ABSTRACT
The human rhinovirus (HRV) 3C and 2A proteases (3Cpro and 2Apro, respectively) are critical in HRV infection, as they are required for viral polyprotein processing as well as proteolysing key host factors to facilitate virus replication. Early in infection, 3Cpro is present as its precursor 3CD, which, although the mechanism of subcellular targeting is unknown, is found in the nucleus as well as the cytoplasm. In this study, we use transfected and infected cell systems to show that 2Apro activity is required for 3CD nuclear localization. Using green fluorescent protein (GFP)-tagged forms of 3Cpro, 3D, and mutant derivatives thereof, we show that 3Cpro is located in the cytoplasm and the nucleus, whereas 3CD and 3D are localized predominantly in the cytoplasm, implying that 3D lacks nuclear targeting ability and that 3Cpro activity within 3CD is not sufficient to allow the larger protein into the nucleus. Importantly, by coexpressing mCherry-2Apro fusion proteins, we demonstrate formally that 2Apro activity is required to allow HRV 3CD access to the nucleus. In contrast, mCherry-3Cpro is insufficient to allow 3CD access to the nucleus. Finally, we confirm the relevance of these results to HRV infection by demonstrating that nuclear localization of 3CD correlates with 2Apro activity and not 3Cpro activity, which is observed only later in infection. The results thus define the temporal activities of 2Apro and 3CD/3Cpro activities in HRV serotype16 infection.
IMPORTANCE The human rhinovirus genome encodes two proteases, 2A and 3C, as well as a precursor protease, 3CD. These proteases are essential for efficient virus replication. The 3CD protein is found in the nucleus early during infection, though the mechanism of subcellular localization is unknown. Here we show that 2A protease is required for this localization, the 3C protease activity of 3CD is not sufficient to allow 3CD entry into the nucleus, and 3D lacks nuclear targeting ability. This study demonstrates that both 2A and 3C proteases are required for the correct localization of proteins during infection and defines the temporal regulation of 2A and 3CD/3C protease activities during HRV16 infection.
INTRODUCTION
Human rhinovirus (HRV), within the Picornaviridae family, causes the majority of common cold episodes and contributes significantly to asthma exacerbations (1, 2). The positive-sense RNA genome of HRV is translated as a single polyprotein that is cotranslationally cleaved by the virally encoded proteases 2Apro and 3Cpro (3). 2Apro and 3Cpro also cleave several host proteins, to effect disruption of host transcription and translation. Host cell targets include poly(A)-binding protein (PABP) and components of the eukaryotic initiation factor 4F complex (eIF4F), such as eIF4G, which effect a considerable reduction in cap-dependent translation (4, 5), the transcription factor OCT1, which results in disruption of host RNA synthesis (6), and nuclear pore complex (NPC) proteins (nucleoporins or Nups), which are required for nucleocytoplasmic trafficking (6–8). Together, the cleavage of these and other proteins (9) enables HRV to redirect host cell machinery for viral processes as well as to interfere with the ability of the host cell to respond to the infection, resulting in efficient virus replication.
HRV completes its life cycle within the cytoplasm of the host cell, with viral replication mediated by the RNA-dependent RNA polymerase, 3Dpol, encoded in the HRV genome. Despite this ability to replicate in the cytoplasm, the HRV viral protease 3Cpro and its precursor 3CD are found within the nucleus during infection (6, 10). While 3Cpro is small enough to diffuse across the nuclear membrane, 3CD and 3Dpol are too large to accumulate in the nucleus in this way. Sequence analysis has predicted a nuclear localization signal (NLS) within the 3Dpol protein (6, 11), but the role of this putative NLS in 3CD entry into the nucleus has not been fully investigated.
Using transfected and HRV-infected cells, we show that the activity of both 2Apro and 3Cpro is required for 3CD entry into the nucleus. Furthermore, the 3Cpro activity of 3CD is not sufficient to enable 3CD entry into the nucleus. Finally, we use HRV serotype 16 (HRV16)-infected cells to demonstrate formally that the timing of 2Apro activity (cleavage of Nup98) correlates with the nuclear localization of 3CD.
MATERIALS AND METHODS
Antibodies.
The primary antibodies for the following proteins were used for Western blot analysis: anti-α/β-tubulin (Cell Signaling Technology catalog no. 2148, used at a 1:1,000 dilution), anti-mCherry (Abcam no. 167453, used at 1:1,000), anti-green fluorescent protein (anti-GFP; Roche no. 11-814 460 001, used at 1:1,000), anti-eIF4G (Santa Cruz no. 11373, used at 1:1,000), anti-hnRNP-A1 (Santa Cruz no. 56700, used at 1:2,000), anti-PABP (Cell Signaling Technology no. 4992, used at 1:1,000), anti-lamin B1 (Invitrogen no. 33-2000, used at 1:1,000), anti-Nup153 (Abcam no. 96462, used at 1:1,000), anti-Nup62 (Santa Cruz no. H122, used at 1:1,000), anti-6×His (Sigma no. H1029, used at 1:2,000), and anti-VP2 (QED Bioscience no. 18758, used at 1:2,000). Antibodies to 3C protease were kindly provided by S. Amineva (Madison, WI) (6) and used at a 1:1,000 dilution.
Cell culture and infection.
Ohio-HeLa cells (provided by Bo Lin, Biota Holdings) and COS-7 cells (CRL-1651; American Type Culture Collection) were grown in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and antibiotics (penicillin, streptomycin, neomycin; Gibco) at 37°C in a humidified atmosphere of 5% CO2. A recombinant HRV16 designed to express an HA-tagged 2Apro was used for all infection experiments (12). Viral stocks were prepared by infecting subconfluent monolayers of Ohio-HeLa cells at a multiplicity of infection (MOI) of 1 by absorption for 1 h with occasional rocking, followed by replacement of the medium with fresh DMEM supplemented with 2% FBS and antibiotics. Once extensive cytopathic effects were observed, infected cultures were frozen at −80°C to release the virus (13). Cultures were thawed, vortexed, and clarified of cellular debris by centrifugation for 15 min at 3,500 rpm. Infectious virus was titrated on Ohio-HeLa cells by a standard 50% tissue culture infective dose (TCID50) protocol, and the titer was calculated using the Spearman-Karber equation (14).
Western blot analysis.
Overnight subconfluent cultures of Ohio-HeLa cells with or without infection with HRV16 at an MOI of 1 were collected for whole-cell lysates or cytoplasmic/nuclear protein extraction at the specified time points. Cytoplasmic and nuclear protein extractions were performed using the NE-PER kit according to the manufacturer's recommendations (ThermoFisher). Whole-cell lysates were collected at the specified time points in RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 50 mM Tris, pH 8) with protease and phosphatase inhibitors (cOmplete, PhosSTOP; Roche) added. Protein extracts were heated at 100°C for 5 min in Laemmli buffer (63 mM Tris-HCl, pH 6.8, 0.1% 2-mercaptoethanol, 0.0005% bromophenol blue, 10% glycerol, 2% SDS) prior to SDS-PAGE. Whole-cell lysates and subcellular fractions were subjected to SDS-PAGE using 10% or 12.5% polyacrylamide gels, followed by protein transfer to nitrocellulose membranes in Tris-glycine-ethanol buffer (25 mM Tris-HCl, 192 mM glycine, 20% ethanol) for 90 min at 400 mA. The blots were stained with Ponceau S (Sigma) to confirm transfer and then blocked for 1 h in 4% skim milk (Diploma) in phosphate-buffered saline (PBS) (10 mM Na2HPO4, 1.7 mM KH2PO4, pH 7.2, 2.7 mM KCl, 137 mM NaCl) prior to incubation with primary antibodies diluted in 1% skim milk in PBS-T (PBS containing 0.1% Tween 20) overnight at 4°C with rocking. After the blots were washed in PBS-T, they were incubated with species-specific secondary antibodies conjugated to horseradish peroxidase diluted at 1:5,000 in 1% skim milk in PBS-T, followed by washing and detection of bound antibodies with enhanced chemiluminescence (ECL; PerkinElmer). Protein bands were detected using the Licor Odyssey Fc, and captured digital images were analyzed using ImageStudio software (Licor). Where required, blots were stripped to remove bound antibodies (2% SDS, 62.5 mM Tris-HCl, pH 6.8, 114.4 mM β-mercaptoethanol) at 50°C for 10 min, followed by washing in PBS-T, blocking in 4% skim milk in PBS, and reprobing using different primary antibodies.
Plasmid constructs.
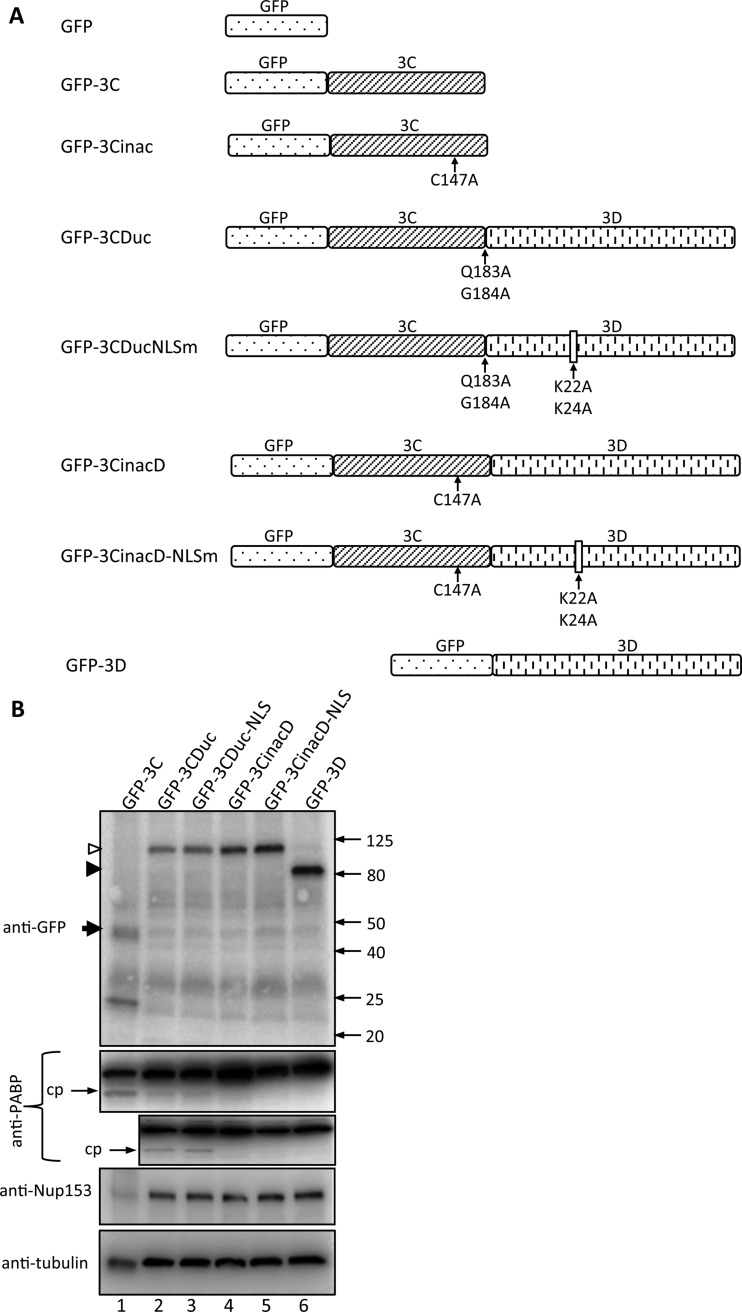
The cloning of 3Cpro into the Gateway-compatible episomally replicating plasmid vector pEPI-DESTC for expression of GFP-3Cpro in mammalian cells has been described previously (10). A mutant form in which the active cysteine in 3Cpro was mutated to alanine (C147A) was generated (GFP-3Cinac). The coding sequence for HRV16 3Dpol was amplified by PCR from the full-length HRV16 genome (15) and recombined into pDONR207 to generate a Gateway-compatible entry clone, which was then recombined into pEPI-DESTC to encode GFP-3Dpol for mammalian cell expression. A form of 3CD that cannot be autocleaved into the component 3Cpro and 3Dpol was generated (GFP-3CDuc); in this construct, the 3Cpro cleavage site is modified such that the P1 and P1′ residues are mutated to alanine (Q183A/G184A). PCR-based site-directed mutagenesis was subsequently performed to mutate the lysine residues in the putative NLS in 3Dpol (6, 11), K22A/K24A, to generate GFP-3CDucNLSm. An additional form of 3CD that cannot be cleaved due to inactivation of the 3Cpro was also generated (GFP-3CinacD). PCR-based site-directed mutagenesis was subsequently performed to mutate the putative NLS in 3Dpol as described above to generate GFP-3CinacD-NLSm. The structure and position of mutations for each GFP-tagged protein are shown below (see Fig. 1A).
FIG 1.
3C in 3CD fusion proteins is proteolytically active. (A) Schematic showing the various GFP fusion protein constructs used in this study. (B) COS-7 cells were transfected to express the indicated GFP fusion proteins and lysed in RIPA buffer containing protease and phosphatase inhibitors 18 h posttransfection (p.t.), and the lysates were subjected to SDS-PAGE and Western blot analysis using the antibodies indicated on the left. The positions of molecular size markers are indicated on the right in kilodaltons; cp, cleavage product of PABP. Empty and filled arrowheads denote GFP-3CDuc and -3D, respectively, with GFP-3C indicated by a filled arrow.
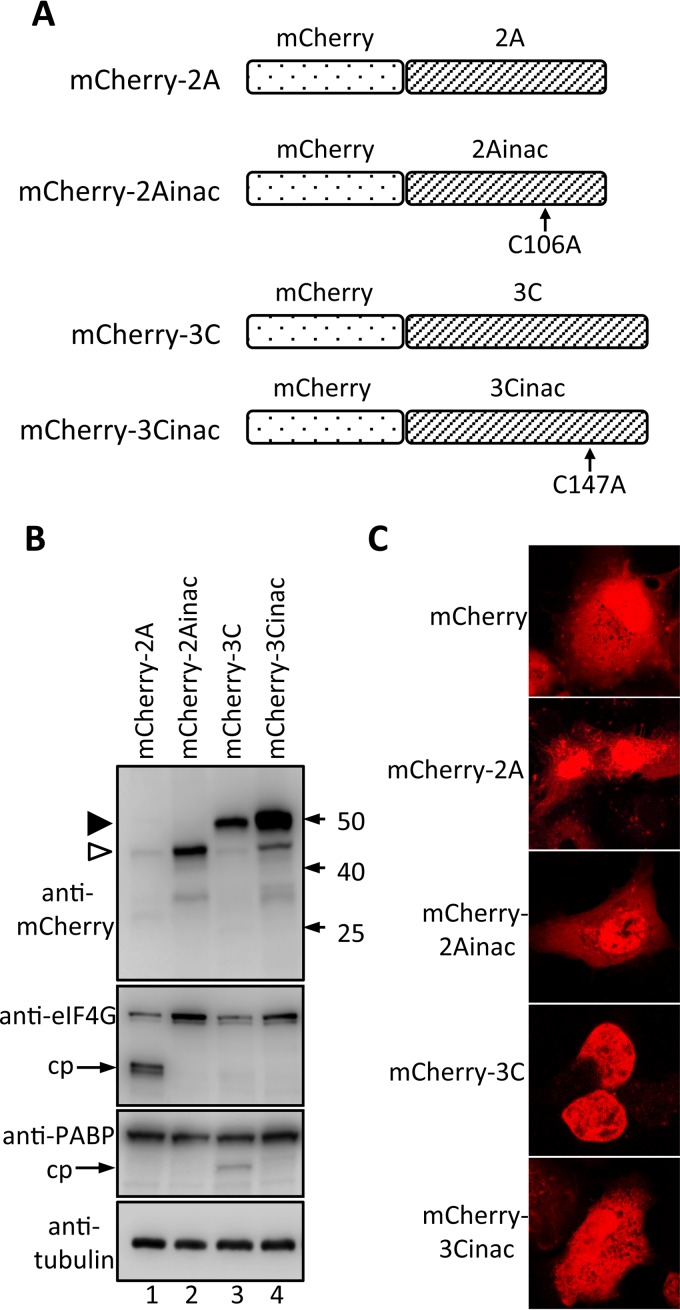
The coding sequence for HRV16 2A was amplified by PCR from the full-length HRV16 genome (15), restriction enzyme digested, and inserted into the mCherry-C1 plasmid vector to encode mCherry-2A. An inactive form of HRV16 2A was also generated; in this construct, the active-site cysteine (Cys 106) (16) was mutated to alanine by site-directed mutagenesis before insertion into plasmid mCherry-C1, to encode mCherry-2Ainac. Likewise, the coding sequence for HRV16 3C was amplified by PCR from the full-length HRV16 genome (15), restriction enzyme digested, and inserted into the mCherry-C1 vector to encode mCherry-3C. An inactive form of HRV 3C was also generated; in this construct, the active-site cysteine (Cys 147) (3) was mutated to alanine by site-directed mutagenesis before insertion into plasmid mCherry-C1, to encode mCherry-3Cinac. The positions of the introduced mutations for each mCherry-tagged protein are shown below (Fig. 3A).
FIG 3.
Proteolytic activity of various mCherry-2A and -3C fusion constructs. (A) Schematic showing the various mCherry fusion protein constructs used in this study. (B) COS-7 cells were transfected to express the indicated mCherry fusion proteins and lysed in RIPA buffer containing protease and phosphatase inhibitors 18 h p.t., and the lysates were subjected to SDS-PAGE and Western blot analysis with the antibodies shown on the left. The positions of molecular mass markers are indicated on the right in kilodaltons; cp, cleavage product. Empty and filled arrowheads denote mCherry-2A and -3C, respectively. (C) COS-7 cells were grown on coverslips and transfected to express mCherry or the indicated mCherry fusion proteins and imaged by CLSM 18 h later.
The cDNA clones for HRV16 2Apro and 3Cpro for expression in Escherichia coli were from Bioclone; the plasmids contained the protease coding sequence inserted into multicloning site of pET-28a for bacterial expression with a hexahistidine (6×His) fusion tag.
Expression of HRV16 proteases in E. coli.
Protease expression was induced in overnight cultures of E. coli using 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h at 37°C, followed by lysis of cells under denaturing conditions (50 mM Tris-HCl pH 7.4, 500 mM NaCl, 5 mM dithiothreitol [DTT], 6 M urea, 5 mM imidazole). Proteases were purified by metal affinity chromatography (BioScale Mini Profinity IMAC columns; Bio-Rad). Briefly, bound proteins were refolded on the column by stepwise removal of urea and eluted under native conditions (50 mM Tris-HCl, pH 7.4, 500 mM NaCl, 5 mM DTT, 500 mM imidazole). Eluted proteins were dialyzed against buffer containing 50 mM Tris-HCl, pH 7.4, 1 mM EDTA, and 5 mM DTT, followed by concentration using Microcon YM-10 columns (Millipore); protein was stored at −20°C in 50% glycerol. The expression and purification of 6×His-tagged proteases were confirmed by SDS-PAGE of samples from bacterial cell lysate and each step in the purification process, followed by staining with Coomassie brilliant blue.
Transfection.
Overnight subconfluent monolayers of COS-7 cells cultured on glass coverslips (Proscitech no. 1) were transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. pEPI-DESTC plasmids encoding various HRV16 proteins were transfected into COS-7 cells, with pEPI-DESTC-GFP as a control. Similarly, mCherry-C1 plasmids encoding 2Apro, 3Cpro, or their protease-inactive mutants were transfected into COS-7 cells, with mCherry-C1 as a control. In certain experiments, mCherry-2A or mCherry-2Ainac was cotransfected with either GFP-3CDuc-, GFP-3CinacD-, GFP-3CDucNLSm-, GFP-3CinacDNLSm-, or GFP-3D-encoding pEPI-DESTC.
CLSM.
At 18 h posttransfection (see above), cells were either lysed in RIPA buffer and subjected to Western blot analysis to confirm protein expression or fixed with 4% formaldehyde in PBS (17). Cells were incubated with Hoechst 33342 (ThermoFisher Scientific) diluted in PBS for 10 min, washed twice in PBS, and mounted using Fluorescence mounting medium (Dako). Digitized fluorescent cell images were collected using a Nikon Ti Eclipse confocal laser-scanning microscope (CLSM) with a Nikon 60×/1.40 oil immersion lens (Plan Apo VC OFN25 DIC N2; optical section of 0.5 μm) and the NIS Elements AR software; data from four individual scans were averaged to obtain the final images for each channel (GFP, mCherry, and Hoechst). Digital images were analyzed as described previously (10, 18, 19), using ImageJ software, to determine the fluorescence intensity above background (Fb) in the nucleus (Fn) compared to that in the cytoplasm (Fc) and to determine the nuclear to cytoplasmic fluorescence ratio (Fn/c).
In vitro protease cleavage assay.
Confluent monolayers of Ohio-HeLa cells were lysed at 107 cells/ml in cold RIPA buffer in the absence of protease inhibitors for 30 min on ice, with rocking, followed by centrifugation at 13,300 rpm for 1 min to remove cell debris. Lysate equivalent to 1.5 × 106 cells was incubated with indicated volumes of purified recombinant HRV16 proteases at 30°C for 6 h prior to heating at 100°C for 5 min in Laemmli buffer to stop the reaction (20), followed by Western blotting as described above.
Statistical analysis.
GraphPad Prism 6 was used for analyses, with a two-tailed Mann-Whitney test to assess statistically significant differences; significance was accepted at a P of ≤0.05.
RESULTS
Nuclear localization of noncleavable 3CD of HRV-infected cells is not due to 3Cpro enzymatic activity or 3D NLS function.
3CD and 3Cpro have been reported to localize in both the host cell nucleus and cytoplasm early in HRV16 infection (6). To begin to analyze the mechanism of 3CD nuclear localization within infected cells, we generated a series of GFP fusion protein constructs for mammalian cell expression (Fig. 1A) of 3Cpro (GFP-3C), 3CD (GFP-3CDuc, in which the 3CD cleavage site Q183/G184 residues are replaced by alanines and hence unable to be cleaved by 3Cpro), 3CD with inactive 3Cpro (GFP-3CinacD, in which the 3Cpro active-site residue C147 is replaced by alanine), 3CD with the alanine substitution mutation of the key residues (K22/K24) within the 3D putative NLS (residues PXKTKLXPS27) (6, 11) (GFP-3CDucNLSm, GFP-3CinacDNLSm), inactive 3Cpro (GFP-3Cinac), and wild-type 3D (GFP-3D).
Initial experiments were performed to confirm fusion protein integrity and 3Cpro activity in transfected cells. Whole-cell lysates were analyzed by Western blotting for GFP, as well as cleavage of PABP (Fig. 1B); although both 2Apro and 3Cpro are able to cleave PABP in vitro (21), 3Cpro is believed to be the main protease responsible in infected cells (22). Anti-GFP antibodies confirmed the correct size of each GFP fusion protein, while probing for PABP revealed a clear cleavage product in lysates from cells expressing GFP-3C, GFP-3CDuc, and GFP-3CDucNLS but not in those expressing GFP-3CinacD, GFP-3CinacD-NLSm, or GFP-3D, as expected (Fig. 1B, compare lanes 1 to 3 with 4 to 6 in the upper PABP blot and lanes 2 and 3 with 4 to 6 in the lower PABP blot). Cleavage of PABP by 3CD was less efficient than that by 3Cpro (Fig. 1B, compare the PABP cleavage product in the lane labeled GFP-3C with that in the lane labeled GFP-3Cuc), consistent with previous reports (23).
CLSM imaging of transfected cells showed active 3Cpro to be both cytoplasmic and nuclear in a similar fashion to GFP alone (Fig. 2A). In contrast, 3CD with or without mutations within the predicted NLS or to inactivate 3Cpro (GFP-3CDuc, -3CDucNLSm, -3CinacD, and -3CinacD-NLSm) was exclusively cytoplasmic (Fig. 2A). GFP-3D, although predominantly cytoplasmic, showed some slight localization to the nucleus. Image analysis (see Materials and Methods) was applied to determine the extent of nuclear accumulation in terms of the nuclear to cytoplasmic fluorescence ratio (Fn/c); multiple transfected cells for each construct were analyzed, with the results supporting the above observations (Fig. 2B). All GFP fusion proteins, including 3D, showed an Fn/c below 0.5, indicative of largely cytoplasmic localization, in contrast to an Fn/c of 1.2 for GFP alone, which is able to diffuse freely and equilibrate in terms of concentration between the nucleus and cytoplasm. GFP-3C, like GFP, showed an Fn/c of around 1.5, indicative of its ability to enter the nucleus by passive diffusion due to its size (ca. 48 kDa) being below the molecular size cutoff (50 to 60 kDa) for passive diffusion into the nucleus (24, 25). The results indicate that 3Cpro activity of 3CDuc is not sufficient to allow 3CDuc into the nucleus and that 3Dpol itself is not intrinsically able to localize strongly in the nucleus in the absence of other HRV components.
FIG 2.
Uncleavable 3CD is unable to target GFP to the nucleus. (A) COS-7 cells were grown on coverslips and transfected to express GFP or the indicated GFP fusion proteins. Cells were imaged by CLSM at 18 h p.t. (B) Images such as those in panel A were subjected to image analysis (see Materials and Methods), where >30 cells for each sample were analyzed to determine Fn/c, the fluorescence in the nucleus relative to that in the cytoplasm. Results shown represent the mean ± standard error of the mean (SEM) (n ≥ 30).
Both HRV 16 2Apro and 3Cpro are required to facilitate 3CD entry into cell nuclei.
Since uncleavable GFP-3CD is largely unable to enter the nucleus in transfected cells (Fig. 2) but is found in the nucleus at early time points during infection (6), we hypothesized that 2Apro activity may be required to enable nuclear localization of 3CD. To test this, we generated a series of mCherry fusion protein constructs for mammalian cell expression (Fig. 3A) of 2Apro (mCherry-2A), inactive 2Apro (mCherry-2Ainac, in which the active-site residue C106 is replaced with alanine), 3Cpro (mCherry-3C), and inactive 3Cpro (mCherry-3Cinac; see above for GFP-3Cinac). Initial experiments were performed to assess fusion protein integrity and 2Apro/3Cpro activity in transfected cells. Whole-cell lysates were analyzed by Western blotting for mCherry, cleavage of eIF4G, a known target of 2Apro, and PABP, a known target of 3Cpro (Fig. 3B). Anti-mCherry antibodies confirmed the correct size of each mCherry fusion protein; the expression levels of mCherry-3C, -3Cinac, and -2Ainac were similar but higher than that of mCherry-2A, in line with observations in other studies, which have attributed the lower levels to the 2Apro-mediated inhibition of cap-dependent translation (26), including that of mCherry-2A. Probing for eIF4G revealed a clear cleavage product only in lysates from cells expressing mCherry-2A (Fig. 3B, eIF4G blot, lane 1), and PABP staining revealed a clear cleavage product only in lysates from cells expressing mCherry-3C (PABP blot, lane 3), as expected. mCherry-2A and -2Ainac were present in both the nucleus and cytoplasm, showing clearly that 2Apro activity is not required for 2Apro nuclear localization (Fig. 3C), which presumably occurs through passive diffusion, as observed for mCherry-3C and mCherry-3Cinac (Fig. 3C).
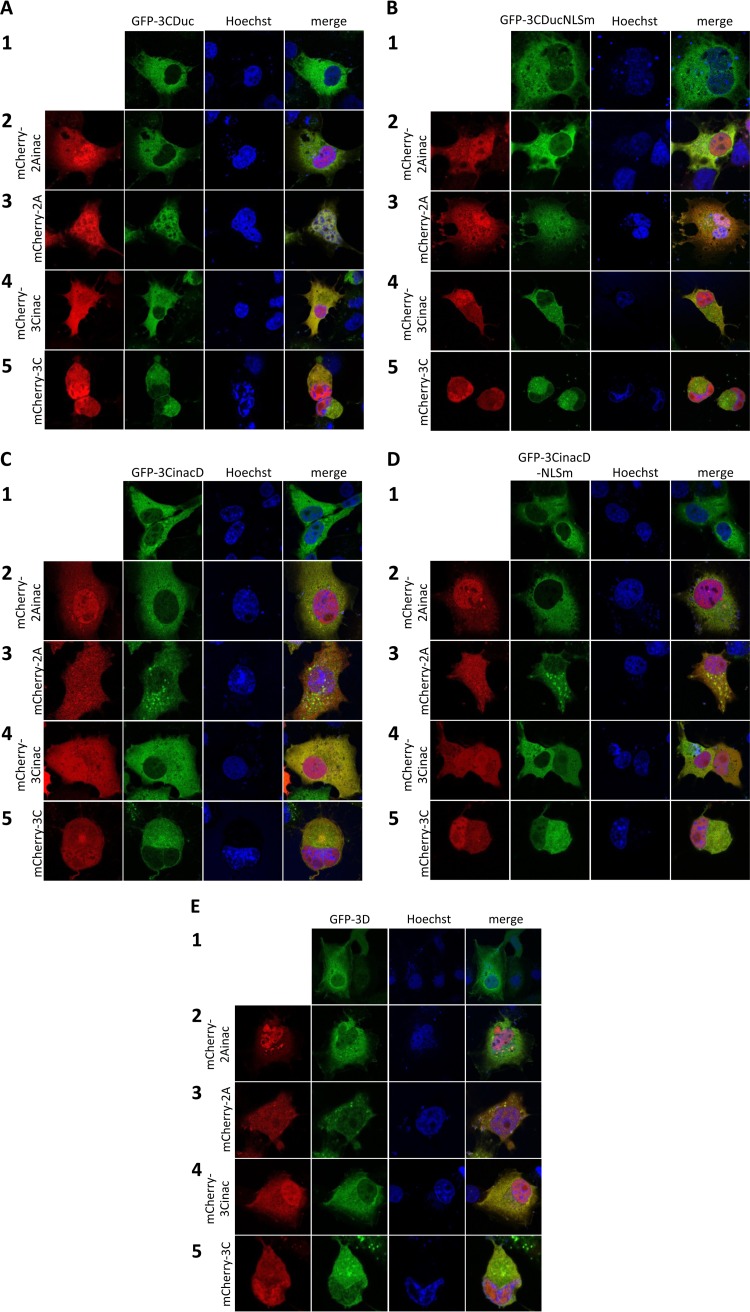
The various mCherry-2A/3C and GFP-3CD/3D fusion constructs were cotransfected, and CLSM was performed to determine the subcellular localization of each fusion protein (Fig. 4). There was no difference in subcellular localization of any of mCherry-2A, -2Ainac, -3C, or -3Cinac in cells coexpressing mCherry and GFP fusion proteins compared to those in which each was expressed alone (compare the localization of each fusion protein in the first column in Fig. 4A to E with those in Fig. 3C). Figure 4A shows the cytoplasmic location of GFP-3CDuc when expressed alone (row 1) or in the presence of mCherry-2Ainac, -3C, or -3Cinac (rows 2, 4, and 5); in the presence of mCherry-2A (row 3), in contrast, GFP-3CDuc clearly localizes in the nucleus as well as the cytoplasm, suggesting that nuclear entry of 3CD can be facilitated by active HRV 2Apro.
FIG 4.
Active HRV16 2Apro and 3Cpro are required for 3CD entry into the nucleus. (A) COS-7 cells grown on coverslips were transfected to express GFP-3CDuc in the absence or presence of the indicated mCherry fusion proteins. CLSM imaging was performed 18 h p.t. Representative images are shown: central images depict localization of GFP-3CDuc (green channel), while the images on the left depict localization of mCherry fusion proteins as labeled (red channel), with the merged image on the right. (B) COS-7 cells grown on coverslips were transfected to express GFP-3CDucNLSm or cotransfected to express GFP-3CDucNLSm and mCherry fusion proteins, as described for panel A. (C) COS-7 cells grown on coverslips were transfected to express GFP-3CinacD or cotransfected to express GFP-3CinacD and mCherry fusion proteins, as described for panel A. (D) COS-7 cells grown on coverslips were transfected to express GFP-3CinacD-NLSm or cotransfected to express GFP-3CinacD-NLSm and mCherry fusion proteins, as described for panel A. (E) COS-7 cells grown on coverslips were transfected to express GFP-3D or cotransfected to express GFP-3D and mCherry fusion proteins, as described for panel A.
Analogously, GFP-3CDuc with a mutated NLS (GFP-3CDucNLSm), in the presence of mCherry-2A but not mCherry-2Ainac, -3C, or -3Cinac, localized in the nucleus as well as the cytoplasm (Fig. 4B). To test the requirement for 3Cpro activity for nuclear localization of GFP-3CDuc, the proteolytically inactive form of 3CD (GFP-3CinacD) was coexpressed with mCherry-2Ainac, -2A, -3Cinac, or -3C (Fig. 4C). The results show that mCherry-2Apro activity is insufficient to enable 3CD access to the nucleus, as none of the coexpression combinations resulted in nuclear accumulation of GFP-3CinacD. Similar results were observed for cotransfections of GFP-3CDucNLSm (Fig. 4D) and GFP-3D (Fig. 4E).
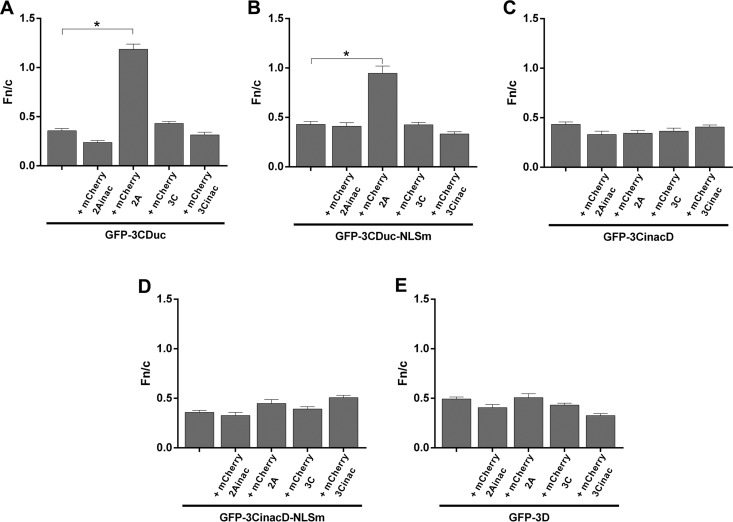
Quantitative analysis to determine the Fn/c ratios for the respective GFP fusion proteins (Fig. 5) confirmed that uncleavable GFP-3CDuc can access only the nucleus when active 2Apro and 3Cpro are both present.
FIG 5.
2Apro and 3Cpro activity is required for 3CD entry into the nucleus based on quantitative imaging. (A to E) Image analysis was performed on cells such as those shown in Fig. 4A to E, respectively, to determine the Fn/c ratios; results represent the mean ± SEM (n ≥ 30). *, P < 0.0001.
Nuclear 3CD in HRV16 infection coincides with 2Apro activity.
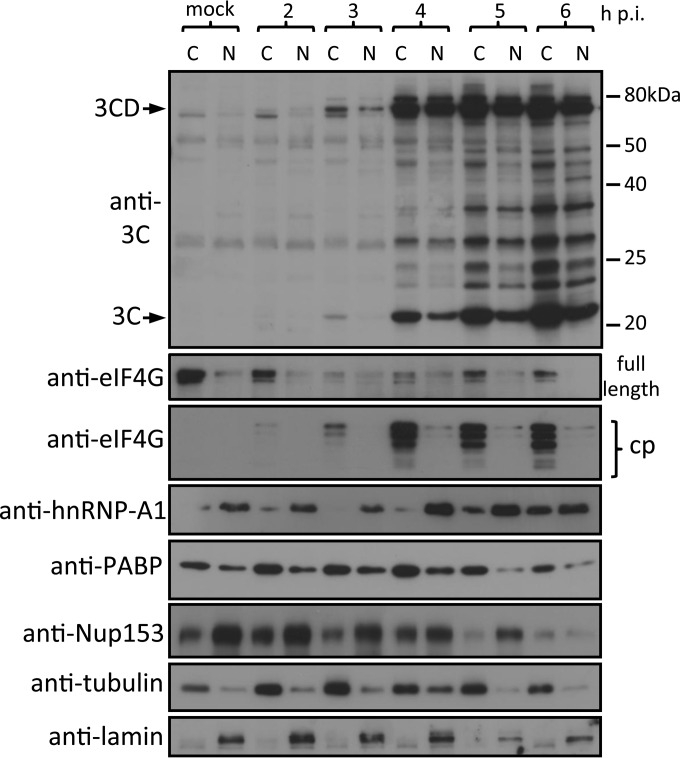
To confirm that HRV16 2Apro is required for the nuclear localization of 3CD during infection, we infected Ohio-HeLa cells with HRV16. As the events that determine localization of 3CD appear to occur early in infection, we collected cytoplasmic and nuclear proteins at 2 to 6 h postinfection (h p.i.); mock-infected cells were collected at 6 h p.i. (Fig. 6). Cellular proteins were analyzed by Western blotting for 3C proteins, eIF4G cleavage products, hnRNP-A1, PABP, and Nup153, with tubulin and lamin as cytoplasmic and nuclear loading controls, respectively. Low levels of 3Cpro were observed exclusively in the cytoplasm at 3 h p.i., simultaneous with the observation of 3CD in both the cytoplasm and nucleus. Depending on the amount of total lysate loaded per lane and exposure time for image capture, we can routinely detect 3CD around 3 and 6 h p.i. (19). These results suggest that nuclear entry of 3CD likely precedes the generation of significant levels of 3Cpro activity in the cell. By 4 h p.i., significant amounts of 3Cpro and 3CD are observed in both the cytoplasmic and nuclear fractions. 2Apro activity is observed 2 to 3 h p.i., as indicated by the presence of eIF4G cleavage products (compare with results for full-length eIF4G). This activity coincides with the presence of 3CD in the nucleus at 3 h p.i., confirming the findings from our transfection experiments indicating that 2Apro activity is required for GFP-3CDuc entry into the nucleus. The effect of 2Apro on nuclear pore integrity is revealed by analysis of hnRNP-A1, a predominantly nuclear protein that shows significant levels of cytoplasmic accumulation from 5 to 6 h p.i. Since we previously showed that the movement of hnRNP-A1 into the cytoplasm is not due to 3Cpro activity, as shown in ectopic expression experiments (19), we can infer that the effect in HRV-infected cells is due to 2Apro. The levels of PABP, a cytoplasmic protein required for cap-dependent translation that is cleaved by 3Cpro, are constant until 6 h p.i.; these results agree with those for whole-cell lysates from later times p.i. (Fig. 7), consistent with significant 3Cpro activity at this time. We previously showed that Nup153 degradation is likely due to 3Cpro activity (19); the results here support this idea in that cleavage of nuclear Nup153 is not observed until 6 h p.i. and occurs after 3CD nuclear entry and movement of hnRNP-A1 to the cytoplasm. Finally, 2Apro activity in the cytoplasm would appear to be maximal at 4 to 5 h p.i., as suggested by the presence of significant levels of eIF4G cleavage products at this time. Nup62 cleavage is observed only later in infection (from 6 h p.i. onwards), paralleling cleavage results for Nup153. Together, these data strongly implicate 2Apro as a necessary mediator of 3CD nuclear entry in infected cells and also as the initial effector of the disruption of nuclear pore function/nuclear transport observed in HRV-infected cells.
FIG 6.
2Apro activity coincides with the presence of 3CD in the nucleus. Ohio-HeLa cells were infected with HRV16 (MOI of 1) or left uninfected (mock), with nuclear (N) and cytoplasmic (C) proteins extracted at the time points shown. Cell lysates were subjected to SDS-PAGE and Western blot analysis. The specificity of the antibodies and bands corresponding to 3C and 3CD are indicated on the left. The positions of molecular size markers are indicated on the right.
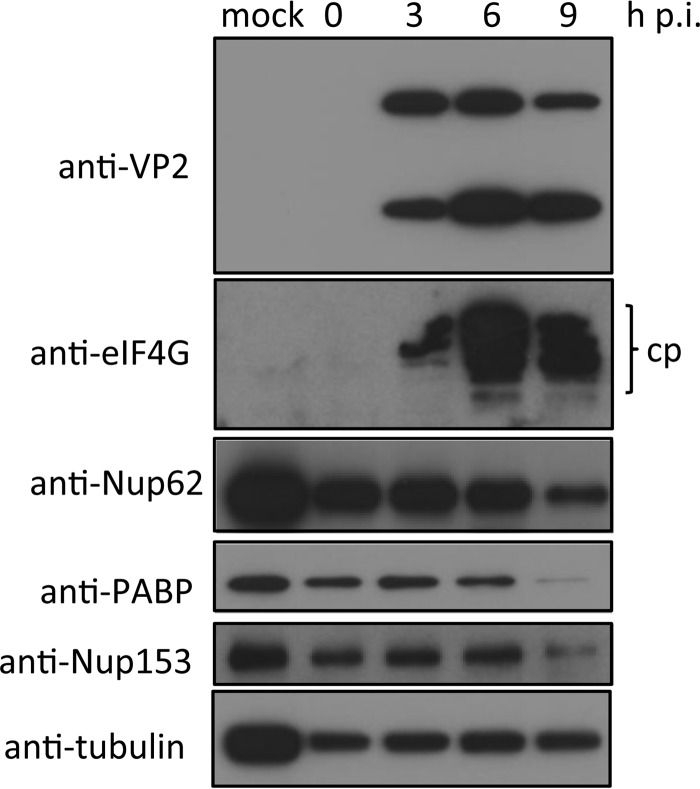
FIG 7.
2Apro activity occurs early in infection, while 3Cpro activity occurs later during infection. Ohio-HeLa cells were infected without (mock) or with HRV16 (MOI of 1), and cells were lysed using RIPA buffer containing protease and phosphatase inhibitors at the time points shown. Cell lysates were subjected to SDS-PAGE and Western blot analysis. cp, cleavage product.
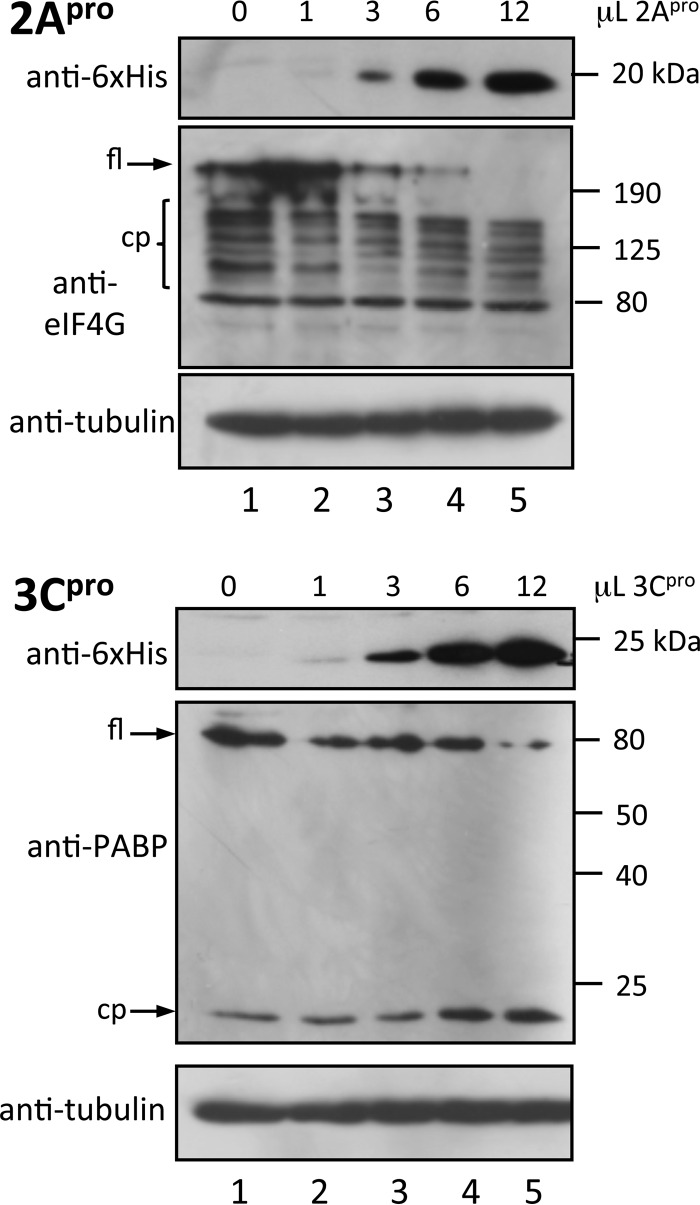
To assess potential intrinsic differences in the protease abilities of 2Apro and 3Cpro, cleavage of PABP and eIF4G, respectively, was examined by adding bacterially expressed 2Apro and 3Cpro to HeLa cell lysates (prepared in the absence of protease inhibitors); in this in vitro system, substrate accessibility, clearly important in infected cells, is not a factor. Some cleavage of both target proteins is evident in the lysates as a result of endogenous protease activity without the addition of exogenous HRV protease (Fig. 8), but it is clear that HRV proteases greatly enhance cleavage of PABP and eIF4G in a dose-dependent fashion. 2Apro and 3Cpro appear to show comparable effects at comparable concentrations, with both proteases cleaving their substrates within a defined period (Fig. 8, compare lane 3 in the two blots with respect to protease levels and loss of full-length substrate), implying very similar intrinsic proteolytic activities in this system with unlimited access to target substrates. The clear implication is that the differential kinetics of cleavage of cellular factors observed in HRV-infected cells is not a function of differences in intrinsic protease activity per se but rather is multifactorial, with key factors including substrate accessibility in the host cell context, polymorphisms in viral proteases, degree of colocalization of viral proteases and cellular protein substrates, and structural and contextual orientation of cleavage sites in proteins.
FIG 8.
Comparable intrinsic activities of recombinant HRV16 2Apro and 3Cpro. The indicated volumes of purified recombinant 2Apro or 3Cpro were incubated with HeLa cell lysate (prepared in the absence of protease inhibitors as described in Materials and Methods) for 6 h at 30°C. The reaction was stopped by heating in Laemmli buffer at 100°C, and samples were analyzed by Western blotting. The antibodies used are indicated on the left of each blot; molecular size markers are shown on the right of each blot. fl, full length; cp, cleavage product.
DISCUSSION
This study demonstrates that HRV 2Apro activity, in conjunction with the 3Cpro activity of the 3CD precursor protein, is required to enable 3CD entry into the nucleus of HRV-infected cells. Importantly, our study also delineates the likely temporal course of events in HRV16 infection, with 2Apro action preceding 3Cpro action. The presence of 3CD in the nuclei of infected cells has been noted for a number of picornaviruses (6, 27, 28), but the mechanism by which this large protein enters the nucleus has been unclear. Together with other recent studies (29–31), the results here suggest that a unique mechanism regulates nuclear localization of picornavirus 3CD/3D, with protease activity being key. When put into the context of the current knowledge of picornaviral protease cleavage of NPC proteins, our data strongly suggest that cleavage of distinct Nups by 2Apro and 3Cpro in a temporally regulated fashion is required to effect the disruption of nucleocytoplasmic trafficking, in part to enable nuclear localization of the large 3CD protein, which cannot otherwise easily access the host nucleus.
Our transfection studies demonstrate that uncleavable GFP-3CD, independent of mutation of the predicted NLS, is largely unable to enter the nucleus when expressed on its own. Proteolytically inactive GFP-3CinacD (with an intact cleavage site between 3C and 3D) is also cytoplasmic, indicating that there is no cryptic sequence within the cleavage site between 3C and 3D that contributes to nuclear targeting. This makes it unlikely that cytoplasmic localization of uncleavable GFP-3CD is due to inactivation of such a sequence, with no evidence for aggregated protein in transfected cells for either protein; the retention of 3Cpro activity by GFP-3CD clearly implies correct folding (Fig. 1). An NLS-like sequence within HRV16 3D (PXKTKLXPS27) similar to sequences from yeast ribosomal proteins was previously predicted by Amineva et al. (6) based on in silico analysis/sequence comparison (and which is similar to the NLS motif of foot-and-mouth disease virus [FMDV] 3Dpol [32]), but mutational analysis had not been previously applied to test NLS functionality and the possibility that HRV protease activity might contribute to nuclear localization had not been assessed. Our data here clearly indicate that mutagenesis of this sequence does not appear to impact nuclear localization of HRV16 3CD under the conditions tested, while GFP-3D itself, although localizing to a slightly greater extent in the nucleus than GFP-3CD (P = 0.03), is predominantly cytoplasmic, completely in line with previous studies for enterovirus 71 (where overexpression can lead to weak nuclear localization in transfected cells) (33) and poliovirus 3Dpol (29). Enterovirus 71 and poliovirus 3Dpol are known to associate with splicing factor Prp8 in the nucleus, leading to inhibition of pre-mRNA splicing, resulting in a decrease in mRNA and accumulation of pre-mRNA intermediates (33). Results for FMDV also suggest an important role for 3Dpol nuclear localization in virus infection, whereby NLS mutations that inhibit it result in attenuated FMDV infection (28), although a link to splicing factor interaction has not been assessed. Although no nuclear function has as yet been assigned to HRV 3Dpol, it seems likely that it plays roles similar to those of the proteins from these other viruses.
What is clear from the present study is that 3Cpro activity in the 3CD context can result in nuclear localization of 3Cpro, helping explain the observations of Amineva et al. (6) and others who used cleavable 3CD constructs in their experimental system. It should be stressed that our approaches do not enable us to exclude the possibility that under certain conditions (e.g., posttranslational modification, etc. [see references 34 and 35]), the NLS may contribute to HRV 3CD nuclear localization. Interestingly, a recent study (31) showed increased nuclear localization of coxsackievirus B3 (CVB3) enhanced GFP (EGFP)-tagged 3Dpol above that of EGFP alone, even though no NLS-like sequence can be identified within the CVB3 3Dpol amino acid sequence, while the nuclear localization of FMDV 3Dpol (28), in contrast to HRV and poliovirus proteins, appears to be dependent on an N-terminal sequence (MRKTKLAPT24) based on mutagenic analysis (28). Clearly, observations with respect to one genus within the Picornaviridae family cannot be assumed to hold true for other genera and, indeed, for serotypes within the same species (16, 36).
GFP-3CD is cytoplasmic when coexpressed with mCherry-2Ainac, -3C, or -3Cinac but can enter the nucleus when coexpressed with active mCherry-2A. Furthermore, the largely cytoplasmic localization of GFP-3CinacD, with or without a mutant NLS, and GFP-3D, even when cotransfected with mCherry-2A or -3C, indicates that the activity of both 3Cpro and 2Apro is required for 3CD nuclear entry, consistent with previous observations for ectopically expressed poliovirus 3CD, which depends on 2Apro activity as well as the 3Dpol NLS (KKKRD129) (27). Clearly, as alluded to above, virus-specific mechanisms are likely to modulate nuclear localization of 3CD/3D in the picornavirus family, but an important role for protease activity is emerging, as confirmed in the present study. The specific substrates that need to be cleaved by 2Apro and 3Cpro to enable HRV 3CD/3D nuclear localization are as yet unclear. However, current literature suggests that cleavage of distinct Nups may be involved (Fig. 6; and see below).
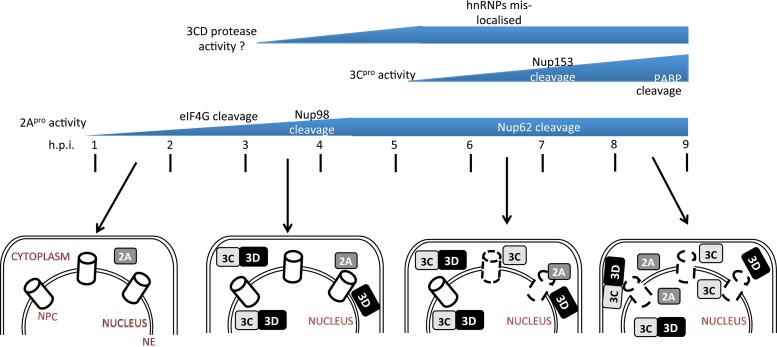
Our infected-cell studies clearly show that 2Apro action in the cytoplasm precedes 3Cpro action early in infection (2 to 3 h p.i.), with the appearance of 3CD in the nucleus importantly coinciding with the 2Apro activity. Figure 9 summarizes our results into a timeline of 2Apro and 3Cpro activity during HRV16 infection and the subcellular localization of the different forms of 3C, making clear the relative timing of 2Apro activity, 3CD and 3Cpro nuclear access, and ultimately NPC breakdown/leakage to the cytoplasm of nuclear proteins such as hnRNP A1. It also incorporates our previous observations with respect to 3Cpro and 2Apro targets (Nup153 and Nup62/Nup98, respectively) (37). 3Cpro activity leads to the redistribution of nucleolin from nucleus to cytoplasm but does not affect localization of hnRNP proteins, implying that complete disruption of nucleocytoplasmic transport as observed in HRV16-infected cells (at 7 to 9 h p.i.) additionally requires 2Apro. Importantly, we have shown that cleavage of Nup98 occurs early (3 h p.i.) in infected cells (37); Nup98 shuttles between the nucleus and cytoplasm and is associated with both the cytoplasmic filaments and the nuclear fibrils of the nuclear pore and, hence, is accessible to the cytoplasmic 2Apro. Importantly, 2Apro-mediated cleavage of Nup62 and 3Cpro-mediated cleavage of Nup153 are detected later in infection (6 h or later p.i.), indicating that access to specific substrates (Nup62 lines the inner channel and Nup153 is in the inner ring of NPC) is a key factor in infected cells. Our in vitro protease assays highlight this; PABP and eIF4G are cleaved, respectively, by 3Cpro and 2Apro with similar efficiencies under in vitro conditions where accessibility is not hindered, in contrast to the situation in infected cells, where eIF4G cleavage is evident from 2 h p.i. while PABP cleavage is only observed later in infection, correlating with its postulated role in the switch from translation to virus replication. The picornaviral polysome must be cleared of initiating or recycling ribosomes before RNA replication can begin, and cleavage of PABP by poliovirus 3Cpro leads to reduced viral as well as cellular translation (22, 38, 39). How PABP cleavage is temporally regulated in virus-infected cells is not clear.
FIG 9.
Timeline of 2A/3Cpro activity and cellular localization during HRV16 infection. Following infection, 2Apro activity is observed from 2 h p.i., as indicated by eIF4G cleavage. By 3 h p.i., 3D and 3CD are present in both the cytoplasm and nucleus, whereas 3Cpro is observed only in the cytoplasm and cleavage of Nup98 by 2Apro is evident, although which specific 3C/3CD protease activity is responsible at this stage is not clear (denoted by a question mark). Since 3CD cannot target efficiently to the nucleus without disruption to nuclear entry points (cleavage of NPC proteins or disruption to other nucleocytoplasmic trafficking proteins), this can be speculated to have occurred by this time. By 4 h p.i., 3CD is seen in the cytoplasm and nucleus, and 2Apro activity against eIF4G is maximal. By 6 to 7 h p.i, breakdown (indicated by broken lines) of the nuclear pore complex (NPC) is clearly evident, as indicated by the relocalization of hnRNP-A1 to the cytoplasm; both 3CD and 3Cpro are present in the nucleus and cytoplasm. By 8 to 9 h p.i., 3CD/3Cpro activity is maximal (as indicated by the loss of full-length PABP and Nup153), as is the effect of 2Apro activity on the NPC, with extensive cleavage of Nup62. 2Apro and 3Cpro are observed in the cytoplasm and the nucleus, 3Dpol is present mostly in the cytoplasm, and 3CD is mostly processed to 3Cpro and 3Dpol. The figure summarizes data for cellular proteins that have been studied to date, but it can be assumed that various other host cell factors will also be impacted as a result of the effect of HRV proteases on the host cell NPC/nucleocytoplasmic trafficking. NE, nuclear envelope.
In conclusion, the present study establishes that nuclear localization of 3CD that occurs early in HRV16 infection is facilitated by the action of 2Apro on the host cell NPC, to render it “leaky,” in concert with 3Cpro activity (in the context of 3CD). The cellular substrates of 3Cpro that are key to 3CD nuclear access and critical targets of 3CD/3D functions in the nucleus remain open questions that are the focus of future studies in this laboratory, along with their importance in the virus life cycle.
ACKNOWLEDGMENTS
This study was supported by the National Health and Medical Research Council (Australia) project grant APP1027312 and Senior Principal Research Fellowship APP1002486/1103050.
We acknowledge access to the Monash Micro Imaging facility at Monash University, Australia.
REFERENCES
- 1.Wark PA, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, Holgate ST, Davies DE. 2005. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med 201:937–947. doi: 10.1084/jem.20041901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gern JE. 2010. The ABCs of rhinoviruses, wheezing, and asthma. J Virol 84:7418–7426. doi: 10.1128/JVI.02290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Racaniello VR. 2001. Picornaviridae: the viruses and their replication, p 685–722. In Knipe DM, Howley PM (ed), Fields virology, 4th ed Lippincott/Raven, Philidelphia, PA. [Google Scholar]
- 4.Lamphear BJ, Yan R, Yang F, Waters D, Liebig HD, Klump H, Kuechler E, Skern T, Rhoads RE. 1993. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human coxsackievirus and rhinovirus. J Biol Chem 268:19200–19203. [PubMed] [Google Scholar]
- 5.Gradi A, Svitkin YV, Sommergruber W, Imataka H, Morino S, Skern T, Sonenberg N. 2003. Human rhinovirus 2A proteinase cleavage sites in eukaryotic initiation factors (eIF) 4GI and eIF4GII are different. J Virol 77:5026–5029. doi: 10.1128/JVI.77.8.5026-5029.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amineva SP, Aminev AG, Palmenberg AC, Gern JE. 2004. Rhinovirus 3C protease precursors 3CD and 3CD′ localize to the nuclei of infected cells. J Gen Virol 85:2969–2979. doi: 10.1099/vir.0.80164-0. [DOI] [PubMed] [Google Scholar]
- 7.Gustin KE, Sarnow P. 2002. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J Virol 76:8787–8796. doi: 10.1128/JVI.76.17.8787-8796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park N, Skern T, Gustin KE. 2010. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J Biol Chem 285:28796–28805. doi: 10.1074/jbc.M110.143404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen LM, Walker EJ, Jans DA, Ghildyal R. 2015. Proteases of human rhinovirus: role in infection. Methods Mol Biol 1221:129–141. doi: 10.1007/978-1-4939-1571-2_10. [DOI] [PubMed] [Google Scholar]
- 10.Ghildyal R, Jordan B, Li D, Dagher H, Bardin PG, Gern JE, Jans DA. 2009. Rhinovirus 3C protease can localize in the nucleus and alter active and passive nucleocytoplasmic transport. J Virol 83:7349–7352. doi: 10.1128/JVI.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weidman MK, Sharma R, Raychaudhuri S, Kundu P, Tsai W, Dasgupta A. 2003. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res 95:75–85. doi: 10.1016/S0168-1702(03)00164-3. [DOI] [PubMed] [Google Scholar]
- 12.Walker EJ, Jensen LM, Ghildyal R. 2015. Reverse genetic engineering of the human rhinovirus serotype 16 genome to introduce an antibody-detectable tag. Methods Mol Biol 1221:171–180. doi: 10.1007/978-1-4939-1571-2_13. [DOI] [PubMed] [Google Scholar]
- 13.Ghildyal R, Ho A, Wagstaff KM, Dias MM, Barton CL, Jans P, Bardin P, Jans DA. 2005. Nuclear import of the respiratory syncytial virus matrix protein is mediated by importin beta1 independent of importin alpha. Biochemistry 44:12887–12895. doi: 10.1021/bi050701e. [DOI] [PubMed] [Google Scholar]
- 14.Mahy BW, Kangro HO. 1996. Virology methods manual. Harcourt Brace, New York, NY. [Google Scholar]
- 15.Lee WM, Wang W. 2003. Human rhinovirus type 16: mutant V1210A requires capsid-binding drug for assembly of pentamers to form virions during morphogenesis. J Virol 77:6235–6244. doi: 10.1128/JVI.77.11.6235-6244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watters K, Palmenberg AC. 2011. Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J Virol 85:10874–10883. doi: 10.1128/JVI.00718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghildyal R, Dagher H, Donninger H, de Silva D, Li X, Freezer NJ, Wilson JW, Bardin PG. 2005. Rhinovirus infects primary human airway fibroblasts and induces a neutrophil chemokine and a permeability factor. J Med Virol 75:608–615. doi: 10.1002/jmv.20315. [DOI] [PubMed] [Google Scholar]
- 18.Poon IK, Oro C, Dias MM, Zhang J, Jans DA. 2005. Apoptin nuclear accumulation is modulated by a CRM1-recognized nuclear export signal that is active in normal but not in tumor cells. Cancer Res 65:7059–7064. doi: 10.1158/0008-5472.CAN-05-1370. [DOI] [PubMed] [Google Scholar]
- 19.Walker EJ, Younessi P, Fulcher AJ, McCuaig R, Thomas BJ, Bardin PG, Jans DA, Ghildyal R. 2013. Rhinovirus 3C protease facilitates specific nucleoporin cleavage and mislocalisation of nuclear proteins in infected host cells. PLoS One 8:e71316. doi: 10.1371/journal.pone.0071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hames BD, Rickwood D. 1998. Gel electrophoresis of proteins: a practical approach, 3rd ed Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 21.Joachims M, Van Breugel PC, Lloyd RE. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol 73:718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuyumcu-Martinez NM, Joachims M, Lloyd RE. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J Virol 76:2062–2074. doi: 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis GJ, Wang QM, Cox GA, Johnson RB, Wakulchik M, Dotson CA, Villarreal EC. 1997. Expression and purification of recombinant rhinovirus 14 3CD proteinase and its comparison to the 3C proteinase. Arch Biochem Biophys 346:125–130. doi: 10.1006/abbi.1997.0291. [DOI] [PubMed] [Google Scholar]
- 24.Shimozono S, Tsutsui H, Miyawaki A. 2009. Diffusion of large molecules into assembling nuclei revealed using an optical highlighting technique. Biophys J 97:1288–1294. doi: 10.1016/j.bpj.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weis K. 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112:441–451. doi: 10.1016/S0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 26.Davies MV, Pelletier J, Meerovitch K, Sonenberg N, Kaufman RJ. 1991. The effect of poliovirus proteinase 2Apro expression on cellular metabolism. Inhibition of DNA replication, RNA polymerase II transcription, and translation. J Biol Chem 266:14714–14720. [PubMed] [Google Scholar]
- 27.Sharma R, Raychaudhuri S, Dasgupta A. 2004. Nuclear entry of poliovirus protease-polymerase precursor 3CD: implications for host cell transcription shut-off. Virology 320:195–205. doi: 10.1016/j.virol.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Briones M, Rosas MF, Gonzalez-Magaldi M, Martin-Acebes MA, Sobrino F, Armas-Portela R. 2006. Differential distribution of non-structural proteins of foot-and-mouth disease virus in BHK-21 cells. Virology 349:409–421. doi: 10.1016/j.virol.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 29.Dougherty JD, Tsai WC, Lloyd RE. 2015. Multiple poliovirus proteins repress cytoplasmic RNA granules. Viruses 7:6127–6140. doi: 10.3390/v7122922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian W, Cui Z, Zhang Z, Wei H, Zhang X. 2011. Poliovirus 2A(pro) induces the nucleic translocation of poliovirus 3CD and 3C′ proteins. Acta Biochim Biophys Sin (Shanghai) 43:38–44. doi: 10.1093/abbs/gmq112. [DOI] [PubMed] [Google Scholar]
- 31.Wu S, Wang Y, Lin L, Si X, Wang T, Zhong X, Tong L, Luan Y, Chen Y, Li X, Zhang F, Zhao W, Zhong Z. 2014. Protease 2A induces stress granule formation during coxsackievirus B3 and enterovirus 71 infections. Virol J 11:192. doi: 10.1186/s12985-014-0192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez-Aparicio MT, Rosas MF, Sobrino F. 2013. Characterization of a nuclear localization signal in the foot-and-mouth disease virus polymerase. Virology 444:203–210. doi: 10.1016/j.virol.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 33.Liu YC, Kuo RL, Lin JY, Huang PN, Huang Y, Liu H, Arnold JJ, Chen SJ, Wang RY, Cameron CE, Shih SR. 2014. Cytoplasmic viral RNA-dependent RNA polymerase disrupts the intracellular splicing machinery by entering the nucleus and interfering with Prp8. PLoS Pathog 10:e1004199. doi: 10.1371/journal.ppat.1004199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jans DA, Xiao CY, Lam MH. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532–544. doi:. [DOI] [PubMed] [Google Scholar]
- 35.Poon IK, Jans DA. 2005. Regulation of nuclear transport: central role in development and transformation? Traffic 6:173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- 36.Chase AJ, Semler BL. 2012. Viral subversion of host functions for picornavirus translation and RNA replication. Future Virol 7:179–191. doi: 10.2217/fvl.12.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker EJ, Jensen LM, Croft S, Ghildyal R. 2015. Variation in the nuclear effects of infection by different human rhinovirus serotypes. Front Microbiol 6:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol Cell Biol 24:1779–1790. doi: 10.1128/MCB.24.4.1779-1790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonderoff JM, Larey JL, Lloyd RE. 2008. Cleavage of poly(A)-binding protein by poliovirus 3C proteinase inhibits viral internal ribosome entry site-mediated translation. J Virol 82:9389–9399. doi: 10.1128/JVI.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]