ABSTRACT
The p12 protein of murine leukemia virus (MLV) Gag is associated with the preintegration complex (PIC), and mutants of p12 (PM14) exhibit defects in nuclear entry/retention. Mutants of the phosphorylated serine 61 also have been reported to have defects in the early life cycle. Here we show that a phosphorylated peptide motif derived from human papillomavirus 8 (HPV-8), the E2 hinge region including residues 240 to 255, can functionally replace the main phosphorylated motif of MLV p12 and can rescue the viral titer of a strain with the lethal p12-PM14 mutation. Complementation with the HPV-8 E2 hinge motif generated multiple second-site mutations in live viral passage assays. Additional p12 phosphorylation sites were detected, including the late domain of p12 (PPPY) as well as the late domain/protease cleavage site of matrix (LYPAL), by mass spectrometry and Western blotting. Chromatin binding of p12-green fluorescent protein (GFP) fusion protein and functional complementation of p12-PM14 occurred in a manner independent of the E2 hinge region phosphorylation. Replacement of serine 61 by alanine within the minimal tethering domain (61SPMASRLRGRR71) maintained tethering, but in the context of the full-length p12, mutants with substitutions in S61 remained untethered and lost infectivity, indicating phosphorylation of p12 serine 61 functions to temporally regulate early and late p12 functions.
IMPORTANCE The p12 protein, required for both early and late viral functions, is the predominant phosphorylated viral protein of Moloney MLV and is required for virus viability. Our studies indicate that the N terminus of p12 represses the early function of the chromatin binding domain and that deletion of the N terminus activates chromatin binding in the wild-type Moloney MLV p12 protein. Mass spectrometry and mutagenesis studies suggest that phosphorylation of both the repression domain and the chromatin binding domain acts to temporally regulate this process at the appropriate stages during infection.
INTRODUCTION
The murine leukemia virus (MLV) (genus Gammaretrovirus, family Retroviridae) Gag protein p12 is a multifunctional protein required in both early and late stages of viral replication. During the late stages of viral assembly, new virions must bud off their host cells, requiring membrane scission. The MLV p12 encodes a late-assembly (L) domain (31PPPY34) (1, 2), reported to interact with the host vesicular trafficking machinery, specifically the E3 ubiquitin ligases in the NEDD4 family, to facilitate Gag ubiquitylation and subsequent budding (3). In addition, the 25DLL27 motif in p12 has been reported to recruit clathrin to budding virions, which is required for correct particle morphogenesis and infectivity (4).
In infected cells, two domains required for early replication were identified by mutational analysis: within the p12 N terminus at residues 10 to 24 (mutants PM5 to PM8) and within the p12 C terminus at residues 60 to 74 (mutants PM13 to PM15) (1, 5). The N terminus of p12 interacts with capsid (CA), as identified via p12 N-terminal mutants failing to produce or stabilize a proper CA core (5, 6). Additional evidence for direct p12-CA interaction has been shown via chimeric viral constructs, in which virus with p12 and CA from different viral species has severely reduced viral DNA production upon infection and failed to produce viable virus (7). The C terminus has been shown to function in binding the host chromatin (5, 8, 9). Both CA and p12 are constituents of the preintegration complex (PIC) (10, 11), and loss of either the N or C terminus blocks the PIC from entering the nucleus (1). Both the p12 N- and C-terminal domains are required to be in cis (5), suggesting that p12 acts as a bridge between the CA within the PIC and the host chromatin. As MLV requires cells to proceed through mitosis for integration (12) and lacks the interactions with host proteins involved in active nuclear transport of HIV (13, 14), p12 is proposed to function in retaining the PIC in the nucleus postmitosis (5, 8, 9). Alternate chromatin tethers functionally replacing the C-terminal function did not alter the MLV integration profile, ruling out the possibility that the p12 chromatin tether is the driving determinant for integration site selection (8).
The role of phosphorylation in retroviruses in the regulation of viral budding (15, 16) remains an open question. These viruses use various late (L) domains to hijack the endosomal sorting complexes required for transport (ESCRT) proteins to aid in viral budding. These ESCRT proteins ubiquitylate Gag, which acts as a cellular signal in the ESCRT pathway to sort cargo into multivesicular bodies, but its exact function in viral budding is unclear (17, 18). The proteins containing the HIV and human T-cell leukemia virus type 1 (HTLV-1) late domains, p6 and matrix, respectively, are both phosphorylated adjacent to their late domains, and loss of this phosphorylation results in diminished viral release and infectivity (15, 19). However, with the exception of the HIV late domain (PTAP), all 11 HIV phosphorylation sites could be mutated simultaneously and were found dispensable for viral release and infectivity in cell culture (16). Ubiquitylation not only is a required step connecting the L domains to budding but also is itself sufficient to replace missing L domains (20). Thus, Gag phosphorylation and intact late domains both facilitate proper ESCRT recruitment and budding.
Many viruses use redundant domains to recruit multiple ESCRT proteins. HIV uses a P(S/T)AP domain to recruit TSG101, an ESCRT I protein, and a redundant LYPxL domain to recruit Alix, an ESCRT III protein (21). HTLV-1 matrix has a third domain, PPxY, used to bind the WW domain of the E3 ubiquitin ligase NEDD4, but also has the P(S/T)AP domain used by HIV (22). MLV has all three domains: the P(S/T)AP domain in matrix, the PPxY domain in p12, and the third domain, LYPxL, overlapping the MA-p12 cleavage site. All three domains were proven functional: the PSAP domain bound TSG101, the PPPY domain bound NEDD4, and the LYPAL domain bound Alix (23). Despite this redundancy, the MLV p12 PPPY domain has been proven to be essential for viral budding, while the PSAP and LYPAL domains increase budding efficiency but are ultimately dispensable (1, 23, 24).
In MLV, the PPPY motif could be functionally replaced with an HIV p6 PTAP motif or an RSV PPPY motif (25). Interestingly, these motifs were able to rescue the budding of the p12 PPPY deletion even when placed into the matrix or nucleocapsid proteins; however, these mutants blocked early viral infection at a stage prior to reaching the nucleus, suggesting a second role for the p12 PPPY domain (25). Recent p12 truncation analysis unveiled the region including the PPPY motif to negatively regulate the C-terminal chromatin binding motif (26).
Previous phosphorylation studies indicated p12 to be the main phosphorylated protein in MLV and that this phosphorylation occurs mainly on serine residues (27–30). All p12 serine residues have been replaced by alanine, and only loss of S61 caused severe replication defects (1, 31). Live passage of an S(61,65)A mutant virus generated second-site mutations restoring replication competence, including M63I. The phosphorylation levels of these suppressor mutants remained low, indicating that these viruses were replicating in the absence of S61 phosphorylation (31). Although loss of phosphorylation sites within p12 decreased viral replication, second-site suppressor mutants that appear to replicate in the absence of the predominant serine phosphorylation sites have been isolated. Thus, the role of this posttranslational modification in the viral life cycle has not been defined. In this report, the role of the p12 M63I phosphorylation-independent mutant in activating p12 chromatin tethering is examined, and the phosphorylation status and function of MLV p12 mutants are analyzed using mass spectrometry, confocal microscopy, and viral titer assays. Novel p12 phosphorylation sites at serine, threonine, and tyrosine are identified and characterized. Here, we discuss the potential role of phosphorylation in the regulation of these late domain functions and the regulation of the p12 chromatin binding sequence.
MATERIALS AND METHODS
Cells, viruses, and infections.
D17 cells carrying the pJET plasmid (32) (encoding the mCAT receptor) and 293T cells (33) were maintained in Dulbecco's modified Eagle medium (DMEM; catalog number 11965; Gibco) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (catalog number S1245OH; Atlanta Biologicals) and 1× antibiotic-antimycotic (catalog number 15240; Gibco). Viral constructs and green fluorescent protein (GFP) fusions were cloned and assayed as previously described (8). Proviral plasmid DNA (pNCA-C) was directly transfected into cells via DEAE-dextran (32), which generates virus capable of multiple rounds of infection. Culture supernatant was collected at each passage to test for the presence of the viral reverse transcriptase enzyme (34). The MLV MA Y130D construct was graciously provided by Alan Rein (35).
p12 mutant cloning.
Cloning of p12 mutants was performed using the pNCA-C plasmid consisting of the Moloney MLV (MMLV) provirus (36). Overlapping PCR with KOD polymerase (Novagen) was used to introduce point mutants throughout p12, using previously cloned p12-PM14 mutants as the templates (8). The overlapping PCR fragment was introduced into pNCA-C (36) using the EcoRI and XhoI sites. Constructs were verified by sequencing p12 with the p20 Rev (5′-GCCCAGACTGGGATTACACCACC-3′) primer. Mutant p12 constructs were cloned into the GFP mammalian expression vector pGIP (37) as previously described (8). p12Δ50–64 (8) has been renamed p12Δ49–64 here to reflect the removal of Gly49 during the substitution of the alternative tethering domains into the p12-Δ constructs.
GFP confocal microscopy.
p12-GFP fusion constructs were generated and imaged as previously described (8). p12 truncations were generated in the same manner, except using Lenti-X 293T cells as both producer and target cells (catalog number 632180; Clontech) and an upright Zeiss Axio Imager Z1 spinning disc confocal microscope with Metamorph Premiere software. All GFP lines were passaged, grown for 2 days, and then imaged after 30 min of Hoechst 33258 DNA staining (catalog number 14530; Sigma). All cells imaged were in mitosis. GFP intensities overlapping the DNA and in the cytoplasm were quantified using Matlab, and the tethering ratio was calculated as the ratio of GFP intensity overlapping the DNA to the intensity of GFP in cytoplasm (8). A tethering ratio correction of −0.1 was applied to every ratio to account for background.
Viral titer assay.
On day 1, 293T cells were transfected with 0.8 μg vesicular stomatitis virus glycoprotein (VSV-G) expression vector pHIT, 0.8 μg lacZ expression plasmid (pRT43.2Tnlsβ-gal) with ψ (retroviral packaging element) (38, 39), and 0.8 μg MMLV viral genome with wild-type p12 and mutants (pNCA-C) using Fugene 6 (Roche) overnight (36). On day 2, cells were treated with 10 mM sodium butyrate for 6 h, virus was collected overnight in fresh medium, and 105 D17 cells were split to 3.5-cm lined plates. On day 3, viral medium was 0.45 μm filtered and normalized for infection by performing an enzyme-linked immunosorbent assay (ELISA) against CA (40). Infections were performed with overnight incubation of filtered viral supernatants corresponding to 230, 23, 2.3, and 0.23 ng of CA, brought to 1 ml total volume with DMEM and 8 μg/ml Polybrene. On day 4, the medium was changed to fresh DMEM. On day 5, cells were fixed in 1 ml 2% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min. Cells were washed twice with PBS and then stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for 3 days (41). On day 8, plates with 1% of lacZ+ cells were scored to avoid high multiplicity of infection (MOI) saturation.
32P labeling of virion proteins.
Confluent 10-cm plates were washed two times in 3 ml phosphate-free DMEM (catalog number 11971-025; Gibco) and starved for 20 min in 3 ml phosphate-free DMEM with 7.5% dialyzed fetal calf serum (FCS); then, 1 mCi of 32P-labeled orthophosphoric acid (catalog number NEX053; PerkinElmer) was added per plate, and the plates were incubated for 5 h at 37°C, 5% CO2. Viral particles were then collected, pelleted, and immunoprecipitated as described previously (42), except that 20 μl of CRL-1890 p12 antiserum was used to form the immune complexes (42).
35S labeling of virion proteins.
Cell lines at near confluence in 10-cm plates were washed twice with 5 ml PBS, starved for 30 min in 5 ml DMEM lacking l-methionine and l-cysteine (catalog number 21013-024; Gibco), and supplemented with 2 mM l-glutamine (catalog number 25030; Gibco). After starvation, 150 μCi of Tran35S-Label (catalog number 510090; MP Biomedicals) was added per plate; 2 h later, 125 μl of dialyzed calf serum was added per plate, and cells were grown 17 h at 37°C and 5% CO2. Viral particles were then collected, pelleted, and immunoprecipitated as described previously (42), except that either 20 μl or 40 μl of CRL-1890 (mouse anti-p12) or 4 μl of 74S000430 (goat anti-p12) antiserum was used to form the immune complexes. The samples electrophoresed on polyacrylamide gel (PAG) represent 36% of the supernatant collected from a 10-cm plate of cells.
Western blots.
Molecular weight markers (Novex Sharp Pre-stained Protein Standards, catalog number S7318; Novex Magic Mark XP Western Standard, catalog number LC5602) were obtained from Invitrogen. TGX gels (4 to 20%) and Tris-glycine (catalog number 161-0732) running buffer were obtained from Bio-Rad, and SDS/PAGE was performed according to the manufacturer's instructions. Transfers were performed using Trans-Blot Turbo transfer packs (0.2 μM polyvinylidene difluoride [PVDF]) and the Bio-Rad Trans-Blot Turbo Mixed MW 2.5A, 7-min protocol. Antibodies used include antiphosphotyrosine antibody, clone 4G10 (catalog number 05-321X; Millipore), phosphoserine/pS antibody (catalog number 61-8100; Novex), rabbit anti-phospho-protein kinase A (anti-phospho-PKA) substrate (RRXS*/T* motif) (catalog number 9624; Cell Signaling Technology), mouse anti-p12 (CRL-1890; ATCC), goat anti-p12 (catalog number 74S000430; Viromed Biosafety Laboratories), goat anti-mouse IgG (H+L) secondary antibody, horseradish peroxidase (HRP) conjugate (catalog number 31430; ThermoFisher Scientific), peroxidase AffiniPure bovine anti-goat IgG (H+L) (catalog number 805-035-180; Jackson ImmunoResearch Inc.), and goat anti-rabbit IgG (H+L) secondary antibody, HRP conjugate (catalog number 31460; ThermoFisher Scientific). Phospho-antibodies that failed to detect phosphorylation of p12 include antiphosphoserine antibody, clone 4A4 (catalog number 05-1000; EMD Millipore), phospho-CDK substrate motif [(K/H)pSP] MultiMab Rabbit (catalog number 9477; Cell Signaling Technology), phospho-mitogen-activated protein kinase/cyclin-dependent kinase (pMAPK/CDK) substrates (PXS*P or S*PXR/K; catalog number 2325; Cell Signaling Technology), phospho-(Ser) Arg-X-Tyr/Phe-X-pSer motif antibody (catalog number 2981; Cell Signaling Technology), and phospho-(Ser) 14-3-3 binding motif (4E2) (catalog number 9606; Cell Signaling Technology).
Mass spectrometry.
Virus analyzed by mass spectrometry was collected from D17 and 293T cells with the mCAT receptor (8). Cell culture supernatant was pelleted at 15,000 × g at 4°C for 1 h and separated by SDS-PAGE. Gel slices correlating with 12 and 15 kDa were extracted, digested with trypsin, chymotrypsin, or protease K, dried, solubilized in 5% acetonitrile–0.1% trifluoroacetic acid (TFA), and analyzed via liquid chromatography-tandem mass spectrometry (LC-MS/MS). Mass spectrometry was performed at the Rutgers Biological Mass Spectrometry Facility. p12-GFP fusion protein mass spectrometry was performed similarly, except that two 10-cm plates of D17 expression cells were lysed and p12-GFP was coimmunoprecipitated with anti-GFP antibody (ThermoFisher catalog number A6455) before SDS-PAGE analysis. Proteins were visualized with Coomassie blue R-250. Ion mass sizes (see Fig. 4) were calculated at http://db.systemsbiology.net:8080/proteomicsToolkit/FragIonServlet.html.
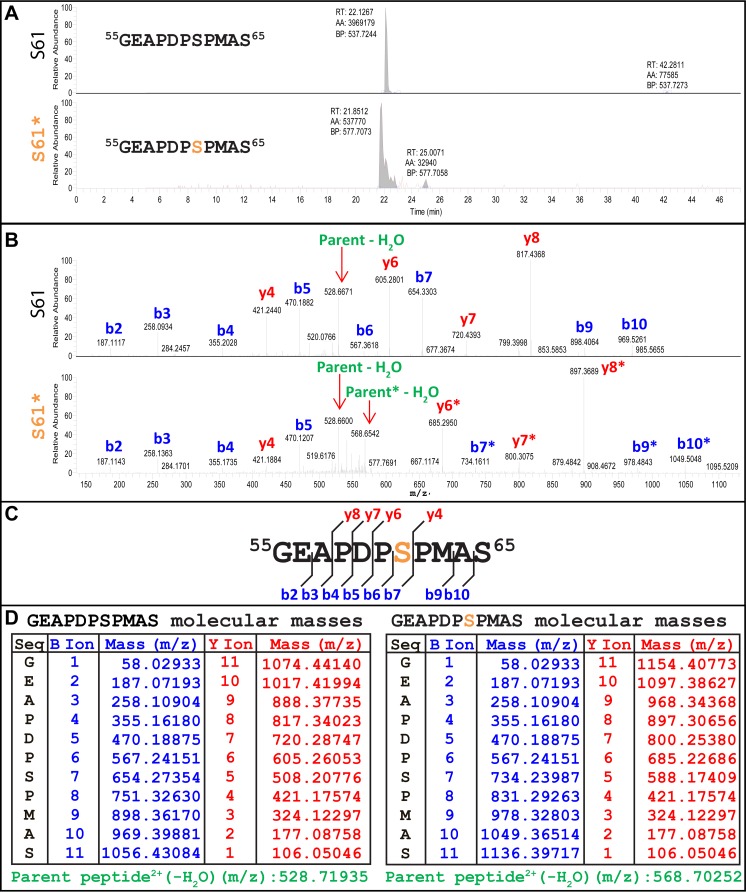
FIG 4.
LC-MS/MS identification of the p12 M63 serine 61 phosphorylation. (A) Peaks of nonphosphorylated (top) and phosphorylated (bottom) MLV p12 M63 55GEAPDPSPMAS65 peptide. The orange serine was determined to be phosphorylated. (B) Fragmentation analysis of nonphosphorylated (top) and phosphorylated (bottom) 55GEAPDPSPMAS65 peptide. Ion peaks were labeled based on ion size calculations shown in panel D. Green labels indicate the full-length, unfragmented parent peptides. Ions with the characteristic 80-Da mass addition signaling phosphorylation are denoted with an asterisk (*). (C) Schematic of detected ions shown in panel B. Red “y” ions include the C terminus, blue “b” ions include the N terminus. (D) Monoisotopic ion mass calculations for the 55GEAPDPSPMAS65 peptide with (right) and without (left) the addition of the 79.96633-Da mass of the phosphorylation group on serine 61. Ions are 1+ charged, and the parent peptide is 2+ charged.
shRNA knockdown of cellular kinases.
293T cells were transfected overnight using Fugene 6 with 2 μg pLKO1 (small hairpin RNA [shRNA] vector), 2 μg pHIT (VSV-G envelope), and 2 μg psPAX2 (lentiviral gag-pol). Cells were then treated with 10 mM sodium butyrate for 6 h to increase plasmid expression. Virus was collected overnight in DMEM, filtered through a 0.45-μm HT Tuffryn membrane, and used to infect fresh 293T cells. Infections were spinoculated 30 min at 1,600 × g with 8 μg/ml Polybrene.
RESULTS
Isoleucine 63 rescues chromatin tethering and viral titer in MLV p12 phosphorylation mutants.
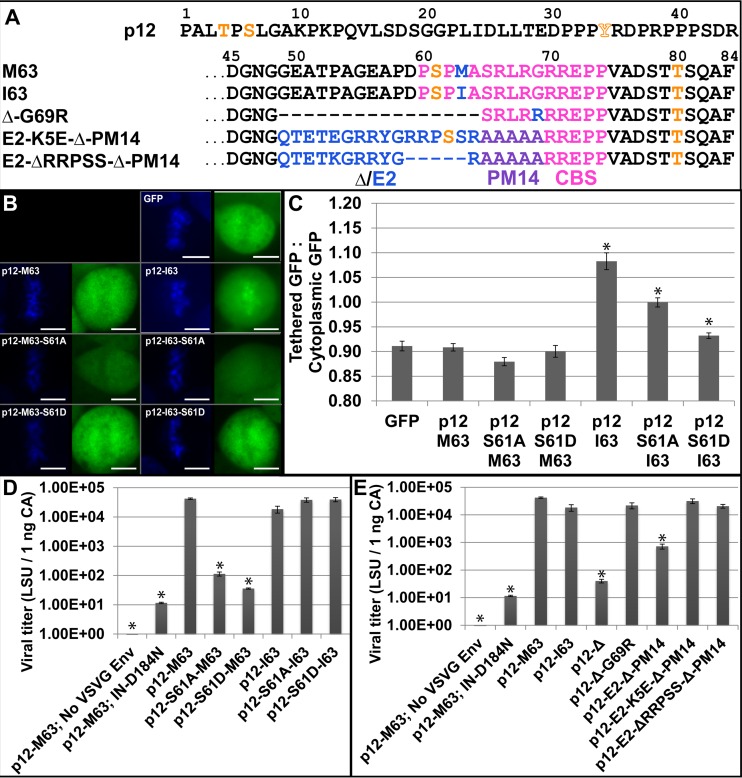
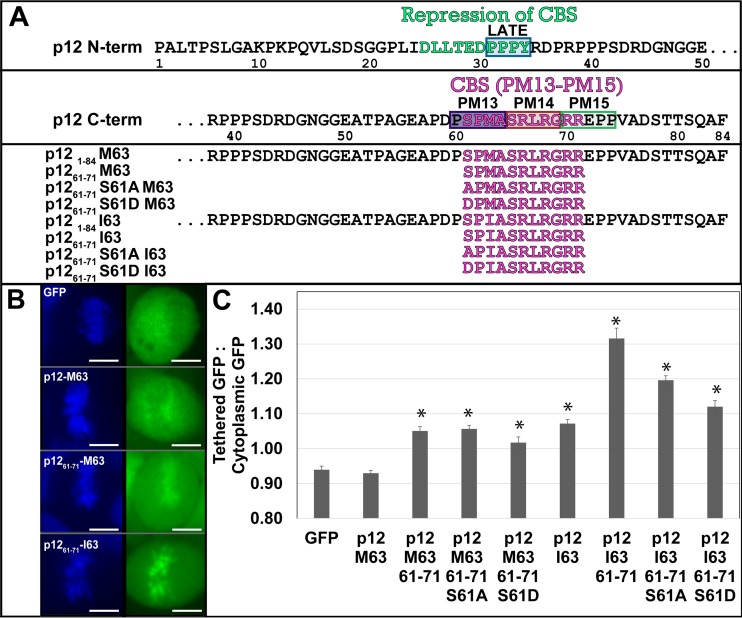
As serine 61 is the key phosphorylated residue of p12 and is located in the chromatin binding sequence (Fig. 1A) (5, 8, 9), the role of phosphorylation in regulating MLV p12 tethering was examined. Previously, we reported that alternative chromatin tethering domains substituted into p12 can replace the function of the PM14 region and can tether p12-GFP fusion proteins to mitotic chromatin (8). Paradoxically, wild-type Moloney MLV (MMLV) p12 M63-GFP had undetectable tethering to chromatin, while p12 I63-GFP did tether (Fig. 1B). This is quantified as a ratio of the intensity of GFP overlapping the mitotic chromatin to the intensity of GFP in the cytoplasm of each cell (Fig. 1C). GFP excluded from the mitotic chromatin scores a ratio of <1.0, while more GFP overlapping the mitotic chromatin versus the cytoplasm scores >1.0. The GFP-only control scores a ratio of 0.9, as does p12 M63-GFP, while p12 I63-GFP scores 1.1, showing increased/detectable levels of mitotic chromatin binding. The p12-I63 sequence variant was previously published as a suppressor mutant that rescued the mutation of serine 61 (9, 31). This suggested that the I63 variant activates the chromatin tethering of p12 in the absence of serine 61 phosphorylation.
FIG 1.
Isoleucine activation of p12 tethering and rescue of viral titer. (A) The 84 residues of p12. M63 and I63 contain the Δ region (residues 49 to 64) that is deleted in the p12-Δ-G69R virus and replaced with the HPV-8 E2 peptide in the last two viruses in the list. Orange indicates phosphorylation sites detected by mass spectrometry (orange outline denotes sites detected only in specific samples), pink denotes the chromatin binding sequence previously defined in p12, blue denotes the sequence variations between viruses, and purple denotes the PM14 mutant backbone. (B) p12-GFP fusion proteins' tethering ability. Left panels, Hoechst stain (DNA); right panels, GFP. Scale bars, 10 μm. (C) Ratio of p12-GFP fusion protein intensity overlapping the mitotic chromatin versus that in the cytoplasm, as seen in panel B. Error bars represent ±1 standard error (SE); n >15. (D) lacZ infection titer of p12 serine 61 mutants in the wild-type p12 M63 context and with the p12 I63 suppressor mutant. Error bars represent ±1 SE; n = 3. (E) lacZ infection titer of all five p12 constructs assayed with mass spectrometry and their parent constructs p12-Δ49–64 and p12-E2-Δ49–64-PM14. Error bars represent ±1 SE; n = 3. All constructs with an asterisk (*) are statistically different from GFP (C) or p12-M63 (D, E) using Student's t test (α = 0.05).
To test this idea, serine 61 was mutated in the p12-GFP constructs. The constructs with S61A and S61D substitutions in M63 remained untethered to the mitotic chromosomes. The S61A substitution in p12-I63-GFP did lower the tethering ratio to 1.0 (the GFP intensity overlapping the chromatin was equal to that in the cytoplasm), which is still significantly above the ratio for GFP alone (0.9) (Student's t test P value, 9 × 10−8) (Fig. 1B and C). The S61D substitution in the p12-I63-GFP background further reduced the tethering ratio (Fig. 1B and C), suggesting that I63 coupled with a phosphomimetic is detrimental to tethering or simply that aspartic acid at position 61 lowers the tethering ratio for reasons unlinked to phosphorylation.
Viral titer assays then confirmed that the p12 M63I substitution, which activated tethering, also rescued the loss of titer in the serine 61 mutants (Fig. 1D). VSV-G Env pseudotyped viruses packaging p12 mutant Gag proteins and a lacZ reporter gene were produced, normalized by ELISA, and infected into D17 cells in a single-round infection assay. Wild-type MMLV (p12 M63) had a viral titer of 4 × 104 lacZ staining units (LSU) per 1 ng CA, while the S61A and S61D mutants had titers over 400-fold lower (1 × 102 and 4 × 101, respectively). Both these phosphorylation mutants were rescued by I63 back to the wild-type titer of 4 × 104, confirming that p12-I63 functions well in the absence of S61 phosphorylation. Interestingly, p12-I63 in the wild-type S61 context decreased titer to 2 × 104, showing a slight fitness cost to the suppressor mutant in a wild-type, phosphorylation-competent setting. The titer of IN D184N, an integrase catalytic core mutant, indicates the background levels of nonvirally mediated insertion into the host genome and lacZ gene expression. These titers confirmed that p12-I63 is able to rescue the viral fitness of MLV p12 serine 61 mutants. This clear requirement of p12 S61 for the fitness of wild-type MLV p12 M63 led to mass spectrometry studies to confirm the phosphorylation states of the various viral and GFP fusion constructs and to probe its requirement for tethering.
Analysis of phosphorylated viral proteins.
Five viral constructs were selected for further analysis, the sequences of which are shown in Fig. 1A. These include the wild-type MMLV (p12-M63), the p12-I63 counterpart, and three constructs bearing exemplary insertions/deletions within the p1249–69 region. Previously, p12 residues glycine 49 through alanine 64 (abbreviated as Δ49–64) were deleted to allow room for alternate chromatin tethering inserts (8). When passaged in cell culture, p12-Δ49–64 virus mutated glycine 69 to arginine (G69R) to improve viral fitness from a titer of 4 × 101 to 2 × 104 (Fig. 1E). The G69R suppressor mutation also activated the p12 chromatin tethering motif to compensate for the loss of serine 61 (8). This p12-Δ49–64-G69R virus showed that MLV can passage in the absence of p12 serine 61 phosphorylation and was useful as a control lacking serine 61 in these phosphorylation studies.
Insertion of a human papillomavirus 8 (HPV-8) E2 sequence into p12-Δ49–64-PM14 construct replaced the p12 S61 phosphorylation motif with an E2 RRXS phosphorylation motif. This HPV-8 E2 sequence is phosphorylated in HPV-8 to activate tethering of the downstream chromatin-binding domain during S phase and mitosis (43). As with p12-Δ49–64, which selected for p12-Δ49–64-G69R, passage of the p12-E2-Δ49–64-PM14 virus produced multiple suppressor mutations in the HPV-8 E2 sequence, including K5E, S15P, ΔRRPSS, and ΔETKGRRY, and one in p12, R71Q (8) (Fig. 1A). The E2 rescue of p12-Δ49–64-PM14 showed that this motif could effectively bind chromatin on its own and could functionally replace the chromatin binding motif of MLV p12 (Fig. 1E). The p12-E2-K5E-Δ49–64-PM14 mutant was chosen for mass spectrometry analysis to detect if the HPV-8 phosphorylation motif would still be phosphorylated in the MLV context. The p12-E2-ΔRRPSS-Δ49–64-PM14 mutant served as a negative control for the deletion of the HPV-8 E2 phosphorylation site. Conveniently, the molecular weight shifts due to these insertions and deletions within p12 allowed for easy identification of p12 and p12 precursor proteins analyzed via SDS polyacrylamide gel electrophoresis.
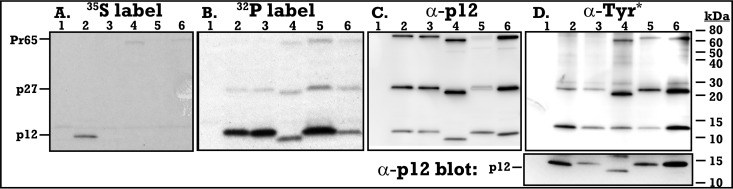
These five viruses were analyzed via 32P labeling, Western blot analysis (Fig. 2 and 3), and mass spectrometry (Fig. 4; Table 1) for the presence of phosphorylation. Virus-associated proteins were first analyzed by 35S labeling to examine maturation. Samples were immunoprecipitated with monoclonal anti-p12 serum (CRL-1890) to focus on p12 and its precursor proteins. The completely processed mature p12 was the predominant product detected in the p12 M63 virus (Fig. 2, lane 2). Labeling with 35S requires the presence of either Met or Cys in the coding sequence. The remaining viral p12 proteins containing I63 or the p12 Δ49–64 deletion are therefore not labeled by 35S. Despite the almost complete cleavage of p12 from Gag precursor observed by immunoprecipitation (Fig. 2A), immature p27 (matrix-p12) and Pr65 (gag) precursor proteins are preferentially detected by anti-p12 Western blotting (Fig. 2C). In contrast, viral protein metabolically labeled with 32P and immunoprecipitated with p12 antibodies detected highly phosphorylated p12 proteins for all five constructs at their predicted molecular masses as well as low levels of phosphorylated precursor p27 and Pr65 proteins (Fig. 2B). p12-M63 (Fig. 2B, lane 2) and p12-I63 (Fig. 2B, lane 3) shared equal levels of phosphorylation, despite p12-I63 functioning independent of S61 phosphorylation (Fig. 1). Surprisingly, deletion of the Δ49–64 region including serine 61 (Fig. 2B, lane 4) showed a diminished but not complete lack of 32P labeling, suggesting that other phosphorylations exist on p12. Addition of the HPV-8 E2 peptide, including the RRPS phosphorylation motif (Fig. 2B, lane 5), returned 32P labeling back to p12-M63 levels, showing that the HPV-8 RRPS phosphorylation motif is being phosphorylated at levels similar to those of p12 S61 in the context of the MLV p12 protein. Lastly, the deletion of this HPV-8 phosphorylation site (Fig. 2B, lane 6) brought 32P labeling back down to a level similar to that of the p12-Δ49–64 virus (Fig. 2B, lane 4). Western blot analysis of the 32P-labeled virus with anti-p12 antiserum shows equal loading of p12 (Fig. 2C). These radiolabeled experiments confirmed the phosphorylation of S61, showed that the MLV-HPV-8 chimeric p12 can be phosphorylated at RRPS, and illuminated the presence of secondary phosphorylation sites in p12 in the absence of S61 and RRPS.
FIG 2.
Maturation and phosphorylation of MLV p12 mutants. (A to C) Viral pellets were labeled with 35S (A) or 32P (B, C), immunoprecipitated against p12, and then analyzed by SDS-PAGE and detected via 35S (A), 32P (B), and anti-p12 antibody (C). p12 and p12-containing precursor polypeptide sizes are marked on the left, while molecular mass standards (in kilodaltons) are marked on the right. (D) Viral pellets run via SDS-PAGE were analyzed by Western blotting with antiphosphotyrosine antibody. A duplicate gel was blotted for p12 (lower panel). Lanes: 1, no virus; 2, p12-M63; 3, p12-I63; 4, p12-Δ49–64-G69R; 5, p12-E2-K5E-Δ49–64-PM14; 6, p12-E2-ΔRRPSS-Δ49–64-PM14.
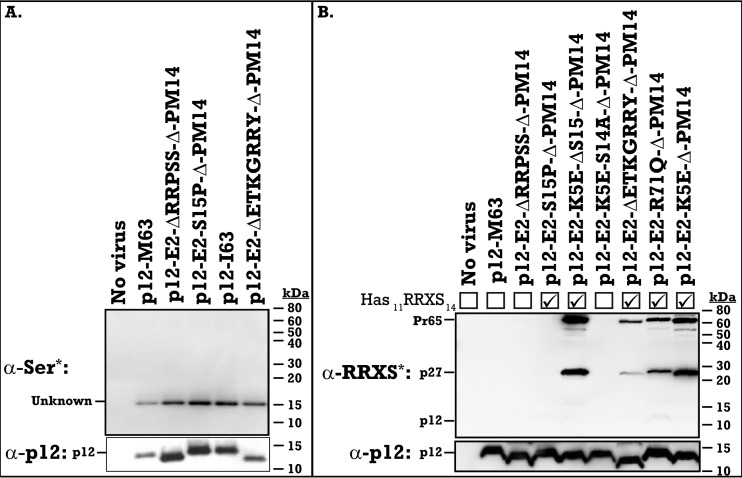
FIG 3.
Phosphorylation of the RRXS motif in the HPV-8 E2 peptide. MLV p12 chimeric viruses with the HPV-8 E2 phosphorylation motif insert and suppressor mutations were pelleted and run by SDS-PAGE on duplicate gels and were analyzed by Western blotting for phosphorylation (top) or for p12 (bottom). (A) Antiphosphoserine Western blot of viral pellets (rabbit anti-phosphoserine, catalog number 61-8100; Novex). (B) Rabbit anti-phospho-PKA substrate (RRXS*/T* motif) (catalog number 9624; Cell Signaling Technology) Western blot of viral pellets. Viruses containing the intact 11RRXS14 motif are indicated by a checkmark. p12 and p12-containing precursor polypeptide sizes are marked on the left, while molecular mass standards (in kilodaltons) are marked on the right.
TABLE 1.
Phosphorylated residues detected on MLV p12 and fitness of mutantsa
| Residue | Viral samples |
p12-GFP | ||
|---|---|---|---|---|
| p12-M/Ib | p12-Δc | Mutant fitnesse | p12-M/Ib | |
| p12 T4 | X | X | Wild type | ND |
| p12 S6 | X | X | Wild type | ND |
| p12 Y34 | ND | X | Nonviablef | ND |
| p12 S61 | X | NA | Revertsf | ND |
| p12 T80 | X | X | Wild type | ND |
| E2 S14/S15 | NA | Xd | Wild type | NA |
X, phosphorylation was detected on the indicated p12 residue; ND, no phosphorylation detected; NA, not applicable.
Includes the p12-M63 and p12-I63 proteins.
Includes the p12-Δ49–64-G69R, p12-E2-K5E-Δ49–64-PM14, and p12-E2-ΔRRPSS-Δ49–64-PM14 proteins.
One phosphorylation was detected on the E2 11RRPSS15 motif, but ion resolution alone could not distinguish between S14 and S15.
Fitness of mutant MLV lacking given phosphorylation sites (T4A, S6A, S61A, T80A, or deletion) via viral spread assay (multiple rounds of infection) done in triplicate.
The identity of the phosphorylated species was analyzed through Western blot assays. Antibodies specific to phosphorylated tyrosine residues detected phosphorylation of the MA-p12 (p27) precursor in all samples, as well as Gag (Pr65) in the three Δ49–64 mutants (Fig. 2D). The strong recognition by the antiphosphotyrosine antibody for the MA-p12 intermediate (p27) and Gag (Pr65) proteins versus the undetected mature p12 implies a functional and conformational epitope present in these polyproteins distinct from the fully processed p12 protein. Unexpectedly, antiphosphoserine Western blotting did not detect p12 or any p12 precursors but did detect a small protein in all five viral samples (Fig. 3A). Five additional phosphoserine antibodies for various motifs (see Materials and Methods) all failed to detect p12 by Western blotting (data not shown). The p12 size shifts due to the Δ49–64 and the E2 inserts are observed in the lower panels of the same samples (anti-p12 Western blots). As these characteristic p12 size shifts are not observed in the low-molecular-mass phosphorylated protein, they are not p12 proteins (Fig. 2D and 3A). The identity of these bands is investigated further via mass spectrometry below.
The HPV-8 E2 RRPS motif is phosphorylated in the chimeric MLV, but its phosphorylation is dispensable for the rescue of the p12-Δ49–64-PM14 mutation.
Despite failing to be recognized by the general antiphosphoserine antibody, phosphorylation of the HPV-8 E2 peptide at 11RRPS14 was detected on p27 and Pr65 precursors. A phospho-PKA substrate antibody recognizing the RRXS*/T* motif detected phosphorylation of the E2 11RRXS14 motif (Fig. 3B). Besides the five original suppressor mutants tested, additional mutations were generated to confirm the phosphorylation site (8). The p12-E2-K5E-S14A-Δ49–64-PM14 virus removes the 11RRXS14 motif and consequently loses the phospho-RRXS signal (Fig. 3B). The p12-E2-K5E-ΔS15-Δ49–64-PM14 virus removes the second overlapping 12RXXS15 motif and has the same phosphorylation signal as p12-E2-K5E-Δ49–64-PM14 (Fig. 3B), confirming that there is no cross-reaction with that RXXS motif. This shows that the 11RRXS14 phosphorylation motif is phosphorylated in a chimeric environment but that this phosphorylation is not essential for the HPV-8 E2 insert to rescue the p12-Δ49–64-PM14 mutation, as both the suppressor mutants, E2 K5E containing serine 14 and E2 ΔRRPSS lacking serine 14, rescued the p12-E2-Δ49–64-PM14 titer back to wild-type levels (Fig. 1E).
Mass spectrometry analysis of viral MLV p12 variants identifies novel phosphorylation sites in p12 and matrix and fails to detect phosphorylation in p12-GFP fusion proteins.
Mass spectrometry was conducted on all five viruses shown in Fig. 1A to identify the residual phosphorylations in the p12-Δ49–64-G69R virus (Fig. 2B), as well as the tyrosine phosphorylations detected by the antiphosphotyrosine antibody (Fig. 2D). Phosphorylations were detected on p12 threonine 4, serine 6, and threonine 80 in all five viruses (Table 1). Serine 61 phosphorylation was detected on p12 M63 and p12 I63 and is deleted from the p12 Δ49–64 viruses (Fig. 4; Table 1). Representative data from MLV p12 M63 55GEAPDPSPMAS65 fragment analysis are displayed in Fig. 4. Two parent peptides, nonphosphorylated (Fig. 4A, top) and single-phosphorylated (Fig. 4A, bottom) 55GEAPDPSPMAS65, were further fragmented to determine the phosphorylated residue (Fig. 4B). Phosphorylation of S61 was determined based on the ions that were detected with the characteristic 80-Da mass addition of the phosphorylation group (H3PO4) (Fig. 4B, bottom). A schematic of all ions detected is provided (Fig. 4C), with b7, b9, b10, y6, y7, and y8 ions all detected as phosphorylated, leading to identification of S61 as phosphorylated. Peptides detected via mass spectrometry (Fig. 4B) were matched to the calculated masses shown in Fig. 4D for identification.
One phosphorylation was detected on serine 14 or 15 of the E2 insert in p12-E2-K5E-Δ49–64-PM14 via mass spectrometry, which was further narrowed to S14 via mutational analysis (Fig. 3B). This E2 S14 phosphorylation is dispensable for tethering and integration, as seen by the wild-type viral titer of p12-E2-ΔRRPSS-Δ-PM14 (Fig. 1E). Interestingly, phosphorylation was also detected by mass spectrometry on p12 tyrosine 34 (Y34), in the PPPY L domain, but only in the three Δ49–64 mutants and not in p12 M63 or p12 I63 (Table 1). Previous studies have shown that p12 serine 61 and p12 tyrosine 34 of the late motif are required for viral fitness in multiple rounds of infection (1). In similar transient expression assays, mutants bearing either p12 T4A, S6A, T80A, E2 S14A, or E2 S15A replicated with kinetics similar to that of wild-type p12-M63 virus, indicating that they are nonessential for viral passage (Table 1).
Mass spectrometry was also performed on the p12-M63-GFP and p12-I63-GFP fusion constructs, in the absence of the other viral proteins. Both proteins were identified with high confidence, but phosphorylation was not detected on either sample. This suggests that serine 61 phosphorylation is dependent on the trafficking of p12 in the viral life cycle and that I63 GFP is able to activate tethering in the absence of serine 61 phosphorylation. This indicates that S61 phosphorylation is important for activating MLV p12 M63 tethering.
To identify the unknown 15-kDa phosphorylated bands, virus was expressed in 293T cells and electrophoresed on SDS-PAG, and the 15-kDa bands were excised for mass spectrometry analysis. Peptides were compared to a database of viral peptides and known human phosphopeptides. In the wild-type virus, the unknown serine phosphorylated band around 15 kDa was not defined using this method. Tyrosine 130 of the MLV matrix protein (15 kDa) was detected as phosphorylated, though, and matches the size of the unknown 15-kDa band detected via Western blotting (Fig. 2D). As p27 contains both MA Y130 and p12 Y34, the position of the tyrosine phosphorylation in this Western blot could not be determined and was further analyzed below.
Destruction of the MA-p12 protease site traps MA-p12 in a highly tyrosine-phosphorylated state.
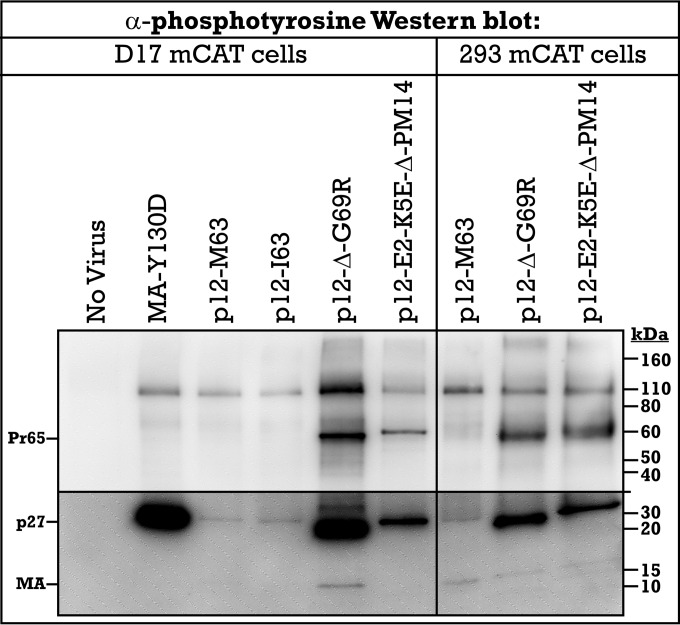
The identity of the p27 tyrosine phosphorylation was analyzed using virus bearing an MA Y130D mutation. MA Y130 was previously characterized as an essential residue for viral protease cleavage between matrix and p12. The matrix Y130D mutant trapped the proteins in the p27 precursor peptide, and this resulted in a 100-fold decrease in infectivity (35). Western blot analysis of viral MA Y130D with antiphosphotyrosine antibody identified that the p27 precursor was trapped in a highly tyrosine-phosphorylated state similar to what was seen with the p12 Δ49–64 mutants, compared to the M63 or I63 virus (Fig. 5). Considering that mass spectrometry detected tyrosine phosphorylation on MA Y130 and p12 Y34, use of MA Y130D indicates that the remaining phosphorylation within p27 should be p12 Y34. It is unknown if the phosphorylated tyrosine precursors represent biologically functional intermediates in the wild-type life cycle.
FIG 5.
Tyrosine phosphorylation of p27 precursor protein. Viral samples collected from D17 and 293 cells, both expressing the mCAT receptor, were run by SDS-PAGE, and phosphorylation of tyrosine residues was detected via antiphosphotyrosine Western blotting. Viral precursor and mature protein sizes are indicated on the left, while molecular mass standards (in kilodaltons) are marked on the right. Bottom panel shows increased exposure to illuminate weak bands.
Activation of p12-M63 chromatin binding.
Phosphorylation of p12 can act directly through activating a tethering competent conformation or indirectly through releasing a repression domain. Deletion studies of p12 indicated that the N terminus of p12 acts as a repression domain of tethering (26). The role of phosphorylation to regulate tethering was examined with the minimal tethering domain 61SPMASRLRGRR71-GFP fusion protein (Fig. 6). Surprisingly, whereas the full-length p12-M63-GFP protein did not bind mitotic chromosomes (Fig. 1), the minimal 61SPMASRLRrRR71-GFP was able to bind mitotic chromosomes. Significantly, substitution of either A or D in p12-S61 maintained chromosomal tethering. This indicates that S61 modification is not directly required for chromatin binding within the context of p1261–71-M63-GFP fusions (Fig. 6). Binding of p1261–71-I63-GFP was significantly higher than that of p1261–71-M63-GFP, with mutations of S61 lowering its binding ability, the level of which are all greater than that observed for p1261–71-M63 (Fig. 6). Mass spectrometry of the p12-I63-GFP construct indicated that the proteins were not phosphorylated (Table 1). Thus, for p12-I63, the effects of the mutations could alter the conformation of the activated p12-I63 state.
FIG 6.
Activation of p12-M63-GFP chromatin binding. Full-length p12-GFP fusion proteins were compared to minimal binding motif constructs containing M63 versus I63. (A) Sequences of fusion proteins. (B) Quantification of GFP intensity overlapping the mitotic chromatin versus that in the cytoplasm; n > 15. Error bars are standard errors. Asterisks (*) indicate ratios that are statistically different from those of GFP using Student's t test (α = 0.001). Studies of GFP and p1261–71-GFP-I63 were duplicated in a companion article in this issue (26) for comparison purposes.
DISCUSSION
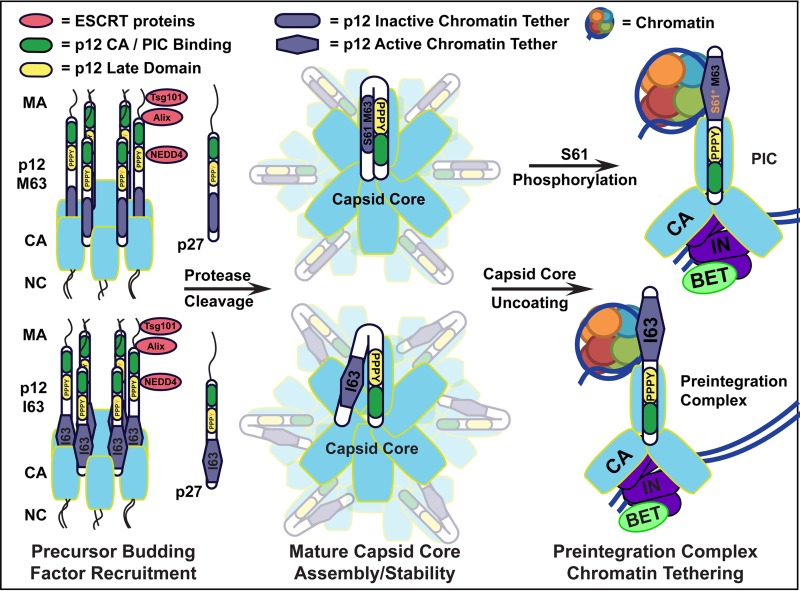
Altogether, p12 functions in viral budding, in capsid core stability and uncoating, and in nuclear retention of the PIC. Tight temporal regulation of the various p12 domains must be required to suppress their functions at inappropriate times, which in many biological systems is regulated by phosphorylation. Previous research has shown that the β-dystroglycan protein is tyrosine phosphorylated at its PPPY domain to disrupt its binding to the WW domain of dystrophin (44). The same phosphorylation offers MLV a mechanism to temporally regulate the PPPY motif binding to the WW domain of NEDD4. MLV Gag precursor recruits NEDD4 via the N-terminal PPPY domain in p12 to function in viral release (Fig. 7, left) (3), while the mature N terminus of p12 must bind the PIC to tether it for integration (Fig. 7, right) (5, 8, 9). As the tethered, mature p12 protein no longer needs to recruit budding proteins and binding them may sterically hinder binding of the PIC and host chromatin, we propose that the phosphorylation of PPPY domain acts to disrupt the binding of NEDD4. This clears the N terminus of p12 to first bind the capsid core (Fig. 7, center) and then bind the preintegration complex, allowing tethering to the host chromatin for nuclear retention (Fig. 7, right). The differential Y34 phosphorylation in p27 versus p12 (Fig. 2D) suggests that p27 may have an important, as-yet-undefined role.
FIG 7.
Model for regulation of p12 chromatin tethering via S61 phosphorylation. (Left) MLV Gag precursor late domains recruit ESCRT proteins for budding. Proteolytic processing proceeds through the p27 (MA-p12) intermediate. PPPY late domain (yellow) phosphorylation is proposed to disrupt NEDD4 binding and allow repression of the chromatin binding motif (blue). (Center) After protease-mediated maturation, the capsid core forms, with the p12 N terminus (green) involved in proper core formation and stability and the p12 C terminus important for core uncoating. (Right) After reverse transcription and capsid core uncoating, the p12 N terminus (green) binds the PIC, containing capsid (light blue), integrase (purple), and BET (lime green), while the C terminus binds mitotic chromatin (represented by an individual nucleosome), to function in nuclear retention of the PIC. (Top right) Phosphorylation of S61 is proposed to disrupt binding of the p12 N-terminal PPPY motif to the p12 C-terminal chromatin binding motif, facilitating capsid core uncoating and relieving chromatin binding repression. (Bottom right) p12 I63, other C-terminal charge shifts, and p12 Δ are proposed to weaken the PPPY interaction with the p12 C-terminal chromatin tethering motif, rescuing the loss of the S61 phosphorylation switch.
Phosphorylation of matrix Y130 may also be used to regulate its late domain. Matrix Y130 is located in the LYPAL late domain at the junction of matrix and p12. During late viral egress, this domain recruits Alix (Fig. 7, left), and then the overlapping protease cleavage site is cleaved during viral maturation. Phosphorylation may help regulate this process correctly, altering the site of Alix binding to allow for subsequent viral protease cleavage. Cleavage of this late domain then relieves the viral particle of the ESCRT budding factor binding domains located in matrix, which are no longer needed during the early stages of infection (Fig. 7, center).
In addition to the PPPY motif's function in late domain budding, recent findings show that the p12 N-terminal 25DLLTEDPPPY34 motif is a repressor of the p12 C-terminal chromatin binding motif, suggesting that these two motifs are binding during infection (26). This dual function for the PPPY motif is supported by previous research indicating that it had both early and late defects in replication (1). The late budding defect in MLV p12 PPPY mutants was rescued with late domains inserted into matrix, an alternate p12 site, and nucleocapsid, but these viruses maintained a block in early infection (25). This early function has a positional effect, which we propose involves interaction with the p12 C terminus. This finding was further confirmed here, as deletion of the N terminus (p1261–71-M63-GFP) activated chromatin tethering in the MMLV wild-type fusion protein (Fig. 6). Furthermore, mutation of S61 in this construct had no effect on the binding of p12-M63-GFP, suggesting that the role of S61 phosphorylation is to disrupt the repressor binding (Fig. 7, center).
Previous research has already established multiple roles for p12 in capsid core stability, first showing that the N terminus of p12 is required for proper capsid core formation and stability (Fig. 7, center) (1, 6). After reverse transcription, C-terminal p12 mutants (PM14 and S61) have been shown to overstabilize the capsid core, blocking appropriate uncoating and nuclear localization of the viral PIC (9). These findings together suggest that the p12 N terminus binding to the C terminus is critical first to form and stabilize the capsid core and that disruption of this interaction is then critical in destabilizing the capsid core and revealing the p12 chromatin tethering motif to function in PIC nuclear retention. S61 phosphorylation by a cell cycle kinase provides a temporal cue for this final capsid core uncoating event. The capsid core must maintain its integrity for antiviral defense until mitosis, at which time p12 S61 phosphorylation can disrupt the binding of the N-terminal DLLTEDPPPY motif, destabilizing the capsid core and revealing the p12 C terminus to tether the PIC to the mitotic chromatin for nuclear retention postmitosis (Fig. 7, right). This was tested with the p12-S61D-M63-GFP construct, which was not found to be tethered to mitotic chromatin. We believe that this is a failure of the phosphomimetic residue to accurately substitute phosphorylation in the context of this assay. The temporal regulation of chromatin binding may have also been lost in the p12-GFP assay, as suggested by the lack of phosphorylation of the p12-GFP fusion proteins. In this model, I63 rescues the loss of S61 by partially disrupting the PPPY interaction, just enough to free the C-terminal chromatin tether when appropriate, but still allowing enough binding to stabilize the capsid core.
In an attempt to identify a cell cycle kinase for this temporal regulation of capsid core uncoating and PIC tethering, NetPhosK 1.0 and GPS 3.0 kinase prediction algorithms were used to predict kinase interactions with the p12 serine 61 motif (45, 46). In addition, a homemade script was used to identify protein interactions with the SPMAS motif. All three methods predicted GSK 3β. Other top hits from one or more methods included Jun N-terminal protein kinase 2 (JNK2), homeodomain-interacting protein kinase 2 (HIPK 2), CDC-like kinase 1 (CLK 1), MAPK 11, CDK 5, MAPK 14, and CDK 1. Expression of these proteins was knocked down using integrated copies of shRNA, expressed in 293T cells on a lentiviral vector. Knockdown of most targets was lethal or crucially slowed cell proliferation, while control shRNAs had no toxicity. This presumably selected for low levels of target knockdown in the surviving and proliferating cells. This low level of knockdown may explain the lack of an effect on viral infection (data not shown). As serine 61 mutation reduces the titer to 0.3% of that of wild-type virus (Fig. 1D), knockdown of the respective kinase was expected to significantly reduce the titer of wild-type p12-M63 virus versus S61 phosphorylation-independent control viruses p12-Δ49–64-G69R and p12-E2-ΔRRPSS-Δ49–64-PM14. Results showed a modest, if any, effect on viral infection (<2-fold change). Either the level of knockdown required to maintain and expand 293T cells was also sufficient to maintain wild-type titer of MLV infection, or the correct kinase target was not predicted by these algorithms.
Kinase inhibitor drugs were also used to test inhibition on wild-type MLV infection. Using the same experimental design as for the shRNA experiment, kinase inhibitors were added to the cells infected by MLV, and the wild-type p12-M63 viral titer was compared to control viruses p12-Δ49–64-G69R and p12-E2-ΔRRPSS-Δ49–64-PM14, which no longer required S61 phosphorylation for viral replication. Published active concentrations of KT5720 (47), roscovitine (48), and valproate (49) (2 μM, 10 μM, and 3 mM, respectively) all had no effect on wild-type MLV replication versus phosphorylation-independent MLV strains. Kinases inhibited by these drugs include the following: phosphorylase kinase (PHK), phosphoinositide-dependent protein kinase 1 (PDK1), MEK, mitogen- and stress-activated protein kinase 1 (MSK1), AKT1, PKBα, and glycogen synthase kinase 3β (GSK 3β) by KT5720; cyclin-dependent kinase 1 (CDK1), CDK2, and CDK5 by roscovitine; and GSK 3β by valproate. Again, either the sublethal drug concentration required to maintain the 293T cells was also sufficient to maintain the wild-type titer of MLV infection, or the correct kinase was not targeted by these drugs.
In conclusion, we have identified novel phosphorylation sites in p12 via mass spectrometry and have probed their role in chromatin tethering and budding. Previous research showed I63 as a viral suppressor mutation to rescue the loss of serine 61 phosphorylation, and we show that I63 activates tethering of p12-GFP in the absence of phosphorylation. We therefore conclude that this activation of p12 tethering rescues the function of p12 S61 phosphorylation and suggest that this S61 phosphorylation activates the chromatin tethering domain in wild-type p12 M63 by displacing the negative regulatory domain located at the p12 N terminus (Fig. 7). p12 I63 overcomes the loss of this regulatory S61 phosphorylation by constitutive activation of the p12 chromatin tethering domain, at the cost of slightly reduced viral fitness.
We found that the exogenous HPV-8 E2 motif QTETKGRRYGRRPSSR can functionally replace the chromatin binding sequence encoded at the p12 C terminus, within residues 49 to 69 (mutated in the p12-Δ49–64-PM14 backbone construct). This alternate viral motif is phosphorylated at levels similar to those of the wild-type 60PSPMA64 motif; however, this phosphorylation is dispensable for the HPV-8 E2-mediated rescue of p12-Δ49–64-PM14 to a wild-type viral titer. These and other suppressor mutants in p12 suggest that phosphorylation of p12 acts to temporally regulate its chromatin tethering but that changes in C-terminal residue charges can compensate for the loss of this mechanism.
Furthermore, we detected phosphorylation within two Gag late domains in specific viral samples at matrix Y130 in the LYPAL motif and within the PPPY late domain. We suggest that these phosphorylations regulate ESCRT factor binding to allow recruitment during viral budding and release during viral entry. The matrix Y130 phosphorylation also overlaps the matrix-p12 protease cleavage site, suggesting that it may also regulate cleavage of p27 into matrix and p12. Release of NEDD4 at the 31PPPY34 motif liberates the p12 N terminus first to bind the C terminus for capsid core stabilization and then to bind the preintegration complex (PIC), required for p12's early functions in nuclear localization of the PIC.
ACKNOWLEDGMENTS
We thank Alan Rein for graciously providing the MLV MA Y130D construct used in this study.
This work was supported by the National Institutes of Health and the New Jersey Commission on Cancer Research (NJCCR) in collaboration with the New Jersey Department of Health (NJDHSS).
Footnotes
For a companion article on this topic, see doi:10.1128/JVI.01084-16.
REFERENCES
- 1.Yuan B, Li X, Goff SP. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J 18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci U S A 97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol 168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang F, Zang T, Wilson SJ, Johnson MC, Bieniasz PD. 2011. Clathrin facilitates the morphogenesis of retrovirus particles. PLoS Pathog 7:e1002119. doi: 10.1371/journal.ppat.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wight DJ, Boucherit VC, Nader M, Allen DJ, Taylor IA, Bishop KN. 2012. The gammaretroviral p12 protein has multiple domains that function during the early stages of replication. Retrovirology 9:83. doi: 10.1186/1742-4690-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wight DJ, Boucherit VC, Wanaguru M, Elis E, Hirst EM, Li W, Ehrlich M, Bacharach E, Bishop KN. 2014. The N-terminus of murine leukaemia virus p12 protein is required for mature core stability. PLoS Pathog 10:e1004474. doi: 10.1371/journal.ppat.1004474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SK, Nagashima K, Hu WS. 2005. Cooperative effect of gag proteins p12 and capsid during early events of murine leukemia virus replication. J Virol 79:4159–4169. doi: 10.1128/JVI.79.7.4159-4169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider WM, Brzezinski JD, Aiyer S, Malani N, Gyuricza M, Bushman FD, Roth MJ. 2013. Viral DNA tethering domains complement replication-defective mutations in the p12 protein of MuLV Gag. Proc Natl Acad Sci U S A 110:9487–9492. doi: 10.1073/pnas.1221736110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elis E, Ehrlich M, Prizan-Ravid A, Laham-Karam N, Bacharach E. 2012. p12 tethers the murine leukemia virus pre-integration complex to mitotic chromosomes. PLoS Pathog 8:e1003103. doi: 10.1371/journal.ppat.1003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prizan-Ravid A, Elis E, Laham-Karam N, Selig S, Ehrlich M, Bacharach E. 2010. The Gag cleavage product, p12, is a functional constituent of the murine leukemia virus pre-integration complex. PLoS Pathog 6:e1001183. doi: 10.1371/journal.ppat.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fassati A, Goff SP. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J Virol 73:8919–8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roe T, Reynolds TC, Yu G, Brown PO. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J 12:2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bin Hamid F, Kim J, Shin CG. 2016. Cellular and viral determinants of retroviral nuclear entry. Can J Microbiol 62:1–15. doi: 10.1139/cjm-2015-0350. [DOI] [PubMed] [Google Scholar]
- 14.Ambrose Z, Aiken C. 2014. HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology 454-455:371–379. doi: 10.1016/j.virol.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemonnot B, Cartier C, Gay B, Rebuffat S, Bardy M, Devaux C, Boyer V, Briant L. 2004. The host cell MAP kinase ERK-2 regulates viral assembly and release by phosphorylating the p6gag protein of HIV-1. J Biol Chem 279:32426–32434. doi: 10.1074/jbc.M313137200. [DOI] [PubMed] [Google Scholar]
- 16.Radestock B, Morales I, Rahman SA, Radau S, Glass B, Zahedi RP, Muller B, Krausslich HG. 2013. Comprehensive mutational analysis reveals p6Gag phosphorylation to be dispensable for HIV-1 morphogenesis and replication. J Virol 87:724–734. doi: 10.1128/JVI.02162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhadina M, Bieniasz PD. 2010. Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog 6:e1001153. doi: 10.1371/journal.ppat.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sette P, Nagashima K, Piper RC, Bouamr F. 2013. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 10:79. doi: 10.1186/1742-4690-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemonnot B, Molle D, Bardy M, Gay B, Laune D, Devaux C, Briant L. 2006. Phosphorylation of the HTLV-1 matrix L-domain-containing protein by virus-associated ERK-2 kinase. Virology 349:430–439. doi: 10.1016/j.virol.2006.02.043. [DOI] [PubMed] [Google Scholar]
- 20.Joshi A, Munshi U, Ablan SD, Nagashima K, Freed EO. 2008. Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic 9:1972–1983. doi: 10.1111/j.1600-0854.2008.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. 2010. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J Virol 84:8181–8192. doi: 10.1128/JVI.00634-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blot V, Perugi F, Gay B, Prevost MC, Briant L, Tangy F, Abriel H, Staub O, Dokhelar MC, Pique C. 2004. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J Cell Sci 117:2357–2367. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- 23.Segura-Morales C, Pescia C, Chatellard-Causse C, Sadoul R, Bertrand E, Basyuk E. 2005. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J Biol Chem 280:27004–27012. doi: 10.1074/jbc.M413735200. [DOI] [PubMed] [Google Scholar]
- 24.Sabo Y, Laham-Karam N, Bacharach E. 2008. Basal budding and replication of the murine leukemia virus are independent of the gag L domains. J Virol 82:9770–9775. doi: 10.1128/JVI.00741-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol 74:7250–7260. doi: 10.1128/JVI.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brzezinski JD, Modi A, Liu M, Roth MJ. 2016. Repression of the chromatin-tethering domain of murine leukemia virus p12. J Virol 90:11197–11207. doi: 10.1128/JVI.01084-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pal BK, Roy-Burman P. 1975. Phosphoproteins: structural components of oncornaviruses. J Virol 15:540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pal BK, McAllister RM, Gardner MB, Roy-Burman P. 1975. Comparative studies on the structural phosphoproteins of mammalian type C viruses. J Virol 16:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikuta K, Luftig RB. 1988. Detection of phosphorylated forms of Moloney murine leukemia virus major capsid protein p30 by immunoprecipitation and two-dimensional gel electrophoresis. J Virol 62:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naso RB, Karshin WL, Wu YH, Arlinghaus RB. 1979. Characterization of 40,000- and 25,000-dalton intermediate precursors to Rauscher murine leukemia virus gag gene products. J Virol 32:187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yueh A, Goff SP. 2003. Phosphorylated serine residues and an arginine-rich domain of the Moloney murine leukemia virus p12 protein are required for early events of viral infection. J Virol 77:1820–1829. doi: 10.1128/JVI.77.3.1820-1829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Reilly L, Roth MJ. 2000. Second-site changes affect viability of amphotropic/ecotropic chimeric enveloped murine leukemia viruses. J Virol 74:899–913. doi: 10.1128/JVI.74.2.899-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol 7:379–387. doi: 10.1128/MCB.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goff S, Traktman P, Baltimore D. 1981. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol 38:239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oshima M, Muriaux D, Mirro J, Nagashima K, Dryden K, Yeager M, Rein A. 2004. Effects of blocking individual maturation cleavages in murine leukemia virus gag. J Virol 78:1411–1420. doi: 10.1128/JVI.78.3.1411-1420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felkner RH, Roth MJ. 1992. Mutational analysis of the N-linked glycosylation sites of the SU envelope protein of Moloney murine leukemia virus. J Virol 66:4258–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C, Rivera A, Ron N, Dougherty JP, Ron Y. 2001. A gene therapy approach for treating T-cell-mediated autoimmune diseases. Blood 97:886–894. doi: 10.1182/blood.V97.4.886. [DOI] [PubMed] [Google Scholar]
- 38.Gemeniano M, Mpanju O, Salomon DR, Eiden MV, Wilson CA. 2006. The infectivity and host range of the ecotropic porcine endogenous retrovirus, PERV-C, is modulated by residues in the C-terminal region of its surface envelope protein. Virology 346:108–117. doi: 10.1016/j.virol.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 39.Ting YT, Wilson CA, Farrell KB, Chaudry GJ, Eiden MV. 1998. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J Virol 72:9453–9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu D-T, Aiyer S, Villanueva RA, Roth MJ. 2013. Development of an enzyme-linked immunosorbent assay based on the murine leukemia virus p30 capsid protein. J Virol Methods 193:332–336. doi: 10.1016/j.jviromet.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fouchier RA, Meyer BE, Simon JH, Fischer U, Malim MH. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J 16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanese N, Roth MJ, Goff SP. 1986. Analysis of retroviral pol gene products with antisera raised against fusion proteins produced in Escherichia coli. J Virol 59:328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekhar V, McBride AA. 2012. Phosphorylation regulates binding of the human papillomavirus type 8 E2 protein to host chromosomes. J Virol 86:10047–10058. doi: 10.1128/JVI.01140-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ilsley JL, Sudol M, Winder SJ. 2001. The interaction of dystrophin with beta-dystroglycan is regulated by tyrosine phosphorylation. Cell Signal 13:625–632. doi: 10.1016/S0898-6568(01)00188-7. [DOI] [PubMed] [Google Scholar]
- 45.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 46.Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. 2008. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics 7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang YY, Martin KC, Kandel ER. 2000. Both protein kinase A and mitogen-activated protein kinase are required in the amygdala for the macromolecular synthesis-dependent late phase of long-term potentiation. J Neurosci 20:6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Azevedo WF, Leclerc S, Meijer L, Havlicek L, Strnad M, Kim SH. 1997. Inhibition of cyclin-dependent kinases by purine analogues: crystal structure of human cdk2 complexed with roscovitine. Eur J Biochem 243:518–526. doi: 10.1111/j.1432-1033.1997.0518a.x. [DOI] [PubMed] [Google Scholar]
- 49.Hall AC, Brennan A, Goold RG, Cleverley K, Lucas FR, Gordon-Weeks PR, Salinas PC. 2002. Valproate regulates GSK-3-mediated axonal remodeling and synapsin I clustering in developing neurons. Mol Cell Neurosci 20:257–270. doi: 10.1006/mcne.2002.1117. [DOI] [PubMed] [Google Scholar]