ABSTRACT
Most double-stranded RNA (dsRNA) viruses are transcribed and replicated in a specialized icosahedral capsid with a T=1 lattice consisting of 60 asymmetric capsid protein (CP) dimers. These capsids help to organize the viral genome and replicative complex(es). They also act as molecular sieves that isolate the virus genome from host defense mechanisms and allow the passage of nucleotides and viral transcripts. Rosellinia necatrix quadrivirus 1 (RnQV1), the type species of the family Quadriviridae, is a dsRNA fungal virus with a multipartite genome consisting of four monocistronic segments (segments 1 to 4). dsRNA-2 and dsRNA-4 encode two CPs (P2 and P4, respectively), which coassemble into ∼450-Å-diameter capsids. We used three-dimensional cryo-electron microscopy combined with complementary biophysical techniques to determine the structures of RnQV1 virion strains W1075 and W1118. RnQV1 has a quadripartite genome, and the capsid is based on a single-shelled T=1 lattice built of P2-P4 dimers. Whereas the RnQV1-W1118 capsid is built of full-length CP, P2 and P4 of RnQV1-W1075 are cleaved into several polypeptides, maintaining the capsid structural organization. RnQV1 heterodimers have a quaternary organization similar to that of homodimers of reoviruses and other dsRNA mycoviruses. The RnQV1 capsid is the first T=1 capsid with a heterodimer as an asymmetric unit reported to date and follows the architectural principle for dsRNA viruses that a 120-subunit capsid is a conserved assembly that supports dsRNA replication and organization.
IMPORTANCE Given their importance to health, members of the family Reoviridae are the basis of most structural and functional studies and provide much of our knowledge of dsRNA viruses. Analysis of bacterial, protozoal, and fungal dsRNA viruses has improved our understanding of their structure, function, and evolution, as well. Here, we studied a dsRNA virus that infects the fungus Rosellinia necatrix, an ascomycete that is pathogenic to a wide range of plants. Using three-dimensional cryo-electron microscopy and analytical ultracentrifugation analysis, we determined the structure and stoichiometry of Rosellinia necatrix quadrivirus 1 (RnQV1). The RnQV1 capsid is a T=1 capsid with 60 heterodimers as the asymmetric units. The large amount of genetic information used by RnQV1 to construct a simple T=1 capsid is probably related to the numerous virus-host and virus-virus interactions that it must face in its life cycle, which lacks an extracellular phase.
INTRODUCTION
Double-stranded RNA (dsRNA) viruses are a diverse group that infect hosts from bacteria to eukaryotes, including fungi, protozoa, plants, and animals (1). Their genome complexity varies from a single segment, like the L-A virus of the yeast Saccharomyces cerevisiae (ScV-L-A) (2), to 12 dsRNA molecules, like the rice dwarf virus (3). Capsid complexity is also quite variable and ranges from a single shell to multilayered concentric capsids. dsRNA viruses nevertheless share many general architectural and functional principles, which indicates parallel strategies in the viral life cycle (4). For example, a specialized T=1 icosahedral capsid that remains undisturbed throughout the dsRNA virus life cycle encompasses its genome and its RNA-dependent RNA polymerase (RdRp).
The T=1 capsid has a dual function, as it provides a platform for RNA transcription and replication and isolates the viral genome from host sentinels to avoid triggering defense mechanisms. The stoichiometry of the T=1 capsid is highly conserved among dsRNA viruses, probably because structural proteins participate in organizing the RdRp complex(es), as well as the dsRNA. Although the simplest icosahedral capsids are built from 60 identical subunits that assemble into 12 pentamers (a T=1 capsid), dsRNA virus T=1 capsids are formed by 60 asymmetric dimers of a single protein (a 120-subunit T=1 capsid) (5).
T=1 capsids have been described in members of the families Reoviridae and Picobirnaviridae (6–10), in the family Cystoviridae (11, 12), and in the families Totiviridae (13–15), Partitiviridae (16, 17), and Megabirnaviridae (18) (Table 1 ). The ubiquitous T=1 capsid is referred to as the inner core in reoviruses and cystoviruses. Members of the family Birnaviridae are exceptions, as they lack the T=1 core (19, 20). Birnaviruses have a single T=13 shell that encapsidates a polyploid dsRNA genome organized as ribonucleoprotein complexes (21, 22). Chrysoviruses, a group of dsRNA mycoviruses with a multipartite genome, have a T=1 capsid with 60 subunits of a single 982-amino-acid capsid protein (CP) (23–25). The CP is formed by a repeated α-helical core, indicative of gene duplication, and the capsid architecture is similar to that of the 120-subunit T=1 layer of reoviruses, cystoviruses, megabirnaviruses, and totiviruses (26).
TABLE 1.
Features of T=1 capsid proteins and genome in dsRNA viruses
| Virusa | dsRNA features |
Capsid features |
||||
|---|---|---|---|---|---|---|
| No. of segmentsb | Size (kbp) | Massc (MDa) | CP (residues) | ϕ/rd (nm) | dsRNA density (bp/100 nm3)e | |
| Reoviridae | ||||||
| Orthoreovirus | 10 | ∼23.5 | 16 | 1,275 | ∼60/24.5 | 38 |
| Rotavirus | 11 | ∼18.5 | 12.6 | 880 | ∼52/23.5 | 34 |
| Orbivirus, BTV | 10 | ∼19.2 | 13.1 | 901 | ∼52/22 | 43 |
| Aquareovirus, GCRV | 11 | ∼23.6 | 16 | 1,027 | ∼60/23 | 46 |
| Phytoreovirus, RDV | 12 | ∼25.7 | 17.5 | 1,019 | ∼57/26 | 35 |
| Cypovirus, CPV | 10 | ∼31.4 | 21.4 | 1,333 | ∼58/24 | 54 |
| Mycoreovirus, MyRV1 | 11 | 23.4 | 16 | |||
| Picobirnaviridae | 2 | ∼4.2 | 2.9 | 590 | ∼35/14 | |
| Cystoviridae, phage ϕ6 | 3 | ∼13.4 | 9.1 | 769 | ∼50/20 | 40 |
| Totiviridae, ScV-L-A | 1 | ∼4.6 | 3.1 | 680 | ∼43/17 | 22 |
| Partitiviridae, PsV-S | 1 (2) | ∼1.7 (3.3) | 1.2 (2.2) | 420 | ∼35/12 | 23 |
| Chrysoviridae, PcV | 1 (4) | ∼3.2 (12.6) | 2.2 (8.6) | 109 | ∼40/16 | 19 |
| Megabirnaviridae, RnMBV1 | 1 (2) | ∼8.1 (16.2) | 5.5 (11) | 135 | ∼52/19 | 28 |
| Quadriviridae, RnQV1 | 1–2 (4) | ∼4.3 (17.1) | 2.9 (11.7) | 1,356 + 1,061 | ∼47/16 | 25 (50)f |
BTV, bluetongue virus; GCRV, grass carp reovirus; RDV, rice dwarf virus; CPV, cytoplasmic polyhedrosis virus; MyRV1, mycoreovirus 1; PsV-S, Penicillium stoloniferum virus S; RnMBV1, Rosellinia necatrix megabirnavirus 1.
For PsV-S, PcV, and RnMBV1 dsRNAs, the genome is formed by two, four, or two dsRNA molecules, respectively, but a mean value was calculated for 1 dsRNA molecule/particle in each column.
Masses were calculated assuming a mass of 682 Da/bp.
ϕ, outer diameter; r, inner radius.
Densities when the volume of a perfect sphere is assumed and any other internal components are ignored.
A value of 25 if there is one dsRNA molecule/particle or 50 if there are two dsRNA molecules/particle.
Here, we used complementary electron microscopy and biophysical analysis to study the capsid structure of Rosellinia necatrix quadrivirus 1 (RnQV1) of the Quadriviridae (27, 28). RnQV1 is associated with latent infections (i.e., it causes no apparent slowing of host growth) and has a multipartite genome consisting of four monocistronic dsRNA segments (genome sizes range from 3.7 to 4.9 kbp). dsRNA-1 (4,942 bp) codes for a protein of unknown function (1,602 amino acid residues), dsRNA-2 (4,352 bp) encodes the P2 capsid protein (1,356 amino acids), dsRNA-3 (4,099 bp) codes for the RdRp (1,117 amino acids), and dsRNA-4 (3,685 bp) codes for the P4 capsid protein (1,061 amino acids). Like many other dsRNA mycoviruses, quadriviruses lack an extracellular phase in their life cycle but have efficient means for both horizontal and vertical transmission (29). Based on multiple alignments of RdRp sequences, quadriviruses appear to be more closely related to totiviruses (with a single genome segment) than to chrysoviruses (with four segments).
The filamentous ascomycete Rosellinia necatrix can be infected by dsRNA viruses belonging to at least five families (30). We analyzed RnQV1 strains 1075 and 1118 (isolated from two independent field strains of R. necatrix) using three-dimensional cryo-electron microscopy (3D cryo-EM) and analytical ultracentrifugation. We found that RnQV1 has a single-shelled T=1 capsid with 60 P2-P4 heterodimers in which one or two dsRNA segments are likely to be encapsidated in a similar particle.
MATERIALS AND METHODS
Virion preparation.
R. necatrix strains infected with Rosellinia necatrix quadrivirus 1 strain W1075 (RnQV1-W1075) or RnQV1-W1118 were used for virion purification, as described previously (31) with modifications. Mycelium was harvested from 14-day stationary-phase cultures grown at room temperature (22 to 25°C) in potato dextrose broth containing 0.5% (wt/vol) yeast extract and homogenized in a Waring blender with 0.1 M sodium phosphate buffer (pH 7.4) containing 0.2 M KCl and 0.5% (vol/vol) β-mercaptoethanol. The homogenate was mixed with an equal volume of chloroform, and the emulsion was broken by centrifugation (8,000 × g; 20 min). The aqueous layer was subjected to two cycles of differential centrifugation (27,000 rpm, 150 min, Beckman type 30 rotor; 10,000 rpm, 10 min, Beckman JA-20 rotor). The pellets were resuspended in buffer A (50 mM Tris-HCl buffer, pH 7.8, 5 mM EDTA, 150 mM NaCl). The final purification step was rate zonal centrifugation in sucrose density gradients (100 to 400 mg/ml in buffer A; 24,000 rpm, 150 min, Beckman SW28 rotor). All centrifugation steps were carried out at 4°C. The two UV-absorbing bands, corresponding to empty particles (upper band) and full particles (lower band), were withdrawn separately with a syringe from the side of the tube, diluted with buffer A, and concentrated by centrifugation (40,000 rpm, 12 h, 4°C, Beckman 50Ti rotor). The pellets were resuspended in buffer A.
Analytical ultracentrifugation of RnQV1-W1075 capsids.
For sedimentation velocity experiments, viral capsids or complete virions in the 0.5- to 1.0-mg/ml range were used. Samples were equilibrated in buffer A. Sedimentation velocity experiments were carried out in an XLI analytical ultracentrifuge (Beckman-Coulter; 10,000 rpm, 20°C), and Rayleigh interference or absorbance at 280 nm was recorded. Sedimentation coefficient distributions were calculated by least-squares boundary modeling of sedimentation velocity data using the c(s) and ls-g*(s) methods (32, 33), as implemented by the SEDFIT program (http://www.analyticalultracentrifugation.com/default.htm). S values were corrected to standard conditions (water, 20°C, and infinite dilution) (34) using the SEDNTERP program (35) to obtain standard S values (S20,w [i.e., 20°C, water]).
Dynamic light scattering.
For dynamic-light-scattering experiments, we used a Protein Solutions DynaPro-MS/X instrument (Protein Solutions, Charlottesville, VA). Viral capsids and complete virions were centrifuged (10,000 × g; 10 min) to remove dust particles; 50-μl samples were loaded in 90° light-scattering cuvettes and measured at 20°C. Translational diffusion coefficients (D) of the viral particles were determined from the scattering data with DYNAMICS autocorrelation analysis software v. 6 (Protein Solutions). Experimental values were buffer and concentration corrected (34) to obtain standard D0 coefficients.
SDS-PAGE.
Concentrated gradient fractions (2 to 5 μl) were added to Laemmli sample buffer to a 1× final concentration (62.5 mM Tris-HCl, 2% SDS, 5% glycerol, 0.012% bromophenol blue, 2 mM dithiothreitol, pH 6.8), heated (3 min; 100°C), and resolved in 11% polyacrylamide gels.
Cryo-electron microscopy.
Samples (5 μl) were applied to acetone-treated Quantifoil R 2/2 holey grids, blotted, and plunged into liquid ethane in a Leica EM CPC cryofixation unit. Micrographs were recorded at a nominal ×50,000 magnification under low-dose conditions (10 e−/Å2) with a FEI Tecnai G2 electron microscope operating at 200 kV and equipped with a field emission gun.
Image processing.
General image-processing operations were performed using Xmipp (36; http://xmipp.cnb.csic.es/) and Spider (37; http://www.wadsworth.org/spider_doc/spider/docs/) software packages. Graphic representations were produced with UCSF Chimera (38; http://www.cgl.ucsf.edu/chimera/).
A Nikon Super CoolScan 9000 ED scanner was used to digitize 423 and 324 micrographs for RnQV1 strains W1075 and W1118, respectively, at a 6.35-μm step size to yield a 1.27-Å pixel size in the specimen. X3d (39) and the Xmipp picking routine were used to manually select 26,729 and 36,582 individual particle images for RnQV1 strains W1075 and W1118, respectively. A 0.7- to 3.8-μm defocus range was determined with CTFfind (40), and contrast transfer function (CTF) phase oscillations were corrected in images by flipping them in the required lobes, and virion particles were extracted and normalized. The Xmipp iterative projection-matching routine was carried out to determine the origin and orientation of each particle using the structure of Penicillium chrysogenum virus (PcV) (EMD-1610 [26]), low-pass filtered to 30 Å, as a starting model. After each refinement iteration, the resolution was assessed from two independent half data sets using the 0.3 criterion of the Fourier shell correlation (FSC). A total of 24,056 (for RnQV1 W1075) and 23,926 (for W1118) particles were included in 3D reconstructions. The amplitude decay profile of the cryo-EM maps was adjusted to match the profile of the X-ray ScV-L-A capsid map (Protein Data Bank [PDB] accession no. 1m1c, [13]). The fitted function was applied to cryo-EM maps in the frequency range from 196 Å to the maximum resolution achieved, and a soft low-pass filter was applied.
The structural asymmetric unit boundaries were established by contouring the map at different σ levels, based on its compactness and contacts with neighboring densities. The asymmetric unit and P2-P4 subunits were segmented using Segger (41) in Chimera and refined iteratively to avoid subunit overlap or loose density. To test whether the selected density corresponded to a single asymmetric unit, the complete capsid map was restored in each cycle after applying icosahedral symmetry to the selected density. The secondary-structure elements (SSE) in the RnQV1 density were identified and modeled using the SSEHunter program (42) integrated in the Gorgon software (43). For difference map calculations, spherically averaged radial density profiles were calculated, normalized, and scaled to match the fit between the cryo-EM map and quasi-atomic model profiles. A difference map was obtained by subtraction.
Secondary-structure predictions.
The Web addresses used for secondary-structure prediction programs were as follows: UniProt, http://www.uniprot.org/; PsiPred, http://bioinf.cs.ucl.ac.uk/psipred/; Jnet, http://www.compbio.dundee.ac.uk/www-jpred/; Porter, http://distill.ucd.ie/porter; Sable, http://sable.cchmc.org; Gor, https://npsa-prabi.ibcp.fr/NPSA/npsa_gor4.html; Yaspin, http://www.ibi.vu.nl/programs/yaspinwww/; Profsec, http://www.predictprotein.org/; and GeneSilico, http://genesilico.pl/meta2/.
Accession number(s).
The 3D reconstructions have been deposited in the EMBL-EBI database (http://www.ebi.ac.uk/) under accession numbers emd-3437 (full RnQV1 strain W1075) and emd-3438 (empty RnQV1 strain W1118).
RESULTS
RnQV1 capsid three-dimensional structure at 8-Å resolution.
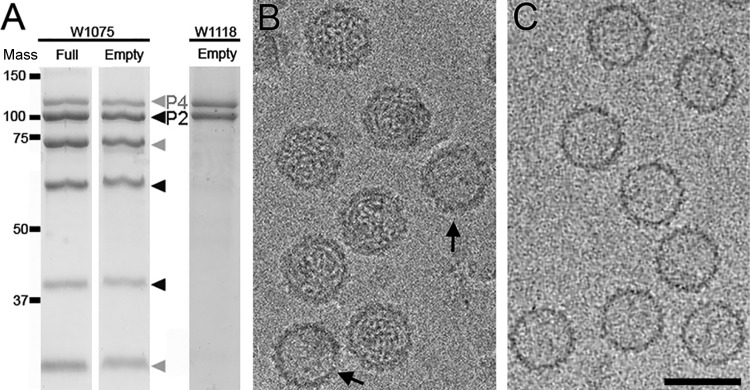
Purified full and empty RnQV1-W1075 particles were analyzed by SDS-PAGE (Fig. 1A, left). Excluding mycoreoviruses (44), RnQV1 is the only mycovirus with several major structural proteins encoded by more than one segment, segments dsRNA-2 and -4. As reported for these particles (27, 28), in addition to lower-mobility bands corresponding to P2 and P4 structural proteins, the proteins were also cleaved in several polypeptides; P2-derived bands had electrophoretic mobilities equivalent to those of proteins with molecular masses of 100, 60, and 40 kDa (Fig. 1A, black arrowheads), whereas P4-related bands were 110, 75, and 30 kDa (Fig. 1A, gray arrowheads). It was unclear whether the P2 and P4 cleavage products were produced in the infected fungal host and/or during virion purification. We also analyzed a second quadrivirus strain, RnQV1-W1118 (which has 81 and 82% P2 and P4 sequence identity with those of RnQV1-W1075), but P2- and P4-related degradation products were basically absent (Fig. 1A, right). RnQV1-W1075 and RnQV1-W1118 P4 and P2 were detected as 110- and 100-kDa bands, which contrasted with the predicted sizes based on the coding capacities of dsRNA-4 and dsRNA-2, which would encode proteins of 1,061 (113,243 Da) and 1,356 (147,420 Da) amino acids, respectively. This discrepancy could be due to anomalous electrophoretic mobilities or, more probably, to further proteolysis at the C terminus.
FIG 1.
Biochemical and cryo-EM analyses of RnQV1 strains W1075 and W1118. (A) Coomassie blue-stained SDS-11% PAGE gels of purified full and empty RnQV1-W1075 (left) and empty RnQV1-W1118 (right) virions used for cryo-EM data acquisition. P4 and P4-related bands are indicated in gray and P2 and P2-related bands in black. Molecular mass markers (10−3 Da) are on the left. (B) Cryo-EM of full RnQV1 strain W1075 (the arrows indicate two empty capsids). (C) Cryo-EM of empty RnQV1 strain W1118. Bar = 50 nm.
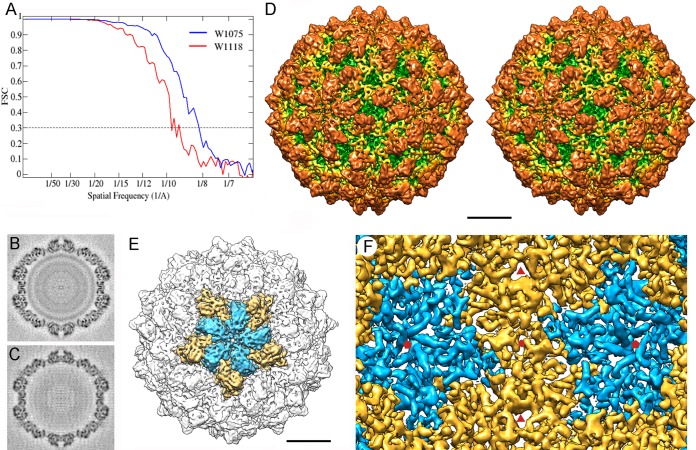
Full RnQV1-W1075 (Fig. 1B) and empty RnQV1-W1118 (Fig. 1C) particles were considered appropriate for independent cryo-EM analysis, and their 3D reconstructions were calculated. Raw cryomicrographs of full and empty RnQV1 particles showed visible surface projections on most particles. The final resolutions of the reconstructions for full W1075 and empty W1118 were estimated to be 8.2 and 9.1 Å, based on a 0.3 FSC threshold (Fig. 2A). The relatively limited resolution could be due to some structural heterogeneity of the purified virions and/or to the imaging conditions used for the study. The central sections of the 3D maps showed no marked structural differences in the protein shell between full and empty capsids (Fig. 2B and C). This interpretation was supported when we used full and empty particles for difference map calculation, which detected only minor differences (see below). In addition, when FSC resolution curves were calculated either for the full and empty maps refined independently or for full and empty particles subjected to joint refinement cycles, the estimated resolutions were 8 to 9 Å, indicating that the two maps were virtually identical (not shown). The data also indicated that the two maps were virtually identical, independently not only of the presence of dsRNA, but also of the processing level of the structural proteins P2 and P4.
FIG 2.
Three-dimensional cryo-EM of RnQV1 virions. (A) Assessment of the resolution of full (W1075) and empty (W1118) RnQV1 reconstructions. FSC resolution curves were calculated for full (blue) and empty (red) capsids. Each set of particle images was subdivided randomly into two subsets, and independent reconstructions were computed from the data. Resolutions for which correlations were <0.3 are indicated. For the 0.3 threshold, the values for full and empty RnQV1 capsids were 8.2 and 9.1 Å, respectively. (B and C) Central sections from the 3D reconstruction of full (B) and empty (C) capsids, viewed along a 2-fold axis. Protein and RNA are dark. The two protein shells are virtually identical, and the RNA density of the full capsid is seen as concentric circles inside the capsid. (D) Stereo view of the radially color-coded outer surface of the full capsid, viewed along a 2-fold axis of icosahedral symmetry. The most prominent features are 120 outward-protruding densities (orange). The map is contoured at 2.5 σ above the mean density. Bar = 100 Å. (E) Surface-shaded virion capsid viewed along an icosahedral 5-fold axis showing the five A (blue) and B (yellow) structural subunits in a pentamer. (F) Inner surface of the RnQV1-W1075 capsid (for clarity, only the density between 145- and 210-Å radii is shown). Icosahedral-symmetry axes are indicated (red symbols).
The outer diameter of the capsid was 470 Å, measured from spherically averaged radial density profiles of maps, and the average capsid shell thickness was 75 Å. The capsid has a T=1 lattice, the most outstanding feature of which is the 120 outward-protruding densities (Fig. 2D, orange). Each pentameric capsomer is formed by an inner ring of five connected, elongated structures (Fig. 2E, blue) surrounded by an outer ring of five slightly larger, partially intercalated structures (Fig. 2E, yellow). Whereas there are marked depressions at the 2- and 3-fold icosahedral-symmetry axes, the capsid is slightly raised at the 5-fold axis positions. The uneven outer surface of the RnQV1 capsid was less defined than the smooth inner surface, where we observed numerous rod densities that probably correspond to α-helices of different lengths (Fig. 2F).
By varying map contour levels, we established structural asymmetric unit boundaries that include the elongated subunits (Fig. 2E, blue and yellow structures). The capsid map was restored computationally after applying icosahedral symmetry to this selected asymmetric unit.
Two structural proteins for the RnQV1 capsid.
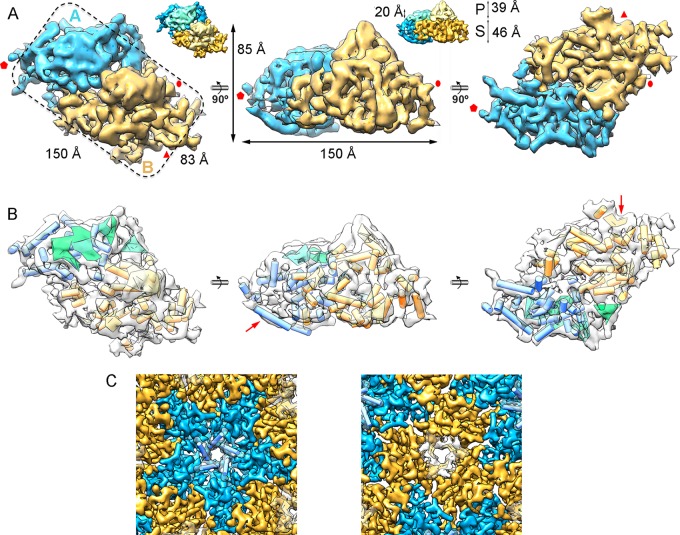
The topographic features of RnQV1-W1075 and RnQV1-W1118 corresponded to the 120-subunit capsid predicted for the ubiquitous T=1 lattice of most dsRNA viruses, in which the asymmetric unit is a CP asymmetric homodimer. Independently of the CP processing level, the RnQV1 capsid was built of P2 and P4 heterodimers, as they were found at similar ratios, as deduced from the biochemical profile of purified RnQV1-W1118 virions (Fig. 1A). The asymmetric unit is a rectangular prism, ∼85-Å maximum height with an ∼150- by ∼83-Å base (Fig. 3A). The two elongated structures that form the asymmetric RnQV1 capsid unit, and which differ slightly in size, correspond to P2 and P4; at this resolution, their concise boundaries cannot be delimited within the asymmetric unit, nor can P2 or P4 be assigned to either of the elongated structures. In addition, P2 and P4 might contribute to both structures by inter- and intradimeric interactions mediated by loops and/or secondary-structure elements that interwine both subunits, as well as domain swapping between the two protomers within each heterodimer. Following established nomenclature (7), we refer to these elongated structures as the A subunit for those that participate directly in interactions at the 5-fold axis (Fig. 2A and 3A, blue) and the B subunit for those intercalated between A subunits (Fig. 2A and 3A, yellow).
FIG 3.
Structure of the RnQV1 capsid and model of the heterodimer fold. (A) Segmented asymmetric unit (A-B heterodimer). The dashed line highlights the rectangular shape. Subunits A (blue) and B (yellow) are indicated. The map is contoured at 2.5 σ above the mean density. Icosahedral-symmetry axes are indicated (red). The insets highlight P and S domains of CP (height is indicated). (B) SSE of subunits A and B, using the color scheme and orientations in panel A; cylinders, α-helices; planks, β-sheets. The arrows indicate the subunit A α-helix that forms the 5-fold axis (middle) and the subunit B β-sheet that forms the 3-fold axis (right). (C) RnQV1 capsid pores at the 5-fold axis (∼16-Å diameter) and the 3-fold axis (∼18-Å side).
RnQV1 A and B subunits, which make up 42% and 58% of the total unit volume, have similar general morphologies consisting of a prominent protruding (P) domain (with similar sizes for PA and PB domains) and a 460-Å-thick shell (S) domain (SB is much larger than SA) (Fig. 3A, insets). SSE in the asymmetric unit were identified with Gorgon software (which includes the SSE Hunter program). We identified 21 α-helices and four β-sheet regions in the A subunit and 25 α-helices and four β-sheet regions in the B subunit (Fig. 3B). Most α-helices face the inner capsid surface. The bundle of five helix-like structures at the 5-fold axis forms a narrow, 16-Å-diameter channel, and three β-sheet-like structures define the triangular pores (∼18-Å sides) at the 3-fold axis (Fig. 3B, red arrows, and C).
The A and B subunits of classic 120-subunit capsids consist of two conformers of the same protein with relatively limited conformational changes, and superposition of the A and B structures matches most SSE. Previous analysis indicated that sequence similarity between P2 and P4 is negligible (27). At the resolution achieved, RnQV1 A and B subunits showed no clear structural resemblance at the SSE level, even in the shell domains, where α-helices are abundant and well defined. When we compared the RnQV1 capsid with other T=1 proteins of dsRNA viruses by superimposing structures that maintain the same orientation in the capsid, we observed no resemblance. SSE prediction based on the sequences of P2 (1,356 amino acids) and P4 (1,061 amino acids) for strains W1075 and W1118 indicated high α-helical content, as we observed directly in the cryo-EM maps (Fig. 4). The SSE predicted for P2 and P4 did not match.
FIG 4.
Sequence alignment and secondary-structure consensus prediction for RnQV1 P2 and P4 CP amino acid sequences. The sequences of P2 (1,356 amino acids for W1075; 1,357 amino acids for W1118) (A) and P4 (1,061 amino acids for W1075; 1,059 amino acids for W1118) (B) were obtained from the UniProt database [H1ACC6, M1VMJ0, H1ACC8, and M1VHN2, respectively). Several SSE prediction methods (PsiPred, Jnet, Porter, Sable, Gor, Yaspin, and Profsec) were used to test correlation with our models of the structural subunits. A consensus SSE prediction was obtained by simple majority at each sequence position. Identical residues (white on red background) and partially conserved residues (red) are indicated. The arrows indicate β-strands, and the spirals indicate α-helices.
dsRNA packaged within the capsid.
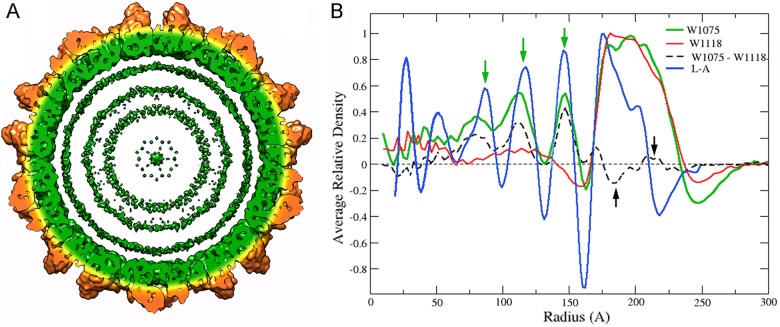
A radial density profile of the full RnQV1 particle showed additional density inside the capsid (r < 161 Å) that corresponds to dsRNA (Fig. 5A and B, green). In the empty capsid, with the same inner and outer shell radii (162 and 235 Å, respectively), the internal density was similar to that of the external solvent (Fig. 5B, red). The difference map calculated by arithmetic subtraction of the density values in both structures showed, in addition to the genome densities, small difference densities at the shell, in the 175- to 195- and 205- to 220-Å radius ranges (Fig. 5B, black arrows). These difference densities, visualized as spurious islands of density rather than large density islands (not shown), can be ascribed to small sequence and structure differences between strains W1075 and W1118 and/or to minor conformational changes in P2 and P4.
FIG 5.
Genomic dsRNA within the RnQV1 virion particle. (A) A 50-Å-thick RnQV1-W1075 slab. Capsid shell coloring is the same as in Fig. 2, contoured at 1.2 σ; dsRNA (green) is represented as three concentric layers contoured at 1.0 σ. (B) Radial density profiles from 3D maps of full (W1075) and empty (W1118) RnQV1 particles. Both profiles are superimposable at the protein shell (radius, ∼162 to 235 Å). A difference map was calculated by arithmetic subtraction of the density values for both structures (full minus empty capsid; dashed line). Small differences in the protein shell are indicated at radii 175 to 195 and 205 to 220 (black arrows); major differences in the genome region are seen as density peaks at radii of 80, 113, and 147 Å (green arrows). The radial density profile from the 3D map of full ScV-L-A virions is also shown (L-A).
After icosahedral symmetry was imposed, the (full) RnQV1-W1075 radial profile showed several concentric shells of RNA density in the particle interior, spaced ∼35 Å apart (Fig. 5B, green arrows). Although it is unlikely that this indicates dsRNA organization, 35 Å is the predominant spacing between dsRNA molecules. The radial profile of dsRNA layers for ScV-L-A (Fig. 5B, blue) (45, 46) was similar to that of RnQV1-W1075; both profiles showed matching dsRNA peaks and indicated almost no protein-RNA interactions between the inner capsid surface and the first underlying dsRNA density shell (Fig. 5B).
Biophysical analysis of RnQV1 particles.
To estimate an independent molecular weight for the RnQV1-W1075 virions, we used analytical ultracentrifugation analysis of empty capsids and complete virions; the sedimentation velocity behavior of these particles is shown in Fig. 6. The main peaks of sedimenting particles for empty and full capsids showed standard sedimentation coefficient values (S20,w) of 132 ± 15 and 189 ± 15 S, respectively. Dynamic-light-scattering experiments with samples from the same viral particle gave standard translational diffusion coefficients (D20,w) for the main scattering species of 7.3 ± 0.5 (empty) and 7.2 ± 0.5 10−12 m2 s−1 (full particles). The combined S20,w and D20,w coefficients, calculated using the Svedberg equation (34), allow the molar mass of the viral particles to be determined in a shape-independent manner. We determined that the main species of empty particles has a molar mass of 15.9 ± 3 MDa, and that of the full virions is 20.6 ± 4 MDa. Considering the molar masses of the P2 and P4 structural CP, the molecular mass for a T=1 capsid with a heterodimer as the asymmetric unit would be ∼15.6 MDa or ∼12.8 MDa, depending on whether P2 was intact or processed, respectively. The experimental values are compatible with a capsid with intact P2 proteins, although processed P2 could not be ruled out.
FIG 6.
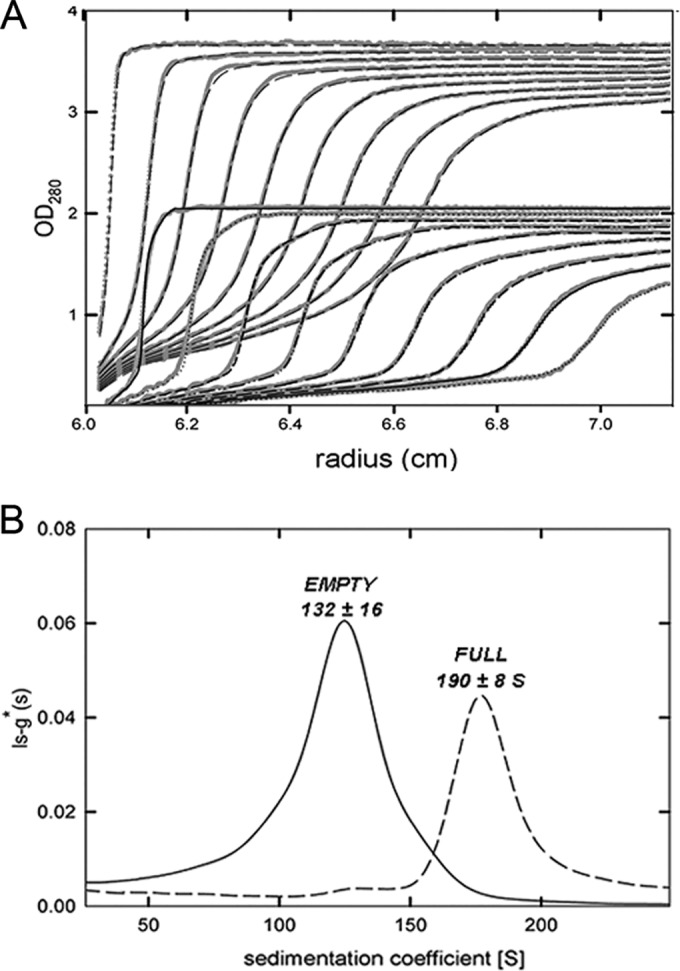
Analytical ultracentrifugation analysis of RnQV1-W1075. Sedimentation velocity experimental data (A) and sedimentation coefficient distribution obtained by c(s) analysis (B) of empty capsids (solid line) and virions or full particles (dashed line).
Given the average size of the dsRNA segments (∼4.3 kbp) and assuming a molar mass of 682 Da/bp, the molar mass of a single genome component is ∼3 MDa. Whereas our biophysical and biochemical analyses showed that the molar mass of the capsid is compatible with a T=1 capsid structure with 60 copies each of P2 and P4, the dsRNA segment/capsid ratio could not be determined unequivocally.
DISCUSSION
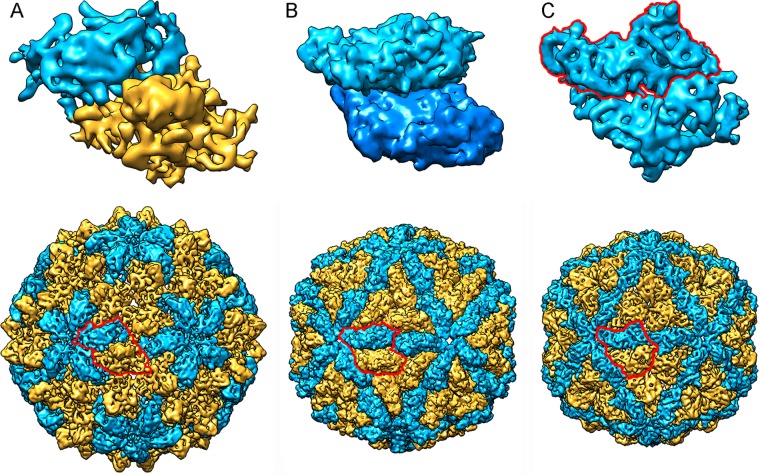
The quasiequivalence theory introduced by Caspar and Klug (47) implies that identical CP subunits interact to form quasiequivalent bonds for the construction of stable icosahedral capsids with multiples of 60 subunits (48, 49). The T=1 capsid, common among dsRNA viruses, is built from 60 copies of a dimer of chemically identical subunits. This 120-subunit capsid has a packing arrangement distinct from that of standard T=1 capsids. It does not follow classical quasiequivalence theory because the two subunits have nonequivalent bonding environments. The PcV T=1 capsid is based on a T=1 lattice (with 60 subunits), but it is a variant of the 120-subunit capsid, as the CP has two motifs with the same fold (26). Our analysis using 3D cryo-EM combined with complementary analytical ultracentrifugation showed that the RnQV1 capsid is a T=1 120-subunit layer composed of 60 heterodimers, a previously unreported organization that allows nonequivalent packing of heterogeneous subunits as dimers. All of these dsRNA virus capsids, exemplified by ScV-L-A (and reovirus inner cores), PcV, and RnQV1, show structural variations of the same framework optimized for RNA metabolism, with 60 asymmetric dimers of a single protein (for ScV-L-A), dimers of similar domains (for PcV), or dimers of two different proteins (for RnQV1) (Fig. 7).
FIG 7.
Comparison of T=1-based inner cores of dsRNA viruses. (Top) Asymmetric units (top views) of RnQV1 (A), ScV-L-A (B), and PcV (C). RnQV1 A and B subunits are shown in blue and yellow, respectively, to indicate that they are distinct proteins; ScV-L-A A and B subunits are shown in blue, since they are conformers of the same protein; and PcV A and B “subunits” are shown in blue (with the boundaries of A outlined in red), as they are two similar covalently bound domains of a single CP. (Bottom) Surface-shaded capsids of RnQV1, ScV-L-A, and PcV viewed along an icosahedral 2-fold axis. To indicate that they are similarly organized capsids, the A subunits (closer to the 5-fold axis) are shown in blue and the B subunits (intercalated between A subunits) in yellow.
Of all the reported T=1 shells, the RnQV1 capsid requires the largest dedication of the viral genome to CP (2,417 amino acid residues; P2 + P4 = ∼270 kDa). This suggests apparent waste of the limited virus coding capacity.
Due to their impact on human and animal health, mammalian viruses are the best-characterized dsRNA virus group. These viruses have a multishelled icosahedral capsid, and each shell has a specific associated role. Whereas the outer shell is protective and is involved in cell entry, the innermost shell (or inner core) has 120 copies of a plate-like protein and is dedicated exclusively to genome and RdRp organization for transcription- and replication-related activities. Except for fungal reoviruses, which like their mammalian counterparts have several protein layers, most fungal dsRNA viruses have a single-shell icosahedral capsid, as they are transmitted by cytoplasmic interchange without leaving the host. The 120-subunit capsids act as a molecular sieve and contain the genome, as well as enzymes involved in dsRNA metabolism (50, 51).
In addition to avoiding dsRNA-mediated intracellular defense mechanisms, the close relationship between the fungal dsRNA virus and its host probably placed many constraints on the virus that it overcame by increasing CP complexity. Fungal hosts are often infected by multiple viruses (52, 53), and virus-virus interactions can also be a major evolutionary determinant. In the filamentous ascomycete R. necatrix, yado-kari virus 1 (YkV1), a positive-sense single-stranded RNA [(+)ssRNA] virus, hijacks the CP of yado-nushi virus 1 (YnV1), a dsRNA virus that resembles totiviruses, to pack its genome and RNA polymerase. The 120-subunit capsids of fungal dsRNA viruses share a corrugated outer surface with protuberances rising above the continuous protein shell. Whereas the average 120-subunit T=1 CP thickness for mammalian dsRNA viruses is 15 to 30 Å, fungal virus CPs are thicker. In ScV-L-A virus, the CP has an extra domain with decapping activity that transfers cap structures from the 5′ end of cellular mRNA to the 5′ end of viral RNA (54, 55). Detailed analysis of the chrysovirus CP (built of two similar domains) showed similar extra domains on the outer capsid surface with unknown functions (26).
Unlike the T=1 core in complex eukaryotic dsRNA viruses, fungal dsRNA viruses consistently show a low degree of genome compaction; in other words, they have spacious capsids (Table 1). Members of the family Reoviridae have an average dsRNA density of ∼40 bp/100 nm3. At an appropriate underfocus, cryo-EM projection images of encapsidated dsRNA in reoviruses and rotaviruses, as well as dsDNA of bacteriophages (56, 57) and herpes simplex virus (58), have a “fingerprint” motif with 25- to 30-Å interstrand spacing, indicative of tight packing (59–62). A high degree of condensation corresponds to a dsRNA density of 40 to 50 bp/100 nm3 or more. ScV-L-A dsRNA is loosely packed, with a genome density of ∼20 bp/100 nm3 and spacing between filaments of 40 to 45 Å (45). Similar values for packed dsRNA were determined for PcV and partitiviruses, all of which also have a single dsRNA molecule (31, 63). The megabirnaviruses contain one segment per capsid that yields a density of ∼28 bp/100 nm3 (18) (Table 1). For RnQV1, the analytical ultracentrifugation/dynamic-light-scattering data were insufficiently accurate and did not allow us to determine the number of dsRNA molecules packaged per capsid. The results nevertheless suggested that a single RnQV1 virion is not replication competent, as the infectious unit would require several viral particles (at least four if there is one dsRNA molecule/particle). Comparison of the (full) RnQV1-W1075 and ScV-L-A radial density profiles suggested one dsRNA molecule per particle, as the predominant spacing between dsRNA molecules is ∼35 Å. Considering the volume available (17,160 nm3 based on an average internal capsid radius of 160 Å) and an ∼4,270-bp average genome size, the density of a single packed dsRNA molecule would be 25 bp/100 nm3 and that of two packed dsRNA molecules would be 50 bp/100 nm3 (Table 1). Images of full RnQV1 particles showed uniformly dense capsids with punctate and/or swirl motifs (as for ScV-L-A and PcV), which indicates loose dsRNA packaging. This relatively low packed genome density is common among fungal viruses that package only a single genomic dsRNA segment per particle and is probably linked to improved template motion during transcription and replication in the more spacious capsids.
The RnQV1 capsid is the T=1 lattice with the largest known capsid proteins (P2 plus P4). Future studies that solve the RnQV1 atomic structure will help to define important aspects of dsRNA virus evolution, i.e., whether P2 and P4 share the folding signature of most dsRNA virus CPs, and of virus-host interaction, i.e., whether the protruding domains have enzyme activity, as for other fungal CPs.
ACKNOWLEDGMENTS
We thank Catherine Mark for editorial assistance.
C.P.M. is a Ph.D. fellow of the La Caixa Foundation International Fellowship Program (La Caixa/CNB). This work was supported by grants from the Spanish Ministry of Economy and Competitivity (BFU2014-52070-C2-2-P to C.A. and G.R. and BFU2014-55475-R to J.R.C.); the Comunidad Autónoma de Madrid (S2013/MIT-2807 to J.R.C.); the Ministry of Education, Culture, Science, Sports and Technology (MEXT) of Japan (16H06436 to N.S.); and the Intramural Research Program of the NIH and the Center for Information Technology (to B.L.T.).
REFERENCES
- 1.Patton JT. 2008. Segmented double-stranded RNA viruses: structure and molecular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 2.Wickner R. 1996. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev 60:250–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu G, Zhou ZH, Baker ML, Jakana J, Cai D, Wei X, Chen S, Gu X, Chiu W. 1998. Structure of double-shelled rice dwarf virus. J Virol 72:8541–8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mertens P. 2004. The dsRNA viruses. Virus Res 101:3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Steven AC, Baumeister W, Johnson LN, OPerham RN. 2016. Molecular biology of assemblies and machines. Garland Science, New York, NY. [Google Scholar]
- 6.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. 2011. Atomic model of an infectious rotavirus particle. EMBO J 30:408–416. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Zientara S, Mertens PP, Stuart DI. 1998. The atomic structure of the Bluetongue virus core. Nature 395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 8.Reinisch KM, Nibert ML, Harrison SC. 2000. Structure of the Reovirus core at 3.6 Å resolution. Nature 404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 9.Duquerroy S, Da Costa B, Henry C, Vigouroux A, Libersou S, Lepault J, Navaza J, Delmas B, Rey FA. 2009. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J 28:1655–1665. doi: 10.1038/emboj.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Jin L, Zhou ZH. 2008. 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature 453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemecek D, Boura E, Wu W, Cheng N, Plevka P, Qiao J, Mindich L, Heymann JB, Hurley JH, Steven AC. 2013. Subunit folds and maturation pathway of a dsRNA virus capsid. Structure 21:1374–1383. doi: 10.1016/j.str.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Omari K, Sutton G, Ravantti JJ, Zhang H, Walter TS, Grimes JM, Bamford DH, Stuart DI, Mancini EJ. 2013. Plate tectonics of virus shell assembly and reorganization in phage phi8, a distant relative of mammalian reoviruses. Structure 21:1384–1395. doi: 10.1016/j.str.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naitow H, Tang J, Canady M, Wickner RB, Johnson JE. 2002. L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol 9:725–728. doi: 10.1038/nsb844. [DOI] [PubMed] [Google Scholar]
- 14.Janssen ME, Takagi Y, Parent KN, Cardone G, Nibert ML, Baker TS. 2015. Three-dimensional structure of a protozoal double-stranded RNA virus that infects the enteric pathogen Giardia lamblia. J Virol 89:1182–1194. doi: 10.1128/JVI.02745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parent KN, Takagi Y, Cardone G, Olson NH, Ericsson M, Yang M, Lee Y, Asara JM, Fichorova RN, Baker TS, Nibert ML. 2013. Structure of a protozoan virus from the human genitourinary parasite Trichomonas vaginalis. mBio 4:e00056–00013. doi: 10.1128/mBio.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan J, Dong L, Lin L, Ochoa WF, Sinkovits RS, Havens WM, Nibert ML, Baker TS, Ghabrial SA, Tao YJ. 2009. Atomic structure reveals the unique capsid organization of a dsRNA virus. Proc Natl Acad Sci U S A 106:4225–4230. doi: 10.1073/pnas.0812071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang J, Ochoa WF, Li H, Havens WM, Nibert ML, Ghabrial SA, Baker TS. 2010. Structure of Fusarium poae virus 1 shows conserved and variable elements of partitivirus capsids and evolutionary relationships to picobirnavirus. J Struct Biol 172:363–371. doi: 10.1016/j.jsb.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyazaki N, Salaipeth L, Kanematsu S, Iwasaki K, Suzuki N. 2015. Megabirnavirus structure reveals a putative 120-subunit capsid formed by asymmetrical dimers with distinctive large protrusions. J Gen Virol 96:2435–2441. doi: 10.1099/vir.0.000182. [DOI] [PubMed] [Google Scholar]
- 19.Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey FA. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Castón JR, Martínez-Torrecuadrada JL, Maraver A, Lombardo E, Rodríguez JF, Casal JI, Carrascosa JL. 2001. C terminus of infectious bursal disease virus major capsid protein VP2 is involved in definition of the T number for capsid assembly. J Virol 75:10815–10828. doi: 10.1128/JVI.75.22.10815-10828.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luque D, Rivas G, Alfonso C, Carrascosa JL, Rodríguez JF, Castón JR. 2009. Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc Natl Acad Sci U S A 106:2148–2152. doi: 10.1073/pnas.0808498106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luque D, Saugar I, Rejas MT, Carrascosa JL, Rodríguez JF, Castón JR. 2009. Infectious Bursal disease virus: ribonucleoprotein complexes of a double-stranded RNA virus. J Mol Biol 386:891–901. doi: 10.1016/j.jmb.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luque D, González JM, Garriga D, Ghabrial SA, Havens WM, Trus B, Verdaguer N, Carrascosa JL, Castón JR. 2010. The T=1 capsid protein of Penicillium chrysogenum virus is formed by a repeated helix-rich core indicative of gene duplication. J Virol 84:7256–7266. doi: 10.1128/JVI.00432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castón JR, Luque D, Gómez-Blanco J, Ghabrial SA. 2013. Chrysovirus structure: repeated helical core as evidence of gene duplication. Adv Virus Res 86:87–108. doi: 10.1016/B978-0-12-394315-6.00004-0. [DOI] [PubMed] [Google Scholar]
- 25.Gómez-Blanco J, Luque D, González JM, Carrascosa JL, Alfonso C, Trus B, Havens WM, Ghabrial SA, Castón JR. 2012. Cryphonectria nitschkei virus 1 structure shows that the capsid protein of chrysoviruses is a duplicated helix-rich fold conserved in fungal double-stranded RNA viruses. J Virol 86:8314–8318. doi: 10.1128/JVI.00802-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luque D, Gómez-Blanco J, Garriga D, Brilot AF, Gonzalez JM, Havens WM, Carrascosa JL, Trus BL, Verdaguer N, Ghabrial SA, Castón JR. 2014. Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage. Proc Natl Acad Sci U S A 111:7641–7646. doi: 10.1073/pnas.1404330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin YH, Chiba S, Tani A, Kondo H, Sasaki A, Kanematsu S, Suzuki N. 2012. A novel quadripartite dsRNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix. Virology 426:42–50. doi: 10.1016/j.virol.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Lin YH, Hisano S, Yaegashi H, Kanematsu S, Suzuki N. 2013. A second quadrivirus strain from the phytopathogenic filamentous fungus Rosellinia necatrix. Arch Virol 158:1093–1098. doi: 10.1007/s00705-012-1580-8. [DOI] [PubMed] [Google Scholar]
- 29.Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N. 2015. 50-plus years of fungal viruses. Virology 479-480:356–368. doi: 10.1016/j.virol.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 30.Kondo H, Kanematsu S, Suzuki N. 2013. Viruses of the white root rot fungus, Rosellinia necatrix. Adv Virus Res 86:177–214. doi: 10.1016/B978-0-12-394315-6.00007-6. [DOI] [PubMed] [Google Scholar]
- 31.Castón JR, Ghabrial SA, Jiang D, Rivas G, Alfonso C, Roca R, Luque D, Carrascosa JL. 2003. Three-dimensional structure of penicillium chrysogenum virus: a double-stranded RNA virus with a genuine T=1 capsid. J Mol Biol 331:417–431. doi: 10.1016/S0022-2836(03)00695-8. [DOI] [PubMed] [Google Scholar]
- 32.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. 2002. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J 82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuck P, Rossmanith P. 2000. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers 54:328–341. doi:. [DOI] [PubMed] [Google Scholar]
- 34.van Holde KE. (ed). 1985. Physical biochemistry. Prentice-Hall, Englewood Cliffs, NJ. [Google Scholar]
- 35.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p 90–125. In Harding SE, Rowe AJ, Horton JC (ed), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, Cambridge, United Kingdom. [Google Scholar]
- 36.Marabini R, Masegosa IM, San Martín MC, Marco S, Fernández JJ, de la Fraga LG, Vaquerizo C, Carazo JM. 1996. Xmipp: an image processing package for electron microscopy. J Struct Biol 116:237–240. doi: 10.1006/jsbi.1996.0036. [DOI] [PubMed] [Google Scholar]
- 37.Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. 2008. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Conway JF, Trus BL, Booy FP, Newcomb WW, Brown JC, Steven AC. 1993. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J Struct Biol 111:222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- 40.Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142:334–347. doi: 10.1016/S1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 41.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. 2010. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol 170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baker ML, Ju T, Chiu W. 2007. Identification of secondary structure elements in intermediate-resolution density maps. Structure 15:7–19. doi: 10.1016/j.str.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker ML, Abeysinghe SS, Schuh S, Coleman RA, Abrams A, Marsh MP, Hryc CF, Ruths T, Chiu W, Ju T. 2011. Modeling protein structure at near atomic resolutions with Gorgon. J Struct Biol 174:360–373. doi: 10.1016/j.jsb.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hillman BI, Supyani S, Kondo H, Suzuki N. 2004. A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the coltivirus genus of animal pathogens. J Virol 78:892–898. doi: 10.1128/JVI.78.2.892-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castón JR, Trus BL, Booy FP, Wickner RB, Wall JS, Steven AC. 1997. Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J Cell Biol 138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng RH, Castón JR, Wang GJ, Gu F, Smith TJ, Baker TS, Bozarth RF, Trus BL, Cheng N, Wickner RB, Steven AC. 1994. Fungal virus capsids, cytoplasmic compartments for the replication of double-stranded RNA, formed as icosahedral shells of asymmetric Gag dimers. J Mol Biol 244:255–258. doi: 10.1006/jmbi.1994.1726. [DOI] [PubMed] [Google Scholar]
- 47.Caspar DLD, Klug A. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol 27:1–24. doi: 10.1101/SQB.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 48.Rossmann M, Johnson J. 1989. Icosahedral RNA virus structure. Annu Rev Biochem 58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 49.Harrison SC. 2007. Principles of virus structure, p 59–98. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Strauss SE (ed), Fields virology 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 50.Zhang X, Ding K, Yu X, Chang W, Sun J, Zhou ZH. 2015. In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus. Nature 527:531–534. doi: 10.1038/nature15767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, Cheng L. 2015. Cryo-EM shows the polymerase structures and a nonspooled genome within a dsRNA virus. Science 349:1347–1350. doi: 10.1126/science.aaa4938. [DOI] [PubMed] [Google Scholar]
- 52.Ghabrial SA, Suzuki N. 2009. Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384. doi: 10.1146/annurev-phyto-080508-081932. [DOI] [PubMed] [Google Scholar]
- 53.Zhang R, Hisano S, Tani A, Kondo H, Kanematsu S, Suzuki N. 2016. A capsidless ssRNA virus hosted by an unrelated dsRNA virus. Nat Microbiol 1:15001. doi: 10.1038/nmicrobiol.2015.1. [DOI] [PubMed] [Google Scholar]
- 54.Tang J, Naitow H, Gardner NA, Kolesar A, Tang L, Wickner RB, Johnson JE. 2005. The structural basis of recognition and removal of cellular mRNA 7-methyl G ‘caps’ by a viral capsid protein: a unique viral response to host defense. J Mol Recognit 18:158–168. doi: 10.1002/jmr.724. [DOI] [PubMed] [Google Scholar]
- 55.Fujimura T, Esteban R. 2011. Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc Natl Acad Sci U S A 108:17667–17671. doi: 10.1073/pnas.1111900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lepault J, Dubochet J, Baschong W, Kellenberger E. 1987. Organization of double-stranded DNA in bacteriophages: a study by cryo-electron microscopy of vitrified samples. EMBO J 6:1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. 1997. Encapsidated conformation of bacteriophage T7 DNA. Cell 91:271–280. doi: 10.1016/S0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 58.Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64:1007–1015. doi: 10.1016/0092-8674(91)90324-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol 122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prasad BV, Rothnagel R, Zeng CQ, Jakana J, Lawton JA, Chiu W, Estes MK. 1996. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 61.Li Z, Baker ML, Jiang W, Estes MK, Prasad BV. 2009. Rotavirus architecture at subnanometer resolution. J Virol 83:1754–1766. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gouet P, Diprose JM, Grimes JM, Malby R, Burroughs JN, Zientara S, Stuart DI, Mertens PP. 1999. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 97:481–490. doi: 10.1016/S0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 63.Ochoa WF, Havens WM, Sinkovits RS, Nibert ML, Ghabrial SA, Baker TS. 2008. Partitivirus structure reveals a 120-subunit, helix-rich capsid with distinctive surface arches formed by quasisymmetric coat-protein dimers. Structure 16:776–786. doi: 10.1016/j.str.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]