Abstract
miR-101 is significantly downregulated in various human cancers, including oral squamous cell carcinoma (OSCC). However, the role of miR-101 in OSCC has not been elucidated. In this study, miR-101 lowly expressed in OSCC tissues and cell lines compared with that in adjacent normal tissues and human normal oral keratinocyte cells. Bioinformatics analysis predicted that miR-101 could potentially target CX chemokine receptor 7 (CXCR7), a promoter of tumor development, to attenuate OSCC progression. Restoring miR-101 expression in OSCC cells suppressed cell proliferation, invasion, and migration. The ectopic expression of CXCR7 in OSCC cells overexpressing miR-101 restored the proliferation and motility capabilities abolished by miR-101. The inhibitory effects of miR-101 on OSCC growth and metastasis were mimicked by CXCR7 knockdown in vivo. CXCR7 expression was upregulated in OSCC tissues. The high expression level of CXCR7 was negatively correlated with miR-101 level and poor prognosis of patients with OSCC. Overall, miR-101 exerts tumor-suppressive functions by targeting CXCR7, leading to inhibition of OSCC cell growth, invasion, and migration. Hence, miR-101 may be a potential target for OSCC diagnosis and therapeutic applications.
Keywords: microRNA-101, oral squamous cell carcinoma, growth, metastasis, CX chemokine receptor 7
Introduction
Oral squamous cell carcinoma (OSCC) is the most common head and neck squamous cell carcinoma, with more than 300,000 new cases diagnosed annually worldwide [1]. The high mortality of OSCC is mainly caused by cervical lymph node metastasis and occasionally by distant organ metastasis [2]. The pathogenesis of OSCC is related to the rates of cell proliferation and apoptosis [3]. The 5-year survival rate of patients with OSCC has not significantly improved, despite progress achieved in OSCC treatment [4]. Therefore, the molecular mechanisms underlying OSCC tumorigenesis must be elucidated to identify tumor-specific biomarkers and therapeutic targets for early diagnosis and treatment of the disease.
MicroRNAs (miRNAs) are a class of small non-coding RNAs that regulate gene function by targeting mRNAs for translational repression or degradation [5]. Abnormalities of miRNAs have been implicated in the occurrence and development of various human cancers, including OSCC [6,7]. Several miRNAs, such as let-7a, let-7d, let-7f, and miR-16 [8], are significantly downregulated in OSCC and are thus considered tumor suppressors. In particular, miR-101 is downregulated in gastric carcinoma [9], hepatocellular carcinoma [10], breast cancer [11], and OSCC [12]. miR-101 promotes apoptosis and inhibits the proliferation and migration of breast cancer cells by downregulating CX chemokine receptor 7 (CXCR7) [11]. However, the role of miR-101 in OSCC remains unknown and needs to be further investigated.
Chemokines, a superfamily of small cytokine-like proteins, can bind to and activate a group of G-protein-coupled seven-span transmembrane receptors, called chemokine receptors. CXCL12, also known as stromal cell derived factor-1, was first detected in murine bone marrow stromal cells. CXCR7, an alternative receptor of CXCL12, plays a key role in cell survival, adhesion, and tumor development [13]. CXCR7 is highly expressed in multiple malignancies; moreover, activation of the CXCR7 signaling pathway by CXCL12 enhances proliferation, metastasis, and invasive activities of tumor cells [14,15]. CXCR7 is concomitantly upregulated with CXCL12 in oral carcinogenesis [16]. Knockdown of CXCR7 significantly inhibits the proliferation, migration, and invasion of breast cancer cells and suppresses tumor growth and lung metastasis in a xenograft model of breast cancer [11]. However, the role of CXCR7 in OSCC growth and metastasis requires further investigation.
In this study, miR-101 was substantially downregulated in OSCC tissues and four OSCC-derived cell lines compared with paired normal tissues and human normal oral keratinocyte cells (hNOKs). miR-101 suppressed OSCC cell growth, invasion, and migration by targeting CXCR7 in vitro and in vivo. Moreover, CXCR7 expression was upregulated in OSCC tissues. The high expression of CXCR7 was negatively correlated with miR-101 expression and poor prognosis of OSCC patients. These findings suggest that miR-101 is a tumor suppressor and miR-101 and CXCR7 are potential targets for OSCC therapy.
Materials and methods
Clinical samples
Tumor samples including paraffin-embedded tissue biopsies and fresh frozen biopsies were obtained from patients receiving surgery for OSCC in School of Stomatology, Fourth Military Medical University from January 2013 to June 2015. This study was approved by the ethics committee of the Fourth Military Medical University (Xi’an, Shaanxi, China). None of the patients received chemotherapy or radiotherapy before surgery. Informed consent was obtained from all participants. Patient followup was performed every 3 months via phone, mail, or outpatient visit. Survival rates were defined as the time between the date of diagnosis and patient death or the end of the study period.
Cell culture
Human OSCC cell lines including Cal-27, Fadu, SCC-9, and SCC-4 as well as human embryonic kidney 293T (HEK293T) cells were obtained from the American Type Culture Collection (Manassas, VA, USA). OSCC cells were maintained in Dulbecco’s modified Eagle’s medium/F12 (DMEM/F12; Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA). The cells were maintained in a humidified incubator at 37°C with 5% CO2. hNOKs were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in an oral keratinocyte medium (ScienCell Research Laboratories). All cells were authenticated based on morphology and growth characteristics and used within 3 months of resuscitation from the frozen stock, with fewer than 20 passages.
Cell stimulation
Cell stimulation was performed as previously described [17]. The cells were serum starved at 37°C for 5 h. The serum-starved cells were stimulated with 100 ng/mL CXCL12 (Peprotech Inc., Princeton, NJ, USA) and incubated at 37°C for the indicated time periods. The cells were harvested for analysis at the end of the stimulation.
Plasmid and lentivirus constructs
The cDNA strands corresponding to the pre-miR-101 sequence were synthesized and cloned into the AgeI and EcoRI sites of pGCsi-H1-CMV-GFP (GeneChem, Shanghai, China) to generate lentivirus-encoding miR-101 (LV-miR-101). The constructs were co-transfected with pHelper 1.0 and pHelper 2.0 Packing Plasmid (GeneChem) into 293T packing cells by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Supernatant was collected 48 h after transfection, centrifuged, filtered, and used for infection. LV-GFP, a control virus, was similarly produced. OSCC cells were transfected with LV-miR-101 or LV-GFP in the presence of 4 mg/mL polybrene (Sigma-Aldrich, St. Louis, MO, USA) at an infection multiplicity of 10. The cells were centrifuged at 1800 rpm for 45 min, cultured for 72 h, and analyzed by fluorescence-activated cell sorting (BD Biosciences, Bedford, MA, USA). shRNA was designed based on the CXCR7 sequence for CXCR7 knockdown. A control vector (NC-shRNA) without significant homology to any mammalian gene sequence was constructed and used as non-silencing control. The resulting lentiviral vector was named CXCR7-lentivirus (LV-shCXCR7). Negative-control lentiviral vector containing NC-shRNA was constructed by a similar process (LV-NC). HEK293T cells were co-transfected with the constructs and lentivirus packaging plasmids, namely, pMD.G and pCMVΔR8.91 (GeneChem). Virus in the conditioned medium was harvested, filtered, and used for infection of OSCC cells. The cells were selected with puromycin (5 μg/mL; Sigma) to generate stable shRNA-expressing clones. The pUNOI-hCXCR7 (open reading frame) plasmid was purchased from InvivoGen (Hong Kong, China).
Cell transfection
Upon reaching 70-80% confluence, the cells were transfected with 80 nM miR-101 mimics or its negative control (miR-NC; RiboBio, Guangzhou, China) by using Lipofectamine® 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Accordingly, 3 μg of DNA was used for plasmid transfection.
Dual-luciferase reporter assay
The CXCR7 3’-UTR region was generated for the dual-luciferase reporter assay by sub-cloning PCR-amplified full-length human CXCR7 cDNA into the Xho1/Xba1 sites of the pGL3 firefly luciferase-expressing vector (Promega, Madison, WI, USA). The miRNA binding sites in the mutation reporters were constructed by site-direct mutagenesis. The primers used to amplify the WT and MUT 3’-UTRs are listed in Table 1. The plasmid pGL3-CXCR7-3’-UTR-wild type (WT) or pGL3-CXCR7-3’-UTR-mutant (MUT) was co-transfected with miR-NC or miR-101 mimics into Fadu or SCC-9 cells. Transfection of chemical-modified RNA molecules or plasmids was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Luciferase assay was conducted 48 h after transfection using a dual-luciferase reporter assay system (Promega). Renilla luciferase was co-transfected as control for normalization.
Table 1.
The primers used to generate the WT and MUT CXCR7 3’-UTRs
| Gene | Primer sequences (5’ to 3’) | |
|---|---|---|
| CXCR7 3’-UTR (WT) | Forward | GAATTCGACGGGTTTACTTGTTTT |
| Reverse | CTGCAGGGAAACAAAAATCTTTAT | |
| CXCR7 3’-UTR (MUT) | Forward | CACCAATAGTGAGAAATATTTCACTTAAAATTTAC |
| Reverse | GTATTTAAATTTTAAGTGAAATATTTCTCACTATTG | |
Quantitative real-time polymerase chain reaction (qPCR)
Total RNAs from the tissues and cells were isolated using TRIzol reagent (Invitrogen) and purified using an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. cDNA was synthesized from the total RNA by using PrimeScript RT reagent kit (TaKaRa, Otsu, Shiga, Japan). The PCR reactions containing SYBR Premix Ex Taq II (TaKaRa) were processed with the following specific primers: for CXCR7, 5’-GCTCACCAAGCTCATCGAT-3’ (forward) and 5’-AAGACCCGAAGCTACTTTGC-3’ (reverse); for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5’-CCTGGATACCGCAGCTAGGA-3’ (forward) and 5’-G CGGCGCAATACGAATGCCCC-3’ (reverse); for miR-101, 5’-CGGCGGTACAGTA CTGTGATAA-3’ (forward) and 5’-CTGGTGTCGTGGAGTCGGCAATTC-3’ (reverse); and for U6, 5’-CTCGCTTCGGCAGCACA-3’ (forward) and 5’-AACGCTT CACGAATTTGCGT-3’ (reverse). The expressions of CXCR7 and miR-101 were based on the 2-ΔΔCt method, using GAPDH and U6 as internal controls, respectively.
Cell proliferation assay
OSCC cells transfected with miR-NC, miR-101 mimics, or miR-101 mimics plus CXCR7-expressing plasmid were seeded in a 96-well plate at a density of 1 × 103 cells/well. After culture for 1, 2, 3, or 4 days, each well was added with 20 μL of 3-(4, 5-dimethylthiazol-2-thiazole)-2, 5-diphenyltetrazolium bromide (MTT; 5 mg/mL; Sigma). The plate was wrapped with foil and incubated at 37°C for 4 h. After removing the medium and the MTT solution, each well was added with 150 μL of dimethyl sulfoxide (Sigma) and incubated for 10 minto resolve the crystals. Optical density (OD) was determined at 490 nm, and each analysis was performed in triplicate.
Colony formation assay
OSCC cells transfected with miR-NC, miR-101, or miR-101 mimics plus CXCR7-expressing plasmid were seeded in a six-well plate at a density of 1 × 103 cells/well. The cells were cultured for 14 days and washed with phosphate-buffered saline (PBS) three times. Each well was added with an ethanol fixation solution (1 mL) for 10 min. After staining with 0.5% crystal violet (Sigma), the colonies were counted under the microscope (Olympus, Tokyo, Japan) and photographed. Five random fields were selected from each well to determine the total number of colonies.
Apoptosis assay
Apoptosis was assessed using an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instructions. After transfection with miR-NC, miR-101 mimics, or miR-101 mimics plus CXCR7-expressing plasmid for 48 h, OSCC cells (1 × 106) were collected, washed, and resuspended in PBS. The cells were added with Annexin V-FITC and PI and incubated for 20 min at 4°C. The cells were analyzed using a FACScan flow cytometer (BD Biosciences) with FlowJo software (Tree Star Inc.).
Transwell assay
The migration and invasion of OSCC cells were detected using the Falcon Cell Culture Inserts with or without Matrigel (BD Biosciences) coating. The upper chamber was pre-coated with 50 μL of 20% growth factor-reduced Matrigel for the invasion assay. The inserts were not coated for the migration assay. After transfection with miR-NC, miR-101 mimics, or miR-101 mimics plus CXCR7-expressing plasmid for 48 h, the cells (5 × 104) were harvested, resuspended in serum-free medium with 0.1% BSA (Sigma), and plated onto the upper chamber. DMEM/F12 containing 10% FBS was added in the lower chamber to act as chemoattractant. Cells in the transwell plates were incubated at 37°C in a humidified atmosphere with 5% CO2 for 24 h. the adherent cells on the upper surface of the insert membrane were carefully removed with cotton tips. Cells on the lower surface of the membrane were fixed with 4% paraformaldehyde for 15 min, permeabilized in 0.1% Triton X-100, and stained with 0.1% crystal violet for 30 min. The invaded or migrated cells were imaged and quantified using a light microscope (Carl Zeiss AG, Germany) in five separate fields per membrane.
Western blot analysis
Proteins from the transfected cells or fresh tissues were extracted with RIPA buffer (Sigma). The proteins were separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis and electro-transferred onto the polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). The membrane was blocked with 5% non-fat milk in TBS containing 0.1% Tween 20 for 2 h at 37°C and incubated with anti-CXCR7 and anti-GAPDH monoclonal antibodies (Abcam, Cambridge, MA, UK) overnight at 4°C. The membrane was then incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at 37°C. The immunoreactive bands were visualized with the enhanced chemiluminescence detection system (Millipore).
Immunohistochemistry
Immunohistochemistry was performed following the manufacturer’s protocols. After deparaffinization and rehydration, the sections were submerged in citrate buffer, treated with 3% H2O2, incubated in 5% goat serum, and incubated with primary antibody against CXCR7 (Abcam) at 4°C overnight. The sections were incubated with a biotinylated secondary antibody by using DAKO Real EnVision Kit (DAKO, Hamburg, Germany) at 37°C for 15 min. The sections were then stained with 3, 3’-diaminobenzidine (DAB; Maixin Biotech., Fuzhou, China) and counterstained with commercial hematoxylin (Maixin Biotech.) prior to microscopic analysis.
Terminal deoxynucleotidyl transferase mediated dUTP nick-end labeling (TUNEL) assay
The tissues were fixed with 10% formalin for 24 h, embedded in paraffin, non-serially sectioned (4 µm thickness), and mounted on polylysine-covered slides. TUNEL staining was performed according to the manufacturer’s protocols (Roche, Mannheim, BW, Germany). The sections were rinsed with PBS, treated with proteinase K, and incubated with terminal deoxynucleotidyl transferase (TdT) reaction mixture at 37°C for 1 h. The sections were then washed with PBS three times, incubated in anti-digoxigenin-peroxidase at 37°C for 30 min, rinsed in PBS, stained with DAB, and counterstained with hematoxylin (Maixin Biotech.). The negative controls were incubated in a medium without TdT enzyme. Apoptotic cells were expressed as ratio of TUNEL-positive cells to the total number of cells.
Animal studies
Female SCID mice (6 weeks old and weighing 20-22 g) were purchased from Institute of Zoology (Chinese Academy of Sciences, Beijing, China) and placed under aseptic conditions with constant humidity and temperature (25°C-28°C) and a 12 h light/dark cycle. All of the experiments were approved by the Institutional Animal Care and Use Committee of the Fourth Military Medical University. For tumor growth assay, the mice were subcutaneously inoculated with Fadu-luciferase cells (1 × 106, PerkinElmer, Santa Clara, CA, USA) stably infected with recombinant lentivirus expressing the miR-101 precursor or shCXCR7 or their control lentivirus on the hind flanks of the mice (n = five/group). Tumor volume was calculated using the following equation: tumor volume = length × width2/2. The mice were euthanized on day 42. Tumors were excised and weighed. For the lung metastasis assay, the mice were injected with Fadu cells (1 × 106) via tail vein with recombinant lentivirus expressing the miR-101 precursor or shCXCR7 or their control lentivirus on the hind flanks of the mice (n = five/group). The mice were monitored for general health and sacrificed by euthanasia 8 weeks post-inoculation. The lung tissues were removed, fixed, paraffin-embedded, serially sectioned, and subjected to hematoxylin and eosin staining. The metastases per section were counted. For the apoptosis assay, the sections were assessed by the TUNEL assay.
Bioluminescence imaging
At 42 days after Fadu-luciferase cell injection, the mice were anesthetized with 2% isoflurane, intraperitoneally injected 150 mg/kg D-luciferin (Promega), and analyzed using an IVIS Imaging System (Caliper LifeScience, MA, USA) to measure the signal intensity of luciferase activity. The signals were expressed as photon flux (photons/s/cm2) by using Living Image software (Xenogen Corporation, Berkeley, CA, USA).
Results
miR-101 is downregulated in OSCC tissues and cell lines
miR-101 expression in 66 pairs of OSCC and adjacent normal oral tissues was determined using qPCR assay to identify the role of miR-101 in OSCC. Figure 1A shows that the expression level of miR-101 in OSCC tissues was significantly lower than that in the adjacent normal tissues. miR-101 levels were assessed in four OSCC cell lines (i.e., Cal-27, Fadu, SCC-9, and SCC-4) and in hNOKs. miR-101 expression in all OSCC cells was downregulated compared with hNOKs (Figure 1B). The Fadu and SCC-9 OSCC cell lines exhibited the lowest levels of miR-101 expression among all the tested OSCC cell lines and were thus selected for further studies. Hence, miR-101 may be an oncosuppressor in OSCC.
Figure 1.
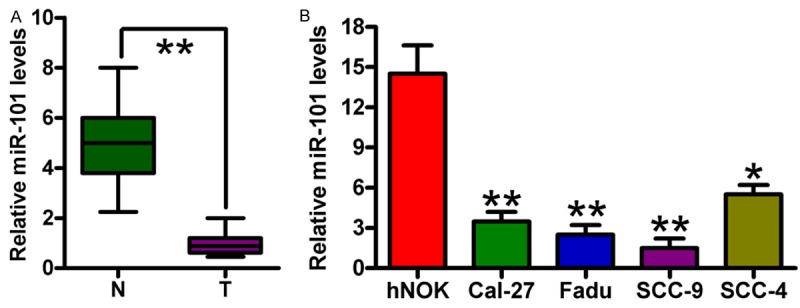
Expression levels of miR-101 in OSCC tissues and cell lines. A. miR-101 expression was analyzed in 66 pairs of OSCC and matched adjacent normal oral tissues through qPCR assay. B. qPCR assay was performed to determine miR-101 expression in the four OSCC cell lines (i.e., Cal-27, Fadu, SCC-9, and SCC-4) and hNOKs. U6 was used as endogenous control. Data are shown as means ± SD of three separate experiments. *P < 0.05, **P < 0.01. N = Normal; T = tumor.
miR-101 directly targets CXCR7 in the OSCC cells
CXCR7 is a putative target of miR-101 in breast cancer [11]. Considering that CXCR7 is involved in the pathogenesis of OSCC [17], we analyzed whether or not it was an function target of miR-101. Analysis using targeting algorithms (TargetScan and microRNA.org) indicated that CXCR7 could be a potential target gene of miR-101. Figure 2A illustrates the predicted miR-101 binding site in the 3’-UTR of CXCR7. The CXCR7 3’-UTR fragment containing the WT- or MUT-miR-101 binding sequence was cloned into the Renilla luciferase reporter and co-transfected with miR-NC or miR-101 mimics into Fadu or SCC-9 cells to validate interactions between miR-101 and CXCR7. miR-101 significantly reduced luciferase activity in WT-3’-UTR of CXCR7 compared with that in miR-NC-transfected cells. However, mutations in the miR-101 binding site almost restored the luciferase activity (Figure 2B). Overexpression of miR-101 markedly reduced the mRNA and protein levels of CXCR7 in the two OSCC cell lines (Figure 2C and 2D). Thus, miR-101 directly targets CXCR7 by downregulating its mRNA and protein expression.
Figure 2.
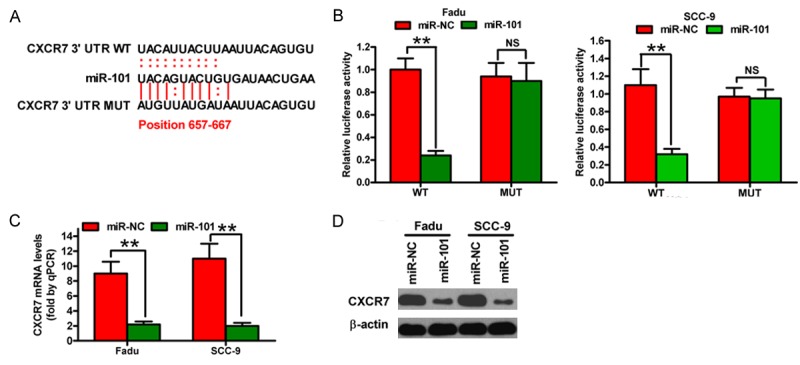
miR-101 directly targets CXCR7. (A) miR-101 binds to the predicted site of the 3’-UTR of CXCR7. (B) Dual-luciferase reporter gene analysis of CXCR7 3’-UTR in FaDu and SCC-9 cells following co-transfection of miR-NC or miR-101 mimics with pGL3 constructs containing WT- or MUT-3’-UTR of CXCR7. Relative luciferase activities are the ratios of Renilla luciferase normalized to the control mimics. (C) and (D) mRNA (C) and protein (D) levels of CXCR7 in the indicated cells transfected with 80 nM miR-NC or miR-101 mimics were measured by qPCR and Western blot assays. GAPDH and β-actin were used as internal controls. Data are shown as means ± SD of three separate experiments. **P < 0.01.
Overexpression of CXCR7 rescues miR-101-induced inhibition of proliferation and motility of OSCC cells
To understand the biological functions of miR-101 in OSCC cells, we transfected miR-NC or miR-101 mimics into OSCC cells. Transfection of miR-101 mimics into OSCC cells significantly inhibited cell viability (Figure 3A). The colony formation decreased in OSCC cells overexpressing miR-101 (Figure 3B and 3C). In addition, miR-101-treated cells induced a higher percentage of apoptotic cells than miR-NC-treated cells (Figure 3D). The transwell assays showed that the migration and invasion of OSCC cells were inhibited by miR-101 overexpression (Figure 3E-H). Furthermore, OSCC cells were co-transferred with miR-101 mimics and CXCR7-expressing plasmids to determine whether or not CXCR7 acts as a functional target of miR-101. As predicted, CXCR7 overexpression significantly restored reduced cell proliferation, migration, and invasion as well as increased apoptosis caused by ectopic expression of miR-101 in OSCC cells (Figure 3A-H). Therefore, miR-101 inhibition on OSCC cell proliferation and motility is dependent on CXCR7 expression.
Figure 3.
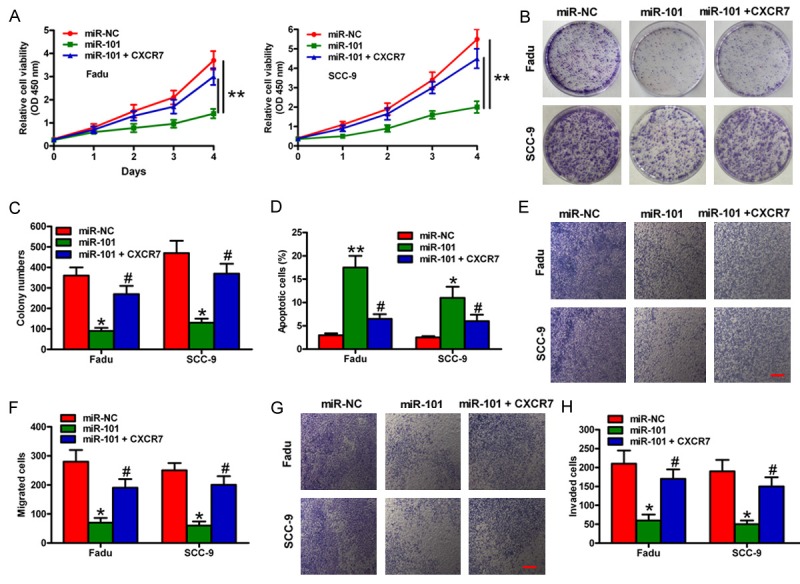
Effects of miR-101 or CXCR7 overexpression on the proliferation and motility of OSCC cells. Fadu and SCC-9 cells stimulated with 100 ng/mL CXCL12 were transfected with miR-NC, miR-101, or miR-101 mimics plus CXCR7-expressing plasmids. (A) MTT assay showing the viability of OSCC cells. (B) Colony formation assay showing the relative proliferation of OSCC cells. (C) Number of colonies was quantified in (B). (D) Cell apoptosis was assessed by flow cytometry assay. (E) and (G) Migration (E) and invasion (G) of OSCC cells were detected by transwell assays. (F) and (H) Numbers of migrated (F) and invaded (H) cells were calculated. Scale bar: 10 μm. Data are shown as means ± SD of three separate experiments. *P < 0.05, **P < 0.01 compared with the miR-NC group, #P < 0.05 compared with the miR-101 group.
miR-101 overexpression or CXCR7 depletion suppresses tumor growth and metastasis of OSCC in vivo
An in vivo xenograft mouse model was subcutaneously injected with miR-101-overexpressing or CXCR7-silencing stable Fadu cells (Figure 4A). The tumor volume (Figure 4B and 4C) and weight (Figure 4D) significantly decreased in both miR-101 overexpression and CXCR7 depletion groups compared with those in the vector control groups. In addition, the number of lung metastatic nodules in the miR-101-overexpressing or CXCR7-silencing stably transfected group was less than that in the control groups (Figure 4E). The miR-101 restoration or CXCR7 depletion also effectively increased the number of apoptotic cells in vivo (Figure 4F). Hence, miR-101 reduces OSCC tumor growth and lung metastasis by inhibiting CXCR7 expression in vivo.
Figure 4.
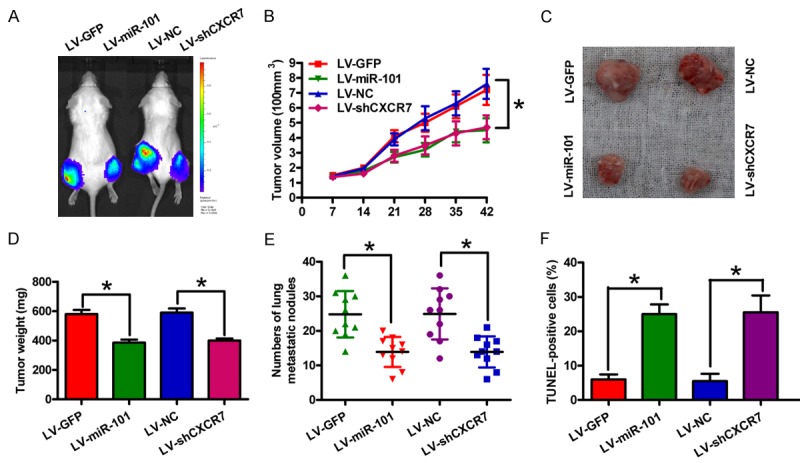
Effects of miR-101 overexpression or CXCR7 depletion on OSCC tumor growth and lung metastasis in vivo. miR-101-overexpressing or CXCR7-silencing stable Fadu cells were used to established an in vivo xenograft or metastasis mouse model by subcutaneous or venous injection. A. In vivo luciferase image detection of the xenograft tumor growth of miR-101 restoration or CXCR7 knockdown Fadu cells. B. Tumor burden was measured and calculated once a week after subcutaneous injection. C. At 6 weeks after subcutaneous inoculation, the tumors were collected and photographed. D. Tumor weight was measured 6 weeks after subcutaneous inoculation. E. Lung metastatic numbers were detected 8 weeks after venous injection. F. Apoptotic cells were assessed by TUNEL assay in tumor tissues from the xenograft models. Data are shown as means ± SD of three separate experiments. *P < 0.05.
CXCR7 is highly expressed and negatively correlated with the prognosis of OSCC
CXCR7 expression was investigated in 66 OSCC specimens through immunohistochemistry staining to explore the potential role of CXCR7 in this disease. Figure 5A shows that CXCR7 was highly expressed in OSCC tissues compared with that in adjacent normal tissues. The upregulation of CXCR7 mRNA and protein levels in OSCC tissues were confirmed using qPCR and Western blot analyses (Figure 5B and 5C). The OSCC tissues were subdivided into the CXCR7-high-expression and -low-expression groups according to the intensity of CXCR7 staining. Patients with high CXCR7 expression levels showed less favorable five-year overall survival (Figure 5D) than patients with low CXCR7 expression. Moreover, CXCR7 expression was significantly negatively correlated with miR-101 expression in OSCC tissues (Figure 5E). Hence, CXCR7 is a key mediator of OSCC progression.
Figure 5.
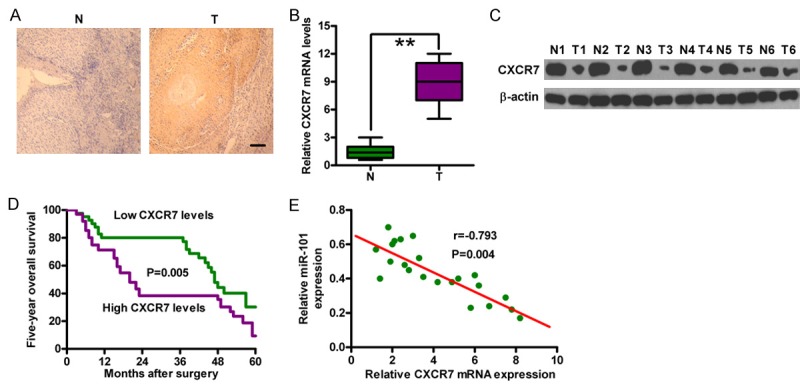
CXCR7 is highly expressed in OSCC tissues and negatively correlated with clinical outcomes. (A) Representative immunohistochemical images of CXCR7 staining in OSCC tissues and the corresponding normal tissues. Scale bar: 10 μm. (B) and (C) mRNA (B) and protein (C) expression of CXCR7 in OSCC tissues and the corresponding normal tissues were detected by qPCR and Western blot analyses. GAPDH and β-actin were used as internal controls. (D) Five-year overall survival of CXCR7 low- and high-expression groups of patients with OSCC. (E) Correlation between CXCR7 levels and miR-101 expression in patients with OSCC (n = 28). **P < 0.01. N = Normal; T = tumor.
Discussion
In this study, miR-101 was found to be a candidate tumor suppressor in OSCC by targeting CXCR7. The key findings are as follows. First, miR-101 was significantly downregulated in OSCC tissues and cell lines compared with the adjacent normal tissues or hNOKs. Second, CXCR7 was a direct target of miR-101. Third, miR-101 inhibited the proliferation, apoptosis resistance, migration, and invasion of OSCC cells in vitro. However, CXCR7 restored the tumor-suppressive effects of miR-101. Fourth, the xenograft and metastasis mouse models confirmed the findings in vitro. Fifth, CXCR7 was upregulated in OSCC tissues, and the high expression of CXCR7 was inversely correlated with miR-101 expression and poor prognosis of patients with OSCC.
miRNAs play critical roles in the regulation of cell proliferation, invasion, and migration. miRNA dysregulation is causally involved in the initiation and progression of human cancers [6,18]. miR-101 is downregulated and acts as a tumor suppressor in multiple cancers [9-12]. In the present study, miR-101 was under-expressed in OSCC tissues and cell lines; hence, miR-101 could function as an oncosuppressor in OSCC. Moreover, miR-101 restoration in Fadu and SCC-9 cells showed significant tumor-suppressing effects by inhibiting cell growth and metastasis in vitro and in vivo. Although the evidence highlighted the important roles of miR-101 in oral tongue squamous cell carcinoma, the underlying mechanism through which miR-101 inhibits OSCC remains unclear. Li et al [11] demonstrated that miR-101 inhibits breast cancer growth and metastasis by targeting CXCR7. Bioinformatics analysis was also performed to elucidate the tumor suppressive effects of miR-101 in OSCC. The results indicated that CXCR7 could be a potential target of miR-101.
CXCR7, initially named as receptor dog cDNA 1 (RDC1), was cloned from a canine thyroid cDNA library and identified as an atypical chemokine receptor [19]. RDC1, which was officially named CXCR7, is a novel receptor of the CXCL12 and initiates a cascade of signal transduction events [20]. Increasing evidence suggests that CXCR7 overexpression plays pivotal roles in malignancies; CXCR7 promotes tumor growth, angiogenesis, and distant metastases to lymph nodes and bone marrow [14,15]. CXCR7 promotes tumor growth and lung metastasis in mouse models of lung and breast cancers [21]. CXCR7 also enhances breast cancer cell proliferation and is correlated with lymph node metastasis and poor prognosis ofbreast cancer [11,22]. Moreover, CXCR7 regulates the tumor growth, angiogenesis, and invasion of human hepatocellular carcinoma cells [23]. CXCR7 is positively expressed in OSCC tissues. Furthermore, CXCR7 knockdown significantly inhibits the proliferation and motilities of OSCC cells [16,17]; hence, CXCR7 plays important roles in OSCC occurrence and development. The mRNA and protein expression levels of CXCR7 were upregulated in OSCC tissues. The high expression level of CXCR7 was negatively correlated with miR-101 expression and poor outcomes of patients with OSCC. The tumor suppressive effects of miR-101 were restored by CXCR7 overexpression in vitro and mimicked by CXCR7 depletion in vivo. Thus, the anti-tumor effects of miR-101 on malignant phenotypes of OSCC are partly mediated by CXCR7 silencing.
In summary, miR-101 was significantly downregulated in OSCC tissues and cell lines. miR-101 inhibited OSCC cell growth, invasion, and migration in vitro and reduced tumor growth and lung metastasis in vivo by targeting CXCR7. miR-101, as a tumor suppressor in OSCC, may thus serve as a potential biomarker for OSCC diagnosis and as a new target for OSCC therapy.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Liu CJ, Liu TY, Kuo LT, Cheng HW, Chu TH, Chang KW, Lin SC. Differential gene expression signature between primary and metastatic head and neck squamous cell carcinoma. J Pathol. 2008;214:489–497. doi: 10.1002/path.2306. [DOI] [PubMed] [Google Scholar]
- 3.Hu J, He Y, Yan M, Zhu C, Ye W, Zhu H, Chen W, Zhang C, Zhang Z. Dose dependent activation of retinoic acid-inducible gene-I promotes both proliferation and apoptosis signals in human head and neck squamous cell carcinoma. PLoS One. 2013;8:e58273. doi: 10.1371/journal.pone.0058273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman HT, Porter K, Karnell LH, Cooper JS, Weber RS, Langer CJ, Ang KK, Gay G, Stewart A, Robinson RA. Laryngeal cancer in the United States: changes in demographics, patterns of care, and survival. Laryngoscope. 2006;116:1–13. doi: 10.1097/01.mlg.0000236095.97947.26. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Wu BH, Xiong XP, Jia J, Zhang WF. MicroRNAs: new actors in the oral cancer scene. Oral Oncol. 2011;47:314–319. doi: 10.1016/j.oraloncology.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 8.Manikandan M, Deva Magendhra Rao AK, Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R, Munirajan AK. Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol Cancer. 2016;15:28. doi: 10.1186/s12943-016-0512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He XP, Shao Y, Li XL, Xu W, Chen GS, Sun HH, Xu HC, Xu X, Tang D, Zheng XF, Xue YP, Huang GC, Sun WH. Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase-2 overexpression and tumor growth. FEBS J. 2012;279:4201–4212. doi: 10.1111/febs.12013. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, Wang J, Mao Y, Zou B, Fan X. MicroRNA-101 suppresses migration and invasion via targeting vascular endothelial growth factor-C in hepatocellular carcinoma cells. Oncol Lett. 2016;11:433–438. doi: 10.3892/ol.2015.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li JT, Jia LT, Liu NN, Zhu XS, Liu QQ, Wang XL, Yu F, Liu YL, Yang AG, Gao CF. MiRNA-101 inhibits breast cancer growth and metastasis by targeting CX chemokine receptor 7. Oncotarget. 2015;6:30818–30830. doi: 10.18632/oncotarget.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng M, Jiang YP, Chen W, Li KD, Liu X, Gao SY, Feng H, Wang SS, Jiang J, Ma XR, Cen X, Tang YJ, Chen Y, Lin YF, Tang YL, Liang XH. Snail and Slug collaborate on EMT and tumor metastasis through miR-101-mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget. 2015;6:6797–6810. doi: 10.18632/oncotarget.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, Miao Z, Penfold ME, Sunshine MJ, Littman DR, Kuo CJ, Wei K, McMaster BE, Wright K, Howard MC, Schall TJ. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun X, Cheng G, Hao M, Zheng J, Zhou X, Zhang J, Taichman RS, Pienta KJ, Wang J. CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010;29:709–722. doi: 10.1007/s10555-010-9256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keeley EC, Mehrad B, Strieter RM. CXC chemokines in cancer angiogenesis and metastases. Adv Cancer Res. 2010;106:91–111. doi: 10.1016/S0065-230X(10)06003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia J, Wang J, Chen N, Dai Y, Hong Y, Chen X, Cheng B. Expressions of CXCR7/ligands may be involved in oral carcinogenesis. J Mol Histol. 2011;42:175–180. doi: 10.1007/s10735-011-9322-x. [DOI] [PubMed] [Google Scholar]
- 17.Chen N, Jiang X, Wang J, Wu T, Cheng B, Xia J. CXCL12-CXCR4/CXCR7 axis contributes to cell motilities of oral squamous cell carcinoma. Tumour Biol. 2016;37:567–575. doi: 10.1007/s13277-015-3803-6. [DOI] [PubMed] [Google Scholar]
- 18.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 19.Libert F, Parmentier M, Lefort A, Dumont JE, Vassart G. Complete nucleotide sequence of a putative G protein coupled receptor: RDC1. Nucleic Acids Res. 1990;18:1917. doi: 10.1093/nar/18.7.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eva C, Sprengel R. A novel putative G protein-coupled receptor highly expressed in lung and testis. DNA Cell Biol. 1993;12:393–399. doi: 10.1089/dna.1993.12.393. [DOI] [PubMed] [Google Scholar]
- 21.Miao Z, Luker KE, Summers BC, Berahovich R, Bhojani MS, Rehemtulla A, Kleer CG, Essner JJ, Nasevicius A, Luker GD, Howard MC, Schall TJ. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci U S A. 2007;104:15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boudot A, Kerdivel G, Habauzit D, Eeckhoute J, Le Dily F, Flouriot G, Samson M, Pakdel F. Differential estrogen-regulation of CXCL12 chemokine receptors, CXCR4 and CXCR7, contributes to the growth effect of estrogens in breast cancer cells. PLoS One. 2011;6:e20898. doi: 10.1371/journal.pone.0020898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng K, Li HY, Su XL, Wang XY, Tian T, Li F, Ren GS. Chemokine receptor CXCR7 regulates the invasion, angiogenesis and tumor growth of human hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2010;29:31. doi: 10.1186/1756-9966-29-31. [DOI] [PMC free article] [PubMed] [Google Scholar]


