ABSTRACT
Hepatitis C virus (HCV) particles are described as lipoviroparticles which are released similarly to very-low-density lipoproteins (VLDLs). However, the release mechanism is still poorly understood; the canonical endoplasmic reticulum-Golgi intermediate compartment (ERGIC) pathway as well as endosome-dependent release has been proposed. Recently, the role of exosomes in the transmission of HCV has been reported. Only a minor fraction of the de novo-synthesized lipoviroparticles is released by the infected cell. To investigate the relevance of multivesicular bodies (MVBs) for viral morphogenesis and release, the MVB inhibitor U18666A was used. Intracellular trafficking was analyzed by confocal microscopy and electron microscopy. Moreover, an mCherry-tagged HCV variant was used. Conditions were established that enable U18666A-dependent inhibition of MVBs without affecting viral replication. Under these conditions, significant inhibition of the HCV release was observed. The assembly of viral particles is not affected. In U18666A-treated cells, intact infectious viral particles accumulate in CD63-positive exosomal structures and large dysfunctional lysosomal structures (multilamellar bodies). These retained particles possess a lower density, reflecting a misloading with lipids. Our data indicate that at least a fraction of HCV particles leaves the cell via the endosomal pathway. Endosomes facilitate the sorting of HCV particles for release or degradation.
IMPORTANCE There are still a variety of open questions regarding morphogenesis and release of hepatitis C virus. The HCV-infected cell produces significant more viral particles that are released, raising the question about the fate of the nonreleased particles. Moreover, the relevance of the endosomal pathway for the release of HCV is under debate. Use of the MVB (multivesicular body) inhibitor U18666A enabled a detailed analysis of the impact of MVBs for viral morphogenesis and release. It was revealed that infectious, fully assembled HCV particles are either MVB-dependently released or intracellularly degraded by the lysosome. Our data indicate that at least a fraction of HCV particles leaves the cell via the endosomal pathway independent from the constitutive secretory pathway. Our study describes a so-far-unprecedented cross talk between two pathways regulating on the one hand the release of infectious viral particles and on the other hand the intracellular degradation of nonreleased particles.
INTRODUCTION
Hepatitis C virus (HCV) is still a major health problem, as approximately 170 million people are chronically infected with HCV worldwide (1). Chronic infection can cause liver cirrhosis and hepatocellular carcinoma, finally leading to death (2).
HCV belongs to the Flaviviridae family. Flaviviridae have a single-stranded positive-sense RNA genome (3). The HCV RNA encompasses about 9,600 bases and is translated into a single polyprotein of about 3,100 amino acids, which is processed by viral and host proteases into the mature viral proteins (4). The members of the structural proteins core, E1, and E2, form the viral particles, and the nonstructural proteins NS2 to NS5B are involved in viral genome replication, particle assembly, and regulation of intracellular processes (5).
The viral genome is replicated in replication complexes which consist of viral and host proteins on the cytoplasmic face of the endoplasmic reticulum (ER) (6). The assembly of the particles takes place at the surface of lipid droplets (LDs) where the core proteins accumulate (7). However, particle assembly and egress are still not fully understood. The particles are associated with various cellular proteins, such as ApoE, ApoB, and TIP47 (8–10), and are called lipoviroparticles (LVPs) because of their loading with lipoproteins and their high similarity to very-low-density lipoproteins (VLDLs) (11).
Autophagy is a conserved cellular process which maintains cell homeostasis by degradation of intracellular components in lysosomes which fuse with autophagosomes (12). HCV is known to induce autophagy that affects its replication, morphogenesis, and release (13–15).
The majority of the de novo-synthesized particles are degraded by lysosomes, whereas only a small fraction is finally released from an infected cell (16, 17). We described recently that HCV is able to shift this equilibrium of particle degradation and release by regulating the cellular proteins TIP47 and syntaxin-17. Expression of TIP47 is upregulated in HCV-infected cells and ensures the trafficking of the particles to late endosomes instead of lysosomes via interaction with Rab9 (10). The soluble N-ethylmaleimide-sensitive-factor attachment receptor (SNARE) protein syntaxin-17, which facilitates the fusion of lysosomes and autophagosomes, is downregulated by HCV to block lysosomal degradation of viral particles (18).
Autophagy is connected to the endosomal pathway which is also important for exocytosis (19). Endocytosis leads to formation of early endosomes that can mature into late endosomes, also called multivesicular bodies (MVBs), by taking up intracellular material and vesicles (20). This process is maintained by the endosomal sorting complexes required for transport (ESCRT) (21, 22). MVBs act as sorting facilities which target their cargo toward different organelles. Moreover, MVBs can fuse with lysosomes or the plasma membrane to cause either degradation or release of their cargo (20).
As the colocalization of HCV structural proteins with markers of late and early endosomes is enhanced by treatment with the MVB transport inhibitor U18666A, MVB-dependent release of HCV particles was postulated (23). Recently, release of infectious HCV was observed to occur via trans-Golgi network (TGN)-endosome trafficking independently from VLDLs (24). Moreover, an inhibitory effect of U18666A on HCV replication by direct inhibition of the enzyme 24-dehydrocholesterol reductase (DHCR24) was observed (25, 26).
U18666A inhibits late endosomes (MVBs) and lipid transport by direct inhibition of the NPC1 protein and thus mimics the lysosomal storage disease Niemann-Pick disease. Niemann-Pick type C relies on a defect in NPC1 that leads to cholesterol accumulation in lysosomal storage structures (27). The same phenotype was observed in U18666A-treated cells. Cholesterol accumulates in large vesicles (multilamellar bodies [MLBs]) positive for lysosomal markers, e.g., LAMP2 (28).
The major cargos of MVBs are exosomes, which are vesicles of 50 to 100 nm and contain different cellular components, such as proteins, RNA, and lipids (29). Hepatitis A virus (HAV) hijacks the exosome machinery to be released and spread, undiscovered from the immune system. In contrast to HAV, which is released as mature particles in exosomes, for HCV so far only the exosome-dependent release of viral genomes has been described (30, 31). However, it is speculated that even whole particles are released by the exosome secretion pathway (32).
The exact mechanism of exosome release is not fully understood. However, exosomes are most likely released by fusion of an exosome-loaded MVB with the plasma membrane which relies on SNAREs (33). Recently, we showed that HCV actively decreases the amount of α-taxilin. α-Taxilin acts as a negative regulator of SNARE functionality by binding to free syntaxin-4 and thereby preventing the formation of SNARE complexes. The decreased amount of α-taxilin in HCV-infected cells promotes SNARE-dependent release (34). Syntaxin-4 is a t-SNARE located on the plasma membrane and has been reported to be involved in exosome release (35) and to accumulate at cholesterol-rich lipid rafts within the plasma membrane to build up exocytosis sites. This localization can be disturbed by interruption of cholesterol transport by U18666A (36).
This study aimed to investigate the release pathway of mature HCV particles via MVBs. For this purpose, the effect of the MVB inhibitor U18666A on the intracellular trafficking and release of HCV particles was characterized. It was revealed that HCV particles are either MVB-dependently released or intracellularly degraded by the lysosome.
MATERIALS AND METHODS
Cell culture.
The Huh7-derived cell clone Huh7.5 (37), which is highly permissive for HCV RNA replication, was used for transfection and infection assays as described previously (38). Isolation of primary human hepatocytes, cultivation, and infection were performed as described previously (38, 39). Huh7 cell clone 9-13, harboring the HCV replicon I377/NS3-3′, has been described previously (40).
Antibodies.
The following commercial primary antibodies were used: anti-NS3 (8G-2; Abcam), anti-β-actin (AC-74; Sigma-Aldrich), anti-core (MA1-080; Thermo Scientific), anti-CD63 (H-193; Santa Cruz Biotechnology), anti-ApoE (A1.4; Santa Cruz Biotechnology), anti-ApoE (PA5-27088; Thermo Scientific), anti-LC3 (PM036; MBL), anti-LAMP2 (555803; BD Pharmingen), and anti-E2 (HCM-091a-5; Austral Biologicals). For detection of NS5A, a polyclonal rabbit derived serum was used (41). The monoclonal antibody anti-HERV-K/GAG is described elsewhere (42).
Alexa Fluor 488- or Alexa Fluor 546-conjugated secondary antibodies were obtained from Invitrogen. Cy3-conjugated antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc., and secondary antibodies for Western blotting were obtained from LI-COR, Inc. Ten-nanometer-gold-particle-conjugated anti-mouse antibody (EM.GAM10) was obtained from BBI Solutions.
Fluorescent dyes.
The following dyes were used in immunofluorescence microscopy: 4′,6-diamidino-2-phenylindole (DAPI; Carl Roth) was used to stain nuclei, filipin III (Sigma-Aldrich) was used to stain cholesterol, and BODIPY 498/503 (Invitrogen) was used to stain lipid droplets.
Cytokines and inhibitors.
Human interferon alpha (tebu-bio) was used at a concentration of 1,000 U/ml. U18666A (Sigma-Aldrich) was used at a concentration of 2 μg/ml for 16 h if not indicated differently. Simvastatin (Selleckchem) was used at a concentration of 10 μM.
Plasmids.
Plasmid pFK-JHF1/J6 has been described previously (43). pFK-Luc-J6 was kindly provided by R. Bartenschlager and has been described previously (44). The J6-E1-mCherry (E1R) construct was generated in a manner similar to that described by Bayer et al. (68). The mCherry sequence was amplified with the primers fwd (AAACGTACGCGATGGTGAGCAA) and rev (AAACGTACGCCTTGTACAGCTCGT) with the plasmid pmCherry (Clontech) as a template. The amplified insert and the plasmid pFK-JHF1/J6 were digested with the enzyme BsiWI (NEB). After dephosphorylation of the plasmid with Antarctic Phosphatase (NEB), the insert was ligated into the plasmid with T4 DNA ligase (Thermo Scientific). The plasmids encoding wild-type or the transdominant negative (tdn) mutants of CHMP3, VPS4A and VPS4B, and for the green fluorescent protein (GFP)-GalT and yellow fluorescent protein (YFP)-Rab7 fusion proteins, were described recently (45) The plasmids encoding the fusion proteins GFP-Grasp65 and YFP-Sec22 were a kind gift from R. Duden, Lübeck, Germany.
RT-PCR.
RNA isolation was performed using peqGOLD TriFast (PEQLAB Biotechnologie GmbH) according to the manufacturer's instructions. cDNA synthesis and real-time PCR (RT-PCR) were performed as described in reference 46. All quantifications were normalized to the amount of RPL27 transcripts. The following primers were used: JFH1_fwd (ATGACCACAAGGCCTTTCG), JFH1_rev (CGGGAGAGCCATAGTGG), hRPL27_fw (AAAGCTGTCATCGTGAAGAAC), and hRPL27_rv (GCTGCTACTTTGCGGGGGTAG).
Quantification of HCV core in the cell culture supernatant.
Cell culture supernatant was centrifuged at 13,300 × g for 10 min to pellet cell debris. A total of 108 μl of the supernatant was used for the Abbott Architect HCV antigen (Ag) chemiluminescent microparticle immunoassay (CMIA) according to the manufacturer's protocol. Measurement was performed with an Architect i2000SR immunoassay analyzer (Abbott). For Western blot analysis, HCV particles were enriched from the supernatant by affinity chromatography on a heparin column as described previously (10).
Quantification of viral genomes in the cell culture supernatant.
Viral RNA was isolated from supernatants using the QIAamp DSP virus kit (Qiagen). Real-time PCR was performed with an artus HCV RG RT-PCR kit (Qiagen) according to the manufacturer's protocol.
Quantification of ApoE in the cell culture supernatant.
Supernatants of 2 × 105 Huh7.5 J6 cells were mixed with 1% Triton X-100 following protein precipitation with 10% trichloroacetic acid (TCA) for 15 min on ice. After centrifugation for 15 min at 14,000 rpm, the protein pellet was washed with −20°C cold acetone. Finally the pellet was solved in Laemmli buffer and analyzed by SDS-PAGE and Western blotting.
In vitro transcription and RNA transfection.
In vitro transcription, electroporation of HCV RNAs, and luciferase assays were performed as described previously (38).
Virus titration.
Virus titers were analyzed based on limited dilution by determination of the 50% tissue culture infective dose (TCID50) as described previously (47, 48). To determine the intracellular TCID50, the cells were washed with phosphate-buffered saline (PBS), trypsinized, and pelleted. The cell pellet was then resuspended in 2 ml of culture medium and was frozen and thawed three times with liquid nitrogen and a 37°C water bath. The cell lysate was centrifuged at 13,300 × g for 10 min, and the virus-containing supernatant was used for TCID50 assay. For the detection of HCV-positive cells, NS5A-specific serum was used (41).
Entry assay.
Confluent Huh7.5 cell monolayers were infected with HCV-containing supernatants for 5 h. After the infection, cells were washed twice with PBS and then incubated with trypsin for 1 min at room temperature to remove attached virus. The cells were washed once with culture medium and twice with PBS before being harvested in order to determine the entered virus by RT-PCR.
Indirect immunofluorescence analysis.
Indirect immunofluorescence analysis was performed as described previously (49). Immunofluorescence staining was analyzed using a confocal laser scanning microscope (CLSM 510 Meta; Carl Zeiss) and ZEN 2009 software. This software was also used to measure fluorescence intensities, Pearson's overlap coefficients, and weighted colocalization coefficients. To analyze the colocalization of CD63 and core, the weighted colocalization coefficients were determined for 19 untreated and U18-treated cells using the software Zen 2009 (Zeiss) and the following formula: M1 = ΣiCh1i,coloc/ΣiCh1i,total; M2 = ΣiCh2i,coloc/ΣiCh2i,total. Total fluorescence per cell was calculated using ImageJ software and the following formula: corrected total cell fluorescence = integrated density − (area of selected cell × mean fluorescence of background readings). In total, a minimum of three cells were measured.
Density gradient centrifugation and infectivity determination.
Iodixanol density gradients were performed as described previously (9). In brief, Huh7.5 J6 cells were treated with 2 μg/ml of U18666A for 16 h. Twenty-milliliter supernatants were concentrated to 1 ml by a Centricon with a 100-kDa cutoff (Millipore). Cell lysates were prepared in 1 ml of culture medium by repetitive freeze-thaw cycles. A density gradient of 10% to 80% Optiprep (Axis-Shield) in PBS was prepared and samples were layered on top. After centrifugation for 18 h at 120,000 × g at 4°C in an SW60Ti rotor, 12 fractions were harvested from top to bottom. The fractions density was determined via the refractive index. To assess the infectivity of each fraction, Huh7.5 cells were infected with an aliquot of each fraction for 16 h, and 72 h postinfection (p.i.), the amount of intracellular RNA was relatively quantified by RT-PCR and normalized to the amount of genomes in the cells infected with the first fraction. To see the ratio of genomes between the fractions, the amount of genomes in one fraction was normalized to the total amount of HCV genomes in all fractions.
Transmission electron microscopy (TEM).
Huh7.5 cells were fixed with 2.5% glutaraldehyde in PBS for 45 min at room temperature. Embedding in Epon and preparation of ultrathin sections were performed as described previously (50), and sections were mounted on freshly glow-discharged carbon-coated nickel grids. For immunogold labeling, cryosections were prepared as described previously (51) and incubated with primary antibody solution for 1 h, followed by incubation with gold-conjugated anti-mouse IgG for 1 h. The specimens were analyzed with a Zeiss EM-109 transmission electron microscope.
Cell viability assay.
Huh7.5 cells were seeded in a 96-well plate (104 cells/well) and treated with different amounts of U18666A or cycloheximide for 16 h. Cell viability was measured using PrestoBlue cell viability reagent (Thermo Scientific) according to the manufacturer's instructions. The reagent's fluorescence was measured after 1 h of incubation in a fluorescence microplate reader (Tecan M200).
mCherry fluorescence measurement.
Huh7.5 cells were electroporated with the J6-E1-mCherry construct and seeded in a 96-well plate (104 cells/well). After 72 h, the cells were washed twice with PBS and covered with 50 μl of PBS. The fluorescence measurement was performed in a fluorescence microplate reader (Tecan M200) with an extinction wavelength of 587 nm and an emission wavelength of 610 nm.
Statistical analyses.
Results are presented as means ± standard errors of the means (SEMs) from at least 3 experiments. The significance of results was analyzed by two-tailed Student's t test, using GraphPad Prism, version 6.07 for Windows (GraphPad Software, San Diego, CA). Error bars in figures represent standard deviations. Statistical significance is represented in figures as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
RESULTS
Time- and dose-dependent inhibition of the viral replication by U18666A.
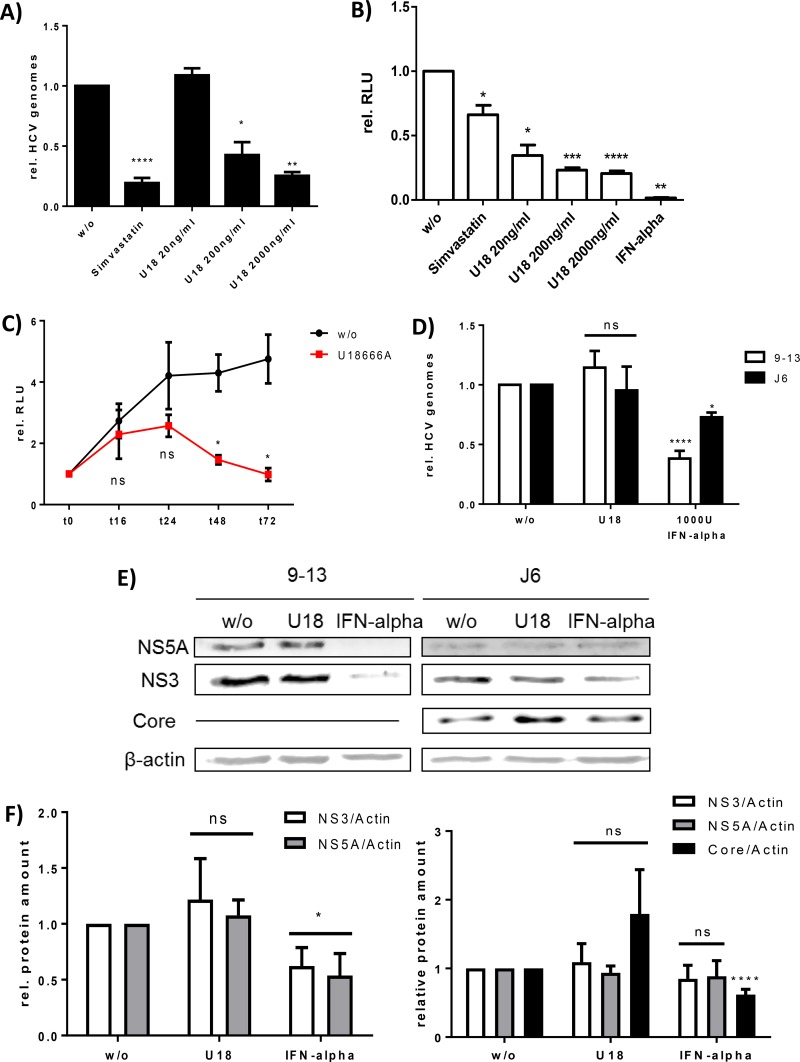
To study the relevance of MVBs for the morphogenesis and release of HCV, the MVB inhibitor U18666A was used. It has been shown that treatment of HCV replicating cells for 72 h with U18666A impairs the replication of HCV due to an inhibition of the enzyme DHCR24 (25). In light of this, it was investigated whether conditions that impair MVB function without affecting HCV genome replication can be defined. Various concentrations of and periods of treatment with U18666A were tested. To analyze the impact of various concentrations of U18666A (20 to 2,000 ng/ml) on viral replication, the subgenomic cell line Huh9-13 and cells replicating the full-length reporter replicon Luc-J6 (44) were incubated with U18666A for 72 h and analyzed by RT-PCR and luciferase reporter gene assay, respectively (Fig. 1A and B). For both systems, a concentration-dependent inhibitory effect on replication was observed after incubation for 72 h. Simvastatin and interferon alpha treatment served as positive controls in these experiments, as both compounds are known to impair HCV replication (52, 53). However, the dual roles of simvastatin in inhibiting viral replication on the one hand and enhancing the release of infectious particles on the other hand (see Fig. S1B at http://download.gsb.bund.de/PEI/U18 supplemental material.pdf) explain the less pronounced effect in the luciferase-based system. To investigate the time dependency of U18666A-mediated inhibition on replication, Huh7.5 cells were electroporated with the Luc-J6 construct. The kinetics of the U18666A treatment, starting 24 h posttransfection (p.t.) (time zero) (Fig. 1C), clearly shows that the replication of HCV is not affected by treatment with 2 μg/ml of U18666A for 16 h. However, after longer treatment, replication is significantly decreased.
FIG 1.
Time- and dose-dependent inhibition of viral replication by U18666A. (A) Huh9-13 cells were treated with 10 μg/ml of simvastatin or the indicated concentrations of U18666A for 72 h. w/o, without treatment. Viral replication was determined by RT-PCR. The graph shows the relative data from three independent experiments, with SEMs. (B) Huh7.5 cells were transfected with the J6-Luc construct and treated with 1,000 U of interferon alpha, 10 μg/ml of simvastatin, or the indicated concentrations of U18666A for 72 h. Viral replication was determined by luciferase reporter gene assay. The graph shows the relative data from three independent experiments, with SEMs. (C) Huh7.5 cells were transfected with the J6-Luc construct and treated with 2 μg/ml of U18666A for 72 h. Viral replication was determined by luciferase reporter gene assay at different time points. The graph shows the relative data from three independent experiments, with SEMs. (D) Huh9-13 and Huh7.5 J6 cells were treated with 2 μg/ml of U18666A or 1,000 U of interferon alpha for 16 h. Viral replication was determined by RT-PCR. The graph shows the relative data from at least three independent experiments, with SEMs. ns, not significant. (E) Western blot analysis of cellular lysates derived from Huh9-13 and Huh7.5 J6 cells which were treated with 2 μg/ml of U18666A or 1,000 U of interferon alpha for 16 h. For detection, specific antibodies against NS3, NS5A, and core were used. Detection of β-actin served as a loading control. (F) Densitometric quantification of Western blots shown in panel E. The graphs show the quantification of at least three independent experiments, with SEMs.
To corroborate the observation that U18666A treatment for 16 h does not affect viral replication, the subgenomic cell line Huh9-13 and Huh7.5 cells electroporated with the full-length HCV J6 genome were used. RT-PCR analysis and Western blot analysis using NS3- and NS5A-specific antisera (Fig. 1D to F) revealed that there is no effect on the genome replication and viral protein amount of U18666A under these conditions. In the case of the cells replicating the full-length HCV J6 genome, a slight increase of the intracellular amount of core protein was detectable (Fig. 1E and F). Again, interferon alpha treatment served as a positive control in this experiment. A cytotoxic effect of U18666A at the concentration used was excluded by cell viability assay (see Fig. S1A at http://download.gsb.bund.de/PEI/U18 supplemental material.pdf). Taken together, these data indicate that incubation of HCV replicating cells for 16 h with the MVB inhibitor U18666A at a concentration of 2 μg/ml does not affect genome replication or viral protein amount and therefore is suitable to study the impact on virus morphogenesis and release.
U18666A inhibits HCV particle release but not assembly.
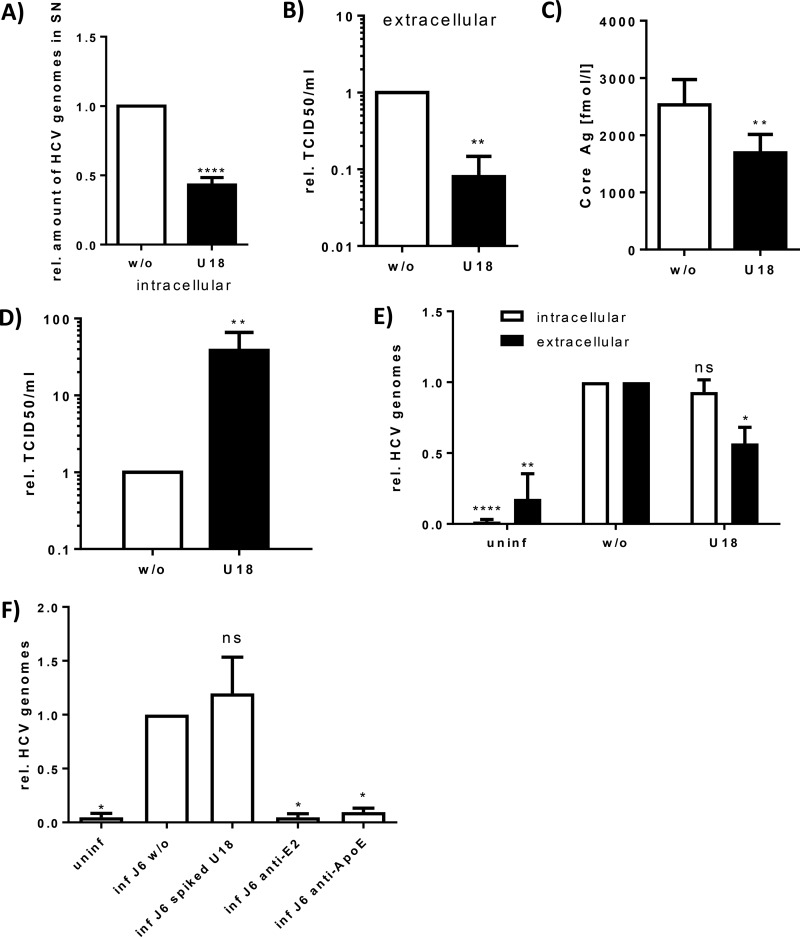
To study whether inhibition of MVB functionality has an impact on HCV morphogenesis and release, HCV-replicating Huh7.5 cells were treated for 16 h with U18666A (2 μg/ml). Quantification of the viral genomes released to the supernatant during this period by RT-PCR revealed a significant decrease in the amount of released viral genomes (Fig. 2A). The amount of viral genomes does not automatically correspond to the amount of infectious viral particles. To test whether this reflects indeed a decrease in the amount of released viral particles, the TCID50 of extracellular viral particles from U18666A-treated cells was determined. In accordance with the RT-PCR data, the extracellular TCID50 of U18666A-treated cells was decreased 10-fold in comparison to that of the untreated controls (Fig. 2B). This was further corroborated by a core-specific chemiluminescent microparticle immunoassay (CMIA): the amount of released HCV core was significantly reduced by U18666A treatment (Fig. 2C). Furthermore, the reduced amount of extracellular core was confirmed by Western blot analysis of cell culture supernatant purified by a heparin column (see Fig. S1C at http://download.gsb.bund.de/PEI/U18 supplemental material.pdf).
FIG 2.
U18666A treatment inhibits HCV particle secretion but not assembly in cell culture and primary cells. HCV-replicating cells were treated with 2 μg/ml of U18666A for 16 h. (A) The amount of released viral genomes in the supernatant was determined by RT-PCR. The graph shows the data from six independent experiments, with SEMs. (B) To determine the amount of released infectious viral particles in the supernatant, a TCID50 assay was performed. The graph shows the relative data from three independent experiments, with SEMs. (C) The amount of released viral particles in the supernatant was determined by core Ag assay. The graph shows the data from seven independent experiments, with SEMs. (D) The amount of intracellular viral particles was assayed by TCID50 of cells lysed by freeze-thawing. The graph shows the relative data from three independent experiments, with SEMs. (E) Primary human hepatocytes were infected over night with supernatant of HCV-replicating cells. Cells were treated 56 h p.i. with 2 μg/ml of U18666A for 16 h. Viral replication and particle release were determined by RT-PCR of cell lysates and supernatants, respectively. The graph shows the relative data from three independent experiments, with SEMs. (F) Huh7.5 cells were infected for 5 h with supernatant of HCV-replicating cells. To determine the entry-inhibiting ability of U18666A, it was added at a concentration of 2 μg/ml. As positive controls for entry inhibition, the supernatant was supplemented with 1 μg/ml of anti-E2 or anti-ApoE antibody. Bound virus was removed by short trypsin treatment before analysis of entered virus by RT-PCR. The graph shows the relative data from four independent experiments, with SEMs.
Vice versa, the amount of intracellular viral particles was quantified by determination of the TCID50; a 50-fold increase after U18666A treatment was observed (Fig. 2D).
The inhibitory effect of U18666A on the release of HCV was further confirmed using HCV-infected primary human hepatocytes (PHHs). PHHs from different donors were infected with HCVcc, and 56 h p.i. the cells were incubated for 16 h with U18666A. In accordance with the data from the Huh7.5 cell line, no effect on the viral replication by U18666A treatment was observed, whereas the amount of released viral genomes was significantly reduced (Fig. 2E).
These data indicate that treatment with U18666A for 16 h does not affect genome replication or HCV particle assembly but inhibits particle release. Assembled infectious particles are retained within the cells.
U18666A does not affect HCV entry.
To investigate whether further steps of the HCV life cycle are affected by U18666A, the effect on the entry of HCV virions into the target cells in the presence or absence of U18666A was analyzed. For this assay, an HCVcc-positive inoculum was spiked with U18666A (2 μg/ml) and compared to the unspiked inoculum. The entry assay (Fig. 2F) revealed that the presence of U18666A does not affect HCV entry. ApoE- and E2-specific antibodies served as controls (10, 54).
This confirms the specificity of the data described above and demonstrates that U18666A has no effect on the entry process of HCV.
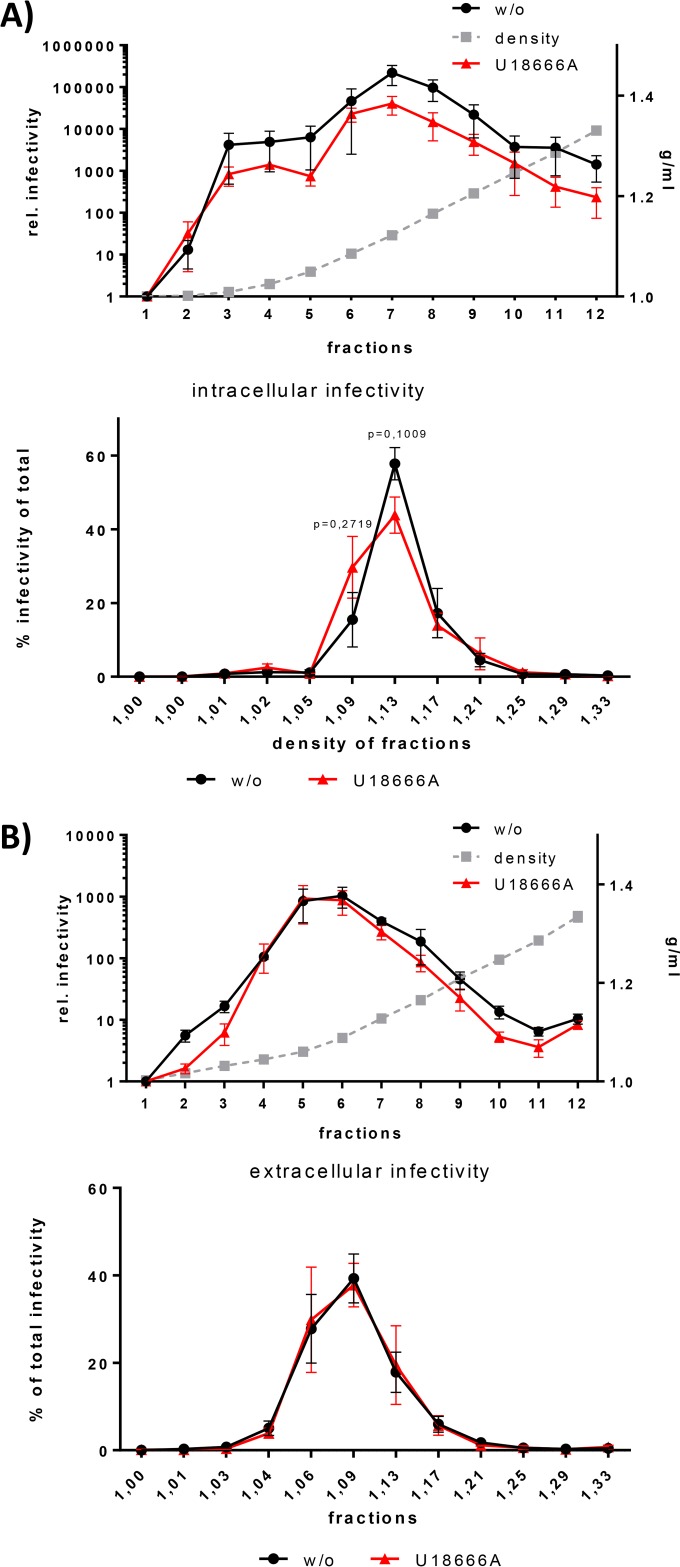
U18666A-dependently retained particles possess a lower density.
In many aspects, HCV LVPs are similar to VLDLs and therefore dependent on the lipid metabolism of the cell. U18666A is known to disturb intracellular lipid transport (55). To investigate whether this is reflected by an improper loading of HCV particles with lipids and lipoproteins, extra- and intracellular HCV particles were concentrated and separated by density gradient centrifugation. The presence of infectious HCV particles in each fraction of the density gradient was assessed by infectivity assays using Huh7.5 cells. When the infectivity of each fraction was normalized to the total infectivity, a shift of the density of the intracellular particles from 1.13 g/ml to 1.09 g/ml by U18666A treatment was observed (Fig. 3A). This shift in the density indicates that U18666A treatment leads to an altered loading of the particles with lipids and lipoproteins. A lower density represents a larger amount of lipids. Therefore, the intracellular retained viral particles derived from U18666A-treated cells seem to have a higher lipid loading than the control particles. The density of the released viral particles does not change after U18666A treatment (Fig. 3B).
FIG 3.
Preferentially, HCV particles with low density are retained intracellularly. (A) HCV J6-replicating Huh7.5 cells were treated with 2 μg/ml of U18666A for 16 h. The cells were lysed by repeated freeze-thaw cycles and loaded on an iodixanol gradient to separate viral particles by their density. Twelve fractions were harvested from top to bottom and were used to infect Huh7.5 cells. The infected cells were harvested 72 h p.i., and viral load was assessed by RT-PCR of the intracellular RNA. The top graph shows the relative infectivity of each fraction, and the bottom graph shows the infectivity as a percentage of the total measured infectivity. The graph shows the relative data from three independent experiments, with SEMs. (B) HCV J6-replicating Huh7.5 cells were treated with 2 μg/ml of U18666A for 16 h. The supernatant was loaded on an iodixanol gradient to separate viral particles by their density. Twelve fractions were harvested from top to bottom and were used to infect Huh7.5 cells. The infected cells were harvested 72 h p.i., and viral load was assessed by RT-PCR of the intracellular RNA. The top graph shows the relative infectivity of each fraction, and the bottom graph shows the infectivity as a percentage of the total measured infectivity. The graph shows the relative data from three independent experiments, with SEMs.
Taken together, these data indicate that that U18666A-mediated inhibition of the release of HCV particles is associated with a block in the secretion of the low-density fraction of HCV particles.
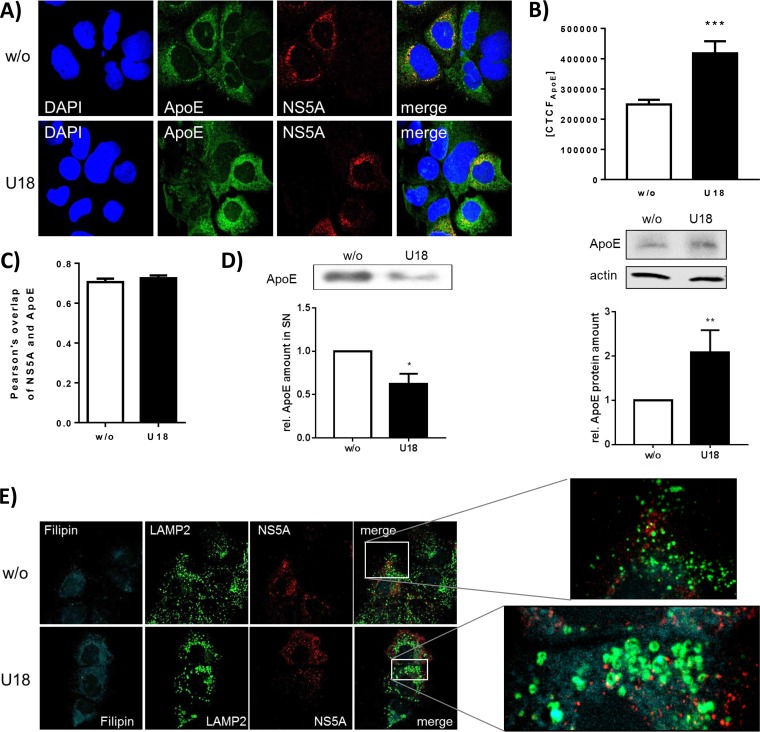
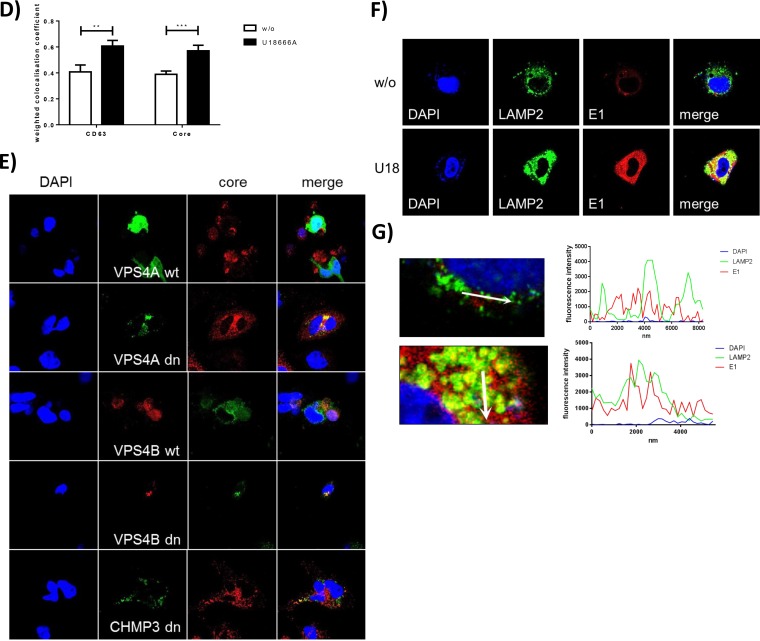
LAMP2 accumulates in U18666A-treated cells.
To study whether the impact of U18666A treatment on HCV release is reflected by a change in the subcellular distribution of viral and cellular proteins, confocal immunofluorescence microscopy was performed. Analyzing the effect of U18666A treatment for 16 h on the localization of NS5A, no change in the amount or localization of NS5A upon U18666A treatment (Fig. 4A) was observed. This was in accordance to the data described above (compare Fig. 1D to F) that indicate that under these conditions, no effect on replication and assembly can be observed. However, a strong accumulation of ApoE, which is normally secreted via exosomes as part of the VLDLs (56, 57), was found by quantification of the immunofluorescence and by Western blotting (Fig. 4B). In accordance with this, the amount of released ApoE is decreased by U18666A treatment (Fig. 4D). Pearson's correlation factor as a value of colocalization of ApoE and NS5A did not change due to U18666A treatment (Fig. 4C), implying that the NS5A-ApoE (58) interaction is not affected by U18666A.
FIG 4.
U18666A has no effect on NS5A localization, but ApoE accumulates and LAMP2 forms ring-like structures. Shown are results of CLSM analysis of HCV-replicating cells which were treated with 2 μg/ml of U18666A for 16 h. (A) NS5A is visualized in red; nuclei were stained with DAPI, in blue, and ApoE is visualized in green. (B) The fluorescence intensity of ApoE was measured in untreated and U18666A-treated cells. The data were confirmed by quantitative Western blot analysis of cellular lysates using ApoE- and actin-specific antibodies. The quantification is based on three independent experiments. (C) Pearson's overlap coefficient of NS5A and ApoE was calculated in untreated and U18666A-treated cells. (D) The amount of secreted ApoE was measured using quantitative Western blotting of TCA-precipitated samples of untreated and U18666A-treated cells. The quantification is based on three independent experiments. (E) NS5A is visualized in red; cholesterol was stained with filipin in cyan, and the lysosomal marker LAMP2 is visualized in green. The enlarged images are shown for treated and untreated cells. (F) The fluorescence intensity of LAMP2 was measured in untreated and U18666A-treated cells. The findings were confirmed by quantitative Western blotting of cell lysates. The quantification is based on three independent experiments. (G) Huh7.5 cells expressing Rab7-YFP were stained for LAMP2, in red. Treatment of U18666A led to the formation of Rab7- and LAMP2-positive rings.
Lysosomes are known to play an essential role in the HCV life cycle (14, 18, 59). Therefore, it was investigated whether lysosomes are affected by U18666A. Costaining of the lysosomal marker LAMP2 in green, NS5A in red, and cholesterol in blue using filipin revealed a significant accumulation of LAMP2 in large ring-like structures (Fig. 4E) after U18666A treatment for 16 h. There is no colocalization between LAMP2 and NS5A detectable, but colocalization between LAMP2 and the cholesterol-binding fluorophore filipin is detectable. As U18666A has been reported to lead to an accumulation of cholesterol in late endosomes (60), it is assumed that the large LAMP2-positive structures represent late endosomes. The accumulation of LAMP2 due to U18666A treatment was quantified by fluorescence intensity and quantitative Western blotting (Fig. 4F). To control whether the formed LAMP2-positive rings really reflect late endosome-derived structures, Huh7.5 cells were transfected with a plasmid coding for a Rab7-YFP fusion protein. U18666A treatment led to the formation of large LAMP2-positive rings which were also positive for the MVB marker Rab7 (Fig. 4G).
The subcellular distribution of ER and Golgi markers is not affected by the presence of U18666A, suggesting that there is no effect on the canonical secretory pathway of this substance (see Fig. S2 at http://download.gsb.bund.de/PEI/U18 supplemental material.pdf).
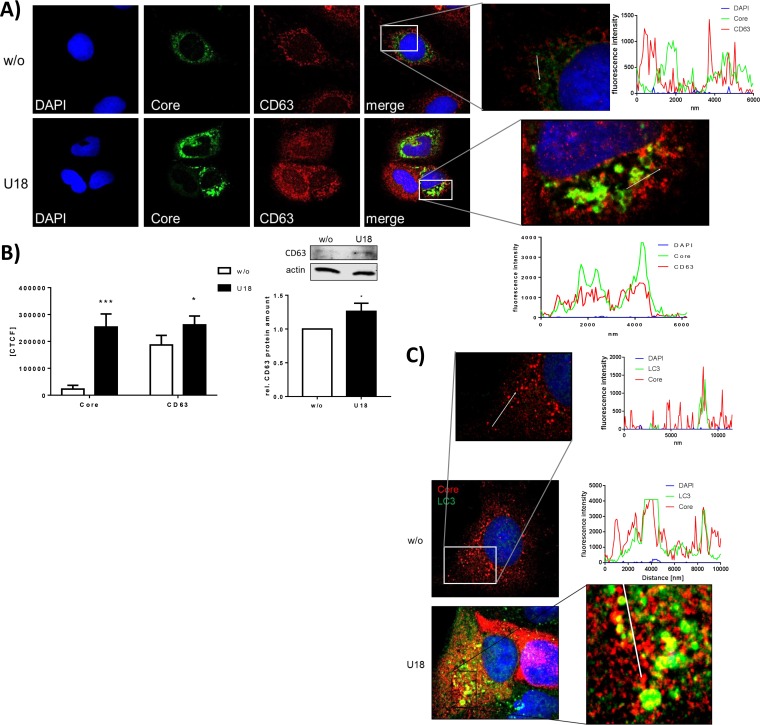
In U18666A-treated cells, core colocalizes within accumulated autophagosomes and exosomes.
The data described above demonstrate that treatment with U18666A for 16 h inhibits particle release without affecting viral replication and particle assembly (compare Fig. 2). The assembled particles are retained in the cell. To characterize the specific place where the particles accumulate, the structural protein core and various subcellular markers were labeled and their distribution was analyzed by confocal immunofluorescence microscopy. In Fig. 5 and in Fig. S3A and B at the URL above, it is clearly visible that HCV core accumulates inside the cells after U18666A treatment. This contributes to the accumulation of core observed by Western blotting (Fig. 2E and F). Also, the exosomal marker CD63 accumulates, as observed by fluorescence intensity measurements and quantitative Western blot analysis (Fig. 5A and B). In addition, the autophagosomal marker LC3 accumulates in U18666A-treated cells, and a fraction of core colocalizes with LC3 (Fig. 5C; see also Fig. S3A at the URL mentioned above). The colocalization of the accumulated core with CD63-positive structures is more pronounced and increases by treatment with U18666A (Fig. 5D; see also Fig. S3B at the URL mentioned above). Taken together, these data show an accumulation of core in autophagosomal and exosomal structures after U18666A treatment, reflecting the involvement of the exosomal pathway for the release of HCV particles. The importance of MVBs for the release of HCV particles is also demonstrated by the accumulation of HCV core in E-class compartments when dominant negative mutants of the ESCRT proteins VPS4A, VPS4B, and CHMP3 are overexpressed (Fig. 5E).
FIG 5.
HCV structural proteins accumulate intracellularly in endosomal structures. (A) CLSM analysis of HCV-replicating cells which were treated with 2 μg/ml of U18666A for 16 h. HCV core is visualized in green; the nuclei were stained with DAPI, in blue, and the exosomal marker CD63 is visualized in red. Shown are the fluorescence intensities of DAPI, core, and CD63 along the white arrows. Enlarged images are shown for treated and untreated cells. (B) The fluorescence intensities of core and CD63 were measured in untreated and U18666A-treated cells (left). The amount of CD63 was also determined by quantitative Western blotting using cell lysates (right). The quantification is based on three independent experiments. (C) CLSM analysis of HCV-replicating cells which were treated with 2 μg/ml ofU18666A for 16 h. HCV core is visualized in red; the nuclei were stained with DAPI, in blue, and the autophagosomal marker LC3 is visualized in green. The graphs show the fluorescence intensities of DAPI, core, and LC3 along the white arrows in the zoomed image. The enlarged images are shown for treated and untreated cells. (D) The weighted colocalization coefficients of core and CD63 were calculated in untreated and U18666A-treated cells. (E) CLSM analysis of Huh7.5 cells that were electroporated with HCV J6 RNA and plasmids encoding the indicated fusion proteins. HCV core was costained with a specific antibody. (F) CLSM analysis of HCV-replicating cells which were treated with 2 μg/ml of U18666A for 16 h. Huh7.5 cells were transfected with an HCV construct harboring an mCherry fusion tag inside the hypervariable region (HVR) of E1, which is visualized in red. The nuclei were stained with DAPI, in blue, and the lysosomal marker LAMP2 is visualized in green. (G) Zoomed image of a part of panel F with intensity profiles showing the fluorescence intensities of DAPI, LAMP2, and E1 along the white arrows.
U18666A leads to accumulation of the glycoprotein E1 in lysosomal structures.
To further corroborate these data, Huh7.5 cells were transfected with the J6-E1-mCherry construct that harbors an mCherry sequence inside the variable region of E1 and thus enables the production of mCherry-labeled viral particles. The functionality of this type of fusion protein and the capacity to form viral particles have been established by Bayer et al. (68) and confirmed for the construct/experimental system used in this study (see Fig. S3C to G at the URL mentioned above). Confocal immunofluorescence microscopy revealed the intracellular accumulation of E1-mCherry fusion protein after U18666A treatment. A more detailed analysis revealed that LAMP2 accumulates (Fig. 5F) in big ring-shaped structures that are filled with E1-mCherry (Fig. 5G) as visualized by a fluorescence intensity profile along the white arrow which depicts two neighbored LAMP2 rings (green) which are filled with E1, visualized by the red fluorescence.
These data indicate that after U18666A treatment, viral particles accumulate in the enlarged LAMP2-positive rings formerly identified to be late endosome derived (Fig. 4G).
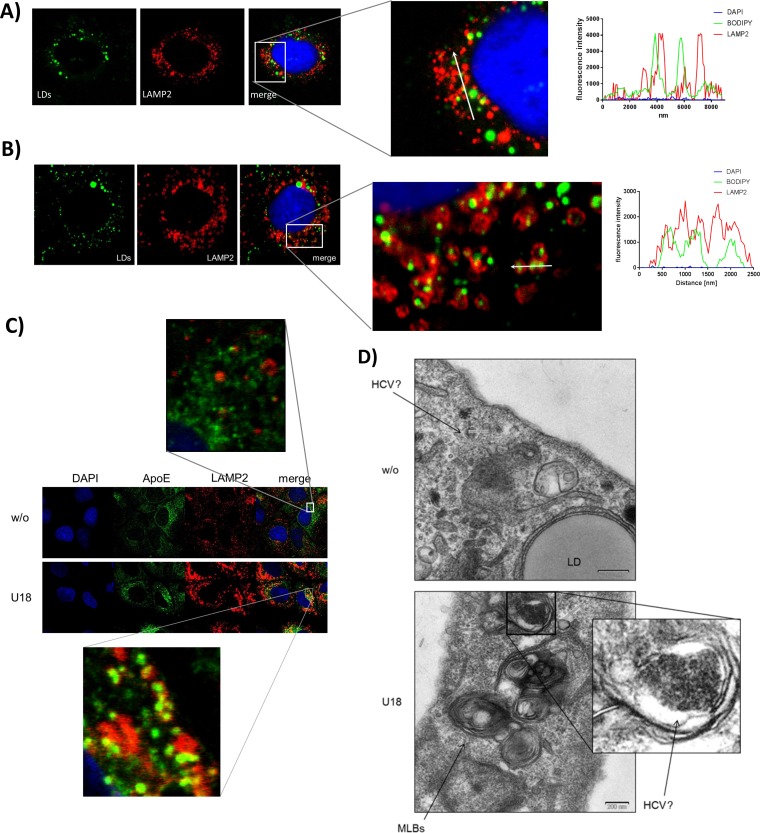
In U18666A-treated cells, lysosomal structures are lipid-filled multilamellar bodies in which HCV particles accumulate.
Lipids are an important key factor in the HCV life cycle. At the surface of lipid droplets, the assembly of HCV particles takes place (5, 61). Furthermore, the lipoviroparticles are heavily associated with lipids and lipoproteins and are considered to resemble VLDLs. Therefore, it was studied whether the lipid distribution changes after U18666A treatment. In confocal laser scanning microscopy (CLSM) images, an accumulation of the lipophilic dye BODIPY inside the LAMP2-positive ring structures was found (Fig. 6B; see also Fig. S4A at the URL mentioned above) compared to the normal distribution in lipid droplets in untreated cells (Fig. 6A). A similar accumulation in the LAMP2-positive rings was observed for the lipoprotein ApoE (Fig. 6C; see also Fig. S4B at the URL mentioned above). For a more detailed view, ultrathin sections of HCV-replicating cells were investigated by electron microscopy (EM). In the untreated cells, some intracellular particles that could represent HCV particles according to the structure and size were identified. Cells treated with U18666A harbor numerous big multilamellar bodies with concentric membrane stacks that represent dysfunctional lysosomes (Fig. 6D). These structures are typical for patients suffering from Niemann-Pick-disease and have been reported for U18666A-treated fibroblasts (62). It can be assumed that these MLBs represent the LAMP2-positive ring structures which are filled with lipids and HCV structural proteins. Indeed, at a higher magnification of these MLBs, electron-dense vesicles are visible that accumulate in the center, surrounded by a light lipid-rich area. These vesicles might reflect HCV particles accumulated in the MLBs according to their size and structure, but this cannot be proven.
FIG 6.
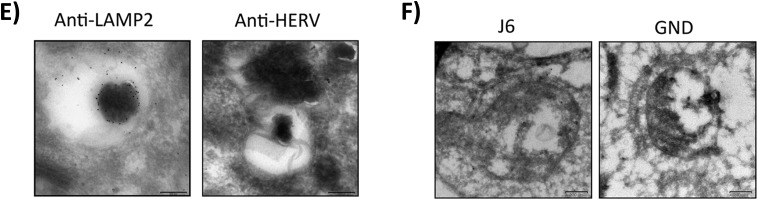
LAMP2-positive rings are filled with lipids and could represent multilamellar bodies filled with HCV particles. (A) CLSM analysis of untreated HCV-replicating cells. The lipid droplets were stained with BODIPY in green, and the lysosomal marker LAMP2 is visualized in red. The nuclei were stained with DAPI in blue. The graph shows the fluorescence intensities of DAPI, BODIPY, and LAMP2 along the white arrow in the zoomed image. (B) CLSM analysis of HCV-replicating cells which were treated with 2 μg/ml of U18666A for 16 h. The lipid droplets were stained with BODIPY in green, and the lysosomal marker LAMP2 is visualized in red. The nuclei were stained with DAPI in blue. The graph shows the fluorescence intensities of DAPI, BODIPY, and LAMP2 along the white arrow in the zoomed image. (C) CLSM analysis of HCV-replicating cells which were left untreated or treated with 2 μg/ml of U18666A for 16 h. ApoE is visualized in green, and the lysosomal marker LAMP2 is visualized in red. The nuclei were stained with DAPI in blue. (D) TEM images of ultrathin sections of HCV-replicating Huh7.5 cells which were treated with 2 μg/ml of U18666A for 16 h. Ultrathin sections were stained with uranyl acetate to identify cellular and viral structures. (E) TEM image of cryosections of HCV-replicating Huh7.5 cells which were treated with 2 μg/ml of U18666A for 16 h. Sections were labeled with 10-nm-gold-conjugated antibodies against LAMP2 or HERV-K as a negative control and stained with uranyl acetate. (F) TEM image of cryosections of Huh7.5 cells electroporated with J6 or GND which were treated with 2 μg/ml of U18666A for 16 h. Sections were labeled with 10-nm-gold-conjugated antibodies against core and E2 and stained with uranyl acetate.
To prove the identity of the MLBs with the LAMP2-positive structures, cryosections of U18666A-treated HCV-replicating Huh7.5 cells were prepared and LAMP2 was labeled with gold-conjugated antibodies. As shown in Fig. 6E, an MLB was labeled with gold particles, whereas no labeling was observed with the control antibody (anti-HERV). There is no fully convincing image of HCV particles in EM images from cryosections published so far. The low abundance and the lipid coat of HCV particles make them hard to visualize. Moreover, the contrast and solution of cryosections are always worse than in ultrathin sections, making it even more unlikely that intact HCV particles will be found. However, we incubated cryosections of Huh7.5 J6 cells treated with U18666A with a mix of antisera against core and E2 and marked both structural proteins with 10-nm gold particles. As shown in Fig. 6F, the structural proteins accumulate inside the MLBs, but there is no virus particle detectable. However, for HCV-negative Huh7.5 GND cells, no labeling inside those structures was found, demonstrating that the staining is specific. This finding clearly indicates that U18666A induces the formation of large LAMP2-positive and lipid-rich MLBs that harbor most likely HCV particles, reflecting the involvement of the lysosomal compartment in the HCV life cycle.
DISCUSSION
The aim of this study was to investigate the relevance of MVBs/late endosomes for the morphogenesis and release of HCV LVPs. To inhibit MVB functionality, the amphiphatic inhibitor U18666A was used. As treatment with U18666A for 72 h inhibits the viral replication by inhibition of DHCR24 (26), the protocol was adapted to conditions that have no inhibitory effect on genome replication. It could be observed that treatment of HCV replicating cells for 16 h with U18666A (2 μg/ml) impairs the release of viral particles but does not affect genome replication as shown for various systems or the entry process. The slight accumulation of core in U18666A-treated cells observed by Western blotting is not statistically significant, whereas it is obvious in immunofluorescence microscopy. Most likely the MLBs cannot be solubilized adequately in SDS buffer, and therefore, the core accumulation is not that strong in Western blotting. Indeed, the membranes of MLBs formed after U18666A treatment have been shown to be detergent resistant (60).
U18666A treatment leads to a significant decrease in the amount of released viral particles, as evidenced by core CMIA, RT-PCR, and TCID50 determination. The TCID50 assay depicts a more pronounced decrease of released viral particles. This could reflect that changes in the lipid composition of the released particles affect their infectivity.
The analysis of intracellular infectivity revealed that U18666A does not affect particle assembly. The amount of intracellular retained particles increases more than 50-fold, which indicates that the particles are assembled and accumulate inside the cells without being released or degraded. In HCV-infected cells, only a minor part of assembled HCV particles are sorted for release, whereas the majority of the de novo-synthesized viral particles is targeted toward the lysosomes for degradation (16, 17). The amount of intracellular particles is more strongly increased as the corresponding amount of extracellular viral particles is decreased. This indicates not only that the particles determined for release are retained but also that these particles that are targeted to the lysosome are prevented from degradation. This reflects that U18666A leads to inhibition of lysosomal activity as shown by the MLBs. An interesting aspect in this context is that these particles that are targeted to the lysosome are infective. This argues against the hypothesis that exclusive defective viral particles are sorted to the lysosomal compartment and raises the question of the sorting criteria.
The density gradient centrifugation revealed a shift of the intracellular particles to a lower density after U18666A treatment. The one-fraction density shift of intracellular particles was reproducibly observed in independent experiments. The density of the released particles does not change, reflecting the secretion competence of the particles with a density of 1.09 g/ml. This secretion-competent fraction of particles increases after U18666A treatment, whereas the fraction of secretion-incompetent particles with a density of 1.13 g/ml decreases intracellularly. Therefore, we conclude that particles can mature to, per se, secretion-competent particles by loading with lipids and lipoproteins inside the MVBs, but the final step of the release is disturbed. Furthermore, we observed an accumulation of lipids and lipoproteins in the intracellular MLBs due to the disturbed intracellular lipid trafficking as visualized by the filipin staining. Remarkably, the filipin staining colocalizes with the LAMP2 signals, reflecting an accumulation of cholesterol in the LAMP2 rings. LAMP2 not only is part of lysosomes but also can be found on late endosomes (63). Based on the results from the CLSM and TEM, it can be concluded that U18666A disturbs the lipid transport, causing functional inhibition of late endosomes and formation of MLBs that leads to an accumulation of the LVPs in late endosomes and MLBs. The ApoE accumulation reflects the impaired endosome-dependent release of VLDLs.
Accumulation of LC3 and of the exosomal marker CD63 due to U18666A treatment was described previously (23, 64). Because of the impaired endosome maturation, exosomes cannot be secreted any longer and accumulate inside the cells. LC3 is known to translocate from autophagosomes to autolysosomes (65). Due to the impaired maturation of endosomes in U18666A-treated cells, LC3 accumulates in big vesicles, which represent nonfunctional lysosomes/MLBs. The accumulation of HCV core in LC3-positive structures further supports the assumption that the HCV life cycle depends on the late endosome machinery. This was further corroborated using an mCherry-tagged E1 HCV variant. J6-E1-mCherry is able to replicate in Huh7.5 cells, and HCV particles are assembled and released comparable to the J6 chimeric construct (Bayer et al., submitted; see also Fig. S2 at the URL mentioned above). Therefore, the mCherry-tagged E1 HCV variant is a suitable tool to study the morphogenesis of HCV particles in electroporated cells. In CLSM studies, a strong accumulation of E1 in U18666A-treated cells was observed. Furthermore, it was found that the LAMP2-positive rings contain E1. As U18666A does not affect viral assembly, we postulate that the E1 fluorescence in LAMP2-positive vesicles represents accumulated particles.
The formation of so-called MLBs that represent defective lysosomal storage structures in cells of patients suffering from Niemann-Pick disease has been described previously (62). The TEM pictures of HCV-replicating Huh7.5 cells exhibit MLBs filled with lipids, reflected by the light areas within these structures. Furthermore, an accumulation of electron-dense, dark vesicles inside the MLBs can be detected, which could reflect HCV particles according to their size (50 nm) and structure. Combining these results with the results of the CLSM analysis, in which large LAMP2-positive rings are filled with lipids and HCV structural proteins and the immunogold labeling in the electron microscopy, we conclude that the MLBs represent the LAMP2-positive structures. Due to U18666A treatment, the lipid transport is blocked by NPC1 inhibition; therefore, lipids and HCV LVPs accumulate in the late endosomes, which later form large nonfunctional MLBs. The importance of early endosomes and members of the ESCRT machinery for HCV particle release has been described previously (23, 66, 67), and our data add further insight in this process. Our data indicate that at least a fraction of HCV particles leaves the cell via the endosomal pathway. Endosomes facilitate the sorting of HCV particles for release or degradation. This is relevant, as only a minor fraction of the de novo-synthesized viral particles are indeed released, while the majority of the particles is finally degraded in the lysosomal compartment. However, as infectious viral particles are found in the lysosomal compartment, the sorting criteria are still enigmatic.
In a recent study, the effect of U18666A on the release of HBV viral and subviral particles was investigated. Due to the relevance of the MVB system for the release of infectious viral particles and of filaments, an inhibitory effect on the release of these particles was observed. In contrast to this, the secretion of spheres that occurs dependent on the ER/Golgi via the constitutive secretory pathway was not affected, indicating that the observed effects of U18666A on the release of HCV are not due to an effect on the Golgi system (45). This was further confirmed by analyzing the subcellular distribution of ER and Golgi markers that is not affected by the presence of U18666A (see Fig. S2 at the URL mentioned above).
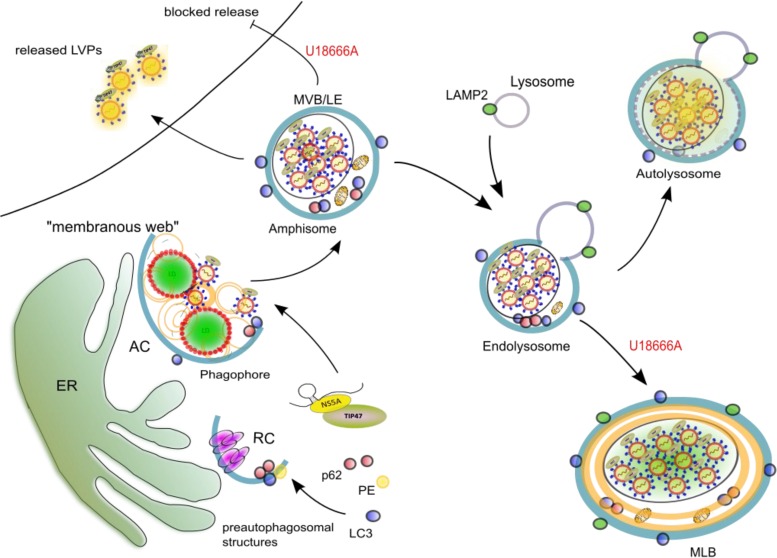
Taken together, our data indicate that treatment with U18666A blocks the release of the viral particles by interference with the functionality of MVBs, which leads to an accumulation of viral particles in exosomes. Moreover, U18666A leads to formation of large dysfunctional lysosomal structures in which the degradation of these viral particles that are targeted toward the lysosome is impaired (Fig. 7). This shows the relevance of the endosomal pathway for the HCV life cycle.
FIG 7.
U18666A treatment leads to the formation of nonfunctional lysosomal MLBs in which the HCV particles accumulate. The viral RNA is transported from the replication complex (RC) to the assembly complex (AC) at the membranous web, where the particles are formed. VLDLs as well as LVPs reach the MVBs, where they are sorted for either release or degradation by fusion with lysosomes. Addition of U18666A disturbs this mechanism by blocking the secretion of cholesterol and exosomes by MVBs. This leads to an accumulation of these HCV particles determined for release in exosomes. As the endolysosomes are overloaded with cholesterol and other lipids, they form nonfunctional MLBs. The HCV particles that are determined for lysosomal degradation accumulate here and are prevented from degradation due to the dysfunctionality of the MLBs.
ACKNOWLEDGMENTS
We thank Andrea Henkes, Gert Carra, Alena Zindel, and Regina Eberle for their excellent technical support and Dagmar Fecht-Schwarz for her critical reading of the manuscript. Furthermore, we thank Malin Finkernagel and the Medizinische Hochschule Hannover for the preparation of human tissues and the isolation of primary cells. Finally, we thank Sigrid Nick and Tanja Westenberger for great help with the core CMIA.
We have no conflicts of interest to declare.
H.R. obtained a grant from the China Scholarship Council. The work was supported by a grant from the German center for infection research (DZIF) to E.H.
REFERENCES
- 1.National Institutes of Health. 2002. National Institutes of Health Consensus Development Conference Statement: management of hepatitis C 2002 (June 10–12, 2002). Gastroenterology; 123:2082–2099. doi: 10.1053/gast.2002.1232082. [DOI] [PubMed] [Google Scholar]
- 2.Alter MJ. 2007. Epidemiology of hepatitis C virus infection. World J Gastroenterol 13:2436. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 4.Appel N, Schaller T, Penin F, Bartenschlager R. 2006. From structure to function: new insights into hepatitis C virus RNA replication. J Biol Chem 281:9833–9836. doi: 10.1074/jbc.R500026200. [DOI] [PubMed] [Google Scholar]
- 5.Dubuisson J, Penin F, Moradpour D. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol 12:517–523. doi: 10.1016/S0962-8924(02)02383-8. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager R, Frese M, Pietschmann T. 2004. Novel insights into hepatitis C virus replication and persistence. Adv Virus Res 63:71–180. doi: 10.1016/S0065-3527(04)63002-8. [DOI] [PubMed] [Google Scholar]
- 7.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 8.Chang KS, Jiang J, Cai Z, Luo G. 2007. Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J Virol 81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J-Y, Acosta EG, Stoeck IK, Long G, Hiet M-S, Mueller B, Fackler OT, Kallis S, Bartenschlager R. 2014. Apolipoprotein E likely contributes to a maturation step of infectious hepatitis C virus particles and interacts with viral envelope glycoproteins. J Virol 88:12422–12437. doi: 10.1128/JVI.01660-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ploen D, Hafirassou ML, Himmelsbach K, Schille SA, Biniossek ML, Baumert TF, Schuster C, Hildt E. 2013. TIP47 is associated with the hepatitis C virus and its interaction with Rab9 is required for release of viral particles. Eur J Cell Biol 92:374–382. doi: 10.1016/j.ejcb.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Bradley D, McCaustland K, Krawczynski K, Spelbring J, Humphrey C, Cook EH. 1991. Hepatitis C virus: buoyant density of the factor VIII-derived isolate in sucrose. J Med Virol 34:206–208. doi: 10.1002/jmv.1890340315. [DOI] [PubMed] [Google Scholar]
- 12.Krámer H. 2013. Route to destruction: autophagosomes SNARE lysosomes. J Cell Biol 201:495–497. doi: 10.1083/jcb.201304065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Ou J-H J. 2015. Hepatitis C virus and autophagy. Biol Chem 396:1215–1222. doi: 10.1515/hsz-2015-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ploen D, Hildt E. 2015. Hepatitis C virus comes for dinner: how the hepatitis C virus interferes with autophagy. World J Gastroenterol 21:8492–8507. doi: 10.3748/wjg.v21.i28.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shrivastava S, Devhare P, Sujijantarat N, Steele R, Kwon Y-C, Ray R, Ray RB, Williams B. 2016. Knockdown of autophagy inhibits infectious hepatitis C virus release by the exosomal pathway. J Virol 90:1387–1396. doi: 10.1128/JVI.02383-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinkert D, Bartenschlager R, Lohmann V. 2005. Quantitative analysis of the hepatitis C virus replication complex. J Virol 79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Post J, Ratnarajah S, Lloyd AR. 2009. Immunological determinants of the outcomes from primary hepatitis C infection. Cell Mol Life Sci 66:733–756. doi: 10.1007/s00018-008-8270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren H, Elgner F, Jiang B, Himmelsbach K, Medvedev R, Ploen D, Hildt E. 20 April 2016. The autophagosomal SNARE protein syntaxin 17 is an essential factor for the hepatitis C virus life cycle. J Virol doi: 10.1128/JVI.00551-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb CA, Dooley HC, Tooze SA. 2013. Endocytosis and autophagy: shared machinery for degradation. Bioessays 35:34–45. doi: 10.1002/bies.201200130. [DOI] [PubMed] [Google Scholar]
- 20.Huotari J, Helenius A. 2011. Endosome maturation. EMBO J 30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. 2002. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol 4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 22.Katzmann DJ, Babst M, Emr SD. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145–155. doi: 10.1016/S0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 23.Lai C-K, Jeng K-S, Machida K, Lai MMC. 2010. Hepatitis C virus egress and release depend on endosomal trafficking of core protein. J Virol 84:11590–11598. doi: 10.1128/JVI.00587-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mankouri J, Walter C, Stewart H, Bentham M, Park WS, Heo WD, Fukuda M, Griffin S, Harris M. 25 May 2016. Release of infectious hepatitis C virus from Huh7 cells occurs via a trans-Golgi network to endosome pathway independent of VLDL. J Virol doi: 10.1128/JVI.00826-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takano T, Tsukiyama-Kohara K, Hayashi M, Hirata Y, Satoh M, Tokunaga Y, Tateno C, Hayashi Y, Hishima T, Funata N, Sudoh M, Kohara M. 2011. Augmentation of DHCR24 expression by hepatitis C virus infection facilitates viral replication in hepatocytes. J Hepatol 55:512–521. doi: 10.1016/j.jhep.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Quan X, Chen X, Sun D, Xu B, Zhao L, Shi X, Liu H, Gao B, Lu X. 2016. The mechanism of the effect of U18666a on blocking the activity of 3β-hydroxysterol Δ-24-reductase (DHCR24): molecular dynamics simulation study and free energy analysis. J Mol Model 22:46. doi: 10.1007/s00894-016-2907-2. [DOI] [PubMed] [Google Scholar]
- 27.Cenedella RJ. 2009. Cholesterol synthesis inhibitor U18666A and the role of sterol metabolism and trafficking in numerous pathophysiological processes. Lipids 44:477–487. doi: 10.1007/s11745-009-3305-7. [DOI] [PubMed] [Google Scholar]
- 28.Lajoie P. 2005. The lipid composition of autophagic vacuoles regulates expression of multilamellar bodies. J Cell Sci 118:1991–2003. doi: 10.1242/jcs.02324. [DOI] [PubMed] [Google Scholar]
- 29.Baixauli F, Lopez-Otin C, Mittelbrunn M. 2014. Exosomes and autophagy. Coordinated mechanisms for the maintenance of cellular fitness. Front Immunol 5:373. doi: 10.3389/fimmu.2014.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longatti A. 2015. The dual role of exosomes in hepatitis A and C virus transmission and viral immune activation. Viruses 7:6707–6715. doi: 10.3390/v7122967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosset F-L, Dreux M. 2014. HCV transmission by hepatic exosomes establishes a productive infection. J Hepatol 60:674–675. doi: 10.1016/j.jhep.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 32.Longatti A, Boyd B, Chisari FV. 2015. Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J Virol 89:2956–2961. doi: 10.1128/JVI.02721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raposo G, Stoorvogel W. 2013. Extracellular vesicles. Exosomes, microvesicles, and friends. J Cell Biol 200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elgner F, Donnerhak C, Ren H, Medvedev R, Schreiber A, Weber L, Heilmann M, Ploen D, Himmelsbach K, Finkernagel M, Klingel K, Hildt E. 2 November 2015. Characterization of α-taxilin as a novel factor controlling the release of hepatitis C virus. Biochem J doi: 10.1042/BJ20150717. [DOI] [PubMed] [Google Scholar]
- 35.Théry C, Zitvogel L, Amigorena S. 2002. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 36.Reverter M, Rentero C, de Muga SV, Alvarez-Guaita A, Mulay V, Cairns R, Wood P, Monastyrskaya K, Pol A, Tebar F, Blasi J, Grewal T, Enrich C. 2011. Cholesterol transport from late endosomes to the Golgi regulates t-SNARE trafficking, assembly, and function. Mol Biol Cell 22:4108–4123. doi: 10.1091/mbc.E11-04-0332R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Himmelsbach K, Sauter D, Baumert TF, Ludwig L, Blum HE, Hildt E. 2009. New aspects of an anti-tumour drug: sorafenib efficiently inhibits HCV replication. Gut 58:1644–1653. doi: 10.1136/gut.2009.182212. [DOI] [PubMed] [Google Scholar]
- 39.Weiss TS, Pahernik S, Scheruebl I, Jauch K-W, Thasler WE. 2003. Cellular damage to human hepatocytes through repeated application of 5-aminolevulinic acid. J Hepatol 38:476–482. doi: 10.1016/S0168-8278(02)00454-3. [DOI] [PubMed] [Google Scholar]
- 40.Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 41.Bürckstümmer T, Kriegs M, Lupberger J, Pauli EK, Schmittel S, Hildt E. 2006. Raf-1 kinase associates with Hepatitis C virus NS5A and regulates viral replication. FEBS Lett 580:575–580. doi: 10.1016/j.febslet.2005.12.071. [DOI] [PubMed] [Google Scholar]
- 42.Bieda K, Hoffmann A, Boller K. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J Gen Virol 82:591–596. doi: 10.1099/0022-1317-82-3-591. [DOI] [PubMed] [Google Scholar]
- 43.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich H-G, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol 80:5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang B, Himmelsbach K, Ren H, Boller K, Hildt E. 2016. Subviral hepatitis B virus filaments, like infectious viral particles, are released via multivesicular bodies. J Virol 90:3330–3341. doi: 10.1128/JVI.03109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masoudi S, Ploen D, Kunz K, Hildt E. 2014. The adjuvant component α-tocopherol triggers via modulation of Nrf2 the expression and turnover of hypocretin in vitro and its implication to the development of narcolepsy. Vaccine 32:2980–2988. doi: 10.1016/j.vaccine.2014.03.085. [DOI] [PubMed] [Google Scholar]
- 47.Steinmann E, Brohm C, Kallis S, Bartenschlager R, Pietschmann T. 2008. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J Virol 82:7034–7046. doi: 10.1128/JVI.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 49.Ploen D, Hafirassou ML, Himmelsbach K, Sauter D, Biniossek ML, Weiss TS, Baumert TF, Schuster C, Hildt E. 2013. TIP47 plays a crucial role in the life cycle of hepatitis C virus. J Hepatol 58:1081–1088. doi: 10.1016/j.jhep.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 50.Kraus B, Boller K, Reuter A, Schnierle BS. 2011. Characterization of the human endogenous retrovirus K Gag protein: identification of protease cleavage sites. Retrovirology 8:21. doi: 10.1186/1742-4690-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stephan O, Schwendemann J, Specke V, Tacke SJ, Boller K, Denner J. 2001. Porcine endogenous retroviruses (PERVs): generation of specific antibodies, development of an immunoperoxidase assay (IPA) and inhibition by AZT. Xenotransplantation 8:310–316. doi: 10.1034/j.1399-3089.2001.00098.x. [DOI] [PubMed] [Google Scholar]
- 52.Kim CS, Jung JH, Wakita T, Yoon SK, Jang SK. 2007. Monitoring the antiviral effect of alpha interferon on individual cells. J Virol 81:8814–8820. doi: 10.1128/JVI.02824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikeda M, Abe K-I, Yamada M, Dansako H, Naka K, Kato N. 2006. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology 44:117–125. doi: 10.1002/hep.21232. [DOI] [PubMed] [Google Scholar]
- 54.Sabo MC, Luca VC, Prentoe J, Hopcraft SE, Blight KJ, Yi M, Lemon SM, Ball JK, Bukh J, Evans MJ, Fremont DH, Diamond MS. 2011. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J Virol 85:7005–7019. doi: 10.1128/JVI.00586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sparrow SM, Carter JM, Ridgway ND, Cook HW, Byers DM. 1999. U18666A inhibits intracellular cholesterol transport and neurotransmitter release in human neuroblastoma cells. Neurochem Res 24:69–77. doi: 10.1023/A:1020932130753. [DOI] [PubMed] [Google Scholar]
- 56.Mensenkamp AR, Havekes LM, Romijn JA, Kuipers F. 2001. Hepatic steatosis and very low density lipoprotein secretion: the involvement of apolipoprotein E J Hepatol 35:816–822. [DOI] [PubMed] [Google Scholar]
- 57.Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G, Luo G. 2014. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog 10:e1004424. doi: 10.1371/journal.ppat.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benga Wagane JA, Krieger SE, Dimitrova M, Zeisel MB, Parnot M, Lupberger J, Hildt E, Luo G, McLauchlan J, Baumert TF, Schuster C. 2010. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology 51:43–53. doi: 10.1002/hep.23278. [DOI] [PubMed] [Google Scholar]
- 59.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. 2009. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5:937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- 60.Sobo K, Le Blanc I, Luyet P-P, Fivaz M, Ferguson C, Parton RG, Gruenberg J, van der Goot Gisou F, Valdivia R. 2007. Late endosomal cholesterol accumulation leads to impaired intra-endosomal trafficking. PLoS One 2:e851. doi: 10.1371/journal.pone.0000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartenschlager R, Penin F, Lohmann V, André P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol 19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Pentchev PG, Comly ME, Kruth HS, Tokoro T, Butler J, Sokol J, Filling-Katz M, Quirk JM, Marshall DC, Patel S. 1987. Group C Niemann-Pick disease: faulty regulation of low-density lipoprotein uptake and cholesterol storage in cultured fibroblasts. FASEB J 1:40–45. [DOI] [PubMed] [Google Scholar]
- 63.Behnke J, Schneppenheim J, Koch-Nolte F, Haag F, Saftig P, Schroder B. 2011. Signal-peptide-peptidase-like 2a (SPPL2a) is targeted to lysosomes/late endosomes by a tyrosine motif in its C-terminal tail. FEBS Lett 585:2951–2957. doi: 10.1016/j.febslet.2011.08.043. [DOI] [PubMed] [Google Scholar]
- 64.Appelqvist H, Nilsson C, Garner B, Brown AJ, Kågedal K, Öllinger K. 2011. Attenuation of the lysosomal death pathway by lysosomal cholesterol accumulation. Am J Pathol 178:629–639. doi: 10.1016/j.ajpath.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hyttinen JMT, Niittykoski M, Salminen A, Kaarniranta K. 2013. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta 1833:503–510. doi: 10.1016/j.bbamcr.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 66.Corless L, Crump CM, Griffin SDC, Harris M. 2010. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol 91:362–372. doi: 10.1099/vir.0.017285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ariumi Y, Kuroki M, Maki M, Ikeda M, Dansako H, Wakita T, Kato N. 2011. The ESCRT system is required for hepatitis C virus production. PLoS One 6:e14517. doi: 10.1371/journal.pone.0014517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bayer K, Banning C, Bruss V, Wiltzer-Bach L, Schindler M. 2016. Hepatitis C virus is released via a noncanonical secretory route. J Virol 90:10558–10573. doi: 10.1128/JVI.01615-16. [DOI] [PMC free article] [PubMed] [Google Scholar]