ABSTRACT
Membrane fusion, which is the key process for both initial cell entry and subsequent lateral spread of herpes simplex virus (HSV), requires the four envelope glycoproteins gB, gD, gH, and gL. Syncytial mutations, predominantly mapped to the gB and gK genes, confer hyperfusogenicity on HSV and cause multinucleated giant cells, termed syncytia. Here we asked whether interaction of gD with a cognate entry receptor remains indispensable for initiating membrane fusion of syncytial strains. To address this question, we took advantage of mutant viruses whose viral entry into cells relies on the uniquely specific interaction of an engineered gD with epidermal growth factor receptor (EGFR). We introduced selected syncytial mutations into gB and/or gK of the EGFR-retargeted HSV and found that these mutations, especially when combined, enabled formation of extensive syncytia by human cancer cell lines that express the target receptor; these syncytia were substantially larger than the plaques formed by the parental retargeted HSV strain. We assessed the EGFR dependence of entry and spread separately by using direct entry and infectious center assays, respectively, and we found that the syncytial mutations did not override the receptor specificity of the retargeted viruses at either stage. We discuss the implications of these results for the development of more effective targeted oncolytic HSV vectors.
IMPORTANCE Herpes simplex virus (HSV) is investigated not only as a human pathogen but also as a promising agent for oncolytic virotherapy. We previously showed that both the initial entry and subsequent lateral spread of HSV can be retargeted to cells expressing tumor-associated antigens by single-chain antibodies fused to a receptor-binding-deficient envelope glycoprotein D (gD). Here we introduced syncytial mutations into the gB and/or gK gene of gD-retargeted HSVs to determine whether viral tropism remained dependent on the interaction of gD with the target receptor. Entry and spread profiles of the recombinant viruses indicated that gD retargeting does not abolish the hyperfusogenic activity of syncytial mutations and that these mutations do not eliminate the dependence of HSV entry and spread on a specific gD-receptor interaction. These observations suggest that syncytial mutations may be valuable for increasing the tumor-specific spreading of retargeted oncolytic HSV vectors.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is an important focus of research as a common human pathogen that often causes mucocutaneous lesions. In addition, HSV has recently shown promise as a tool for the development of novel therapeutic modalities against human cancers (1).
Membrane fusion is the key process required for both initial entry of the virion into cells and subsequent lateral spread of HSV-1. HSV-1 entry depends on the interaction of gD with one of its cognate receptors: herpesvirus entry mediator (HVEM), nectin-1, or 3-O-sulfated heparan sulfate (3-OS-HS) (2–4). Receptor binding triggers a conformational change in gD that in turn activates the fusion mechanism executed by other envelope glycoproteins (5–7); fusion merges the viral envelope with cell membranes, resulting in capsid release into the cytoplasm. The lateral spread of HSV-1 typically occurs through release of progeny virions into spaces between infected and juxtaposed uninfected cells, and it causes cell rounding and aggregation, with limited cell-cell fusion (8). However, certain HSV mutants can rapidly spread to adjacent cells by mediating fusion between infected and surrounding uninfected cells, leading to the formation of multinucleated giant cells, termed syncytia (9–12). Mutations responsible for this hyperfusogenic phenotype, referred to as syncytial mutations, have been mapped to at least four viral genes, i.e., gB (11, 13–18), gK (12, 19–21), UL20 (22, 23), and UL24 (24), but are typically encountered as a single point mutation in the gB or gK gene.
The envelope glycoprotein gB is a type I membrane protein composed of 904 amino acids and is believed to execute membrane fusion during HSV entry and cell-cell fusion, based on the presence of fusion loops that mediate membrane interaction (25, 26) and its structural similarity to vesicular stomatitis virus glycoprotein G, a well-characterized fusion protein (27). From the results of their bimolecular fluorescence complementation studies, Atanasiu and colleagues suggested that activation of gB is accomplished through the coordinated, sequential activities of the 4 glycoproteins gB, gD, gH, and gL, which constitute the so-called fusion machinery, as follows (28): (i) a conformational change in gD is induced by receptor binding, (ii) receptor-activated gD alters the conformation of gH/gL, and (iii) altered gH/gL stimulates or unmasks the fusogenic activity of gB. All of the gB syncytial mutations identified to date have been mapped to the C-terminal cytoplasmic domain (cytodomain) (11, 13–18). This strongly suggests that the gB cytodomain restricts the fusion activity of gB and that the cytodomain mutations lead to relaxation of this restriction.
HSV-1 gK is a glycosylated membrane protein that consists of 338 amino acids and has several hydrophobic domains (12). It is known that gK is required for virus-induced cell-cell fusion (29, 30). However, the transient coexpression of gB, gD, gH, and gL is both necessary and sufficient to cause virus-free cell-cell fusion (31), while gK is not required. Thus, this transient coexpression system does not precisely recapitulate virus-induced cell-cell fusion. Interestingly, the transient coexpression of gK with the four other glycoproteins has been reported to inhibit cell-cell fusion (32), indicating that gK has a restrictive function in membrane fusion. In contrast to gB, syncytial mutations in gK have been predominantly, though not exclusively, mapped to the N-terminal extracellular domain (33). Since these mutations are thus located on opposite sides of the membrane, their mechanisms are probably distinct.
Considering that syncytial mutations confer hyperfusogenic potential upon the HSV fusion machinery, the question of whether or not they eliminate the typical dependence of HSV-induced membrane fusion on receptor recognition by gD arises. Silverman and colleagues reported that some gB syncytial mutations enabled a degree of virus-free cell-cell fusion in the absence of gD (34), although this was not confirmed by additional experiments using a different setup (35). Cocchi and colleagues reported that plaque formation by the syncytial strains HSV-1(MP) and HFEM-syn was not blocked by an anti-nectin-1 monoclonal antibody that efficiently blocked the formation of plaques by a nonsyncytial strain. This finding suggests that syncytial strains spread by a gD-receptor interaction-independent mechanism (36). However, Even and colleagues observed that transfection of nectin-1-transduced cells with viral DNAs of the same syncytial strains, HSV-1(MP) and HFEM-syn, produced large syncytia, whereas no plaques were observed in cultures of cells that had not been transduced with nectin-1. This finding indicates that spread of these strains remains dependent on a gD receptor (37).
We recently reported that entry of HSV can be retargeted to cells that express tumor-associated antigens, including epidermal growth factor receptor (EGFR), carcinoembryonic antigen (CEA), and epithelial cell adhesion molecule (EpCAM). Our method involves mutational detargeting of gD from its canonical receptors and retargeting by insertion of a receptor-specific single-chain antibody (scFv) (38–40). We also reported that not only direct entry but also subsequent lateral spread of the EpCAM-retargeted HSV strain is strictly dependent on cellular expression of the target receptor (40).
In the present study, we took advantage of the unique receptor specificities of our retargeted viruses in an attempt to resolve the contradictory evidence regarding whether or not syncytial mutations override the dependence of HSV-induced membrane fusion on gD interaction with a receptor. We reasoned that it should be advantageous to use gD-retargeted viruses for this purpose because wild-type gD can react with multiple receptors, including not only its authentic receptors, HVEM, nectin-1, and 3-OS-HS, but also some cryptic receptors, such as nectin-3 (38, 41), which would complicate the analysis and interpretation of the results. We introduced well-known syncytial mutations into gB and/or gK of our retargeted viruses and report here that gD retargeting does not prevent the formation of syncytia by cells that express the target receptor. We present the results of separate analyses of direct virus entry and lateral spread, which showed in turn that the syncytial mutations did not impair the specificity of entry of retargeted viruses at either stage. These data clearly demonstrate that the hyperfusogenic activity conferred by gB and gK syncytial mutations still requires an activating signal from receptor-engaged gD. We discuss the implications of this conclusion for the design of improved oncolytic HSV vectors.
MATERIALS AND METHODS
Cells.
The details on the following cell lines were described previously: African green monkey kidney Vero cells (ATCC CCL-81), Vero/Cre cells (42) (provided by David Leib, Dartmouth Medical School, NH), Vero-EpCAM cells (40), Chinese hamster ovary CHO-K1 cells (ATCC CCL-61), HVEM-transduced CHO-HVEM cells (38), nectin-1-transduced CHO-nectin-1 cells (3) (provided by Patricia Spear, Northwestern University, IL), EGFR-transduced CHO-EGFR cells (43) (provided by Stephen Russell, Mayo Clinic, MN), baby hamster kidney J1-1.2 cells (44) (provided by Gabriella Campadelli-Fiume, University of Bologna, Italy), HVEM-transduced J-HVEM cells (45), nectin-1-transduced J-nectin-1 cells (46), and EGFR-transduced J-EGFR cells (47). Human pancreatic carcinoma PANC-1 cells (ATCC CRL-1469) were cultured in Dulbecco's modified Eagle's medium (DMEM; Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, MA). Human pancreatic carcinoma AsPC-1 cells (ATCC CRL-1682), PK-8 cells (RBRC RCB2700), BxPC-3 cells (ATCC CRL-1687), human bile duct carcinoma HuCCT1 cells (RBRC RCB1960), and murine colon carcinoma CT26 cells (ATCC CRL-2638) were cultured in RPMI 1640 medium (Nacalai Tesque) supplemented with 10% FBS. CT26-EGFR cells were established by infection of CT26 cells with a human EGFR-expressing retroviral vector that was produced by transfection of PLAT-A cells (provided by Toshio Kitamura, University of Tokyo, Japan) with the plasmid pMXc-puro-hEGFR, followed by selection for resistance to 10 μg/ml puromycin (Invivogen, San Diego, CA); pMXc-puro-hEGFR was created by inserting the coding sequence for human EGFR into the multicloning site of pMXc-puro (also provided by Toshio Kitamura). Human renal cell carcinoma ACHN cells (ATCC CRL-1611) were cultured in Eagle's minimal essential medium (Wako, Osaka, Japan) supplemented with 10% FBS. All the cell lines were negative for mycoplasma contamination.
HSV-BAC recombineering and viruses.
All HSV-bacterial artificial chromosome (BAC) constructs generated in this study were derived from KOS-37 BAC (42) (provided by David Leib). All BAC recombinations were performed using scarless gene modification based on the Red recombination system (48) and the plasmids pRed/ET (Gene Bridges, Heidelberg, Germany) and pBAD-I-SceI (48) (provided by Nikolaus Osterrieder, Free University of Berlin, Germany). All constructs were confirmed by PCR analysis, pulsed-field gel electrophoresis analysis of restriction enzyme digests, and targeted DNA sequencing. Targeting plasmids for Red recombination were constructed as described previously (48). Briefly, the kanamycin resistance gene flanked by an I-SceI restriction site (I-SceI–aphAI fragment) was amplified from pEPkan-S2 (48) (also provided by Nikolaus Osterrieder) by PCR with different targeting primers as specified below. All targeting fragments for Red recombination were purified using a Qiagen gel extraction kit (Qiagen, Hilden, Germany) or Qiagen PCR purification kit (Qiagen). The BAC constructs pKGN, pKGNE, and pKGNEp were described previously (40). The BAC construct pKGNE-Kt was generated by exchanging the codon for alanine (GCG) at residue 40 in the gK gene of pKGNE with a codon for threonine (ACC). The targeting fragment containing gK-40T-I-SceI-aphAI-40T-gK was obtained by a PCR using pEPkan-S2 as the template, with the primers 5′-ACCGTCTTCGGTGCCAGTCCGCTGCACCGATGTATTTACACCGTACGCCCCACCGGCACCAGGATGACGACGATAAGTAGGG-3′ and 5′-ACGAGGGCGGTGTCGTTGTTGGTGCCGGTGGGGCGTACGGTGTAAATACATCGGTGCAGCCTACAACCAATTAACCAATTCTGATTAG-3′, and was used for recombination with the gK region of pKGNE, followed by aphAI gene removal, resulting in pKGNE-Kt. The BAC constructs pKGNE-Bh and pKGNE-BhKt were generated by exchanging the codon for arginine (CGC) at residue 858 in the gB genes of pKGNE and pKGNE-Kt, respectively, with a codon for histidine (CAT). The targeting fragment containing gB-858H-I-SceI-aphAI-858H-gB was obtained by a PCR using pEPkan-S2 as the template, with the primers 5′-GAGATGATACGGTACATGGCCCTGGTGTCGGCCATGGAGCATACGGAACACAAGGCCAAGAGGATGACGACGATAAGTAGGG-3′ and 5′-CAGCGCGCTCGTGCCCTTCTTCTTGGCCTTGTGTTCCGTATGCTCCATGGCCGACACCAGCTACAACCAATTAACCAATTCTGATTAG-3′, and was used for recombination with the gB region of pKGNE or pKGNE-Kt followed by aphAI gene removal, resulting in pKGNE-Bh or pKGNE-BhKt, respectively. The BAC construct pKGNEp-BhKt was generated by exchanging the EGFR-retargeted gD allele (gD:224/38C-scEGFR) (39) of pKGNE-BhKt with an EpCAM-retargeted gD allele (gD:224/38C-scEpCAM) (40); the scEGFR (528) gene was provided by Izumi Kumagai, Tohoku University, Japan. The targeting fragment containing I-SceI-aphAI-scEpCAM-gD was obtained by a PCR using pgD:224/38C-scEpCAMkan (40) as the template, with the primers 5′-AAGCAGGGGTTAGGGAGTTG-3′ and 5′-TCCGGACGTCTTCGGAGGCCCC-3′, and was used for recombination with the gD region of pKGNE-BhKt followed by aphAI gene removal, resulting in pKGNEp-BhKt.
Recombinant viruses KGN, KGNE, and KGNEp were described previously (40). KGNE-Bh, KGNE-Kt, and KGNE-BhKt were established by transfection of Vero/Cre cells with pKGNE-Bh, pKGNE-Kt, and pKGNE-BhKt, respectively, followed by two rounds of limiting dilution on Vero cells. KGNEp-BhKt was established by cotransfection of Vero-EpCAM cells with pKGNEp-BhKt and pxCANCre (provided by Izumu Saito, University of Tokyo, Japan), followed by two rounds of limiting dilution on Vero-EpCAM cells. Confirmation of BAC deletion was carried out as described previously (49). Propagation, purification, and titration of viruses were performed essentially as described previously (45).
Plaque morphology.
Vero cells were infected with viruses at 30 PFU per well in multiwell plates for 24 h, fixed with 4% paraformaldehyde (Nacalai Tesque), and observed after a 10-min incubation at room temperature (RT) with 5 μg/ml wheat germ agglutinin-Alexa Fluor 594 (Thermo Fisher Scientific) and 1 μM Hoechst 33342 (Thermo Fisher Scientific). For immunofluorescence assay, Vero cells infected for 24 h were incubated with a 1:400 dilution of mouse anti-gD monoclonal antibody (MAb) DL6 (Santa Cruz Biotechnology, Santa Cruz, CA) in phosphate-buffered saline (PBS) at RT for 1 h, fixed with 4% paraformaldehyde at RT for 30 min, incubated with 10% horse serum (Thermo Fisher Scientific) in PBS (HS-PBS) at 37°C for 1 h, and finally incubated with a 1:400 dilution of Cy3-conjugated sheep anti-mouse IgG (Sigma, St. Louis, MO) in 1% HS-PBS at RT for 1 h.
Flow cytometric analysis.
Flow cytometric analyses were performed using a FACSCalibur instrument (BD Biosciences, Franklin Lakes, NJ). Mouse anti-EGFR MAb 528 (Santa Cruz Biotechnology) and an isotype (IgG2a, κ)-matched control antibody, MOPC-173 (Biolegend, San Diego, CA), were used as primary antibodies. Alexa Fluor 488–goat anti-mouse IgG(H+L) (Thermo Fisher Scientific) was used as a secondary antibody.
Plaque formation, cell killing, entry, and infectious center assays.
Plaque formation (40), cell killing (39), entry (40), and infectious center assays (45) were described previously. Fifty percent lethal doses (LD50) in cell killing assays were estimated from the sigmoid-like dose-response curves drawn by ImageJ logistic curve-fitting software (50). For entry-blocking assays, cells were incubated with 100 μg/ml mouse anti-EpCAM MAb MY24 (40, 51) or an isotype (IgG1, κ)-matched control antibody, MG1-45 (Biolegend), for 1 h at RT, infected for 2 h at 37°C, and washed with acidic buffer as described previously (38), and cells expressing enhanced green fluorescent protein (EGFP) were counted in each well at 12 h postinfection.
Statistical analysis.
Comparative analyses of the experimental data were performed using the Steel-Dwass test (for comparing four groups) or the Welch t test (for comparing two groups). Differences were considered statistically significant if the P value was <0.05.
RESULTS
Introduction of syncytial mutations into an EGFR-retargeted HSV strain.
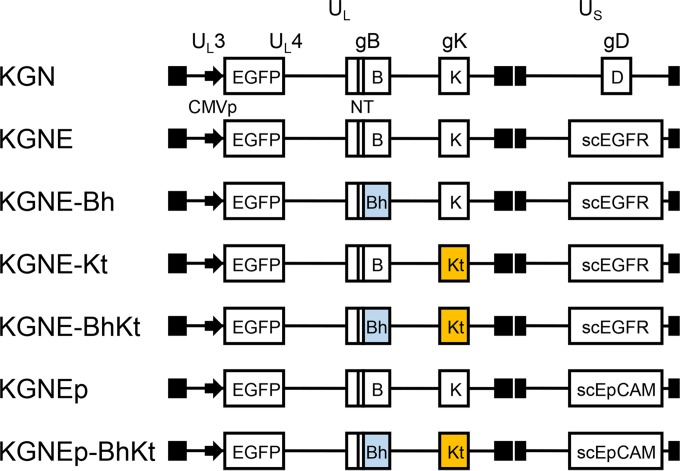
We used the Red recombination system in bacteria (48) to introduce one or two well-known syncytial (syn) mutations, i.e., an arginine-to-histidine substitution at residue 858 of gB (gB:R858H) (11) or an alanine-to-threonine substitution at residue 40 of gK (gK:A40T) (12), into pKGNE, a previously described, EGFR-retargeted HSV-1–BAC construct containing an EGFP reporter gene (40). We converted these HSV-BAC constructs into BAC-deleted infectious viruses by transfection of Cre recombinase-expressing Vero cells; the BAC region in these constructs is flanked by loxP sequences on both sides (42). Limiting dilution of representative viral clones was performed to exclude the copresence of BAC-bearing viruses, in consideration of the possibility that the presence of BAC in the viral genome could affect the efficiency of virus cell-to-cell spread. We refer to the mutant viruses as KGNE-Bh (encoding gB:R858H), KGNE-Kt (encoding gK:A40T), and KGNE-BhKt (encoding gB:R858H and gK:A40T) (Fig. 1).
FIG 1.
Genomic structures of recombinant HSVs. The schematics show representations of the genomes of the recombinant viruses used in this study. UL, unique long segment; US, unique short segment; CMVp, human cytomegalovirus immediate early (IE) promoter; scEGFR, anti-EGFR scFv-fused gD; scEpCAM, anti-EpCAM scFv-fused gD; NT, D285N/A549T mutations in gB that increase the rate of virus entry (38); Bh, R858H mutation in gB; Kt, A40T mutation in gK. Closed boxes show terminal and internal inverted repeats.
Syncytial mutations alter the plaque morphology of EGFR-retargeted HSV.
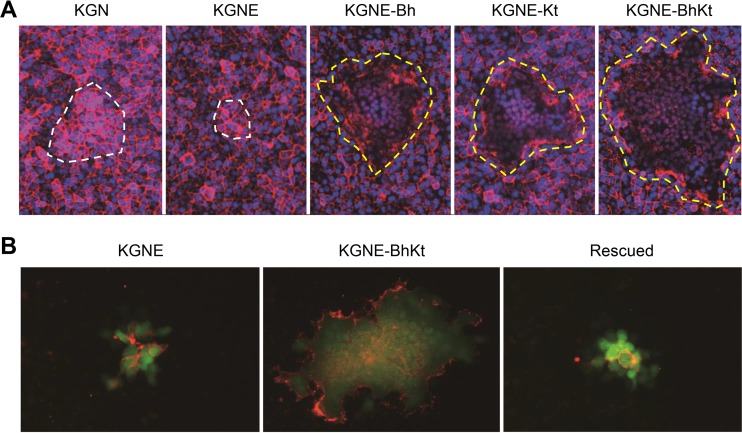
We infected Vero cells with the EGFR-retargeted viruses containing the syncytial mutations and compared their plaques to those formed by the parental EGFR-retargeted virus, KGNE (40), and the wild-type-gD version, KGN (40). The cell membranes and nuclei were stained by red fluorescence-labeled wheat germ agglutinin and Hoechst 33342, respectively. As shown in Fig. 2A, KGNE-Bh, KGNE-Kt, and KGNE-BhKt all developed large syncytia at 24 h postinfection. In contrast, KGN and KGNE did not form syncytia but caused rounding of the infected cells after several days (data not shown), as is typical of wild-type HSV-1 infection (8). To visualize the cell membranes solely of infected cells, we carried out an immunofluorescence assay using an anti-gD MAb that recognizes membrane-proximal residues of the gD ectodomain distal to the amino-terminal portion that was modified for the EGFR retargeting. As shown in Fig. 2B, KGNE yielded plaques composed of dozens of infected (green) cells that in multiple instances were visibly separated from each other by gD-positive plasma membranes, while KGNE-BhKt produced plaques that appeared as single continuous green areas, with gD outlining limited to the edges, consistent with a single plasma membrane characteristic of syncytium formation. A rescued virus, derived by replacement of the syncytial mutations in the gB and gK genes of the KGNE-BhKt BAC with the respective wild-type residues, revealed a plaque morphology similar to that of KGNE (Fig. 2B). These results indicate that the altered receptor specificity of gD was compatible with the typical hyperfusogenic activity of the syncytial mutants.
FIG 2.
Plaque morphologies of syncytial, EGFR-retargeted HSVs. (A) Vero cells infected for 24 h with the viruses indicated above the panels were stained with wheat germ agglutinin-Alexa Fluor 594 (red) and Hoechst 33342 (blue) for detection of cell membranes and nuclei, respectively. Dotted lines indicate the margins of plaques (white, nonsyncytial plaques; yellow, syncytial plaques). (B) Vero cells infected for 24 h with the viruses indicated above the panels were incubated with the anti-gD MAb DL6 and stained with a Cy3-conjugated secondary antibody. Red, Cy3 signals; green, EGFP signals. Rescued, a rescued virus derived by replacement of the gB:R858H and gK:A40T mutations of KGNE-BhKt BAC with the respective wild-type residues.
Syncytial mutations in EGFR-retargeted HSV enable extensive formation of syncytia and augment cell killing on human tumor cell lines.
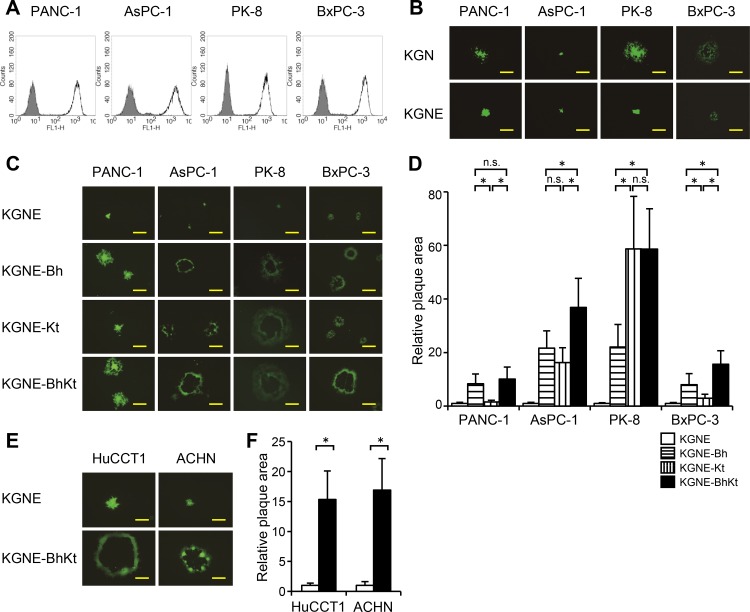
A number of reports have demonstrated that the antitumor activity of various types of oncolytic HSV can be enhanced by syncytial mutations (52–55). To determine if these effects are preserved in the context of our EGFR-retargeted HSV strain, we infected a panel of human pancreatic carcinoma cell lines (PANC-1, AsPC-1, PK-8, and BxPC-3) and examined the lateral spread of virus and cytocidal efficiency. Each of these cell lines highly expressed EGFR (Fig. 3A). However, when they were infected by KGNE, PK-8 and BxPC-3 cells showed reduced cell-to-cell spread compared to that for infection by the wild-type-gD version, KGN, whereas PANC-1 and AsPC-1 cells showed similar levels of spread by the two viruses (Fig. 3B), suggesting that cellular factors affecting the efficiency of virus cell-to-cell spread might not be limited to the level of EGFR expression. As shown in Fig. 3C, while KGNE yielded plaques or foci mostly composed of fewer than 100 individual infected cells at 72 h postinfection, all three syn mutant viruses, KGNE-Bh, KGNE-Kt, and KGNE-BhKt, formed syncytia on each cell line at this point. These results demonstrate that gD retargeting does not abolish the hyperfusogenic activity of syncytial mutants on human cancer cells.
FIG 3.
Lateral spread of syncytial, EGFR-retargeted HSVs on human carcinoma cell lines. (A) Surface expression of EGFR as analyzed by flow cytometry. Closed histograms represent staining using an isotype-matched negative-control IgG as the primary antibody. Open histograms represent staining using the anti-EGFR MAb 528 as the primary antibody. (B, C, and E) The cell lines listed above the panels were infected for 2 h with the viruses indicated to the left and then overlaid with methylcellulose-containing medium. EGFP signals were recorded at 3 days postinfection. Photographs of representative plaques are shown. Bars, 500 μm (B), 500 μm (C), and 1 mm (E). (D and F) Mean areas of plaques (n = 15) in panels C and E, normalized to the respective means of KGNE plaque areas. Error bars represent standard deviations. White bars, KGNE; horizontally striped bars, KGNE-Bh; vertically striped bars, KGNE-Kt; black bars, KGNE-BhKt. *, P < 0.05 by the Steel-Dwass test (D) or the Welch t test (F); n.s., not significant.
Measurement of plaque areas revealed that KGNE-Bh formed significantly larger syncytia than those of KGNE-Kt on the PANC-1 and BxPC-3 cell lines (Fig. 3D); the differences in plaque area between KGNE and KGNE-Kt on these two cell lines were not remarkable, albeit they were statistically significant. Conversely, KGNE-Kt formed significantly larger syncytia than those of KGNE-Bh on PK-8 cells (Fig. 3D). Of particular interest, while the double mutant KGNE-BhKt virus developed syncytia on PANC-1 and PK-8 cells that were similar in size to those of each of the more efficient of the single mutant viruses on these lines (KGNE-Bh and KGNE-Kt, respectively), it formed larger syncytia than those of either KGNE-Bh or KGNE-Kt on both the AsPC-1 and BxPC-3 cell lines (Fig. 3D). Thus, the two syncytial mutations appear to affect distinct events in the cell-cell fusion process induced by EGFR-retargeted HSV.
To determine the extent of syncytium formation by the double mutant KGNE-BhKt virus on other types of human cancer cells, we performed plaque assays on bile duct carcinoma HuCCT1 and renal cell carcinoma ACHN cells that express EGFR on the surface (data not shown). As shown in Fig. 3E and F, KGNE-BhKt generated large syncytia on both cell lines, while KGNE produced small plaques without evidence of cell-cell fusion. These findings indicate that EGFR-retargeted HSV with combined syncytial mutations can produce increased lateral spread in a variety of tumor cell types.
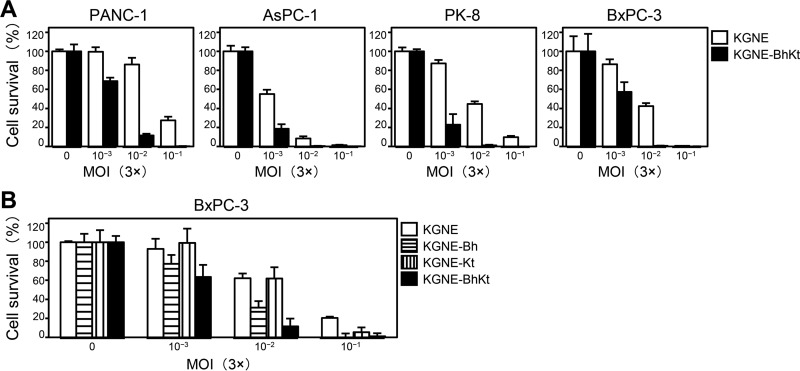
To assess the cell killing potential of KGNE-BhKt, the pancreatic cell lines described above were infected with increasing amounts of virus, and cell viability was determined by a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay at 4 days postinfection. As shown in Fig. 4A, KGNE-BhKt showed significantly increased cell killing activity compared to that of KGNE on all four lines; the difference in 50% lethal dose (LD50) between the two viruses was 19-fold for PANC-1 cells, 2.6-fold for AsPC-1 cells, 18-fold for PK-8 cells, and 6.5-fold for BxPC-3 cells (data not shown). Furthermore, we compared the cell killing efficiencies of the single mutant viruses, KGNE-Bh and KGNE-Kt, to that of the double mutant KGNE-BhKt on BxPC-3 cells, because the plaque formation assay (Fig. 3D) showed that the robustness of syncytium formation was significantly different among the three viruses on this cell line. As shown in Fig. 4B, KGNE-BhKt showed increased cell killing activity over that of KGNE-Bh and KGNE-Kt. In addition, KGNE-Bh showed more robust cell killing activity than KGNE-Kt (Fig. 4B); the LD50 multiplicities of infection (MOIs) were 5.3 × 10−2 for KGNE, 1.3 × 10−2 for KGNE-Bh, 3.9 × 10−2 for KGNE-Kt, and 4.9 × 10−3 for KGNE-BhKt (data not shown). These results demonstrate that the relative robustness of cell killing activities among these four viruses paralleled their plaque sizes on BxPC-3 cells and that the two syncytial mutations together can increase the in vitro efficiency of cancer cell killing by our gD-retargeted HSV strain.
FIG 4.
Cell killing activity of syncytial, EGFR-retargeted HSVs. (A and B) The cell lines indicated above the panels were infected at MOIs ranging from 0.003 to 0.3 for 96 h, and percent cell viability relative to that of uninfected cells was measured by MTT assay. Means for 6 replicates are shown, and error bars represent standard deviations. White bars, KGNE; horizontally striped bars, KGNE-Bh; vertically striped bars, KGNE-Kt; black bars, KGNE-BhKt.
Syncytial mutations do not impair the specificity of EGFR-retargeted HSV entry and lateral spread.
Since HSV syn mutations appear to act by lowering the kinetic barrier to membrane fusion (35), it is conceivable that they can cause off-target fusion of retargeted HSV at the similar but mechanistically distinct stages of initial virus entry and subsequent lateral spread. Therefore, we separately assessed the EGFR dependence of entry and spread by performing direct entry and infectious center assays, respectively; infectious center assays specifically examine viral cell-to-cell spread by comparing different “acceptor” cells for plaque formation induced by a single source of intracellular virus (donor cells) (56, 57).
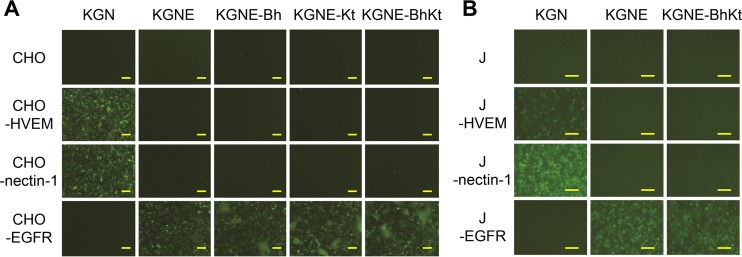
We analyzed the specificity of initial virus entry by using the Chinese hamster ovary CHO-K1 (CHO) cell line, which is resistant to HSV infection because of the absence of gD receptors, and derivative lines which had been transduced with human HVEM (CHO-HVEM), nectin-1 (CHO-nectin-1), or EGFR (CHO-EGFR). Viral entry was identified qualitatively by EGFP fluorescence at 12 h postinfection. As shown in Fig. 5A, KGN at an MOI of 3 showed minimal entry into CHO-K1 or CHO-EGFR cells but efficiently entered CHO-HVEM and CHO-nectin-1 cells, which express authentic gD receptors. In contrast, efficient entry of KGNE at the same MOI was observed only for CHO-EGFR cells (Fig. 5A), consistent with our previous work (39, 40). All three syn mutant viruses entered exclusively into CHO-EGFR cells (Fig. 5A). Similar EGFR-specific entry results were obtained for KGNE-BhKt in another gD receptor-deficient cell line, i.e., baby hamster kidney J1.1-2 (J), and its derivatives, J-HVEM, J-nectin-1, and J-EGFR (Fig. 5B). These results clearly demonstrate that the syncytial mutations did not impair the specificity of primary entry by the EGFR-retargeted HSV strain. Thus, gD-receptor interaction stringently dictates the tropism of HSV entry, even in the presence of syncytial mutations.
FIG 5.
Specificity of entry by syncytial, EGFR-retargeted HSVs. The cell lines indicated to the left of the photographs were infected for 12 h (A) or 8 h (B) with the viruses indicated above the panels at an MOI of 3, and EGFP fluorescence was visualized. Bars, 125 μm.
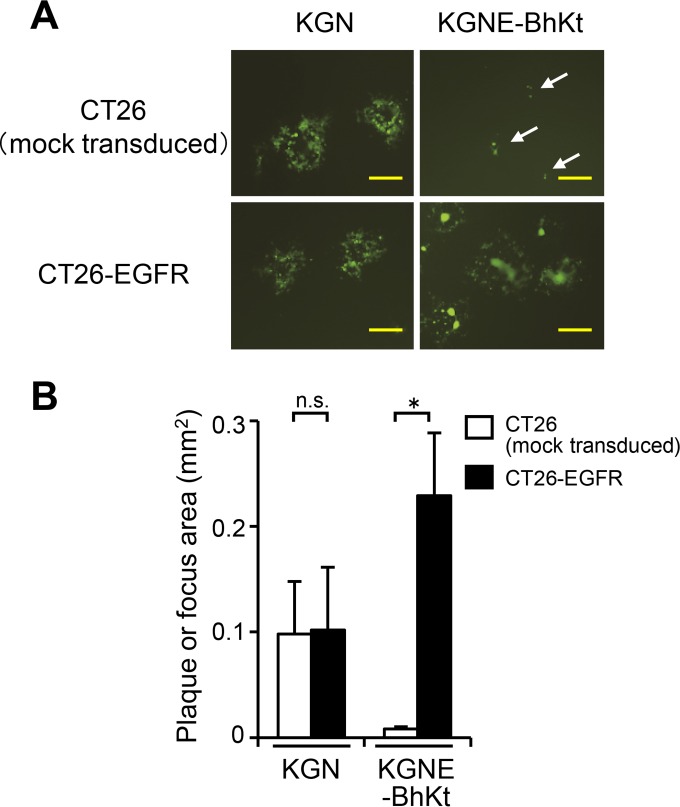
To perform infectious center assays that examined the EGFR selectivity of KGNE-BhKt-induced lateral spread, we used murine colon carcinoma CT26 cells, which have been reported to lack EGFR expression (58), and a derivative cell line that we created by transduction with human EGFR (CT26-EGFR cells). CT26 cells are susceptible to wild-type HSV and have been used by others to test the efficacy of oncolytic HSVs (59, 60). CT26-EGFR cells were infected with KGN or KGNE-BhKt at an MOI of 10 to achieve 100% infection. After residual extracellular virions were inactivated by an acidic wash, equal numbers of infected (donor) cells were seeded onto monolayers of uninfected (acceptor) cells, either CT26-EGFR cells or mock-transduced CT26 cells. The cultures were overlaid with 1% methylcellulose, and plaque formation was assessed 2 days later by examination of EGFP fluorescence. As shown in Fig. 6A and B, plaques formed by KGN bearing wild-type gD were readily detected and similar in size, irrespective of cellular EGFR expression, in accordance with the reported HSV susceptibility of CT26 cells (59, 60). In contrast, KGNE-BhKt developed syncytia on CT26-EGFR acceptor cells, while only single infected (presumably donor) cells or very small foci of single infected cells were detected in mock-transduced CT26 acceptor cells (Fig. 6A and B). Similar results were obtained in additional experiments that used a different CT26-EGFR clone (data not shown), arguing against a clone-specific effect for these outcomes. These observations indicate that cellular EGFR expression is required for syncytium formation by KGNE-BhKt. Taken together, these results led us to conclude that introduction of syncytial mutations does not impair the specificity of either entry or spread of our EGFR-retargeted HSV strain.
FIG 6.
Specificity of lateral spread by KGNE-BhKt. (A) CT26-EGFR cells were infected with the viruses indicated above the panels (MOI of 10). Extracellular viruses were inactivated by an acidic wash, and equal numbers of infected (donor) cells were added to monolayers of the uninfected (acceptor) cells indicated to the left. The mixed cultures were overlaid with methylcellulose-containing medium, and EGFP signals were recorded at 2 days postinfection. Bars, 500 μm. Arrows show single green cells or small foci. (B) Mean areas of plaques or foci (n = 15) in the wells examined for panel A. Error bars represent standard deviations. White bars, CT26 (mock-transduced) cells; black bars, CT26-EGFR cells. *, P < 0.05 by the Welch t test; n.s., not significant.
Syncytial mutations enable the formation of large syncytia by EpCAM-retargeted HSV.
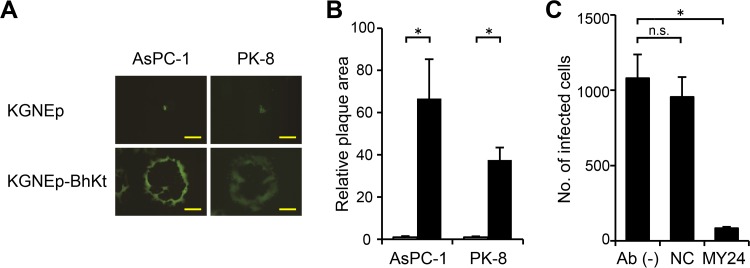
To extend our observations, we tested whether the combined syncytial mutations would confer hyperfusogenic activity on an HSV strain retargeted to a different tumor-associated antigen, EpCAM. We replaced the anti-EGFR scFv fused to detargeted gD in the KGNE-BhKt BAC construct with an anti-EpCAM scFv (40) to generate the recombinant virus KGNEp-BhKt (Fig. 1), and we performed plaque assays on AsPC-1 and PK-8 cells, which express EpCAM on the surface (data not shown). As shown in Fig. 7A, KGNEp-BhKt developed large syncytia on both cell lines at 3 days postinfection, whereas KGNEp (40), an EpCAM-retargeted virus without syncytial mutations, formed only small foci mostly composed of fewer than several dozen individual infected cells. The differences in mean plaque areas were 67-fold for AsPC-1 and 37-fold for PK-8 cells (Fig. 7B). We examined whether KGNEp-BhKt infection is also dependent on the cellular expression of its target receptor. We pretreated AsPC-1 cells with the anti-EpCAM MAb MY24 and then performed a virus entry assay. As shown in Fig. 7C, entry of KGNEp-BhKt was efficiently inhibited by MY24, while it was not inhibited by the isotype-matched control antibody. These data further supported our conclusion that introduction of syncytial mutations does not impair the specificity of our gD-retargeted HSV strains. Taken together, these results indicate that the type of receptor recognized by retargeted gD is not a critical determinant for establishment of syncytia. In addition, they suggest that syncytial mutations can be used to enhance the spreading efficiency of different receptor-retargeted HSV vectors.
FIG 7.
Lateral spread of syncytial, EpCAM-retargeted HSV on human cancer cells. (A) The cell lines listed above the panels were infected for 2 h with the viruses indicated to the left and then overlaid with methylcellulose-containing medium. EGFP signals were recorded at 3 days postinfection. Photographs of representative plaques are shown. Bars, 500 μm. (B) Mean areas of the plaques (n = 15) from panel A normalized to the respective means of KGNE plaque areas. Error bars represent standard deviations. White bars, KGNEp; black bars, KGNEp-BhKt. *, P < 0.05 by the Welch t test. (C) Inhibition of KGNEp-BhKt entry by pretreatment of AsPC-1 cells with 100 μg/ml anti-EpCAM MAb MY24 or an isotype-matched negative-control antibody, as indicated below the columns. Pretreated cells were incubated with KGNEp-BhKt at an MOI of 0.01 for 2 h, extracellular viruses were inactivated, and cells expressing EGFP were counted at 12 h postinfection. Means for 3 replicates are shown, and error bars represent standard deviations. Ab (−), no antibody; NC, isotype-matched negative-control antibody (MG1-45). *, P < 0.05 by the Welch t test; n.s., not significant.
DISCUSSION
Our investigations have shown that the syncytial mutations gB:R858H and gK:A40T do not compromise the specificities of EGFR-retargeted HSV entry and lateral spread. Furthermore, our results illustrate that gD retargeted to either of two noncanonical receptors (EGFR and EpCAM) does not abolish the hyperfusogenic activity of gB and gK syncytial mutants. These results support the conclusion that syncytial HSV strains remain dependent on the interaction of gD with a specific receptor for both entry and spread, although the nature of the receptor appears to be less consequential.
We have shown that the relative robustness of syncytium formation by the EGFR-retargeted gB and gK syn mutants varied among several human pancreatic carcinoma cell lines tested and that the two mutations combined were more effective than each individual mutation at inducing cell-cell fusion and killing cells in certain cell lines. Thus, the efficiencies of syncytium formation and cell killing were shown to be dependent on both the syncytial mutation(s) introduced and unknown host cell characteristics. These observations are consistent with the supposition that the locations of the gB and gK mutations opposite each other on the cell or viral membrane indicate that they act by different mechanisms (the gB:R858H mutation is located in the cytodomain, whereas the gK:A40T mutation is in the ectodomain).
Rogalin and Heldwein recently used virus-free cell-cell fusion experiments to demonstrate that gB syn mutations reduce the kinetic barrier to fusion, possibly by rendering gB more sensitive to activation by gH/gL or by increasing the rate of fusion after the activating event (35). From the results of structural and biochemical studies, the same group also proposed that the gB cytodomain may restrict the fusogenic activity of gB by shielding certain key residues in a structure that involves components of the cell membrane (34). This would be consistent with any effects of host cell characteristics, such as membrane lipid composition or surface charge distribution, on the phenotype of the R858H mutation.
How gK syncytial mutations mediate a hyperfusogenic phenotype remains unclear. Chouljenko and colleagues reported that an N-terminal gK peptide that encompassed the positions of common syncytial mutations can directly bind to the ectodomain of gB, which suggests that this domain in gB plays a role in the regulation of the fusion activity of gB and is responsive to changes in gK (61). They used a computer-assisted prediction of structure to suggest that the gK N terminus may bind predominantly to gB domain I (61), which contains fusion loops (26, 27); the gK mutations may affect the insertion of the fusion loops into the membrane of the target cell. Additionally, Chouljenko et al. showed that the N terminus of native gK can bind to gH as well, which suggests an alternative scenario in which mutations in this domain may facilitate the activation of the fusion activity of gB by gH (61). It should be noted that our receptor-retargeted HSVs contain two mutations in the gB ectodomain, D285N and A549T, that increase the rate of virus entry (38). However, since these mutations do not cause syncytium formation or enhance lateral spread (38, 57), it is unlikely that they directly influence the hyperfusogenic effects of syncytial mutants. Nevertheless, since the D285N mutation is positioned in gB domain I, it may potentially modify the gB-gK interface, and thereby the effects of gK syncytial mutations, a scenario that remains to be addressed.
HSV-1 has recently become a promising tool for oncolytic virotherapy (1). Many of the oncolytic HSVs that have been tested clinically, to date, have been modified genetically to inactivate or delete viral genes, such as the infected cell polypeptide 34.5 (ICP34.5) gene and the ICP6 gene, which are essential for replication in normal but not tumor cells (62–68). Although the results of clinical trials have shown that these types of oncolytic HSV can safely be administered to humans, there is significant room for improvement in treatment efficacy, which is hampered, at least in part, by the reduced capacity of these viruses to replicate in tumor cells (69). Several groups of investigators have sought to take advantage of cell-cell fusion mechanisms to overcome the problem and have reported that syncytial mutations can enhance the potency of oncolytic HSV either or both in vitro and in vivo (52–55). Since transductional targeting diminishes the dependence of oncolytic HSV on attenuating mutations for safety, our observations that gD retargeting does not interfere with syncytium formation by gB and gK syn mutants and that even a combination of syn mutations does not abolish the strict dependence of infection on the target receptor suggest that retargeted syn mutant HSV may offer oncolytic activity superior to that of the previous oncolytic syn mutant HSV strain. While gD retargeting alone may not be sufficient to fully restrict replication of the virus to tumor cells, we previously showed that the incorporation of specific microRNA-responsive elements (miR-T) into the viral genome enhances tumor cell specificity, and thereby safety, without reducing oncolytic activity (70). Future direct comparisons of different combinations of these features may clarify which oncolytic HSV design best serves the dual demands of safety and effective oncolytic activity.
ACKNOWLEDGMENTS
We thank Izumi Kumagai, David Leib, Nikolaus Osterrieder, Patricia Spear, Stephen Russell, Gabriella Campadelli-Fiume, Toshio Kitamura, and Izumu Saito for reagents. We also thank Tetsuro Watabe, Kiyoko Fukami, Kenichiro Asano, Takeshi Fukuhara, and Yasuhiro Yoshimatsu (Tokyo University of Pharmacy and Life Sciences) for helpful discussions. This study was supported by the Imaging Core Laboratory, The Institute of Medical Science, The University of Tokyo.
H.U., J.B.C., and J.C.G. are coinventors of intellectual property licensed to Oncorus, Inc. (U.S. patent applications 13/641,649 and 15/032,958). J.C.G. is a founder of Oncorus, Inc., and Chairman of the Scientific Advisory Board.
Funding Statement
This work, including the efforts of Hiroaki Uchida, was funded by Japan Society for the Promotion of Science (JSPS) (25290059 and 15K15144), Takeda Science Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, Daiwa Securities Health Foundation, and Sumitomo Foundation. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Peters C, Rabkin SD. 2015. Designing herpes viruses as oncolytics. Mol Ther Oncolytics 2:15010. doi: 10.1038/mto.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436. doi: 10.1016/S0092-8674(00)81363-X. [DOI] [PubMed] [Google Scholar]
- 3.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 4.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13–22. doi: 10.1016/S0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 5.Carfi A, Willis SH, Whitbeck JC, Krummenacher C, Cohen GH, Eisenberg RJ, Wiley DC. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell 8:169–179. doi: 10.1016/S1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 6.Fusco D, Forghieri C, Campadelli-Fiume G. 2005. The pro-fusion domain of herpes simplex virus glycoprotein D (gD) interacts with the gD N terminus and is displaced by soluble forms of viral receptors. Proc Natl Acad Sci U S A 102:9323–9328. doi: 10.1073/pnas.0503907102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, Eisenberg RJ, Cohen GH, Wiley DC, Carfi A. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J 24:4144–4153. doi: 10.1038/sj.emboj.7600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 9.Ruyechan WT, Morse LS, Knipe DM, Roizman B. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol 29:677–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Read GS, Person S, Keller PM. 1980. Genetic studies of cell fusion induced by herpes simplex virus type 1. J Virol 35:105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bzik DJ, Fox BA, DeLuca NA, Person S. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- 12.Debroy C, Pederson N, Person S. 1985. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology 145:36–48. doi: 10.1016/0042-6822(85)90199-0. [DOI] [PubMed] [Google Scholar]
- 13.Cai WH, Gu B, Person S. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol 62:2596–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baghian A, Huang L, Newman S, Jayachandra S, Kousoulas KG. 1993. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J Virol 67:2396–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel JP, Boyer EP, Goodman JL. 1993. Two novel single amino acid syncytial mutations in the carboxy terminus of glycoprotein B of herpes simplex virus type 1 confer a unique pathogenic phenotype. Virology 192:112–120. doi: 10.1006/viro.1993.1013. [DOI] [PubMed] [Google Scholar]
- 16.Gage PJ, Levine M, Glorioso JC. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J Virol 67:2191–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster TP, Melancon JM, Kousoulas KG. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18–29. doi: 10.1006/viro.2001.1004. [DOI] [PubMed] [Google Scholar]
- 18.Diakidi-Kosta A, Michailidou G, Kontogounis G, Sivropoulou A, Arsenakis M. 2003. A single amino acid substitution in the cytoplasmic tail of the glycoprotein B of herpes simplex virus 1 affects both syncytium formation and binding to intracellular heparan sulfate. Virus Res 93:99–108. doi: 10.1016/S0168-1702(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 19.Bond VC, Person S. 1984. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology 132:368–376. doi: 10.1016/0042-6822(84)90042-4. [DOI] [PubMed] [Google Scholar]
- 20.Pogue-Geile KL, Spear PG. 1987. The single base pair substitution responsible for the Syn phenotype of herpes simplex virus type 1, strain MP. Virology 157:67–74. doi: 10.1016/0042-6822(87)90314-X. [DOI] [PubMed] [Google Scholar]
- 21.Dolter KE, Ramaswamy R, Holland TC. 1994. Syncytial mutations in the herpes simplex virus type 1 gK (UL53) gene occur in two distinct domains. J Virol 68:8277–8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baines JD, Ward PL, Campadelli-Fiume G, Roizman B. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol 65:6414–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLean CA, Efstathiou S, Elliott ML, Jamieson FE, McGeoch DJ. 1991. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J Gen Virol 72:897–906. doi: 10.1099/0022-1317-72-4-897. [DOI] [PubMed] [Google Scholar]
- 24.Tognon M, Guandalini R, Romanelli MG, Manservigi R, Trevisani B. 1991. Phenotypic and genotypic characterization of locus Syn 5 in herpes simplex virus 1. Virus Res 18:135–150. doi: 10.1016/0168-1702(91)90014-M. [DOI] [PubMed] [Google Scholar]
- 25.Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J Virol 81:4858–4865. doi: 10.1128/JVI.02755-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hannah BP, Cairns TM, Bender FC, Whitbeck JC, Lou H, Eisenberg RJ, Cohen GH. 2009. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J Virol 83:6825–6836. doi: 10.1128/JVI.00301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 28.Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J Virol 84:12292–12299. doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melancon JM, Luna RE, Foster TP, Kousoulas KG. 2005. Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and UL20p function redundantly in virion de-envelopment. J Virol 79:299–313. doi: 10.1128/JVI.79.1.299-313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chouljenko VN, Iyer AV, Chowdhury S, Chouljenko DV, Kousoulas KG. 2009. The amino terminus of herpes simplex virus type 1 glycoprotein K (gK) modulates gB-mediated virus-induced cell fusion and virion egress. J Virol 83:12301–12313. doi: 10.1128/JVI.01329-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner A, Bruun B, Minson T, Browne H. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol 72:873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avitabile E, Lombardi G, Campadelli-Fiume G. 2003. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J Virol 77:6836–6844. doi: 10.1128/JVI.77.12.6836-6844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foster TP, Alvarez X, Kousoulas KG. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J Virol 77:499–510. doi: 10.1128/JVI.77.1.499-510.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silverman JL, Greene NG, King DS, Heldwein EE. 2012. Membrane requirement for folding of the herpes simplex virus 1 gB cytodomain suggests a unique mechanism of fusion regulation. J Virol 86:8171–8184. doi: 10.1128/JVI.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogalin HB, Heldwein EE. 2015. Interplay between the herpes simplex virus 1 gB cytodomain and the gH cytotail during cell-cell fusion. J Virol 89:12262–12272. doi: 10.1128/JVI.02391-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cocchi F, Menotti L, Dubreuil P, Lopez M, Campadelli-Fiume G. 2000. Cell-to-cell spread of wild-type herpes simplex virus type 1, but not of syncytial strains, is mediated by the immunoglobulin-like receptors that mediate virion entry, nectin1 (PRR1/HveC/HIgR) and nectin2 (PRR2/HveB). J Virol 74:3909–3917. doi: 10.1128/JVI.74.8.3909-3917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Even DL, Henley AM, Geraghty RJ. 2006. The requirements for herpes simplex virus type 1 cell-cell spread via nectin-1 parallel those for virus entry. Virus Res 119:195–207. doi: 10.1016/j.virusres.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Uchida H, Chan J, Goins WF, Grandi P, Kumagai I, Cohen JB, Glorioso JC. 2010. A double mutation in glycoprotein gB compensates for ineffective gD-dependent initiation of herpes simplex virus type 1 infection. J Virol 84:12200–12209. doi: 10.1128/JVI.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchida H, Marzulli M, Nakano K, Goins WF, Chan J, Hong CS, Mazzacurati L, Yoo JY, Haseley A, Nakashima H, Baek H, Kwon H, Kumagai I, Kuroki M, Kaur B, Chiocca EA, Grandi P, Cohen JB, Glorioso JC. 2013. Effective treatment of an orthotopic xenograft model of human glioblastoma using an EGFR-retargeted oncolytic herpes simplex virus. Mol Ther 21:561–569. doi: 10.1038/mt.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibata T, Uchida H, Shiroyama T, Okubo Y, Suzuki T, Ikeda H, Yamaguchi M, Miyagawa Y, Fukuhara T, Cohen JB, Glorioso JC, Watabe T, Hamada H, Tahara H. 2016. Development of an oncolytic HSV vector fully retargeted specifically to cellular EpCAM for virus entry and cell-to-cell spread. Gene Ther 23:479–488. doi: 10.1038/gt.2016.17. [DOI] [PubMed] [Google Scholar]
- 41.Cocchi F, Menotti L, Di Ninni V, Lopez M, Campadelli-Fiume G. 2004. The herpes simplex virus JMP mutant enters receptor-negative J cells through a novel pathway independent of the known receptors nectin1, HveA, and nectin2. J Virol 78:4720–4729. doi: 10.1128/JVI.78.9.4720-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gierasch WW, Zimmerman DL, Ward SL, Vanheyningen TK, Romine JD, Leib DA. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J Virol Methods 135:197–206. doi: 10.1016/j.jviromet.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura T, Peng KW, Vongpunsawad S, Harvey M, Mizuguchi H, Hayakawa T, Cattaneo R, Russell SJ. 2004. Antibody-targeted cell fusion. Nat Biotechnol 22:331–336. doi: 10.1038/nbt942. [DOI] [PubMed] [Google Scholar]
- 44.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J Virol 72:9992–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uchida H, Shah WA, Ozuer A, Frampton AR Jr, Goins WF, Grandi P, Cohen JB, Glorioso JC. 2009. Generation of herpesvirus entry mediator (HVEM)-restricted herpes simplex virus type 1 mutant viruses: resistance of HVEM-expressing cells and identification of mutations that rescue nectin-1 recognition. J Virol 83:2951–2961. doi: 10.1128/JVI.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frampton AR Jr, Stolz DB, Uchida H, Goins WF, Cohen JB, Glorioso JC. 2007. Equine herpesvirus 1 enters cells by two different pathways, and infection requires the activation of the cellular kinase ROCK1. J Virol 81:10879–10889. doi: 10.1128/JVI.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakano K, Kobayashi M, Nakamura K, Nakanishi T, Asano R, Kumagai I, Tahara H, Kuwano M, Cohen JB, Glorioso JC. 2011. Mechanism of HSV infection through soluble adapter-mediated virus bridging to the EGF receptor. Virology 413:12–18. doi: 10.1016/j.virol.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 49.Miyagawa Y, Marino P, Verlengia G, Uchida H, Goins WF, Yokota S, Geller DA, Yoshida O, Mester J, Cohen JB, Glorioso JC. 2015. Herpes simplex viral-vector design for efficient transduction of nonneuronal cells without cytotoxicity. Proc Natl Acad Sci U S A 112:E1632–E1641. doi: 10.1073/pnas.1423556112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaguchi M, Nishii Y, Nakamura K, Aoki H, Hirai S, Uchida H, Sakuma Y, Hamada H. 2014. Development of a sensitive screening method for selecting monoclonal antibodies to be internalized by cells. Biochem Biophys Res Commun 454:600–603. doi: 10.1016/j.bbrc.2014.10.133. [DOI] [PubMed] [Google Scholar]
- 52.Fu X, Zhang X. 2002. Potent systemic antitumor activity from an oncolytic herpes simplex virus of syncytial phenotype. Cancer Res 62:2306–2312. [PubMed] [Google Scholar]
- 53.Takaoka H, Takahashi G, Ogawa F, Imai T, Iwai S, Yura Y. 2011. A novel fusogenic herpes simplex virus for oncolytic virotherapy of squamous cell carcinoma. Virol J 8:294. doi: 10.1186/1743-422X-8-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Israyelyan AH, Melancon JM, Lomax LG, Sehgal I, Leuschner C, Kearney MT, Chouljenko VN, Baghian A, Kousoulas KG. 2007. Effective treatment of human breast tumor in a mouse xenograft model with herpes simplex virus type 1 specifying the NV1020 genomic deletion and the gBsyn3 syncytial mutation enabling high viral replication and spread in breast cancer cells. Hum Gene Ther 18:457–473. doi: 10.1089/hum.2006.145. [DOI] [PubMed] [Google Scholar]
- 55.Israyelyan A, Chouljenko VN, Baghian A, David AT, Kearney MT, Kousoulas KG. 2008. Herpes simplex virus type-1 (HSV-1) oncolytic and highly fusogenic mutants carrying the NV1020 genomic deletion effectively inhibit primary and metastatic tumors in mice. Virol J 5:68. doi: 10.1186/1743-422X-5-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roller RJ, Rauch D. 1998. Herpesvirus entry mediator HVEM mediates cell-cell spread in BHK(TK−) cell clones. J Virol 72:1411–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uchida H, Chan J, Shrivastava I, Reinhart B, Grandi P, Glorioso JC, Cohen JB. 2013. Novel mutations in gB and gH circumvent the requirement for known gD receptors in herpes simplex virus 1 entry and cell-to-cell spread. J Virol 87:1430–1442. doi: 10.1128/JVI.02804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castle JC, Loewer M, Boegel S, de Graaf J, Bender C, Tadmor AD, Boisguerin V, Bukur T, Sorn P, Paret C, Diken M, Kreiter S, Tureci O, Sahin U. 2014. Immunomic, genomic and transcriptomic characterization of CT26 colorectal carcinoma. BMC Genomics 15:190. doi: 10.1186/1471-2164-15-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toda M, Rabkin SD, Kojima H, Martuza RL. 1999. Herpes simplex virus as an in situ cancer vaccine for the induction of specific anti-tumor immunity. Hum Gene Ther 10:385–393. doi: 10.1089/10430349950018832. [DOI] [PubMed] [Google Scholar]
- 60.Esaki S, Goshima F, Kimura H, Murakami S, Nishiyama Y. 2013. Enhanced antitumoral activity of oncolytic herpes simplex virus with gemcitabine using colorectal tumor models. Int J Cancer 132:1592–1601. doi: 10.1002/ijc.27823. [DOI] [PubMed] [Google Scholar]
- 61.Chouljenko VN, Iyer AV, Chowdhury S, Kim J, Kousoulas KG. 2010. The herpes simplex virus type 1 UL20 protein and the amino terminus of glycoprotein K (gK) physically interact with gB. J Virol 84:8596–8606. doi: 10.1128/JVI.00298-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu BL, Robinson M, Han ZQ, Branston RH, English C, Reay P, McGrath Y, Thomas SK, Thornton M, Bullock P, Love CA, Coffin RS. 2003. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther 10:292–303. doi: 10.1038/sj.gt.3301885. [DOI] [PubMed] [Google Scholar]
- 63.MacLean AR, ul-Fareed M, Robertson L, Harland J, Brown SM. 1991. Herpes simplex virus type 1 deletion variants 1714 and 1716 pinpoint neurovirulence-related sequences in Glasgow strain 17+ between immediate early gene 1 and the ‘a’ sequence. J Gen Virol 72:631–639. doi: 10.1099/0022-1317-72-3-631. [DOI] [PubMed] [Google Scholar]
- 64.Meignier B, Longnecker R, Roizman B. 1988. In vivo behavior of genetically engineered herpes simplex viruses R7017 and R7020: construction and evaluation in rodents. J Infect Dis 158:602–614. doi: 10.1093/infdis/158.3.602. [DOI] [PubMed] [Google Scholar]
- 65.Mineta T, Rabkin SD, Yazaki T, Hunter WD, Martuza RL. 1995. Attenuated multi-mutated herpes simplex virus-1 for the treatment of malignant gliomas. Nat Med 1:938–943. doi: 10.1038/nm0995-938. [DOI] [PubMed] [Google Scholar]
- 66.Todo T, Martuza RL, Rabkin SD, Johnson PA. 2001. Oncolytic herpes simplex virus vector with enhanced MHC class I presentation and tumor cell killing. Proc Natl Acad Sci U S A 98:6396–6401. doi: 10.1073/pnas.101136398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parker JN, Gillespie GY, Love CE, Randall S, Whitley RJ, Markert JM. 2000. Engineered herpes simplex virus expressing IL-12 in the treatment of experimental murine brain tumors. Proc Natl Acad Sci U S A 97:2208–2213. doi: 10.1073/pnas.040557897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chase M, Chung RY, Chiocca EA. 1998. An oncolytic viral mutant that delivers the CYP2B1 transgene and augments cyclophosphamide chemotherapy. Nat Biotechnol 16:444–448. doi: 10.1038/nbt0598-444. [DOI] [PubMed] [Google Scholar]
- 69.Kramm CM, Chase M, Herrlinger U, Jacobs A, Pechan PA, Rainov NG, Sena-Esteves M, Aghi M, Barnett FH, Chiocca EA, Breakefield XO. 1997. Therapeutic efficiency and safety of a second-generation replication-conditional HSV1 vector for brain tumor gene therapy. Hum Gene Ther 8:2057–2068. doi: 10.1089/hum.1997.8.17-2057. [DOI] [PubMed] [Google Scholar]
- 70.Mazzacurati L, Marzulli M, Reinhart B, Miyagawa Y, Uchida H, Goins WF, Li A, Kaur B, Caligiuri M, Cripe T, Chiocca EA, Amankulor N, Cohen JB, Glorioso JC, Grandi P. 2015. Use of miRNA response sequences to block off-target replication and increase the safety of an unattenuated, glioblastoma-targeted oncolytic HSV. Mol Ther 23:99–107. doi: 10.1038/mt.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]