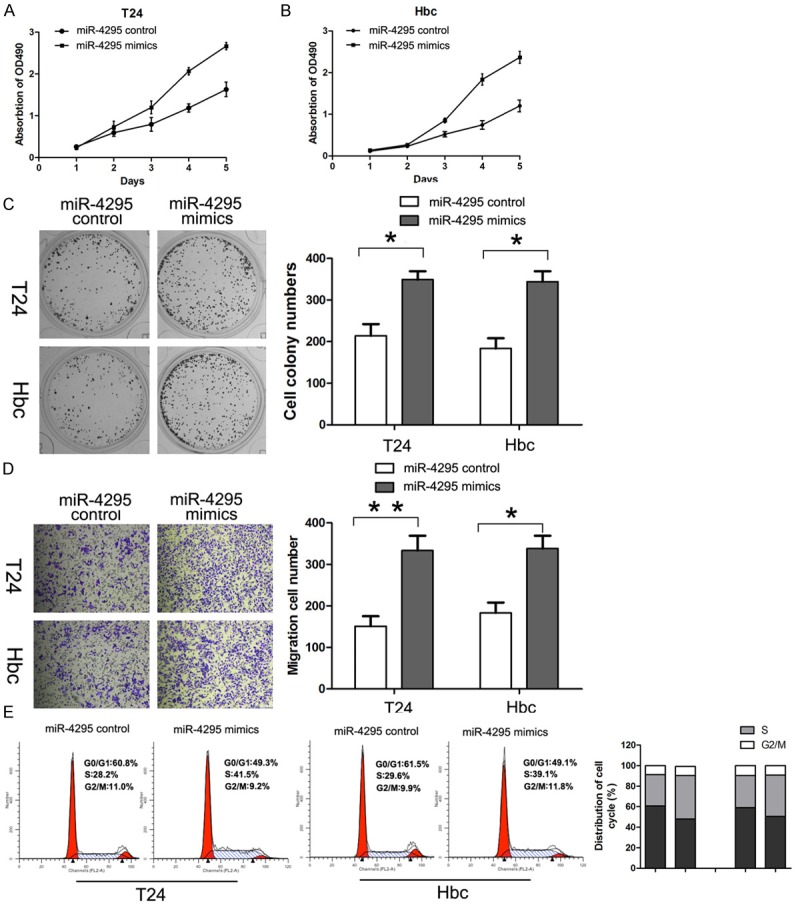
MiR-4295 promotes bladder cancer cell growth and cell cycle progression. A, B. CCK-8 assay was performed every 24 h until 96 h after transfection, and the proliferation curves of T24 and Hbc cells were plotted. C. Representative micrographs (left) and quantifications (right) of colonies formed by the indicated cells and stained with crystal violet. D. Cell migration of the indicated cells in a transwell cell migration assay, as shown by the quantification and representative images. E. PI staining and flow cytometry were used to analyze the cell cycle distribution of T24 and Hbc cells. The bars represent the mean ± SD of three independent experiments. **P <0.01, *P <0.05.
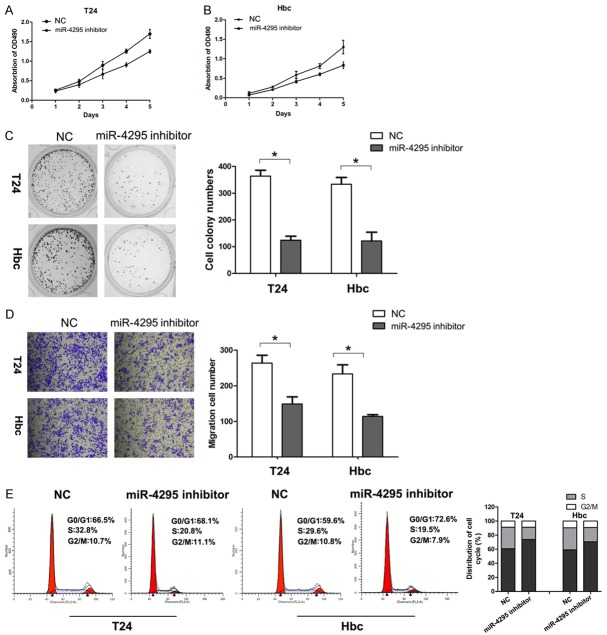
Inhibition of miR-4295 attenuates the proliferative capacity of bladder cancer cells
To further confirm whether the inhibition of miR-4295 reduces bladder cancer cell proliferation, loss-of-function studies were performed using a miR-4295 inhibitor. The CCK-8 and colony formation assays showed that the inhibition of miR-4295 significantly decreased the proliferative capacity of T24 and 5637 cells, compared with cells transfected with NC (Figure 3A-C). The migration assay revealed that the suppression of miR-4295 led to a decrease in the number of cells that had migrated (Figure 3D). Flow cytometry showed that the cells transfected with the miR-4295 inhibitor had a significant increase in the percentage of cells in the G1/G0 phase and a decrease in the percentage of cells in S phase, compared with NC transfected cells (Figure 3E). These results suggest that the downregulation of miR-4295 suppresses proliferation and cell cycle progression of bladder cancer cells.
Figure 3.
The inhibition of miR-4295 suppresses bladder cancer cell growth and cell cycle progression. A, B. CCK-8 assay was performed every 24 h until 96 h after transfection, and the proliferation curves of T24 and Hbc cells were plotted. C. Representative micrographs (left) and quantifications (right) of crystal violet stained colonies formed by the indicated cells. D. Cell migration of the indicated cells in a transwell cell migration assay, as shown by the quantification and representative images. E. PI staining and flow cytometry were used to analyze the cell cycle distribution of T24 and Hbc cells. The bars represent the mean ± SD of three independent experiments. **P <0.01, *P <0.05.
MiR-4295 directly targets BTG1
MiRNAs execute post-transcriptional regulation, mostly by binding to the 3’-UTR of the downstream genes. Accordingly, we used the bioinformatics prediction software, Targetscan, to identify possible targets that are involved in the regulation of cell growth (http://www.targetscan.org/). Among these thousands of candidates, we focused on BTG1. Luciferase reporter assays were performed to verify the interaction between miR-4295 and the 3’UTR of BTG1. The 3’UTR of BTG1 was cloned downstream of the pmirGLO Dual-Luciferase miRNA target expression vector. In addition, a mutated BTG1 3’-UTR vector was also constructed (Figure 4A). Cells that were co-transfected with the wt-3’-UTR-reporter and miR-4295 showed significantly decreased relative luciferase activity compared with cells transfected with the miR-4295 control. However, the luciferase activity of the reporter carrying the 3’-UTR with mutated binding sites was unaffected by the co-transfection with miR-4295 (Figure 4B). Western Blot analysis was used to confirm the regulation of BTG1 by miR-4295 (Figure 4C, 4D). These results strongly indicate that the BTG1 is a direct target of miR-4295.
Figure 4.

Mir-4295 directly targets the 3’UTR of BTG1 mRNA. A. Bioinformatics analyses of the binding of miR-4295 to the 3’-UTR of BTG1 mRNA. B. A luciferase reporter assay was performed to determine whether BTG1 is a target of miR-4295. C, D. Western blot analysis was used to assess the protein level of BTG1 in T24 and Hbc cells transfected with miR-4295 mimic or miR-4295 inhibitor. GAPDH was used as an endogenous reference. The bars represent the mean ± SD of three independent experiments. **P <0.01, *P <0.05.
Overexpression of BTG1 reverses the oncogenic effect of miR-4295 on cell proliferation
Next, a rescue experiment was conducted to further confirm the role of BTG1 in miR-4295 induced bladder cancer proliferation. An overexpression vector, pcDNA3.1(+)/BTG1, was constructed to enhance endogenous BTG1 expression. The results of the CCK-8 and colony formation assays indicated that the proliferation suppressing effect of miR-4295 was reversed by BTG1 in both T24 and Hbc cells (Figure 5A-C). Additionally, the migration assay indicated that BTG1 significantly reversed the effect of miR-4295 on the migration capability of T24 and Hbc cells (Figure 5D). Furthermore, cell cycle analysis demonstrated that BTG1 increased the proportion of cells in S phase, and inhibited the effect of miR-4295 on cell cycle progression (Figure 5E). These results suggested that the overexpression of BTG1 could reverse the effect of miR-4295 on the proliferation and cycle progression of bladder cancer cells.
Figure 5.
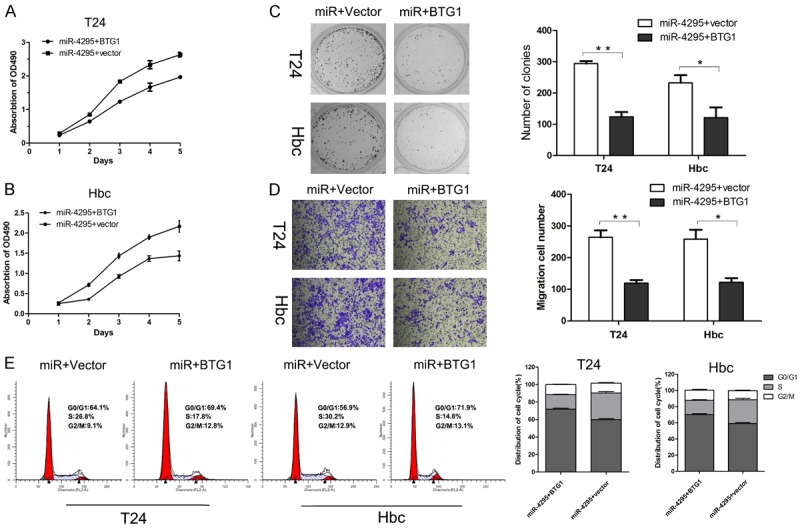
A rescue experiment was conducted to confirm that BTG1 is a target of Mir-4295. A, B. The CCK-8 assay was performed every 24 h until 96 h after co-transfection with miR-4295 mimics and BTG1 overexpression vector or with miR-4295 and control vector. The proliferation curves of the various conditions are shown. C. Representative micrographs (left) and quantifications (right) of crystal violet stained colonies formed by the T24 and Hbc cells co-transfected with miR-4295 mimics and BTG1 overexpression vector or with miR-4295 and control vector. D. Cell migration of T24 and Hbc cells that were treated as indicated in a transwell cell migration assay, as shown by the quantification and representative images. E. PI staining and flow cytometry were used to analyze the cell cycle distribution of T24 and Hbc cells that were treated as indicated. The bars represent the mean ± SD of three independent experiments. **P <0.01, *P <0.05.
Discussion
Various miRNAs play important roles in the invasion and metastases of malignant tumor cells. An increasing amount of evidence indicates that the dysregulation of various miRNAs is involved in bladder carcinogenesis and progression [12,13]. It has been demonstrated that miRNAs exert their biological function in bladder cancer by multiple ways. For instance, miRNAs regulate the proliferation, invasion and metastasis of bladder cancer cells by targeting numerous genes, such as STMN1, BMI-1, LASS2 and c-FOS [14-17]. However, despite developments in bladder cancer diagnosis and treatment, knowledge regarding the molecular mechanisms underlying bladder carcinogenesis and progression still remain limited. Additionally, specific tumor biomarkers that can be used as therapeutic targets are lacking. As such, there is an urgent need to identify novel biomarkers that have potential clinical significance as therapeutic targets.
MiR-4295 is a newly identified miRNA, and only a few studies on it exist. However, it has been shown to have an essential role in tumor carcinogenesis and progression. MiR-4295 has been found to inhibit cell growth, causing G0/G1 arrest and apoptosis in glioma cells, and to attenuate non-small cell lung cancer growth by targeting USP28. In the present study, we determined that miR-4295 expression was significantly higher in bladder cancer tissues and cell lines than in normal bladder tissue, indicating that miR-4295 may be involved in bladder carcinogenesis or progression. Functional studies were conducted to determine the role of miR-4295 in bladder cancer. Our findings revealed that the overexpression of miR-4295 significantly promoted the proliferation, colony formation and migration of bladder cancer cells, as well as cell cycle progression. Additionally, to verify our hypothesis, we knocked down the expression of miR-4295 by transfecting cells with a miR-4295 inhibitor. As expected, the knock down of miR-4295 attenuated the proliferative capacity of bladder cancer cells.
BTG1 was initially identified in B lymphoblastic leukemia, and its expression appears to be highest in the G0/G1 phases of the cell cycle [18]. BTG proteins can shuttle between the nucleus and the cytoplasm, though their function is dependent on their nuclear localization, which enables them to bind to the nuclear receptor, TRα, and the myogenic factor, MyoD [19,20]. Weak BTG1 expression has recently been reported in various other types of cancer, including thyroid, lung, prostate, renal, and breast cancer [21-28], and was reported to be positively correlated with invasion depth, lymphatic and venous invasion, and lymph node metastasis [29]. These findings indicate that BTG1 expression might be considered a valuable biomarker for predicting the aggressive behaviors of gastric cancer. Additionally, BTG1 expression has been found to be regulated by miRs, such as miR-19, miR22, miR-17-92 and miR-miR-454-3p [30-32], in prostate cancer, colorectal cancer and lung cancer. In this work, we first found that the expression of BTG1 was significantly downregulated in bladder carcinomas and cell lines. Next, BTG1 was predicted to be a potential target of miR-4295. In addition, in bladder carcinomas, the expression of BTG1 and miR-4295 were shown to correlate inversely, which supports a physiological role for miR-4295 in regulating BTG1 expression. To verify this hypothesis, a luciferase assay was performed, and the results were as expected.
In conclusion, our study indicated that miR-4295 is significantly upregulated in bladder cancer cells and carcinomas. MiR-4295 promotes cell proliferation and cell cycle progression of bladder cancer cells by targeting BTG1. These findings may suggest miR-4295 as a novel potential biomarker for cancer diagnosis and therapy. Further studies into other mechanisms underlying the function of miR-4295 are warranted.
Disclosure of conflict of interest
None.
References
- 1.Micheli A, Francisci S, Krogh V, Rossi AG, Crosignani P. Cancer prevalence in Italian cancer registry areas: the ITAPREVAL study. ITAPREVAL Working Group. Tumori. 1999;85:309–369. doi: 10.1177/030089169908500502. [DOI] [PubMed] [Google Scholar]
- 2.Sager CC, Benamran DA, Wirth G, Iselin CE. [New approaches for the treatment of superficial bladder cancer] . Rev Med Suisse. 2015;11:2281–2284. [PubMed] [Google Scholar]
- 3.Jin Y, Lu J, Wen J, Shen Y, Wen X. Regulation of growth of human bladder cancer by miR-192. Tumour Biol. 2015;36:3791–3797. doi: 10.1007/s13277-014-3020-8. [DOI] [PubMed] [Google Scholar]
- 4.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282–289. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Zhao X, Li J, Huang S, Wan X, Luo H, Wu D. MiRNA-29c regulates cell growth and invasion by targeting CDK6 in bladder cancer. Am J Transl Res. 2015;7:1382–1389. [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Wu Q, Xu B, Wang P, Fan W, Cai Y, Gu X, Meng F. miR-124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine-tuning UHRF1. FEBS J. 2015;282:4376–4388. doi: 10.1111/febs.13502. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S, Zhang C, Liu W, Zheng W, Zhang Y, Wang S, Huang D, Liu X, Bai Z. MicroRNA-24 upregulation inhibits proliferation, metastasis and induces apoptosis in bladder cancer cells by targeting CARMA3. Int J Oncol. 2015;47:1351–1360. doi: 10.3892/ijo.2015.3117. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Li Q, Wang S, Zhang J. miR4855p inhibits bladder cancer metastasis by targeting HMGA2. Int J Mol Med. 2015;36:1136–1142. doi: 10.3892/ijmm.2015.2302. [DOI] [PubMed] [Google Scholar]
- 10.Lei M, Xie W, Sun E, Sun Y, Tian D, Liu C, Han R, Li N, Liu M, Han R, Liu L. microRNA-21 Regulates Cell Proliferation and Migration and Cross Talk with PTEN and p53 in Bladder Cancer. DNA Cell Biol. 2015;34:626–632. doi: 10.1089/dna.2015.2868. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Xu B, Qiang Y, Huang H, Wang C, Li D, Qian J. Overexpression of deubiquitinating enzyme USP28 promoted non-small cell lung cancer growth. J Cell Mol Med. 2015;19:799–805. doi: 10.1111/jcmm.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou XU, Qi L, Tong S, Cui YU, Chen J, Huang T, Chen Z, Zu XB. miR-128 downregulation promotes growth and metastasis of bladder cancer cells and involves VEGF-C upregulation. Oncol Lett. 2015;10:3183–3190. doi: 10.3892/ol.2015.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Zhang Y, Liu X, Fang A, Wang J, Yang Y, Wang L, Du L, Wang C. Direct quantitative detection for cell-free miR-155 in urine: a potential role in diagnosis and prognosis for non-muscle invasive bladder cancer. Oncotarget. 2016;7:3255–66. doi: 10.18632/oncotarget.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Cao J, Zhao X. miR-221 facilitates the TGFbeta1-induced epithelial-mesenchymal transition in human bladder cancer cells by targeting STMN1. BMC Urol. 2015;15:36. doi: 10.1186/s12894-015-0028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Y, Yang X, Deng X, Zhang X, Li P, Tao J, Lu Q. MicroRNA-218 inhibits bladder cancer cell proliferation, migration, and invasion by targeting BMI-1. Tumour Biol. 2015;36:8015–8023. doi: 10.1007/s13277-015-3532-x. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Zhang W, Zuo Y, Ding M, Ke C, Yan R, Zhan H, Liu J, Wang J. miR-9 promotes cell proliferation and inhibits apoptosis by targeting LASS2 in bladder cancer. Tumour Biol. 2015;36:9631–9640. doi: 10.1007/s13277-015-3713-7. [DOI] [PubMed] [Google Scholar]
- 17.Lan G, Yang L, Xie X, Peng L, Wang Y. MicroRNA-490-5p is a novel tumor suppressor targeting c-FOS in human bladder cancer. Arch Med Sci. 2015;11:561–569. doi: 10.5114/aoms.2015.52359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang R, Hu W, Sun G, Wang J, Han X. [Expression of BTG1 protein in laryngeal squamous cell carcinoma and its clinical significance] . Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2015;29:1447–1450. [PubMed] [Google Scholar]
- 19.Busson M, Carazo A, Seyer P, Grandemange S, Casas F, Pessemesse L, Rouault JP, Wrutniak-Cabello C, Cabello G. Coactivation of nuclear receptors and myogenic factors induces the major BTG1 influence on muscle differentiation. Oncogene. 2005;24:1698–1710. doi: 10.1038/sj.onc.1208373. [DOI] [PubMed] [Google Scholar]
- 20.Kawamura-Tsuzuku J, Suzuki T, Yoshida Y, Yamamoto T. Nuclear localization of Tob is important for regulation of its antiproliferative activity. Oncogene. 2004;23:6630–6638. doi: 10.1038/sj.onc.1207890. [DOI] [PubMed] [Google Scholar]
- 21.Lu YF, Sun GG, Liu Q, Yang CR, Cheng YJ. BTG1 expression in thyroid carcinoma: diagnostic indicator and prognostic marker. Int J Oncol. 2014;45:1574–1582. doi: 10.3892/ijo.2014.2543. [DOI] [PubMed] [Google Scholar]
- 22.Zhu R, Li W, Xu Y, Wan J, Zhang Z. Upregulation of BTG1 enhances the radiation sensitivity of human breast cancer in vitro and in vivo. Oncol Rep. 2015;34:3017–3024. doi: 10.3892/or.2015.4311. [DOI] [PubMed] [Google Scholar]
- 23.Zheng HC, Li J, Shen DF, Yang XF, Zhao S, Wu YZ, Takano Y, Sun HZ, Su RJ, Luo JS, Gou WF. BTG1 expression correlates with pathogenesis, aggressive behaviors and prognosis of gastric cancer: a potential target for gene therapy. Oncotarget. 2015;6:19685–19705. doi: 10.18632/oncotarget.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C, Tao T, Xu B, Lu K, Zhang L, Jiang L, Chen S, Liu D, Zhang X, Cao N, Chen M. BTG1 potentiates apoptosis and suppresses proliferation in renal cell carcinoma by interacting with PRMT1. Oncol Lett. 2015;10:619–624. doi: 10.3892/ol.2015.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26:262–272. doi: 10.1016/j.ccr.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito Y, Suzuki T, Yoshida H, Tomoda C, Uruno T, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Kuma K, Yamamoto T, Miyauchi A. Phosphorylation and inactivation of Tob contributes to the progression of papillary carcinoma of the thyroid. Cancer Lett. 2005;220:237–242. doi: 10.1016/j.canlet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Iwanaga K, Sueoka N, Sato A, Sakuragi T, Sakao Y, Tominaga M, Suzuki T, Yoshida Y, K-Tsuzuku J, Yamamoto T, Hayashi S, Nagasawa K, Sueoka E. Alteration of expression or phosphorylation status of tob, a novel tumor suppressor gene product, is an early event in lung cancer. Cancer Lett. 2003;202:71–79. doi: 10.1016/j.canlet.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 28.Yoneda M, Suzuki T, Nakamura T, Ajima R, Yoshida Y, Kakuta S, Katsuko S, Iwakura Y, Shibutani M, Mitsumori K, Yokota J, Yamamoto T. Deficiency of antiproliferative family protein Ana correlates with development of lung adenocarcinoma. Cancer Sci. 2009;100:225–232. doi: 10.1111/j.1349-7006.2008.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun GG, Wang YD, Cheng YJ, Hu WN. The expression of BTG1 is downregulated in nasopharyngeal carcinoma and possibly associated with tumour metastasis. Mol Biol Rep. 2014;41:5979–5988. doi: 10.1007/s11033-014-3475-0. [DOI] [PubMed] [Google Scholar]
- 30.Lu K, Liu C, Tao T, Zhang X, Zhang L, Sun C, Wang Y, Chen S, Xu B, Chen M. MicroRNA-19a regulates proliferation and apoptosis of castration-resistant prostate cancer cells by targeting BTG1. FEBS Lett. 2015;589:1485–1490. doi: 10.1016/j.febslet.2015.04.037. [DOI] [PubMed] [Google Scholar]
- 31.Alanazi I, Hoffmann P, Adelson DL. MicroRNAs are part of the regulatory network that controls EGF induced apoptosis, including elements of the JAK/STAT pathway, in A431 cells. PLoS One. 2015;10:e0120337. doi: 10.1371/journal.pone.0120337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Tang J, Li C, Kong J, Wang J, Wu Y, Xu E, Lai M. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2015;356:781–790. doi: 10.1016/j.canlet.2014.10.029. [DOI] [PubMed] [Google Scholar]