Abstract
Recent evidence has demonstrated that A kinase interacting protein 1 (AKIP1), a molecular regulator of protein kinase A, was overexpressed in breast cancer. However, the prognostic and biological role of AKIP1 in breast cancer is still elusive. The purpose of our study was to elucidate the role and molecular mechanism of AKIP1 in breast cancer development. The mRNA levels of AKIP1 in breast cancer and paired normal breast tissues were examined by quantitative real-time PCR. The relationship of AKIP1 expression with clinicopathological characteristics and clinical prognosis of breast cancer patients was investigated. In vitro migration and invasion assays were performed in MCF-7 and SK-BR-3 cells to determine its role in metastasis and the possible mechanism. The result showed that AKIP1 expression was up-regulated in breast cancer tissues compared with that in normal breast tissues. High expression of AKIP1 was associated significantly with advanced tumor stage (P<0.001), tumor size (P=0.029), and lymph node metastasis (P=0.004). Moreover, overexpression of AKIP1 was significantly correlated with poor overall survival and recurrence-free survival (P=0.038 and P=0.005, respectively). Furthermore, down-regulation of AKIP1 remarkably inhibited breast cancer cell motility and invasion through inhibiting the Akt/GSK-3β/Snail pathway. Therefore, AKIP1 may represent a prospective prognostic indicator and a potential therapeutic target of breast cancer.
Keywords: AKIP1, breast cancer, prognosis, metastasis
Introduction
Breast cancer represents the most common cancer in women in the United States and about 246,660 new cases of invasive breast cancer are expected to be diagnosed, and 40,450 women will die from breast cancer in the United States in 2016 [1]. Over the past decades, diagnosis and management of breast cancer have improved through combined efforts in surgery, radiotherapy and chemotherapy, but the clinical survival rate for patients with advanced stage diseases is still unfavorable [2]. For patients with distant metastasis, the 5-year survival rate is less than 20% [3]. Therefore, it is crucial to develop more effective screening and enhance our ability to predict the progression and survival of the disease. In addition to conventional clinicopathological parameters, molecular markers may provide an alternative approach.
A kinase interacting protein 1 (AKIP1) is a 23 kDa protein originally identified in mRNA screens of breast and prostate cancer cell lines [4]. AKIP1 has three splice variants, the full length protein (AKIP1a), one that lacks the third exon (AKIP1b), and one that lacks the third and fifth exon (AKIP1c). As it has no significant homologies to other described proteins or particular catalytic domains, it has been supposed to have a role as an adaptor or structural intracellular protein [5]. AKIP1 has been shown to localize to the cytoplasm, nucleus, and mitochondria and associate with proteins with different sub-cellular localizations [6]. The literature on the biochemical and biological roles of AKIP1 is quite limited. Recently, it has identified as a potential factor controlling stress adaptation in the heart, as overexpression of AKIP1 in the heart protected against ischemia/reperfusion and improved cardiac function [7]. Besides, AKIP1a has been shown to scaffold NF-κB in a PKA phosphorylation dependent manner and enhance transcription [8]. In contrast, AKIP1b was shown to recruit the histone deacetylase silent mating type information regulation 2 homolog (SIRT1) in a NEDDylation dependent manner and repress transcription [9]. AKIP1 has also been shown as a novel PKAc binding protein that targets PKAc to specific locations within cells, and is therefore hypothesized to be a putative molecular integrator regulating myocyte death and survival [6]. Recent work demonstrated that AKIP1 behaved as an oncogene leading to the tumorigenesis and invasiveness. In particular, it acts as a molecular regulator of protein kinase A and nuclear factor kappa B signaling in breast cancer [10]. However, the clinicopathological and biological roles of AKIP1 in breast cancer remain largely unknown.
In the present study, we investigated AKIP1 protein expression and its correlation with clinicopathologic features and clinical outcomes in breast cancer samples, and reported that AKIP1 might induce breast cancer metastasis through regulating Akt/GSK-3β/Snail signaling pathway. These data might provide information for the prediction of breast cancer prognosis and the establishment of targeted therapies.
Materials and methods
Cell lines
Breast cancer cell lines MCF-7 and SK-BR-3 were grown in the DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 100 μg/μL streptomycin and 100 μg/μL penicillin in a humidified incubator containing 5% CO2 at 37°C.
Patients and samples
Fresh tumor tissue with paired non-cancerous tissue samples of 10 breast cancer patients were obtained in operation from the Maternal and Child Health Hospital of Guangxi Zhuang Autonomous Region. A total of 150 paraffin-embedded breast cancer samples, which were histologically and clinically diagnosed in patients with radical surgery in Children’s Hospital of Guangxi Zhuang Autonomous Region, between 1998 and 2004, were also included in this study. The clinical and pathologic parameters were reviewed from impatient medical records and presented in Table 1. All of the enrolled participants had intact follow-up information and were not exposed to radiotherapy or chemotherapy prior to surgical resection. Clinical and pathological data of the 150 patients with breast cancer were collected, such as age, tumor stage, differentiation grade, lymph node metastases, pathologic characteristics and recurrence. Clinical stage of breast cancer was classified according to the American Joint Committee on Cancer guidelines. This study was reviewed and approved by the Institutional Medical Ethics Committee of Maternal and Child Health Hospital of Guangxi Zhuang Autonomous Region and written informed consent from each participant was obtained.
Table 1.
Patients characteristics and AKIP1 expression
| Characteristic | Total | AKIP1 | p | ||
|---|---|---|---|---|---|
|
| |||||
| Low | High | ||||
| Age | ≤50 y | 50 | 28 | 22 | 0.728 |
| >50 y | 100 | 53 | 47 | ||
| Side | Left | 79 | 46 | 33 | 0.990 |
| Right | 71 | 35 | 36 | ||
| Tumor stage | I | 70 | 50 | 20 | <0.001 |
| II-IV | 80 | 31 | 49 | ||
| Tumor size | ≤2 cm | 71 | 45 | 26 | 0.029 |
| >2 cm | 79 | 36 | 43 | ||
| Differentiation | 1/2 | 45 | 26 | 19 | 0.543 |
| 3 | 105 | 55 | 50 | ||
| Histological type | DC | 128 | 71 | 57 | 0.384 |
| others | 22 | 10 | 12 | ||
| LN metastasis | No | 71 | 47 | 24 | 0.004 |
| Yes | 79 | 34 | 45 | ||
DC: ductal carcinoma; LN: lymph nodes.
RNA extraction and real-time PCR
Total RNA from human cancer/non-cancerous tissues and breast cancer cells was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized from 1 mg of total RNA by use of the SuperScriptH III First-Strand Synthesis System (Invitrogen). Real-time PCR was carried out using an CFX96 Real-Time System (BIO-RAD). SYBR green 2X master mixture (Invitrogen) was used in a total volume of 10 mL. The primer sequences were as follows: AKIP1 sense, 5’-GAAGGATCCGTCGACATGGAATACTGCCTGGCGGC-3’; antisense, 5’-GAACTCGAGTCATACGGGGAACACCAAGTCCAC-3’; Snail sense, 5’-ACCACTATGCCGCGCTCTT-3’, antisense, 5’-GGTCGTAGGGCTGCTGGAA-3’; GAPDH sense, 5’-GAAGGTGAAGGTCGGAGTC-3’, antisense, 5’-GAAGATGGTGATGGGATTTC-3’. GAPDH was used as an internal control. All reactions were run in triplicate in three independent experiments.
Small RNA transfection
Stealth small interfering RNAs (siRNAs) targeting AKIP1 and control siRNA were designed by Invitrogen. Cells were transfected with siRNA with Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Final concentrations of siRNAs were 100 nM, and the cells were harvested 72 h after transfection.
Immunohistochemical analysis
Immunohistochemical staining was carried out on 4-mm thick sections of the paraffin-embedded tissues, and the staining process was strictly performed according to the streptavidin-biotinperoxidase complex method. After deparaffinisation and rehydration, the sections were treated with 3% hydrogen peroxide to block endogenous peroxidase. Non-specific binding was blocked with normal goat serum for 30 min at 37°C, then the sections were incubated with rabbit polyclonal anti-AKIP1 (1:100; Sigma, USA) antibody overnight at 4°C. After washing with PBS, the sections were incubated with a horseradish peroxidase-labeled polymerconjugated anti-mouse secondary antibody (Zymed) at 37°C for 30 min. Then the sections were stained with 3, 3-diaminobenzidine tetrahydrochloride for 5 min and nuclei were counterstained with hematoxylin for 3 min, and then mounted with neutral balsam. PBS was used to replace the antibody as a negative control.
The immunostaining results were evaluated independently by two pathologists blinded to the clinical parameters, and the final score was the average of the scores by two observers. We used the intensity and extent of the staining to evaluate the expression of AKIP1. The staining intensity was scored as 0 (negative), 1 (weak staining exhibited as light yellow), 2 (moderate staining exhibited as yellow brown), 3 (strong staining exhibited as brown). Extent of staining was scored as: 1 (<10% positive tumor cells); 2 (10-50% positive tumor cells); 3 (51-80% positive tumor cells), and 4 (>80% positive tumor cells), according to the percentages of the positive staining areas relative to the whole cancer area or entire section for the normal samples. The sum of intensity and extent score was used as the final staining scores (0 to 7) for AKIP1. For the purpose of statistical evaluation, tumors having a final staining score of <4 classified tumors with low AKIP1 expression and score ≥4 classified as high AKIP1 expression.
Western blotting
Cell lysates were prepared by a SDS lysis buffer. Protein concentration was measured using the BCA protein assay kit (Pierce, Rockford, IL). Equal amounts of protein were separated by electrophoresis on a 12% sodium dodecyl sulfate-PAGE gel. The proteins were electro-transferred from gel to polyvinylidene difluoride membrane (Millipore, Bedford, MA). The membrane was blocked with 5% dry milk solution for 30 min, and then incubated with p-GSK-3β, GSK-3β, p-Akt, Akt (Cell Signaling Technology, USA), and Snail (sigma, USA) antibodies for 2 hours at room temperature. After washing, the membrane was incubated with Horseradish Peroxidase conjugating secondary antibodies. Finally, membrane was detected with an enhanced chemiluminescence kit (Amersham Bioscience, Piscataway, NJ) according to the manufacturer’s instructions.
Wound healing assay
Cells were seeded in 6-well culture plates and cultured under permissive conditions until 90% confluence. Then the confluent cell monolayer was scratched with a sterile plastic pipette tip to produce a straight line. The debris was removed and the edge of the scratch was smoothed with PBS washing. Photographic images were taken at 0 and 24 h along the scrape line by microscope. Results were expressed as relative scratch width, based the distance migrated relative to the original scratched distance. Experiments were repeated a minimum of three times.
Cell migration and invasion assay
Cell migration ability was measured in Corning Transwell insert chambers. Briefly, cells in serum-free medium were plated in the upper chamber of the insert without Matrigel. Medium with 10% fatal bovine serum was added into the lower chambers as a chemoattractant. After 24 h incubation, the non-invading cells attached to the upper surface of the membrane were removed with a sterile cotton swab, and the migration cells penetrated through the membrane were stained with 0.1% crystal violet for 20 min at room temperature. Cell invasion ability was conducted in a modified 24-well Boyden chamber. The inserts were coated with 1 mg/mL Matrigel matrix according to the manufacturer’s recommendations. Methods used in cell invasion assay were similar to those in the cell migration assay. Three independent experiments were performed and the data are presented as the mean ± SD.
Statistical analysis
Statistical analyses were performed using a statistical software package (SPSS 18.0, SPSS Inc, Chicago, IL, USA). The significance of AKIP1 levels was determined by Student’s t-test. The chi-square test was used to analyze the relationship between AKIP1 expression and clinicopathological characteristics. Survival times were evaluated using the Kaplan-Meier survival curves, and compared by the log-rank test. Multivariate analysis was performed using a Cox’s proportional hazards regression model. P<0.05 were considered to be statistically significant.
Results
AKIP1 is overexpressed in breast cancer
To determine the AKIP1 expression pattern in breast cancer tissues, real-time RT-PCR was performed in 10 pairs of breast cancer and adjacent non-cancerous tissues. In all the 10 patients, increased levels of AKIP1 mRNA expression were detected in all the cancer tissues in comparison to the normal breast tissues (Figure 1). The representative immunostaining of AKIP1 in breast cancer tissues was shown in Figure 2. These data suggested that AKIP1 might serve as an oncogene in breast cancer.
Figure 1.
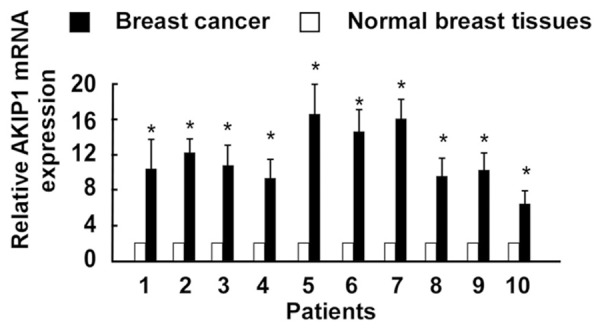
Expression of AKIP1 is overexpressed in breast cancer tissues compared with normal breast tissues through quantitative real-time PCR. (*P<0.05).
Figure 2.
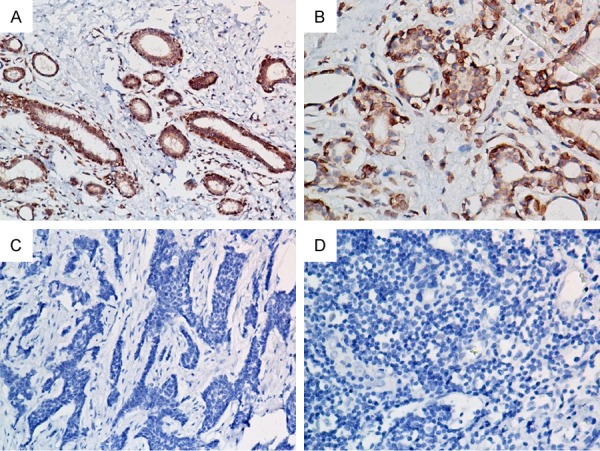
Representative immunohistochemical staining of AKIP1 in breast cancer tissues. (A, B) High AKIP1 expression in breast cancer tissues. (C, D)Low AKIP1 expression in breast cancer tissues. Magnifications: 200× for (A and C), 400× for (B and D).
Correlation between AKIP1 expression and clinicopathological features
The clinical significances of AKIP1 overexpression in patients with breast cancer were summarized in Table 1. The results showed that AKIP1 expression levels were significantly different between groups in regards to tumor stage (P<0.001), tumor size (P=0.029), and lymph node metastasis (P=0.004). However, there was no significant association between AKIP1 expression and age, tumor side, histological grade, and histological type.
Correlation of AKIP1 expression with clinical prognosis
To evaluate the impact of AKIP1 expression and clinicopathological features on clinical outcomes of the patients, we used Kaplan-Meier analysis and the log-rank test for censored survival data. As displayed in Figure 3, Kaplan-Meier survival curves showed that patients with high AKIP1 expression had poorer overall survival compared to those with low AKIP1 expression (P=0.038), recurrence-free survival also gradually declined with increasing AKIP1 expression scores (P=0.005). Multivariate Cox proportional hazards model analysis showed that AKIP1 could be used as a potential prognostic factor for overall and recurrence-free survival in breast cancer patients (P=0.022 and P=0.018, respectively) (Table 2).
Figure 3.
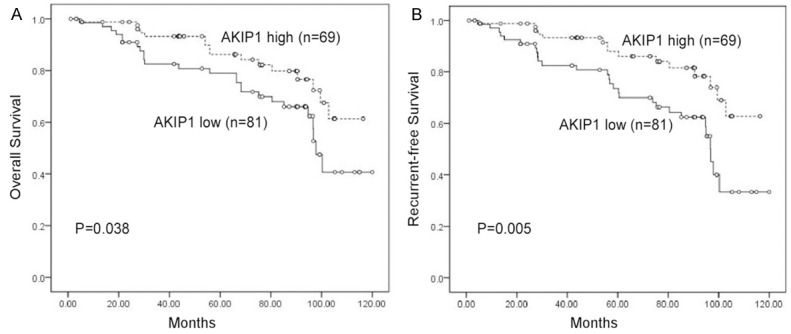
Kaplan-Meier survival curves of patients with breast cancer according to AKIP1 expression. A. Overall survival (OS). B. Recurrence-free survival (RFS).
Table 2.
Multivariate analyses of OS and RFS in relation to clinical parameters
| Prognostic variables | OS | RFS | ||
|---|---|---|---|---|
|
| ||||
| Hazard Ratio (95% CI) | P | Hazard Ratio (95% CI) | P | |
| Age (>50 y vs ≤50 y) | 0.547 (0.309-1.759) | 0.535 | 0.735 (0.207-1.488) | 0.487 |
| Side (Left vs Right) | 1.235 (0.246-1.147) | 0.481 | 1.153 (0.167-1.391) | 0.447 |
| Tumor Stage (II-IV vs I) | 2.194 (0.589-5.383) | 0.016 | 2.208 (0.797-5.424) | 0.059 |
| Tumor size (>2 cm vs ≤2 cm) | 2.736 (0.516-6.767) | 0.065 | 2.838 (0.572-7.868) | 0.094 |
| Tumor grade (Grade 3 vs 1/2) | 1.840 (0.084-2.979) | 0.205 | 1.760 (0.206-2.838) | 0.345 |
| Histological type (DC vs others) | 1.266 (0.265-4.225) | 0.578 | 1.465 (0.726-4.276) | 0.513 |
| LN metastasis (yes vs no) | 3.343 (1.173-7.618) | 0.007 | 3.375 (1.109-7.7) 43 | 0.015 |
| AKIP1 expression (high vs low) | 2.891 (1.097-4.216) | 0.022 | 2.925 (1.136-4.357) | 0.018 |
DC: ductal carcinoma; LN: lymph nodes.
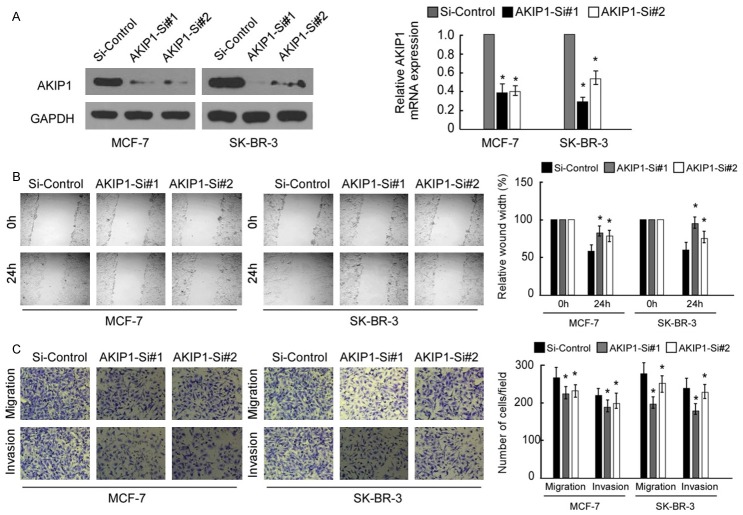
Downregulation of AKIP1 inhibits the migration and invasion of breast cancer cell lines
To evaluate the impact of AKIP1 on the migration and invasion abilities of breast cancer cell, we knocked down the expression of AKIP1 using its specific siRNA(s) in breast cancer cells MCF-7 and SK-BR-3. Efficient silencing of AKIP1 expression in these cells was verified by qRT-PCR (Figure 4A). Both the si-AKIP1-transfected MCF-7 and SK-BR-3 cells had significantly slower closure of the wound area compared with their respective control cells (Figure 4B, both P<0.05). This result was confirmed using Boyden’s chamber assay. The migration potentials of both cells were both significantly reduced (Figure 4C, both P<0.05), and suppression of AKIP1 in both cell lines inhibited the cell invasion abilities in the Matrigel substrate as well (Figure 4C, both P<0.05).
Figure 4.
Knock down of AKIP1 inhibits the migration and invasion capacities of breast cancer cells. A. The transfection efficiency of AKIP1 was analyzed by measuring transcript levels using qRT-PCR analyses in MCF-7 and SK-BR-3 cells. B. Effect of AKIP1 knockdown in cell migration of MCF-7 and SK-BR-3 was determined by wound healing assay (left panels), the uncovered areas in the wound healing assays were quantified as a percentage of the original wound area (right panels). C. Transwell cell migration assay and Matrigel invasion assay of breast cancer cells transfected with Si-Control, AKIP1-Si#1, and AKIP1-Si#2 (left panels). Quantifications of migrated cells through the membrane and invaded cells through Matrigel of each cell line are shown as proportions of their vector controls (right panels). Bar graphs show the statistical analysis of three independent experiments (*P<0.05).
AKIP1 knockdown inhibits Akt/GSK-3β/Snail signaling pathway
In our study, we first examined the expression levels of relative genes of the Akt/GSK-3β/Snail signaling in MCF-7 and SK-BR-3 cell lines with AKIP1 knockdown by real-time PCR and western blot. As shown in Figure 5A, silencing of AKIP1 reduced the expression of phosphorylated Akt, but not total Akt, while the expression of Snail and phosphorylated GSK-3β, but not total GSK-3β, were also down-regulated. Additionally, the transcriptional level of Snail was decreased by AKIP1 suppression (Figure 5B). Taken together, our results suggest that AKIP1 mediates migration and invasion in breast cancer cells via regulation of the Akt/GSK-3β/Snail signaling pathway.
Figure 5.
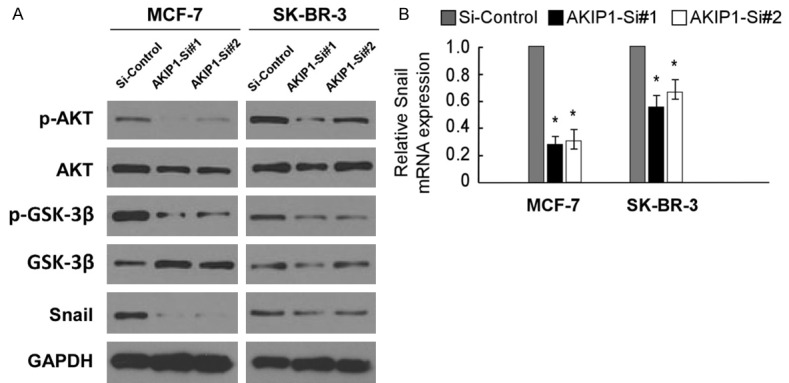
Down-regulation of AKIP1 inhibited Akt/GSK-3β/Snail signaling pathway. A. MCF-7 and SK-BR-3 cells were treated with AKIP1 si-RNA, and control si-RNA, following which cells were harvested for analysis of AKIP1, p-GSK-3β, GSK-3β, p-Akt, Akt, and Snail level. B. Real-time PCR analysis shows AKIP1 expression in AKIP1-silenced breast cancer cells. Bar graphs show the statistical analysis of 3 independent experiments (*P<0.05).
Discussion
In this study, we found that the expression of AKIP1 was elevated in breast cancer tissues in compared with normal breast tissues by Real-time PCR. We also found that the expression of AKIP1 was associated with poor overall survival and recurrent-free survival in breast cancer patients. Moreover, overexpression of AKIP1 was correlated with clinical stage and lymph nodes status. Furthermore, suppression of AKIP1 repressed cellular motility and invasion through inhibiting Akt/GSK-3β/Snail signal pathway. The current study suggests that AKIP1 may contribute to the progression of breast cancer progression.
As a kinase interacting protein, AKIP1 localizes to the cytoplasm, nucleus, and mitochondria and associates with several cell signaling factors, including PKA, NFκB, apoptin, RAC1, TAP73 and AIF [11], suggesting that AKIP1 might have multiple functions in cells. Gao demonstrated that AKIP1 suppressed NFkappaB-dependent transcription through its ability to bind to p65 and the cyclin D1 promoter in a neddylation-dependent manner, which influenced life span in yeast and worms [9]. Yu et al. observed perinuclear co-localization of AKIP1 and Rac1 in CSF-1-treated neonatal rat osteoclasts but not in resting osteoclasts, indicating that AKIP1 may influence the ability of Rac1 to remodel the actin cytoskeleton [12]. In the subsequent study, they revealed that AKIP1 is induced in hypertrophic hearts and can stimulate adaptive cardiomyocyte growth, which involves Akt signaling [13]. They also reported that AKIP1 overexpression improves mitochondrial function to enhance respiration without excess superoxide generation, thereby implicating a role for AKIP1 in mitochondrial stress adaptation [11]. Recently, AKIP1 has become a focus in tumor research. It is worth noting that Lin et al. found that Nkx2-8 repressed NF-κB activity by restraining nuclear localization of NF-κB p65 via downregulation of AKIP1 in esophageal squamous cell carcinoma cells [14]. They also found that overexpressing AKIP1 induces, whereas silencing AKIP1 reduces, ESCC angiogenesis and lymphangiogenesis, and that it transcriptionally upregulates vascular endothelial growth factor-C (VEGF-C) via interaction with its promoter through cooperation with multiple transcriptional factors [15]. Early evidence showed that AKIP1 is mainly overexpressed in breast cancer cells, suggesting that AKIP1 might act as a potent oncogenic protein in breast cancer development. However, the expression of AKIP1 in breast cancer tissues and its potential clinical and biological significances have not been described.
In the present study, we characterized the expression pattern of AKIP1 via real-time PCR in 10 pairs of breast cancers and normal breast tissues. The results showed that AKIP1 was highly expressed in breast cancer tissues, but relatively low expression was found in normal tissues, suggesting that AKIP1 expression was significantly associated with the carcinogenesis and the development of breast cancer. Moreover, the data of present study showed that the expression of AKIP1 protein was tightly related with clinical stage, tumor size, lymphatic metastasis, and clinical prognosis. Previous studies have described an oncogenic role of AKIP1 in tumor development. Leung et al. demonstrated that AKIP1 is a protein partner of TAp73, and that they cooperate with each other to exert tumor suppressive functions and sensitize the response of cervical cancer cells to radiotherapy [16]. Zimmerman et al. showed that AKIP1 interacts with the apoptosis-inducing protein Apoptin in triggering apoptin-induced tumor-selective cell death in a large panel of human tumors [17]. The presented data herein is consistent with previous reports suggest that AKIP1 may play a substantial role in tumor biological aggressiveness.
As an essential regulator of cell growth, AKIP1 has been shown to be involved in the regulation of cancer cell metastasis, while the mechanism by which AKIP1 contributes to tumor cell migration is not well understood. In the present study, we found that knockdown of AKIP1 with specific siRNA in MCF-7 and SK-BR-3 cell lines can decrease cancer cell migration and invasion. Moreover, we for the first time, found that downregulation of AKIP1 remarkably repressed cellular motility and invasion by inhibiting Akt/GSK-3β/Snail pathway. Our results was supported by Yu et al., who demonstrated that Akt kinase signaling pathways have been linked to hypertrophic growth and AKIP1 specifically induced phosphorylation of Akt [13]. Likewise, Lin et al. indicated that AKIP1 might promote angiogenesis and lymphangiogenesis of esophageal squamous cell carcinoma via interaction with its promoter through cooperation with multiple transcriptional factors, including SP1, AP2 and nuclear factor-κB (NF-κB) [15]. These findings highlighted the underlying molecular mechanism of AKIP1 in cancer metastasis.
In conclusion, we have demonstrated that the elevated expression of AKIP1 in breast cancer contributes to aggressiveness and metastasis through Akt/GSK-3β/Snail pathway. Meanwhile, high AKIP1 expression in breast cancer significantly correlates with patients’ shorter survival time. Therefore, AKIP1 has potential as an attractive prognostic marker and a new target for breast cancer treatment.
Disclosure of conflict of interest
None.
References
- 1.Machida Y, Tozaki M, Shimauchi A, Yoshida T. Breast density: the trend in breast cancer screening. Breast Cancer. 2015;22:253–261. doi: 10.1007/s12282-015-0602-2. [DOI] [PubMed] [Google Scholar]
- 2.Saad ED, Katz A, Buyse M. Overall survival and post-progression survival in advanced breast cancer: a review of recent randomized clinical trials. J. Clin. Oncol. 2010;28:1958–1962. doi: 10.1200/JCO.2009.25.5414. [DOI] [PubMed] [Google Scholar]
- 3.Donohoe J, Marshall V, Tan X, Camacho FT, Anderson R, Balkrishnan R. Predicting Late-stage Breast Cancer Diagnosis and Receipt of Adjuvant Therapy: Applying Current Spatial Access to Care Methods in Appalachia. Med Care. 2015;53:980–988. doi: 10.1097/MLR.0000000000000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitching R, Li H, Wong MJ, Kanaganayakam S, Kahn H, Seth A. Characterization of a novel human breast cancer associated gene (BCA3) encoding an alternatively spliced proline-rich protein. Biochim Biophys Acta. 2003;1625:116–121. doi: 10.1016/s0167-4781(02)00562-6. [DOI] [PubMed] [Google Scholar]
- 5.Leon DA, Canaves JM. In silico study of breast cancer associated gene 3 using LION Target Engine and other tools. Biotechniques. 2003;35:1222–1226. 1228, 1230–1231. [PubMed] [Google Scholar]
- 6.Sastri M, Barraclough DM, Carmichael PT, Taylor SS. A-kinase-interacting protein localizes protein kinase A in the nucleus. Proc Natl Acad Sci U S A. 2005;102:349–354. doi: 10.1073/pnas.0408608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sastri M, Haushalter KJ, Panneerselvam M, Chang P, Fridolfsson H, Finley JC, Ng D, Schilling JM, Miyanohara A, Day ME, Hakozaki H, Petrosyan S, Koller A, King CC, Darshi M, Blumenthal DK, Ali SS, Roth DM, Patel HH, Taylor SS. A kinase interacting protein (AKIP1) is a key regulator of cardiac stress. Proc Natl Acad Sci U S A. 2013;110:E387–396. doi: 10.1073/pnas.1221670110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao N, Asamitsu K, Hibi Y, Ueno T, Okamoto T. AKIP1 enhances NF-kappaB-dependent gene expression by promoting the nuclear retention and phosphorylation of p65. J Biol Chem. 2008;283:7834–7843. doi: 10.1074/jbc.M710285200. [DOI] [PubMed] [Google Scholar]
- 9.Gao F, Cheng J, Shi T, Yeh ET. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat Cell Biol. 2006;8:1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 10.Gao N, Hibi Y, Cueno M, Asamitsu K, Okamoto T. A-kinase-interacting protein 1 (AKIP1) acts as a molecular determinant of PKA in NF-kappaB signaling. J Biol Chem. 2010;285:28097–28104. doi: 10.1074/jbc.M110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H, Tigchelaar W, Koonen DP, Patel HH, de Boer RA, van Gilst WH, Westenbrink BD, Sillje HH. AKIP1 expression modulates mitochondrial function in rat neonatal cardiomyocytes. PLoS One. 2013;8:e80815. doi: 10.1371/journal.pone.0080815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu KP, Itokawa T, Zhu ML, Syam S, Seth A, Insogna K. Breast cancer-associated gene 3 (BCA3) is a novel Rac1-interacting protein. J Bone Miner Res. 2007;22:628–637. doi: 10.1359/jbmr.070105. [DOI] [PubMed] [Google Scholar]
- 13.Yu H, Tigchelaar W, Lu B, van Gilst WH, de Boer RA, Westenbrink BD, Sillje HH. AKIP1, a cardiac hypertrophy induced protein that stimulates cardiomyocyte growth via the Akt pathway. Int J Mol Sci. 2013;14:21378–21393. doi: 10.3390/ijms141121378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C, Song L, Gong H, Liu A, Lin X, Wu J, Li M, Li J. Nkx2-8 downregulation promotes angiogenesis and activates NF-kappaB in esophageal cancer. Cancer Res. 2013;73:3638–3648. doi: 10.1158/0008-5472.CAN-12-4028. [DOI] [PubMed] [Google Scholar]
- 15.Lin C, Song L, Liu A, Gong H, Lin X, Wu J, Li M, Li J. Overexpression of AKIP1 promotes angiogenesis and lymphangiogenesis in human esophageal squamous cell carcinoma. Oncogene. 2015;34:384–393. doi: 10.1038/onc.2013.559. [DOI] [PubMed] [Google Scholar]
- 16.Leung TH, Ngan HY. Interaction of TAp73 and breast cancer-associated gene 3 enhances the sensitivity of cervical cancer cells in response to irradiation-induced apoptosis. Cancer Res. 2010;70:6486–6496. doi: 10.1158/0008-5472.CAN-10-0688. [DOI] [PubMed] [Google Scholar]
- 17.Zimmerman R, Peng DJ, Lanz H, Zhang YH, Danen-Van Oorschot A, Qu S, Backendorf C, Noteborn M. PP2A inactivation is a crucial step in triggering apoptin-induced tumor-selective cell killing. Cell Death Dis. 2012;3:e291. doi: 10.1038/cddis.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]