ABSTRACT
The glycoprotein of Ebola virus (EBOV GP), a member of the family Filoviridae, facilitates viral entry into target cells. In addition, EBOV GP antagonizes the antiviral activity of the host cell protein tetherin, which may otherwise restrict EBOV release from infected cells. However, it is unclear how EBOV GP antagonizes tetherin, and it is unknown whether the GP of Lloviu virus (LLOV), a filovirus found in dead bats in Northern Spain, also counteracts tetherin. Here, we show that LLOV GP antagonizes tetherin, indicating that tetherin may not impede LLOV spread in human cells. Moreover, we demonstrate that appropriate processing of N-glycans in tetherin/GP-coexpressing cells is required for tetherin counteraction by EBOV GP. Furthermore, we show that an intact receptor-binding domain (RBD) in the GP1 subunit of EBOV GP is a prerequisite for tetherin counteraction. In contrast, blockade of Niemann-Pick disease type C1 (NPC1), a cellular binding partner of the RBD, did not interfere with tetherin antagonism. Finally, we provide evidence that an antibody directed against GP1, which protects mice from a lethal EBOV challenge, may block GP-dependent tetherin antagonism. Our data, in conjunction with previous reports, indicate that tetherin antagonism is conserved among the GPs of all known filoviruses and demonstrate that the GP1 subunit of EBOV GP plays a central role in tetherin antagonism.
IMPORTANCE Filoviruses are reemerging pathogens that constitute a public health threat. Understanding how Ebola virus (EBOV), a highly pathogenic filovirus responsible for the 2013-2016 Ebola virus disease epidemic in western Africa, counteracts antiviral effectors of the innate immune system might help to define novel targets for antiviral intervention. Similarly, determining whether Lloviu virus (LLOV), a filovirus detected in bats in northern Spain, is inhibited by innate antiviral effectors in human cells might help to determine whether the virus constitutes a threat to humans. The present study shows that LLOV, like EBOV, counteracts the antiviral effector protein tetherin via its glycoprotein (GP), suggesting that tetherin does not pose a defense against LLOV spread in humans. Moreover, our work identifies the GP1 subunit of EBOV GP, in particular an intact receptor-binding domain, as critical for tetherin counteraction and provides evidence that antibodies directed against GP1 can interfere with tetherin counteraction.
INTRODUCTION
Infection with Ebola virus (EBOV) (formerly Zaire ebolavirus), a member of the genus Ebolavirus within the family Filoviridae, causes severe and frequently fatal disease. The Ebola virus disease (EVD) epidemic in Western Africa in 2013 to 2016 was associated with 11,316 deaths and entailed secondary cases in the United States and Spain (1, 2), indicating that EVD constitutes a global public health threat. The interferon (IFN) system, an important component of innate immunity, is a first-line defense against infection by EBOV and other viruses (3, 4). Sensors of the IFN system detect viral invaders and trigger the production and release of IFN. Binding of IFN to receptors on neighboring cells, in turn, induces the expression of roughly 300 to 400 proteins, many of which exert antiviral activity (5). As a consequence, IFN-exposed cells transit into an antiviral state. Understanding how IFN-induced antiviral factors reduce EBOV infection and how the virus evades this process might yield insights into viral pathogenesis and might help to establish targets for intervention.
The IFN-induced antiviral factor tetherin (CD317, BST-2, or HM1.24) restricts the release of progeny virions from infected cells (6, 7). Tetherin's particular membrane topology is pivotal to this activity. The protein has an N-terminal transmembrane domain and a C-terminal glycosylphosphatidylinositol (GPI) anchor, which permit tetherin to simultaneously insert into the viral and the plasma membranes. As a consequence, tetherin forms a physical tether between newly formed virus particles and the host cell (8). Several viruses encode tetherin antagonists that allow robust viral spread in tetherin-positive target cells (9). The Vpu protein of HIV-1 is the prototype tetherin antagonist, and it is well established that specific interactions between the transmembrane domains of these proteins are required for tetherin antagonism (10–13). Antagonism encompasses Vpu-dependent removal of tetherin from the site of viral budding—the plasma membrane—and rerouting of the protein for endosomal degradation (14–16).
The glycoprotein (GP) of filoviruses is inserted into the viral envelope and facilitates viral entry into target cells, a process that depends on the interactions of the receptor-binding domain (RBD) in GP with the cellular protein Niemann-Pick disease type C1 (NPC1) (17, 18). Moreover, EBOV GP counteracts tetherin (19) by a novel mechanism (19–22), which might involve GP-dependent inhibition of tetherin association with the viral matrix protein VP40 (23). Tetherin antagonism by GP might be required for efficient EBOV spread in the host, since macrophages, central viral target cells (24), express tetherin (25, 26). In contrast, it is unknown whether the GP of a related filovirus, Lloviu virus (LLOV) (genus Cuevavirus) (27), counteracts tetherin. In addition, it is poorly understood which domains in EBOV GP contribute to tetherin counteraction. EBOV GP was found to interact with tetherin via its transmembrane unit, GP2 (20), and evidence was provided that the transmembrane domain (TM) within GP2 is necessary but not sufficient for tetherin counteraction (28, 29). However, the EBOV GP TM mutant that was unable to counteract tetherin was also defective in mediating viral entry (28) and thus might have been partially misfolded. In addition, a separate study revealed that EBOV GP counteracts an artificial tetherin molecule (21), suggesting that GP binding to tetherin may not be required for antagonism. More recent work indicated that deletion of the glycan cap of EBOV GP, an N-glycosylated region displayed at the top of GP, might be incompatible with tetherin antagonism (29), but the underlying mechanism was not investigated. In sum, it is at present unknown how EBOV GP antagonizes tetherin, and it is incompletely understood which determinants in the viral GP control tetherin antagonism.
Here, we analyze whether LLOV GP antagonizes tetherin, and we examine the role of the surface unit, GP1, of EBOV GP in tetherin antagonism. We show that LLOV GP counteracts the antiviral activity of tetherin, indicating that tetherin might not pose an effective barrier against LLOV spread in human cells. Moreover, we demonstrate that appropriate processing of N-glycans, as well as an intact RBD, is required for tetherin counteraction by EBOV GP, although inhibition of the RBD interaction partner NPC1 has no effect. Finally, we identified an antibody directed against the GP1 subunit of EBOV GP that may block tetherin antagonism. These results indicate a central role of the GP1 subunit of EBOV GP in tetherin counteraction and suggest that antibodies directed against this subunit can interfere with viral release by blocking GP-dependent tetherin antagonism.
MATERIALS AND METHODS
Cell culture, plasmids, and antibodies.
Human embryonal kidney 293T (HEK293T) cells, N-acetylglucosamine transferase I-deficient (GnTI−) HEK293S cells (30), and HeLa cells were maintained in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS); 1% penicillin-streptomycin; and, in the case of GnTI− cells, 10 μM sodium pyruvate. Mouse hybridoma cells secreting anti-vesicular stomatitis virus glycoprotein (VSV G) antibody (I1-hybridoma; CRL-2700; ATCC) were cultivated in DMEM supplemented with 20% FBS and 1% penicillin-streptomycin. The cells were cultured at 37°C in a humidified atmosphere containing 5% CO2. Plasmids encoding the following proteins were described previously: tetherin (31); EBOV GP wild type (wt) and EBOV GP with mutations in the RBD (32), a deleted mucin-like domain (MLD) (33), LLOV GP (35); HIV Vpu (20); VSV G (36); murine leukemia virus (MLV) vector encoding luciferase (36); MLV Gag-Pol (36); HIV-1 p55-Gag (28); and EBOV VP40 harboring a myc tag (37). Tetherin with an N-terminal AU1 antigenic tag was generated by PCR-based mutagenesis and inserted into plasmid pcDNA3.1 using EcoRV and NheI restriction sites. The integrity of the PCR-amplified sequence was confirmed by automated sequence analysis. The following antibodies have also been previously described: EBOV GP1-specific monoclonal antibodies (38, 39), anti-Gag monoclonal antibody (40), and a polyclonal rabbit antiserum raised against EBOV GP1 (41). The following antibodies were purchased from commercial providers: monoclonal anti-V5 antibody (Invitrogen), mouse anti-AU1 antibody (Covance), monoclonal rabbit anti-tetherin antibody (Abcam), anti-tetherin monoclonal antibody (B02P; Abnova), polyclonal anti-β-actin antibody (Abnova; Sigma), and horseradish peroxidase (HRP)-coupled secondary antibodies directed against mouse and rabbit immunoglobulin (Dianova) and fluorescein isothiocyanate (FITC)-coupled secondary antibodies against mouse immunoglobulin (Dianova).
Analysis of viral glycoprotein-mediated transduction.
Transduction was analyzed as described previously (36). In brief, for the production of MLV vectors bearing filovirus GPs, 293T control or GnTI− cells were seeded in T25 cell culture flasks and cotransfected with plasmids encoding MLV Gag-Pol, an MLV vector encoding firefly luciferase, and a viral glycoprotein or empty plasmid, employing calcium phosphate as the transfection reagent. At 16 h posttransfection, the cells were washed and supplemented with fresh medium. At 48 h posttransfection, the culture supernatants were collected, sterile filtered through a 0.45-μm filter, aliquoted, and stored at −80°C. For transduction of target cells, 293T cells seeded in a 96-well plate were incubated with 50 μl/well of vector preparation for 6 h at 37°C. Thereafter, 50 μl/well of fresh DMEM culture medium was added. At 72 h postransduction, the culture supernatants were removed, the cells were lysed, and luciferase activity was measured in the cell lysates by employing a commercially available kit (PJK) and the Hidex Chameleon V luminometer with Microwin 2000 software.
For analysis of the antiviral activities of U18666A and cationic amphiphiles, VSV-based pseudotypes were used for consistency with previous work (42). The pseudotypes were generated and used for transduction as described previously (43). In brief, 293T cells seeded in 6-well-plates were calcium phosphate transfected with plasmids encoding VSV G or EBOV GP or with empty plasmid (pCAGGS) as a negative control. At 18 h posttransfection, the cells were inoculated with VSV*ΔG-Luc (44, 45) at a multiplicity of infection (MOI) of 3 for 1 h at 37°C. Thereafter, the cells were washed with phosphate-buffered saline (PBS) and incubated for 1 h at 37°C with a 1:1,000 dilution of hybridoma supernatant containing anti-VSV G antibody in order to neutralize residual virus. Finally, fresh culture medium was added to the cells, and the supernatants were collected at 18 to 20 h postransduction, clarified from the cell debris by centrifugation, aliquoted, and stored at −80°C. To assess the blockade of viral entry by cationic amphiphiles, 293T target cells seeded in 96-well plates were preincubated with each compound or diluent for 3 h at 37°C. Subsequently, the cells were inoculated with equal volumes of pseudotypes and incubated for 18 h at 37°C in the presence of inhibitor. Finally, luciferase activities in cell lysates were measured as described for cells transduced with MLV pseudotypes.
Inhibition of virus-like-particle release by tetherin and tetherin antagonism by filoviral glycoproteins.
Release of virus-like particles (VLPs) and its inhibition by tetherin were examined as described previously (20, 28). In brief, 293T control cells or GnTI− cells were seeded in 48-well plates and cotransfected with plasmids encoding HIV-1 p55-Gag, tetherin, and a potential tetherin antagonist or with empty plasmid, using the calcium phosphate method. For experiments with EBOV VP40, a plasmid encoding VP40 instead of HIV Gag was used. At 16 h posttransfection, the transfection medium was replaced by fresh culture medium. For blockade of EBOV GP-dependent tetherin antagonism, GP1-specific monoclonal antibodies were added to the culture medium at a final concentration of 20 μg/ml, or cationic amphiphiles (U18666A, clomifene, and terconazole; all purchased from Sigma) were added at the indicated concentrations. At 48 h posttransfection, the supernatants were collected, and the cells were lysed in 50 μl of 2× SDS-containing lysis buffer (30 mM Tris [pH 6.8], 10% glycerol, 2% SDS, 5% β-mercaptoethanol, 0.1% bromphenol blue, 1 mM EDTA). The lysates were incubated at 95°C for 30 min. The supernatants were cleared of remaining cell debris by centrifugation, and VLPs were pelleted from the cleared supernatants by centrifugation through a 20% sucrose cushion. The concentrated VLPs were lysed in 30 μl 2× SDS loading buffer and incubated at 95°C for 30 min. Subsequently, the cell lysates and lysed supernatants were investigated for the presence of Gag or VP40, respectively, employing Western blot analysis.
Immunoblotting.
For immunoblotting, the proteins were separated via SDS-polyacrylamide gel electrophoresis using a 12.5% polyacrylamide gel and transferred onto nitrocellulose membranes (GE Lifesciences; 0.2 μm). The membranes were blocked in 5% milk powder in PBS with 0,1% Tween 20, and Gag protein was detected using 1:100-diluted supernatants of hybridoma cells secreting a mouse anti-Gag antibody. If murine antibodies against EBOV GP were added to inhibit tetherin antagonism, Gag expression was detected using a human monoclonal anti-Gag antibody at a dilution of 1:5,000. VP40 was detected using 1:3-diluted supernatants of a hybridoma cell line that secretes anti-myc antibody. Expression of EBOV GP wt and mutants was detected by employing a GP1-specific rabbit serum at a dilution of 1:1,000. For the detection of LLOV GP, a V5-tagged version of the protein was employed, and its expression was detected by employing an anti-V5 antibody at a dilution of 1:5,000. Expression of β-actin was detected after stripping the membranes (Tris-HCl, SDS, and β-mercaptoethanol; 50°C; 30 min) employing anti-β-actin antibodies at a dilution of 1:10,000. HRP-coupled anti-mouse, anti-rabbit, and anti-human secondary antibodies were used at a final concentration of 0.1 μg/ml. Bound secondary antibodies were detected using a commercially available enhanced chemiluminescence (ECL) kit (GE Healthcare), and signals were visualized using the ChemoCam imaging system and ChemoStarProfessional software (Intas). For quantification of the signal intensity, the program ImageJ was used (46). For normalization, Gag/VP40 signals measured in culture supernatants were divided by the respective signals detected in cell lysates.
Analysis of Ebola virus glycoprotein expression at the cell surface.
For analysis of the surface expression of EBOV GP and mutants, 293T cells were transfected with the respective plasmids and washed and harvested in PBS at 48 h posttransfection. Expression of EBOV GP at the cell surface was detected by employing GP-specific mouse monoclonal antibody 5E6 and an FITC-conjugated anti-mouse secondary antibody. Staining of cells fixed with 2% paraformaldehyde (PFA) was analyzed by employing an LSR II flow cytometer (BD Biosciences) and FACS Diva software (BD Biosiences). The data were further analyzed using FCS Express 4 Flow research software (De Novo Software).
Coimmunoprecipitation.
For the analysis of EBOV GP interactions with tetherin by coimmunoprecipitation (co-IP), 293T cells were cotransfected with plasmids encoding EBOV GP wt or EBOV GP with mutations in the RBD and a plasmid encoding tetherin with an AU1 antigenic tag added to the N terminus. In parallel, antibody-agarose conjugates for immunoprecipitation were generated. For this, agarose beads (A/G Plus Agarose; Santa Cruz) were washed two times with co-IP buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 5 mM EDTA, 0,5% IGEPAL), blocked with cold-water fish gelatin at 4°C for 2 h on a rotating shaker, washed again with co-IP buffer, and incubated with anti-AU1 antibody for 2 h at 4°C on a rotating shaker. At 48 h posttransfection, the 293T cells were harvested, washed with PBS, and resuspended in co-IP buffer. After lysis for 20 min at 4°C, the solutions were cleared from cellular debris by centrifugation at 600 × g and aliquoted. The aliquots were stored for subsequent analysis by immunoblotting or were incubated with agarose-bound anti-AU1 antibody for 20 min at room temperature. After washing eight times with co-IP buffer, the agarose beads were resuspended in 20 μl of 2× SDS loading dye and analyzed by immunoblotting.
Proximity ligation assay.
For analysis of EBOV GP interactions with tetherin via proximity ligation assay (PLA), 100,000 HeLa cells per well were seeded in 12-well plates containing coverslips and then transfected with the indicated EBOV GP expression plasmids using Lipofectamine 2000 according to the manufacturer's protocol (Thermo Fisher). At 24 h posttransfection, the cells were fixed for 20 min with 2% PFA at 4°C, permeabilized for 10 min with 1% saponin, and blocked for 1 h with 10% fetal calf serum (FCS) at room temperature. The primary antibodies, an anti-tetherin monoclonal antibody (B02P; Abnova) and a rabbit anti-EBOV GP serum raised against the GP1 subunit (41), were diluted 1:100 and 1:500 in 1% FCS, respectively, and the cells were subsequently incubated in the primary antibody solution for 1 h at room temperature. Incubation with PLA probes, the ligation reaction, the amplification reaction, and mounting of the coverslips were performed according to the manufacturer's protocol (Duolink; Sigma-Aldrich). Finally, staining was analyzed, employing spinning-disc microscopy and image analysis as described previously (47).
Sequence alignment.
The alignment of a portion of the filovirus RBDs was performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Sequences were obtained from the NCBI (National Center for Biotechnology Information) database, including consensus sequences for Zaire ebolavirus (EBOV) (n = 172), Sudan ebolavirus (SUDV) (n = 20), Bundibugyo ebolavirus (BDBV) (n = 8), Taï Forest ebolavirus (TAFV) (n = 4), Reston ebolavirus (RESTV) (n = 13), and Marburg virus (MARV) (n = 84). In contrast, only a single sequence was available for LLOV.
Statistical analysis.
Statistical significance was calculated using an unpaired two-tailed t test employing GraphPad software.
RESULTS
The Lloviu virus glycoprotein is a tetherin antagonist.
We employed a previously described HIV Gag-based VLP assay (20, 28) to assess inhibition of viral budding by tetherin and its counteraction by EBOV GP, EBOV GP mutants, and LLOV GP. HIV Gag was chosen for this endeavor because expression of filovirus GPs does not modulate release of Gag VLPs from tetherin-negative cells. In contrast, release of EBOV VP40-based VLPs from tetherin-negative cells is augmented by EBOV GP (20), which complicates the analysis of tetherin antagonism. Therefore, a VP40-based assay was used only for confirmatory purposes.
We commenced our analysis by asking whether LLOV GP counteracts tetherin. As a prerequisite to these studies, we determined LLOV GP expression and facilitation of viral entry. Analysis of epitope-tagged proteins revealed that LLOV GP and EBOV GP were appreciably expressed in transfected 293T cells (Fig. 1A), with EBOV GP expression being more efficient. Moreover, both proteins mediated host cell entry when incorporated into retroviral vectors (Fig. 1B), although EBOV GP-driven entry was more robust than LLOV GP-mediated entry, in keeping with published data (35). Thus, under the conditions chosen, LLOV GP was expressed and functional and could be examined for tetherin counteraction. For this, HIV-1 Vpu and EBOV GP were employed as positive controls, while transfection of cells with empty plasmid served as a negative control. Tetherin expression reduced Gag VLP release, and this effect was counteracted by EBOV GP and Vpu, as expected, and by LLOV GP (Fig. 1C and D). This observation adds LLOV GP to the list of viral tetherin antagonists and, jointly with previous work (19, 20), suggests that filoviruses of all three genera, Ebolavirus, Marburgvirus, and Cuevavirus, can antagonize tetherin via their GPs. In addition, this finding raises the question of which features conserved between filovirus GPs control tetherin antagonism.
FIG 1.
LLOV GP is a tetherin antagonist. (A) Plasmids encoding V5-tagged versions of the indicated glycoproteins were transiently transfected into 293T cells. Transfection of empty plasmid (Mock) served as a negative control. Glycoprotein expression in cell lysates was detected by Western blotting, using anti-V5 antibody. Detection of β-actin served as a loading control. The results were confirmed in two separate experiments. (B) MLV vectors bearing the indicated glycoproteins were used to transduce 293T cells, and luciferase activities in cell lysates were measured at 72 h postransduction. Transduction mediated by EBOV GP wt was set as 100%. The averages and standard errors of the mean (SEM) of five independent experiments are shown. (C) 293T cells were transiently transfected with plasmids encoding HIV Gag, tetherin, and the indicated viral glycoproteins or with empty plasmid as a negative control (Mock). HIV-1 Vpu served as a positive control for tetherin antagonism. The presence of Gag in supernatants and cell lysates was determined by Western blotting using an anti-Gag antibody. Detection of β-actin in cell lysates served as a loading control. (D) Averages of four independent experiments conducted as described for panel C and quantified via the ImageJ program. Release of Gag from cells coexpressing EBOV GP and tetherin was set as 100%.
Processing of N-glycans is required for tetherin antagonism.
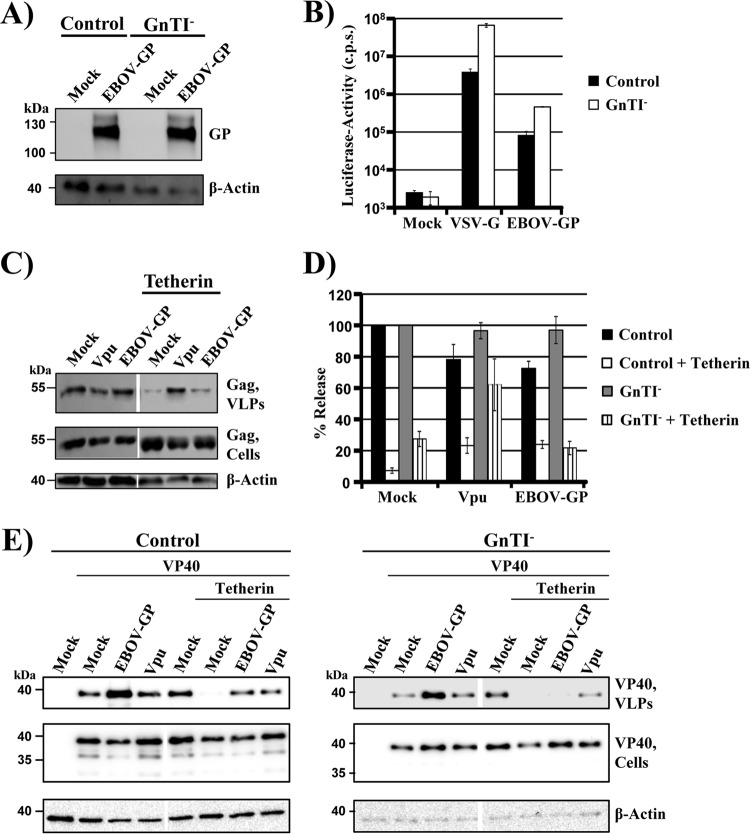
A hallmark of all filovirus glycoproteins is their extensive N-linked glycosylation, raising the question of whether N-glycans contribute to tetherin counteraction. We employed 293S GnTI− cells, in which processing of N-glycans is stalled at the high-mannose stage (30), to examine whether appropriate N-glycosylation is a prerequisite for tetherin counteraction by EBOV GP. Expression levels of EBOV GP in transfected control and GnTI− cells were comparable (Fig. 2A), and pseudotypes produced in both cell lines were readily able to transduce target cells (Fig. 2B). Moreover, tetherin expression restricted Gag-VLP release from both control and GnTI− cells, with restriction in GnTI− cells being less effective (Fig. 2C and D), potentially due to a modest accumulation of tetherin in cytoplasmic compartments of these cells (data not shown). Thus, tetherin and GP are expressed in biologically active forms in control and GnTI−cells. However, EBOV GP failed to rescue Gag VLP release from blockade by tetherin in GnTI− cells, while tetherin counteraction by GP was efficient in control cells and levels of tetherin antagonism by Vpu were comparable in the two cell lines (Fig. 2C and D). Similar results were obtained when release of VP40 VLPs was examined (Fig. 2E), indicating that adequate processing of N-glycans is a prerequisite for tetherin counteraction by EBOV GP.
FIG 2.
Processing of N-glycans is required for tetherin antagonism by EBOV GP, but not Vpu. (A) A plasmid encoding EBOV GP was transiently transfected into control 293T or GnTI− cells. Transfection of empty plasmid (Mock) served as a negative control. Glycoprotein expression in cell lysates was detected by Western blotting, using serum raised against the GP1 subunit of EBOV GP. Detection of β-actin served as a loading control. Three separate experiments yielded similar results. (B) Equal volumes of MLV vectors produced in control or GnTI− cells and bearing the indicated viral glycoproteins were used to transduce 293T cells. At 72 h postransduction, luciferase activities were measured in cell lysates. The results of a single representative experiment carried out with triplicate samples are shown. The error bars indicate standard deviations (SD). Similar results were obtained in three separate experiments. c.p.s., counts per second. (C) GnTI− cells were transfected with plasmids encoding HIV Gag, the indicated viral glycoproteins, and tetherin or with empty plasmid (Mock). HIV-1 Vpu served as a positive control for tetherin antagonism. The presence of Gag protein in culture supernatants and cell lysates was determined by Western blotting. Detection of β-actin in cell lysates served as a loading control. (D) Averages of five independent experiments with control and GnTI− cells conducted as described for panel C and quantified via the ImageJ program. The release of Gag from cells expressing only Gag without any antagonist and without tetherin was set as 100%; the error bars indicate SEM. (E) Control and GnTI− cells were transfected with plasmids encoding VP40 harboring a myc tag, the indicated viral glycoproteins, and tetherin or with empty plasmid (Mock). HIV-1 Vpu served as a positive control for tetherin antagonism. The presence of VP40 in culture supernatants and cell lysates was determined by Western blotting using an anti-myc antibody. The results of single blots are shown, from which irrelevant lanes were cut out. Detection of β-actin in cell lysates served as a loading control. Similar results were obtained in three separate experiments.
An intact receptor-binding domain is required for tetherin counteraction by the Ebola virus glycoprotein.
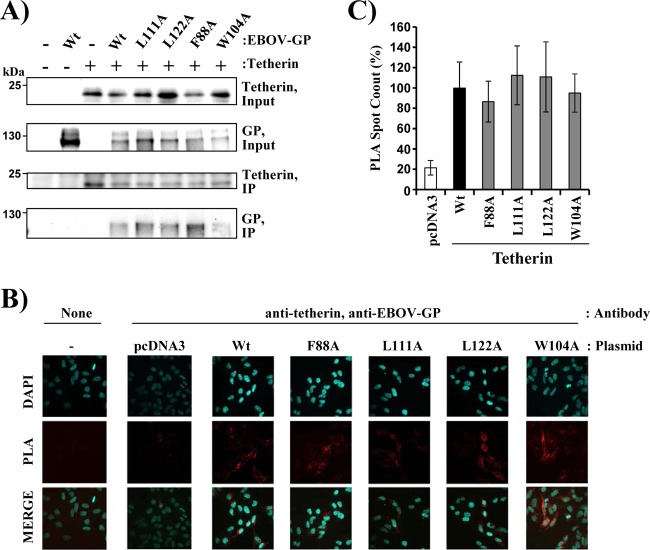
We next investigated whether two conserved elements in the GP1 subunit, the MLD and the RBD, are required for tetherin antagonism. Deletion of the MLD was compatible with robust GP expression (Fig. 3A) and slightly increased GP-driven entry (Fig. 3B), in agreement with published data (48). Moreover, the MLD was dispensable for tetherin antagonism (Fig. 3C and D), in keeping with a previous study (19). In order to determine the role of the RBD in tetherin counteraction, we characterized four point mutations in the RBD, three of which (F88A, L111A, and L122A) were previously reported to abrogate GP-driven host cell entry (32), while the fourth (W104A) was shown not to impede the entry process (50). The three amino acid residues essential for entry are fully conserved between members of the genera Ebolavirus and Cuevavirus, and two out of three are also present in the RBD of marburgviruses (the third was replaced by a conservative substitution, L111I) (Fig. 4A). All the mutants were comparably expressed in transfected 293T cells (Fig. 4B and Table 1), and F88A, L111A, and L122A mutants indeed failed to mediate efficient entry into target cells, while entry driven by the W104A mutant was robust (Fig. 4C). Notably, the abilities of these mutants to drive host cell entry correlated with their capacities to antagonize tetherin: the F88A, L111A, and L122A mutants exhibited strongly reduced tetherin antagonism in the Gag VLP assay, while the W104A mutant was active (Fig. 4D and E). Similar results were obtained in the VP40 VLP assay: F88A, L11A, and L122A mutants were unable to appreciably counteract tetherin, while tetherin counteraction by the W104A mutant was robust (Fig. 4F). Finally, expression of all the mutants augmented release of VP40 VLPs from tetherin-negative control cells (Fig. 4F), suggesting that augmentation of VLP release and tetherin antagonism can be genetically separated. In sum, these results show that an intact RBD is required for tetherin antagonism by EBOV GP.
FIG 3.
The MLD in EBOV GP is dispensable for tetherin antagonism. (A) Plasmids encoding the indicated viral glycoproteins were transiently transfected into 293T cells. Transfection of empty plasmid (Mock) served as a negative control. Glycoprotein expression in cell lysates was detected by Western blotting using serum raised against the GP1 subunit of EBOV GP. Detection of β-actin served as a loading control. Three separate experiments yielded similar results. (B) Equal volumes of MLV vectors bearing the indicated glycoproteins were used for transduction of 293T target cells. Luciferase activities in cell lysates were measured at 72 h postransduction. Transduction mediated by EBOV GP wt was set as 100%. The averages and SEM of five independent experiments are shown. (C) 293T cells were transiently transfected with plasmids encoding HIV Gag, the indicated viral glycoproteins, and tetherin or with empty plasmid (Mock). HIV-1 Vpu served as a positive control for tetherin antagonism. The presence of Gag protein in supernatants and cell lysates was determined by Western blotting using an anti-Gag antibody. Detection of β-actin in cell lysates served as a loading control. (D) Averages of at least five independent experiments conducted as described for panel C and quantified via the ImageJ program. The release of Gag from cells expressing GP and tetherin was set as 100%; the error bars indicate SEM. ***, P < 0.0001.
FIG 4.
EBOV GP requires an intact receptor-binding domain for tetherin antagonism. (A) Amino acid sequence alignment of portions (residues 85 to 125 in EBOV GP) of filovirus RBDs that harbor the amino acid residues investigated for tetherin antagonism (green; numbering according to EBOV GP). *, positions which have a single, fully conserved residue; :, conservation between groups of strongly similar properties (scoring >0.5 in the Gonnet PAM 250 matrix); ., conservation between groups of weakly similar properties (scoring ≤0.5 in the Gonnet PAM 250 matrix). (B) Plasmids encoding the indicated viral glycoproteins were transiently transfected into 293T cells. Transfection of empty plasmid (Mock) served as a negative control. Glycoprotein expression in cell lysates was detected by Western blotting, using serum raised against GP1 of EBOV GP. Detection of β-actin served as a loading control. Four independent experiments yielded highly comparable results. (C) Equal volumes of MLV vectors bearing the indicated viral glycoproteins were used to transduce 293T cells. Luciferase activity in cell lysates was measured at 72 h postransduction. Transduction mediated by EBOV GP wt was set as 100%. The averages and SEM of five independent experiments are shown. (D) 293T cells were transfected with plasmids encoding HIV Gag, tetherin, and the indicated viral glycoproteins or empty plasmid (Mock). HIV-1 Vpu served as a positive control for tetherin antagonism. The presence of HIV Gag in culture supernatants and cell lysates was determined by Western blotting. Detection of β-actin served as a loading control. (E) Averages of at least five independent experiments conducted as described for panel C and quantified via the ImageJ program. The release of Gag from cells coexpressing GP and tetherin was set as 100%; the error bars indicate SEM. (F) 293T cells were transiently transfected with plasmids encoding VP40 harboring a myc tag, tetherin, and the indicated viral glycoproteins or with empty plasmid (Mock). HIV-1 Vpu served as a positive control for tetherin antagonism. The presence of VP40 in culture supernatants and cell lysates was determined by Western blotting. Detection of β-actin in cell lysates served as a loading control. *, P < 0.05; ***, P < 0.0001.
TABLE 1.
Expression of the Ebola virus glycoprotein mutants analyzed
| Glycoprotein | Expression (%) |
|
|---|---|---|
| Cell lysates (Western blotting)a | Cell surface (FACS)b | |
| Wt | 100 | 100 |
| F88A | 120.0 ± 10.1 | 67.7 ± 14.6 |
| L111A | 108.6 ± 14.5 | 45.1 ± 8.4 |
| L122A | 120.5 ± 20.5 | 87.3 ± 12.0 |
| W104A | 123.4 ± 15.7 | 86.7 ± 12.4 |
Averages of three experiments; expression was analyzed with rabbit serum raised against GP1.
Averages of four experiments; expression was analyzed with antibody 5E6. FACS, fluorescence-activated cell sorter.
Inhibition of NPC1 does not interfere with tetherin antagonism by the Ebola virus glycoprotein.
During viral entry, proteolytic processing of GP in endosomes exposes the RBD for subsequent binding to NPC1 (17, 18). A recent study reported evidence for the presence of proteolytically processed GP on the surfaces of 293T cells transfected to express GP (51). Moreover, low levels of endogenous NPC1 were detected at the plasma membranes of 293 cells (51). These observations suggest that NPC1 might be required not only for EBOV GP-driven host cell entry but also for tetherin antagonism. To address this possibility, we employed the compound U18666A, a cationic amphiphile (52). U18666A binds to the sterol-sensing domain of U18666A (53), induces cholesterol accumulation in endosomes, and blocks EBOV entry (42). The compound robustly inhibited EBOV GP- but not VSV G-driven entry (Fig. 5A), as expected. A modest inhibition of VSV G-dependent entry was observed in the presence of 20 μM U18666A (Fig. 5A) and coincided with modestly reduced cell viability (33% reduction, as determined by a CellTiter-Glo Luminescent Cell Viability assay [Promega]) (data not shown), suggesting that this effect was nonspecific. Despite efficient blockade of GP-driven entry, U18666A treatment did not interfere with tetherin antagonism by EBOV GP (Fig. 5B and C), indicating that NPC1 functions required for viral entry are dispensable for GP-mediated tetherin antagonism. Several cationic amphiphiles other than U18666A were also found to raise endosomal cholesterol levels and to block EBOV entry in an NPC1-dependent fashion (42). Therefore, we asked whether two of these compounds, clomifene and terconazole, interfere with tetherin antagonism. Both compounds efficiently reduced viral entry, as expected, but did not inhibit tetherin antagonism by GP (Table 2), confirming that biological properties of NPC1 required for GP-driven cell entry are dispensable for tetherin antagonism.
FIG 5.
U18666A does not block tetherin antagonism by the Ebola virus glycoprotein. 293T cells were treated with the indicated concentrations of compound, incubated with equal volumes of VSV pseudotypes bearing VSV G or EBOV GP, and luciferase activities in the cell lysates were determined at 16 h postransduction. The averages of two independent experiments performed with triplicate samples are shown; the error bars indicate SEM. Transduction in the absence of inhibitor was set as 100%. (B) 293T cells were cotransfected with plasmids encoding HIV Gag, tetherin, and EBOV GP or with empty plasmid (Mock). HIV-1 Vpu served as a positive control for tetherin antagonism. At 12 h posttransfection, the indicated concentrations of U18666A were added to cultures expressing EBOV GP. The presence of HIV Gag in culture supernatants and cell lysates was determined by Western blotting. Detection of β-actin served as a loading control. (C) Averages of three independent experiments conducted as described for panel B and quantified via the ImageJ program. The release of Gag from untreated, tetherin-negative control cells was set as 100%; the error bars indicate SEM.
TABLE 2.
Inhibition of Ebola virus glycoprotein-driven entry and tetherin antagonism by cationic amphiphiles
| Inhibitor | Concn (μM) | Cell viabilitya | Entry inhibitiona | Release inhibitiona |
|---|---|---|---|---|
| Chlomifene | 5 | ++ | +++ | − |
| Terconazole | 9.4 | +++ | +++ | − |
| U18666A | 20 | ++ | +++ | − |
+++, ≥75%; ++, ≥50%; −, <25%.
Mutations in the receptor-binding domain of the Ebola virus glycoprotein that inhibit tetherin antagonism do not interfere with tetherin binding.
It has been reported that EBOV GP interacts with tetherin (19). Therefore, we investigated whether mutations in the RBD that inhibit tetherin antagonism also block tetherin binding. For this, we first employed coimmunoprecipitation. Expression of EBOV GP wt, EBOV GP mutants, and tetherin was readily detectable in cotransfected cells, and pulldown of tetherin resulted in coprecipitation of wt EBOV GP (Fig. 6A), as expected. Notably, wt EBOV GP and GP mutants with exchanges in the RBD that inhibit tetherin antagonism were coprecipitated with comparable efficiencies (Fig. 6A), suggesting that lack of tetherin antagonism by the RBD mutants tested was not due to lack of tetherin binding. We next investigated whether differences in tetherin binding of EBOV GP wt and RBD mutants became apparent when endogenous tetherin expression was examined. For this, we transfected HeLa cells, which constitutively express high levels of endogenous tetherin, with plasmids encoding EBOV GP wt and mutants and determined interactions with tetherin via a proximity ligation assay. We observed comparable tetherin binding of wt and mutant GPs (Fig. 6B and C), confirming that lack of tetherin antagonism by the RBD mutants is not due to lack of tetherin binding.
FIG 6.
Mutations in the receptor-binding domain of the Ebola virus glycoprotein that interfere with tetherin antagonism are compatible with tetherin binding. (A) 293T cells were cotransfected with plasmids encoding tetherin with an N-terminal AU1 tag and EBOV GP wt or the indicated EBOV GP mutants. Coimmunoprecipitation was performed with anti-AU1 antibody coupled to agarose beads, and proteins in cell lysates and in precipitates were detected by Western blotting, employing rabbit serum raised against the GP1 subunit of EBOV GP and a rabbit monoclonal antibody directed against tetherin. The results of a single representative experiment, confirmed in a separate experiment, are shown. (B and C) HeLa cells were transfected with plasmids encoding EBOV GP or the indicated EBOV GP mutants or with empty plasmid as a control. For the PLA, the cells were stained with anti-tetherin and anti-EBOV GP primary antibodies. The images were analyzed by automatically counting the red spots of 20 transfected cells per sample using Volocity software (version 6.3). (B) Representative microscopy images. (C) Mean values and standard deviations of the relative numbers of PLA spots per cell (n = 20). The PLA spot count for cells transfected with EBOV GP wt was set as 100%. DAPI, 4′,6-diamidino-2-phenylindole.
Evidence that an antibody directed against the GP1 subunit can block tetherin counteraction by the Ebola virus glycoprotein.
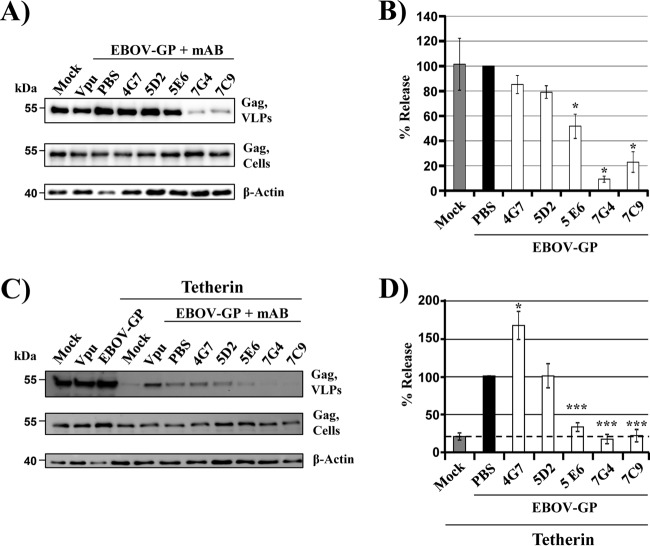
The results obtained so far pointed toward an important role of the GP1 subunit in tetherin counteraction by EBOV GP. GP1 is a central target for the humoral immune response, and thus, we examined whether antibodies directed against GP1 might block tetherin antagonism. For this, we made use of previously described monoclonal antibodies that bind to the MLD in naive GP and protect mice and guinea pigs from a lethal challenge with EBOV (38, 39). We first assessed whether these antibodies interfere with VLP release from control cells. Antibodies 4G7 and 5D2 did not impact VLP release, while antibody 5E6 modestly and antibodies 7G4 and 7C9 strongly inhibited particle release (Fig. 7A and B). Blockade of release might be due to cross-linking of GP on the cell surface and GP on the virion surface, resulting in a tetherin-like restriction of particle release. When the antibodies were tested on cells coexpressing tetherin and GP, similar results were obtained, with the exception of antibody 5E6 (Fig. 7C and D). This antibody had a modest impact on VLP release from tetherin-negative cells (Fig. 7A and B) but reduced particle release from tetherin-positive cells close to background level (Fig. 7D, dashed line), indicating that it interferes with GP-mediated tetherin antagonism. Finally, it is noteworthy that none of the antibodies inhibited VLP release from tetherin-positive or tetherin-negative cells expressing GP without MLD (not shown), indicating that the above-described effects were specific. In sum, our findings suggest that antibody 5E6 can interfere with tetherin antagonism by GP, although part of its release-restricting activity is tetherin independent.
FIG 7.
Evidence that an antibody directed against the GP1 subunit of EBOV GP can block tetherin counteraction. (A) 293T cells were transiently transfected with plasmids encoding HIV Gag and EBOV GP or with empty plasmid (Mock). HIV-1 Vpu served as a positive control for tetherin antagonism. At 16 h posttransfection, the medium was replaced with fresh culture medium supplemented with the indicated antibodies at a final concentration of 20 μg/ml or with PBS. The presence of Gag in culture supernatants and cell lysates was determined by Western blotting. Detection of β-actin served as a loading control. mAb, monoclonal antibody. (B) Averages of two to five independent experiments conducted as described for panel A and quantified via the ImageJ program. The release of Gag from cells expressing GP and treated with PBS was set as 100%; the error bars indicate SEM. (C) The experiment was conducted as described for panel A but cells coexpressing tetherin were examined. (D) Averages of three independent experiments conducted as described for panel A and quantified via the ImageJ program. The release of Gag from cells expressing GP and tetherin and treated with PBS was set as 100%; the error bars indicate SEM. The dashed line indicates the assay background, which was defined by VLP release from tetherin-positive cells, which do not express a tetherin antagonist. *, P < 0.05; ***, P < 0.0001.
DISCUSSION
Tetherin is expressed in macrophages (25, 26) and mature dendritic cells (54), which are important filovirus targets (24, 55, 56), and tetherin counteraction by GP might be essential for robust viral spread in the host. However, it is largely unclear how GP antagonizes tetherin and which domains in GP are required. The present study shows that an intact RBD, as well as appropriate N-glycosylation of GP, is essential for tetherin antagonism and confirms that the MLD is dispensable. Moreover, we demonstrate that an antibody against GP1, which protects against fatal EBOV challenge in a mouse model (38), can block tetherin antagonism by GP. These results indicate that the GP1 subunit plays a central role in tetherin antagonism and suggest that blockade of GP-dependent tetherin antagonism might contribute to the protective activity of certain anti-GP1 antibodies.
Previous work demonstrated that EBOV GP (19) and the GPs of other members of the genera Ebolavirus and Marburgvirus counteract tetherin (20, 28), although these analyses were semiquantitative and subtle differences in the efficiency of tetherin counteraction might have been missed. In contrast, it was unknown whether the GP of LLOV, which was detected in dead Schreiber's bats (Minioperus schreibersii) in Northern Spain (27), also counteracts tetherin. Absence of tetherin counteraction by LLOV GP would suggest that LLOV might not be able to spread efficiently in the human host and that LLOV GP could potentially be used as a tool for mutagenic analysis designed to identify domains required for tetherin antagonism. However, the findings of the present study indicate that LLOV GP robustly counteracts tetherin, suggesting that tetherin antagonism is conserved among all filoviruses known to date. This finding raises the question of which determinants in filovirus GPs are required for tetherin antagonism.
A conserved feature of all filovirus GPs is their extensive N- and O-linked glycosylation. Several N-glycans are located in a surface-exposed area, the glycan cap (57), while the mucin-like domain is extensively modified with O-linked and N-linked glycans. N-glycans limit access to the RBD and are required for binding to cellular lectins and for protection against antibodies (58, 59), since glycans can shield underlying epitopes from binding of neutralizing antibodies. N-glycosylation of proteins starts in the endoplasmic reticulum (ER), where precursor glycans consisting mainly of mannose residues are transferred en bloc onto certain asparagine residues. Upon glycoprotein import into the Golgi apparatus, these high-mannose-type N-glycans are processed into hybrid and complex forms. Processing of N-glycans in the Golgi apparatus can be blocked by inactivating GnTI and results in the trapping of N-glycans in their high-mannose form. The present study shows that exclusive modification of GP with high-mannose N-glycans is compatible with efficient GP expression and GP-driven host cell entry, as expected from a previous analysis (60), but may be incompatible with efficient tetherin antagonism. Such a scenario would be in keeping with a recent study reporting that the glycan cap is essential for tetherin antagonism (29), a finding that was confirmed by the present analysis (not shown). However, the possibility that the absence of tetherin antagonism by GP in GnTI− cells is due to altered N-glycosylation of tetherin or a cellular factor involved in tetherin antagonism (see below) rather than inappropriate glycosylation of GP itself cannot be excluded.
Another hallmark of filovirus GPs, apart from their extensive glycosylation, is the presence of an RBD, which binds to host cell factors involved in viral entry (50, 61, 62). Our study shows that mutations in the RBD that abrogate viral entry also inhibit tetherin antagonism. In contrast, an RBD mutation that did not interfere with viral entry was compatible with tetherin antagonism. A straightforward interpretation of these findings is that GP might need to engage the same cellular factor for entry and tetherin counteraction. The cholesterol transporter NPC1 has been identified as a receptor for filoviruses that is bound by the RBD and that is essential for entry into cultured cells and for viral spread in the host (17, 18, 42, 63). GP binds to NPC1 upon viral uptake into host cell endosomes and processing of GP by the endosomal cysteine proteases cathepsin B and L (17, 18). It is thus not obvious how NPC1 could contribute to tetherin antagonism by GP, which probably occurs at the cell surface or during transport of tetherin to the cell surface. However, a recent study provided evidence that newly expressed GP can be proteolytically processed and transported to the cell surface (51). Moreover, small amounts of NPC1 were detected at the plasma membrane (51). As a consequence, one can speculate that both GP-driven viral entry and tetherin antagonism might depend on NPC1. However, inhibition studies with U18666A and related cationic amphiphiles that induce cholesterol accumulation in endosomes (all compounds) (42), bind NPC1 (U18666A) (53), and block EBOV GP-driven entry in an NPC1-dependent fashion (all compounds) (42) revealed that blockade of NPC1 functions required for viral entry does not interfere with tetherin antagonism. Notably, the RBD has initially been identified as an element in GP1 that is required for efficient binding of soluble GP1 to the surfaces of susceptible cells (62), a process believed to be independent of NPC1, due to its predominantly endosomal localization. Subsequent studies provided evidence that cell adherence and susceptibility to GP-driven entry are correlated and that adherent (and thus susceptible) cells express an RBD binding partner at the cell surface that is present only in intracellular pools within nonadherent (and thus nonsusceptible) cells (64, 65). Although the nature of this cellular factor is at present unknown, it is tempting to speculate that it might be required not only for viral entry, but also for tetherin antagonism.
Wild-type tetherin and artificial tetherin, which was designed in silico, exhibit the same domain organization and exert antiviral activity but share no sequence homology (8). Previous studies reported that EBOV GP antagonizes the antiviral activities of both proteins (21) and interacts with wt tetherin (19). One can speculate that EBOV GP may not bind to artificial tetherin, which suggests that interactions of GP with tetherin might not be required for tetherin antagonism. Alternatively, GP might bind wt and artificial tetherin, and these interactions might be necessary but not sufficient for tetherin counteraction. Both scenarios are in agreement with our finding that RBD mutants largely defective in tetherin counteraction still bind to transfected and endogenously expressed tetherin, as determined by coimmunoprecipitation and proximity ligation assays.
The GP1 subunit is an important target for the antibody response, and it is conceivable that antibody binding blocks GP-dependent tetherin antagonism. Indeed, we obtained evidence that one out of five GP1-directed antibodies, which were previously shown to protect mice from lethal EBOV infection (38, 39), may inhibit tetherin antagonism by EBOV GP, suggesting that this process occurs at the cell surface. However, analysis and interpretation of the activity are complicated by the observation that the antibody (and two others) also interferes with VLP release from tetherin-negative GP-expressing cells. The release of Gag-based VLPs is not modulated by GP; the finding that antibodies directed against GP can block this process was therefore unexpected. One explanation could be that these antibodies simultaneously bind to GP on the particle and on the cell surface, resulting in a tetherin-like restriction of particle release. Moreover, the antibody potentially interfering with tetherin antagonism recognizes an epitope in the MLD, a domain that is dispensable for tetherin antagonism. Thus, one must postulate that its interference with tetherin antagonism is not due to the inhibition of MLD interactions with potential cellular binding partners. Instead, the antibody might inhibit conformational changes in GP, which could be required for tetherin counteraction, or might limit the accessibility of epitopes located close to the MLD due to steric effects. Alternatively, the antibody might reduce GP stability, as has recently been reported for an inhibitor targeting a cavity between GP1 and GP2 (66). In sum, our data suggest that antibodies generated against EBOV GP1 may interfere with tetherin antagonism and/or inhibit particle release from tetherin-negative cells in a GP-dependent manner.
Collectively, our study demonstrates a central role of the GP1 subunit, in particular the RBD, in tetherin counteraction and identifies a GP1-specific antibody that may block this process. It will be interesting to investigate whether GP1-specific antibodies generated in EVD patients block tetherin antagonism and whether a previously reported (62, 64, 65) but so far unidentified cellular interaction partner of the RBD contributes to tetherin antagonism.
ACKNOWLEDGMENTS
We thank C. Basler, Department of Microbiology, Mount Sinai School of Medicine, for the kind gift of plasmids encoding EBOV GP RBD mutants. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: anti-HIV-1 p24 monoclonal (71-31) from Susan Zolla-Pazner and p96ZM651gag-opt from Yingying Li, Feng Gao, and Beatrice H. Hahn.
Funding Statement
This work, including the efforts of Michael Schindler, was funded by Deutsche Forschungsgemeinschaft (DFG) (SCHI1073/4-1). This work, including the efforts of Stefan H. Pöhlmann, was funded by Deutsche Forschungsgemeinschaft (DFG) (PO 716/8-1). This work, including the efforts of Mariana González Hernández, was funded by Deutscher Akademischer Austauschdienst (DAAD) country-related cooperation programme with Mexico: CONACYT (Consejo Nacional de Ciencia y Tecnología) (Stipend).
REFERENCES
- 1.Ebola Response Team WHO. 2016. Ebola virus disease among male and female persons in West Africa. N Engl J Med 374:96–98. doi: 10.1056/NEJMc1510305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. 17 February 2016. Ebola situation report-17 February 2016. http://apps.who.int/ebola/current-situation/ebola-situation-report-17-february-2016.
- 3.Basler CF. 2015. Innate immune evasion by filoviruses. Virology 479-480:122–130. doi: 10.1016/j.virol.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misasi J, Sullivan NJ. 2014. Camouflage and misdirection: the full-on assault of ebola virus disease. Cell 159:477–486. doi: 10.1016/j.cell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 7.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neil SJ. 2013. The antiviral activities of tetherin. Curr Top Microbiol Immunol 371:67–104. doi: 10.1007/978-3-642-37765-5_3. [DOI] [PubMed] [Google Scholar]
- 10.McNatt MW, Zang T, Bieniasz PD. 2013. Vpu binds directly to tetherin and displaces it from nascent virions. PLoS Pathog 9:e1003299. doi: 10.1371/journal.ppat.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwabu Y, Fujita H, Kinomoto M, Kaneko K, Ishizaka Y, Tanaka Y, Sata T, Tokunaga K. 2009. HIV-1 accessory protein Vpu internalizes cell-surface BST-2/tetherin through transmembrane interactions leading to lysosomes. J Biol Chem 284:35060–35072. doi: 10.1074/jbc.M109.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vigan R, Neil SJ. 2010. Determinants of tetherin antagonism in the transmembrane domain of the human immunodeficiency virus type 1 Vpu protein. J Virol 84:12958–12970. doi: 10.1128/JVI.01699-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banning C, Votteler J, Hoffmann D, Koppensteiner H, Warmer M, Reimer R, Kirchhoff F, Schubert U, Hauber J, Schindler M. 2010. A flow cytometry-based FRET assay to identify and analyse protein-protein interactions in living cells. PLoS One 5:e9344. doi: 10.1371/journal.pone.0009344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Fruh K, Moses AV. 2009. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {beta}TrCP-dependent mechanism. J Virol 83:7931–7947. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, Piguet V. 2009. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog 5:e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC. 2009. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog 5:e1000450. doi: 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal CP, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. 2009. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci U S A 106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kühl A, Banning C, Marzi A, Votteler J, Steffen I, Bertram S, Glowacka I, Konrad A, Sturzl M, Guo JT, Schubert U, Feldmann H, Behrens G, Schindler M, Pöhlmann S. 2011. The Ebola virus glycoprotein and HIV-1 Vpu employ different strategies to counteract the antiviral factor tetherin. J Infect Dis 204:S850–S860. doi: 10.1093/infdis/jir378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez LA, Yang SJ, Hauser H, Exline CM, Haworth KG, Oldenburg J, Cannon PM. 2010. Ebola virus glycoprotein counteracts BST-2/Tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J Virol 84:7243–7255. doi: 10.1128/JVI.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez LA, Yang SJ, Exline CM, Rengarajan S, Haworth KG, Cannon PM. 2012. Anti-tetherin activities of HIV-1 Vpu and Ebola virus glycoprotein do not involve removal of tetherin from lipid rafts. J Virol 86:5467–5480. doi: 10.1128/JVI.06280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustin JK, Bai Y, Moses AV, Douglas JL. 2015. Ebola virus glycoprotein promotes enhanced viral egress by preventing Ebola VP40 from associating with the host restriction factor BST2/tetherin. J Infect Dis 212(Suppl 2):S181–S190. doi: 10.1093/infdis/jiv125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez O, Leung LW, Basler CF. 2012. The role of antigen-presenting cells in filoviral hemorrhagic fever: gaps in current knowledge. Antiviral Res 93:416–428. doi: 10.1016/j.antiviral.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindler M, Rajan D, Banning C, Wimmer P, Koppensteiner H, Iwanski A, Specht A, Sauter D, Dobner T, Kirchhoff F. 2010. Vpu serine 52 dependent counteraction of tetherin is required for HIV-1 replication in macrophages, but not in ex vivo human lymphoid tissue. Retrovirology 7:1. doi: 10.1186/1742-4690-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrensch F, Karsten CB, Gnirss K, Hoffmann M, Lu K, Takada A, Winkler M, Simmons G, Pöhlmann S. 2015. Interferon-induced transmembrane protein-mediated inhibition of host cell entry of ebolaviruses. J Infect Dis 212(Suppl 2):S210–S218. doi: 10.1093/infdis/jiv255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negredo A, Palacios G, Vazquez-Moron S, Gonzalez F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, de la Cruz MM, Herrera JE, Pizarro M, Hutchison SK, Echevarria JE, Lipkin WI, Tenorio A. 2011. Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog 7:e1002304. doi: 10.1371/journal.ppat.1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gnirss K, Fiedler M, Kramer-Kuhl A, Bolduan S, Mittler E, Becker S, Schindler M, Pöhlmann S. 2014. Analysis of determinants in filovirus glycoproteins required for tetherin antagonism. Viruses 6:1654–1671. doi: 10.3390/v6041654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vande Burgt NH, Kaletsky RL, Bates P. 2015. Requirements within the Ebola viral glycoprotein for tetherin antagonism. Viruses 7:5587–5602. doi: 10.3390/v7102888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeves PJ, Callewaert N, Contreras R, Khorana HG. 2002. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A 99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz P, Hahn BH, Hatziioannou T, Kirchhoff F. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez O, Ndungo E, Tantral L, Miller EH, Leung LW, Chandran K, Basler CF. 2013. A mutation in the Ebola virus envelope glycoprotein restricts viral entry in a host species- and cell-type-specific manner. J Virol 87:3324–3334. doi: 10.1128/JVI.01598-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marzi A, Akhavan A, Simmons G, Gramberg T, Hofmann H, Bates P, Lingappa VR, Pöhlmann S. 2006. The signal peptide of the ebolavirus glycoprotein influences interaction with the cellular lectins DC-SIGN and DC-SIGNR. J Virol 80:6305–6317. doi: 10.1128/JVI.02545-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reference deleted.
- 35.Maruyama J, Miyamoto H, Kajihara M, Ogawa H, Maeda K, Sakoda Y, Yoshida R, Takada A. 2014. Characterization of the envelope glycoprotein of a novel filovirus, lloviu virus. J Virol 88:99–109. doi: 10.1128/JVI.02265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wrensch F, Winkler M, Pöhlmann S. 2014. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses 6:3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin-Serrano J, Zang T, Bieniasz PD. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med 7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 38.Qiu X, Alimonti JB, Melito PL, Fernando L, Stroher U, Jones SM. 2011. Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clin Immunol 141:218–227. doi: 10.1016/j.clim.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 39.Qiu X, Fernando L, Melito PL, Audet J, Feldmann H, Kobinger G, Alimonti JB, Jones SM. 2012. Ebola GP-specific monoclonal antibodies protect mice and guinea pigs from lethal Ebola virus infection. PLoS Negl Trop Dis 6:e1575. doi: 10.1371/journal.pntd.0001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorny MK, Gianakakos V, Sharpe S, Zolla-Pazner S. 1989. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc Natl Acad Sci U S A 86:1624–1628. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marzi A, Wegele A, Pöhlmann S. 2006. Modulation of virion incorporation of Ebolavirus glycoprotein: effects on attachment, cellular entry and neutralization. Virology 352:345–356. doi: 10.1016/j.virol.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 42.Shoemaker CJ, Schornberg KL, Delos SE, Scully C, Pajouhesh H, Olinger GG, Johansen LM, White JM. 2013. Multiple cationic amphiphiles induce a Niemann-Pick C phenotype and inhibit Ebola virus entry and infection. PLoS One 8:e56265. doi: 10.1371/journal.pone.0056265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann M, Gonzalez HM, Berger E, Marzi A, Pöhlmann S. 2016. The glycoproteins of all filovirus species use the same host factors for entry into bat and human cells but entry efficiency is species dependent. PLoS One 11:e0149651. doi: 10.1371/journal.pone.0149651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berger Rentsch M, Zimmer G. 2011. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One 6:e25858. doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffmann M, Muller MA, Drexler JF, Glende J, Erdt M, Gutzkow T, Losemann C, Binger T, Deng H, Schwegmann-Wessels C, Esser KH, Drosten C, Herrler G. 2013. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS One 8:e72942. doi: 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagen N, Bayer K, Rosch K, Schindler M. 2014. The intraviral protein interaction network of hepatitis C virus. Mol Cell Proteomics 13:1676–1689. doi: 10.1074/mcp.M113.036301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medina MF, Kobinger GP, Rux J, Gasmi M, Looney DJ, Bates P, Wilson JM. 2003. Lentiviral vectors pseudotyped with minimal filovirus envelopes increased gene transfer in murine lung. Mol Ther 8:777–789. doi: 10.1016/j.ymthe.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Reference deleted.
- 50.Manicassamy B, Wang J, Jiang H, Rong L. 2005. Comprehensive analysis of ebola virus GP1 in viral entry. J Virol 79:4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Markosyan RM, Miao C, Zheng YM, Melikyan GB, Liu SL, Cohen FS. 2016. Induction of cell-cell fusion by Ebola virus glycoprotein: low pH is not a trigger. PLoS Pathog 12:e1005373. doi: 10.1371/journal.ppat.1005373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cenedella RJ. 2009. Cholesterol synthesis inhibitor U18666A and the role of sterol metabolism and trafficking in numerous pathophysiological processes. Lipids 44:477–487. doi: 10.1007/s11745-009-3305-7. [DOI] [PubMed] [Google Scholar]
- 53.Lu F, Liang Q, Abi-Mosleh L, Das A, De Brabander JK, Goldstein JL, Brown MS. 2015. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife 4:e12177. doi: 10.7554/eLife.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coleman CM, Spearman P, Wu L. 2011. Tetherin does not significantly restrict dendritic cell-mediated HIV-1 transmission and its expression is upregulated by newly synthesized HIV-1 Nef. Retrovirology 8:26. doi: 10.1186/1742-4690-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. 2003. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol 163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryabchikova EI, Kolesnikova LV, Luchko SV. 1999. An analysis of features of pathogenesis in two animal models of Ebola virus infection. J Infect Dis 179(Suppl 1):S199–S202. doi: 10.1086/514293. [DOI] [PubMed] [Google Scholar]
- 57.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jeffers SA, Sanders DA, Sanchez A. 2002. Covalent modifications of the ebola virus glycoprotein. J Virol 76:12463–12472. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lennemann NJ, Rhein BA, Ndungo E, Chandran K, Qiu X, Maury W. 2014. Comprehensive functional analysis of N-linked glycans on Ebola virus GP1. mBio 5:e00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin G, Simmons G, Pöhlmann S, Baribaud F, Ni H, Leslie GJ, Haggarty BS, Bates P, Weissman D, Hoxie JA, Doms RW. 2003. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J Virol 77:1337–1346. doi: 10.1128/JVI.77.2.1337-1346.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brindley MA, Hughes L, Ruiz A, McCray PB Jr, Sanchez A, Sanders DA, Maury W. 2007. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J Virol 81:7702–7709. doi: 10.1128/JVI.02433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuhn JH, Radoshitzky SR, Guth AC, Warfield KL, Li W, Vincent MJ, Towner JS, Nichol ST, Bavari S, Choe H, Aman MJ, Farzan M. 2006. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J Biol Chem 281:15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- 63.Wang H, Shi Y, Song J, Qi J, Lu G, Yan J, Gao GF. 2016. Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1. Cell 164:258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dube D, Schornberg KL, Stantchev TS, Bonaparte MI, Delos SE, Bouton AH, Broder CC, White JM. 2008. Cell adhesion promotes Ebola virus envelope glycoprotein-mediated binding and infection. J Virol 82:7238–7242. doi: 10.1128/JVI.00425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dube D, Schornberg KL, Shoemaker CJ, Delos SE, Stantchev TS, Clouse KA, Broder CC, White JM. 2010. Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc Natl Acad Sci U S A 107:16637–16642. doi: 10.1073/pnas.1008509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Y, Ren J, Harlos K, Jones DM, Zeltina A, Bowden TA, Padilla-Parra S, Fry EE, Stuart DI. 2016. Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature 535:169–172. doi: 10.1038/nature18615. [DOI] [PMC free article] [PubMed] [Google Scholar]