ABSTRACT
The entry of avian metapneumovirus (aMPV) into host cells initially requires the fusion of viral and cell membranes, which is exclusively mediated by fusion (F) protein. Proteolysis of aMPV F protein by endogenous proteases of host cells allows F protein to induce membrane fusion; however, these proteases have not been identified. Here, we provide the first evidence that the transmembrane serine protease TMPRSS12 facilitates the cleavage of subtype B aMPV (aMPV/B) F protein. We found that overexpression of TMPRSS12 enhanced aMPV/B F protein cleavage, F protein fusogenicity, and viral replication. Subsequently, knockdown of TMPRSS12 with specific small interfering RNAs (siRNAs) reduced aMPV/B F protein cleavage, F protein fusogenicity, and viral replication. We also found a cleavage motif in the aMPV/B F protein (amino acids 100 and 101) that was recognized by TMPRSS12. The histidine, aspartic acid, and serine residue (HDS) triad of TMPRSS12 was shown to be essential for the proteolysis of aMPV/B F protein via mutation analysis. Notably, we observed TMPRSS12 mRNA expression in target organs of aMPV/B in chickens. Overall, our results indicate that TMPRSS12 is crucial for aMPV/B F protein proteolysis and aMPV/B infectivity and that TMPRSS12 may serve as a target for novel therapeutics and prophylactics for aMPV.
IMPORTANCE Proteolysis of the aMPV F protein is a prerequisite for F protein-mediated membrane fusion of virus and cell and for aMPV infection; however, the proteases used in vitro and vivo are not clear. A combination of analyses, including overexpression, knockdown, and mutation methods, demonstrated that the transmembrane serine protease TMPRSS12 facilitated cleavage of subtype B aMPV (aMPV/B) F protein. Importantly, we located the motif in the aMPV/B F protein recognized by TMPRSS12 and the catalytic triad in TMPRSS12 that facilitated proteolysis of the aMPV/B F protein. This is the first report on TMPRSS12 as a protease for proteolysis of viral envelope glycoproteins. Our study will shed light on the mechanism of proteolysis of aMPV F protein and pathogenesis of aMPV.
INTRODUCTION
Avian metapneumovirus (aMPV), a member of the genus Metapneumovirus in the subfamily Pneumovirinae of the family Paramyxoviridae, causes acute rhinotracheitis (TRT) in turkeys and swollen head syndrome (SHS) in chickens (1–3). Thus, aMPV is a major disease threat to the turkey and chicken industries (4, 5). However, no effective vaccine or drug is currently available for large commercial poultry producers to prevent or treat aMPV (6). Based on a phylogenetic analysis, aMPV is divided into four subtypes: aMPV/A, aMPV/B, aMPV/C, and aMPV/D (7, 8).
The entry of paramyxoviruses into host cells initially requires the fusion of viral and cell membranes, which is mediated by one or more viral glycoproteins on the exterior of the viral envelope (9–12). For metapneumoviruses like aMPV, fusion (F) protein alone, without the help of an attachment protein, can induce membrane fusion (13–16). Consistent with this observation, recombinant aMPV lacking the small hydrophobic (SH) and attachment (G) glycoproteins replicates efficiently in vivo and vitro (17, 18). Collectively, these results suggest that aMPV F protein alone can mediate membrane fusion and viral infection.
The prerequisite for paramyxovirus F protein mediation of membrane fusion is cleavage of the F protein, followed by a conformational change (19–22). The paramyxovirus F protein is synthesized as a full-length precursor (F0) that is subsequently cleaved by cellular proteases to generate F1 and F2. Cleavage of F0 exposes a fusion peptide at the N terminus of F1 that is responsible for mediating membrane fusion (19, 23–25).
Many enveloped virus glycoproteins that mediate virus-cell membrane fusion, and therefore virus infection, are cleaved by type II transmembrane serine proteases (TTSPs) (25–27). The TTSP family contains more than 20 members that share a number of structural features, including an N-terminal transmembrane domain and a C-terminal extracellular serine protease domain (28–33).
In summary, a prerequisite for aMPV infection is proteolysis of the aMPV F protein; however, the proteases that cleave aMPV F protein remain unknown. Here, we have assembled evidence that the transmembrane serine protease TMPRSS12 facilitates the cleavage of aMPV/B F protein.
MATERIALS AND METHODS
Cells, virus, and plasmids.
Vero cells, BHK-21 cells, DF-1 cells, and chicken embryo fibroblasts (CEF) were grown in Dulbecco's modified Eagle's medium (DMEM; Thermo Scientific, Rockford, IL) supplemented with 10% fetal bovine serum (Gibco, Invitrogen/Life Technologies, Australia) and 1% penicillin and streptomycin (Summus, Beijing, China). The aMPV/B strain aMPV/f and Newcastle disease virus (NDV), LaSota vaccine strain, were maintained in our laboratory. F genes of aMPV/B strain VCO3/60616 (GenBank accession number AB548428.1) and human metapneumovirus (hMPV) Canada 97-83 strain (GenBank accession number AY297749.1) were subcloned into a pCAGGS expression vector carrying a Flag tag and were named aMPV/B-F and hMPV-F, respectively. Monkey hepsin (Mo_hepsin; GenBank accession number XM_005588816.1), monkey TMPRSS12 (Mo_TMPRSS12; GenBank accession number XM_005570851.1), human TMPRSS2 (Hu_TMPRSS2; GenBank accession number NM_005656.3), and chicken TMPRSS12 (Ch_TMPRSS12; GenBank accession number XM_424480.3) were subcloned into a pCAGGS vector with a hemagglutinin (HA) tag. Mutations were introduced into aMPV/B-F, hepsin, and TMPRSS12 using an In-Fusion HD cloning kit (Clontech, Mountain View, CA, USA). F, hepsin, and TMPRSS12 genes used were confirmed by DNA sequencing and Western blot analysis.
Syncytium formation assay to measure fusogenicity of the F protein.
The syncytium formation assay was performed to analyze F protein fusogenic activity as previously described (34–36). Vero cells grown in 6-well plates to 85% confluence were transfected with 1 μg plasmid carrying the F gene with ExFect transfection reagent (Vazyme Biotech, Nanjing, China) according to the manufacturer's protocol. Forty-eight hours after transfection, syncytia induced by the F protein in transfected Vero cells were observed and photographed using a digital microscope. The areas forming syncytia were analyzed with a Magnetic Lasso tool and the histogram function of Adobe Photoshop software. The ratio of the area forming syncytia to the total area was calculated to estimate the fusogenicity of F protein as previously described (34–36). To study the effect of TMPRSS12 on fusogenic activity of F protein, TMPRSS12 was selectively overexpressed or knocked down in Vero cells before transfection with the plasmid carrying the F gene.
Reporter gene assay to measure fusogenicity of the F protein.
The reporter gene assay was performed to estimate the fusogenic activity of F protein according to previous reports (11, 21). To study the effect of TMPRSS12 on fusogenic activity, TMPRSS12 was selectively overexpressed or knocked down in Vero cells. At 6 h posttransfection, the transfected Vero cells also were transfected with 1 μg of plasmid carrying the F gene and 0.5 μg of T7 control plasmid with luciferase cDNA (Promega, Madison, WI, USA). At 24 h posttransfection, the transfected Vero cells were detached, resuspended, and overlaid onto BHK-21 cells that were infected with MVA-T7 at a multiplicity of infection (MOI) of 1. At 48 h postinfection, luciferase activity of the cell lysates was quantitated using an Envision multilabel reader (PerkinElmer, Waltham, MA, USA) to estimate the fusogenic activity of the F protein.
Knockdown of TMPRSS12 with siRNA.
Confluent monolayers of Vero cells, DF-1 cells, or CEF cells in 6-well plates were transfected with small interfering RNAs (siRNAs) targeting Mo_TMPRSS12 or Ch_TMPRSS12 or with control siRNAs synthesized by GenePharma (Shanghai, China) using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). At 6 h of treatment with siRNAs, the cells were infected with 50 μl of 106 50% tissue culture infective doses (TCID50)/ml aMPV/f or transfected with aMPV/B-F.
To assess whether trypsin improved aMPV/B infection in TMPRSS12 siRNA-treated cells, TMPRSS12 siRNA-treated cells were infected with aMPV/B and then maintained in medium with 10 μg/ml trypsin. To assess whether overexpression of TMPRSS12 restored aMPV/B susceptibility to TMPRSS12 siRNA-treated cells, TMPRSS12 was overexpressed in siRNA-treated cells before they were infected with aMPV/B. At 48 h postinfection (p.i.), the supernatant of infected cells was collected to measure the virus titer.
Biotinylation and Western blot assay.
The extent of F protein cleavage and cell surface expression levels of TMPRSS12 were determined by biotinylation and Western blot assay as previously described (37, 38). Vero cells or DF-1 cells at 85% confluence in 6-well plates were transfected with aMPV/B-F/hMPV-F carrying a Flag tag or TMPRSS12 carrying an HA tag. At 48 h posttransfection, membrane-impermeable EZ-Link Sulfo-NHS-SS-Biotin (Thermo Scientific, Rockford, IL) was used to biotinylate surface proteins. The biotinylated Vero cells or DF-1 cells were lysed with cell lysis buffer (Thermo Scientific) for 30 min at 4°C. The lysates were centrifuged at 12,000 × g for 10 min at 4°C to collect the supernatants, and the cell surface proteins were immunoprecipitated using streptavidin beads (Thermo Scientific). Proteins were separated by 12% SDS-PAGE (GenScript, Nanjing, China), and the separated proteins were transferred to a nitrocellulose membrane (Hybond-C Super; GE Healthcare, Piscataway, NJ). The blot was probed with polyclonal anti-Flag (GenScript) or anti-HA antibody (Sigma) diluted 1:1,000, followed by incubation with IRDye 800CW goat anti-rabbit IgG (H+L) (1:10,000 dilution; LI-COR Biosciences, Bad Homburg, Germany) in a dark place at room temperature for 1 h. Membranes were exposed to LI-COR Odyssey (LI-COR Biosciences) for visualization of proteins. aMPV/B F protein expression was evaluated quantitatively to determine F protein cleavage levels using Gelpro32 software.
Determination of viral titers.
Mo_TMPRSS12 or Ch_TMPRSS12 was selectively overexpressed or knocked down in confluent monolayers of Vero cells, DF-1 cells, or CEF cells in 6-well plates. The transfected cells were infected with 50 μl of 106 TCID50/ml aMPV/f at 6 h posttransfection. The supernatant samples from Vero cells, DF-1 cells, or CEF cells infected with aMPV/f were collected at 24, 48, 72, and 96 h p.i. and maintained at −80°C until use. To determine aMPV/f titers, 100 μl of each 10-fold serial dilution of supernatant was cultured with Vero cells at 80% confluence in 96-well plates. At 4 days postinfection, syncytium formation was observed under a digital microscope to calculate the 50% tissue culture infective dose (TCID50) using the Reed and Muench method (39).
RT-PCR analysis of TMPRSS12 mRNA expression.
Chicken tissue samples were homogenized and centrifuged at 8,000 × g for 2 min. RNA was extracted from the samples using TRIzol (TaKaRa, Dalian, China) according to the manufacturer's instructions. The extracted RNA was resuspended in 15 μl of diethyl pyrocarbonate (DEPC)-treated water. To obtain cDNA, a reverse transcription (RT) reaction was performed with a QuantScript RT kit (Tiangen, Beijing, China) according to the manufacturer's instructions. The PCR was performed using Premix Taq (TaKaRa Taq, version 2.0, plus dye) (TaKaRa) with the following primer pairs: chTMPRSS12-F, 5′-CCGATGTCTGTAACGGCTCTGATGC-3′ (forward); chTMPRSS12-R, 5′-CATACAGGAGCACCATCCCTAC-3′ (reverse); ChGAPDH-F, 5′-CCCTGAAAATTGTCAGCAATGCATC-3′ (forward); ChGAPDH-R, 5′-CATCAAAGGTGGAGGAATGGCTGTC-3′ (reverse). Amplification was performed using the following cycling conditions: 95°C for 5 min; 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min; and elongation at 72°C for 7 min. Five microliters of each PCR product was analyzed by 2% agarose gel electrophoresis and visualized with a Gel Doc XR+ gel imaging system (Bio-Rad Laboratories, CA, USA).
qRT-PCR analysis of TMPRSS12 mRNA expression.
Quantitative RT-PCR (qRT-PCR) was performed with the following primer pairs and probes using a PrimeScript one-step RT-PCR kit (TaKaRa) on a LightCycler 480 (Roche, Rotkreuz, Switzerland): chTMPRSS12-F, 5′-TTGGGAGGCGTTGATTCCT-3′ (forward); chTMPRSS12-R, 5′-GACCCCCATCATGTAGTATTTGTTG-3′ (reverse); chTMPRSS12-Probe, 5′-TTGGCGTGCCACCACCCCA-3′ (probe); Chactin-F, 5′-CTGGCATTGCTGACAGGAT-3′ (forward); Chactin-R, 5′-GCCTCCAATCCAGACAGAGT-3′ (reverse); Chactin-probe, 5′-AGAAGGAGATCACAGCCCTGGCACC-3′ (probe). The following temperature program was used for the qRT-PCR assay: 42°C for 5 min, 95°C for 10 s, and 40 cycles of 95°C for 5 s and 60°C for 30 s. Fluorescent signals were collected during the elongation step.
Animal experiments and virus isolation.
Sixty 1-day-old aMPV-free specific-pathogen-free (SPF) chickens were randomly divided into two groups. Thirty chickens in the infected group were inoculated intranasally and by eye drop with 0.2 ml of 106.5 TCID50/ml aMPV/f per chicken, and 30 chickens, used as the control group, were inoculated with 0.2 ml of DMEM per chicken. The two groups were kept in separate rooms. At 3, 7, 10, 14, 21, and 28 days postinfection, three chickens from the infected group and three from the control group were randomly selected and euthanized to collect tissues. All tissue samples were homogenized and centrifuged at 8,000 × g for 2 min. These samples were passaged approximately six times in Vero cells until syncytium formation, when aMPV/B was isolated. Isolation was confirmed by qRT-PCR according to a previous report (40). All animal studies were approved by the Review Board of Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
Statistical analysis.
All experiments were repeated three to five times separately. Data were statistically analyzed by multiple one-way comparisons using SPSS 13.0 software. A P value of less than 0.05 was considered statistically significant.
RESULTS
aMPV/B grows in vitro without the addition of trypsin.
In a previous study, we demonstrated that the cleavage of aMPV/B F protein is independent of exogenous trypsin (34). Here, we investigated the effect of adding trypsin on replication of aMPV/B. As shown in Fig. 1, in the absence of trypsin, aMPV/B can be propagated in Vero and DF-1 cells (Fig. 1A and C), and aMPV/B titers without trypsin treatment approached those observed with trypsin treatment at 72 h postinfection (Fig. 1B and D), whereas exogenous trypsin was essential for NDV growth in vitro (Fig. 1E and F). Moreover, in the absence of trypsin, we propagated aMPV/B continuously in Vero cells for 20 passages and in DF-1 cells for another five passages, and the titer at each passage was approximately 107 and 106 TCID50/ml, respectively. Briefly, these results demonstrated that aMPV/B grew independently of exogenous trypsin in vitro, indicating that aMPV/B F protein is cleaved by endogenous proteases.
FIG 1.
Addition of trypsin has no effect on the replication of aMPV/B. (A and C) aMPV/B replication kinetics in Vero cells (A) and DF-1 cells (C) left untreated or treated with 10 μg/ml of trypsin (TPCK) were tested for up to 96 h p.i. Viral titers were determined using Vero cells in the absence of trypsin. (B and D) At 72 h p.i., the supernatant of infected Vero cells (B) and DF-1 cells (D) was collected to determine titer. (E) The hemagglutination titers of NDV (LaSota strain) treated with trypsin or left untreated were tested for up to 96 h p.i. (F) At 72 h p.i., the supernatant of freeze-thawing infected CEF cells was collected to determine hemagglutination titer. CEF cells, chicken embryo fibroblast cells. **, P < 0.05; ***, P < 0.005; #, P > 0.05 (nonsignificant).
Overexpression of TMPRSS12 and hepsin increases cleavage and fusogenic activity of aMPV/B F protein and aMPV/B replication.
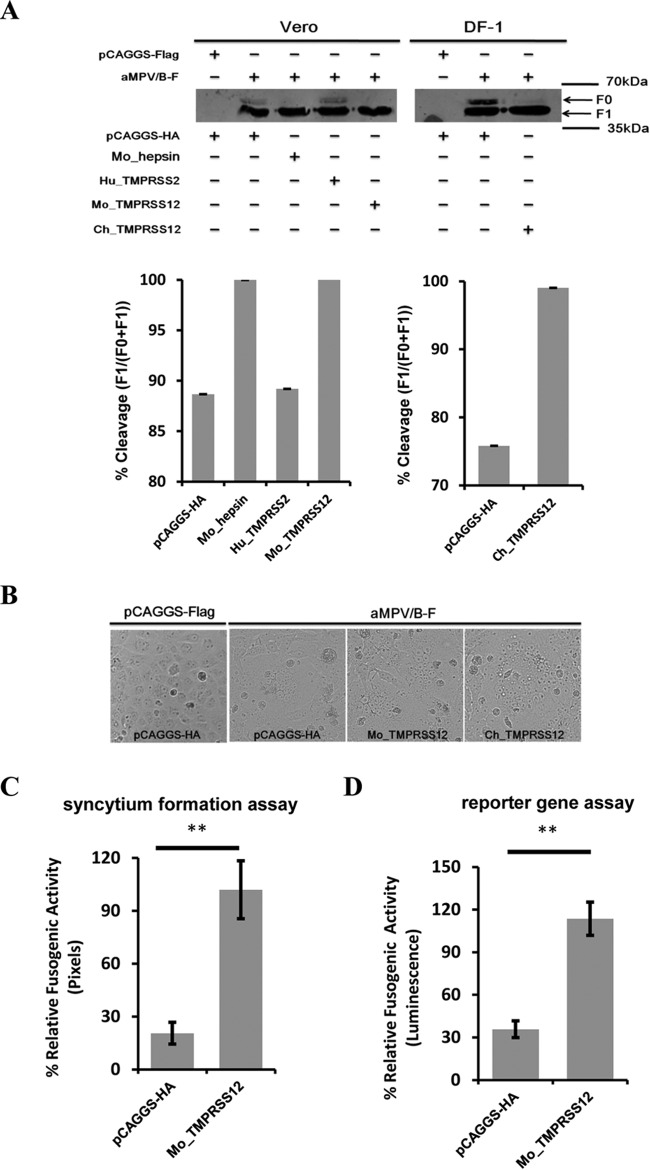
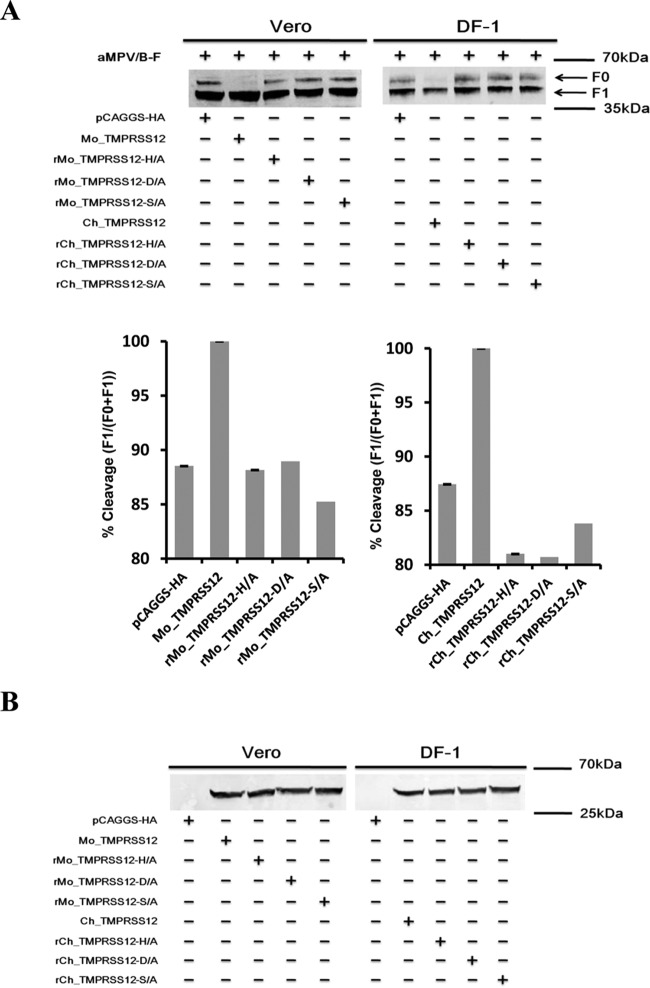
Some glycoproteins of viruses are cleaved by the transmembrane serine protease hepsin or TMPRSS2 (25, 27, 41–43). The protein sequences of hepsin and TMPRSS12 are highly similar. Hence, we investigated the role of hepsin, TMPRSS2, and TMPRSS12 in aMPV/B F protein cleavage. We overexpressed Mo_hepsin, Hu_TMPRSS2, or Mo_TMPRSS12 in Vero cells, as well as Ch_TMPRSS12 in DF-1 cells, before transfection with aMPV/B-F. As shown in Fig. 2A, transfection of Mo_hepsin, Mo_TMPRSS12, and Ch_TMPRSS12 slightly increased the extent of aMPV/B F protein cleavage compared to transfection of the pCAGGS vector carrying an HA tag, whereas transfection with Hu_TMPRSS2 did not.
FIG 2.
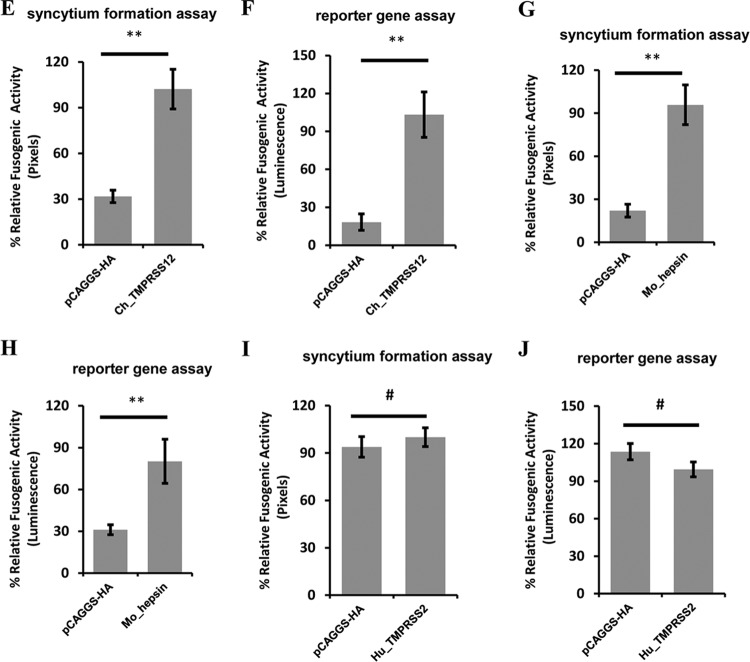
Overexpression of TMPRSS12 enhances cleavage level and fusogenic activity of aMPV/B F protein. (A) The effect of hepsin, TMPRSS2, and TMPRSS12 expression on the extent of aMPV/B F protein cleavage. The cleavage of aMPV/B F protein was analyzed by biotinylation and Western blot assay. (B) Effect of TMPRSS12 expression on syncytium formation in Vero cells. Syncytium formation in Vero cells was observed and photographed (magnification, ×100; 8.5 cm by 9.1 cm, as shown). (C to J) The effect of TMPRSS12, hepsin, and TMPRSS2 expression on aMPV/B F protein fusogenic activity was analyzed by syncytium formation (C, E, G, and I) and reporter gene assays (D, F, H, and J).
These findings prompted us to ascertain whether overexpression of TMPRSS12 promoted aMPV/B F protein fusogenic activity. Our results showed that syncytium formation in Vero cells overexpressing TMPRSS12 was greater than that in empty vector-treated cells (Fig. 2B). We further investigated the effect of Mo_TMPRSS12 and Ch_TMPRSS12 overexpression on aMPV/B F protein fusogenic activity with quantitative assays. These results indicated that transient expression of Mo_TMPRSS12 and Ch_TMPRSS12 enhanced the fusogenicity of aMPV/B F protein approximately 4- to 6-fold (Fig. 2C, D, E, and F). In addition, we found that overexpression of Mo_hepsin enhanced the fusogenicity of aMPV/B F protein, whereas overexpression of Hu_TMPRSS2 did not (Fig. 2G, H, I, and J).
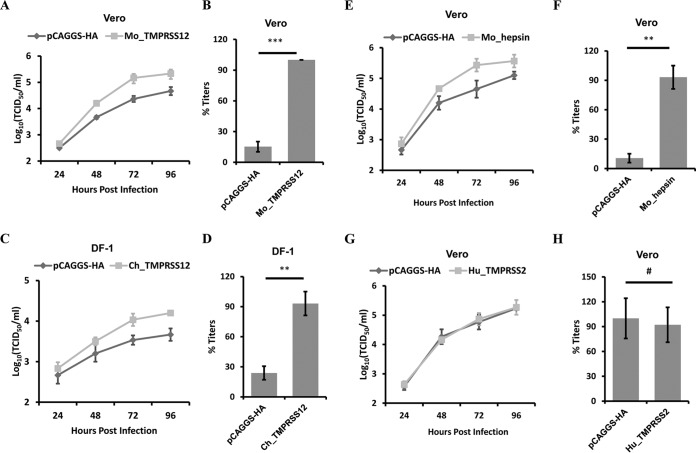
To investigate whether TMPRSS12 facilitated the replication of aMPV/B, we transfected Vero cells with Mo_TMPRSS12 and DF-1 cells with Ch_TMPRSS12 before the cells were infected with aMPV/B. As shown in Fig. 3A, transient expression of Mo_TMPRSS12 in Vero cells enhanced aMPV/B replication compared to that following transfection with the vector pCAGGS, and it increased aMPV/B titers approximately 9-fold relative to the expression of vector pCAGGS at 72 h p.i. (Fig. 3B). Additionally, we discovered that aMPV/B replication was promoted in DF-1 cells overexpressing Ch_TMPRSS12 compared to that in empty vector-treated controls (Fig. 3C). At 72 h p.i., aMPV/B titers were increased by Ch_TMPRSS12 approximately 5-fold (Fig. 3D). Furthermore, overexpression of Mo_hepsin increased replication of aMPV/B, whereas Hu_TMPRSS2 did not (Fig. 3E, F, G, and H). Overall, overexpression of TMPRSS12 and hepsin increased aMPV/B F protein cleavage and fusogenicity and aMPV/B replication.
FIG 3.
Overexpression of TMPRSS12 enhances aMPV/B replication. (A and C) aMPV/B replication kinetics in different cells overexpressing TMPRSS12 were tested up to 96 h p.i. (B and D) At 72 h p.i., the supernatant in infected cells overexpressing TMPRSS12 was collected to determine titer. (E and G) aMPV/B replication kinetics in Vero cells overexpressing hepsin or TMPRSS2 were tested up to 96 h p.i. (F and H) At 72 h p.i., the supernatant in infected cells overexpressing hepsin or TMPRSS2 was collected to determine titer. **, P < 0.05; ***, P < 0.005; #, P > 0.05 (nonsignificant).
Knockdown of TMPRSS12 reduces aMPV/B F protein cleavage and fusogenic activity and aMPV/B replication.
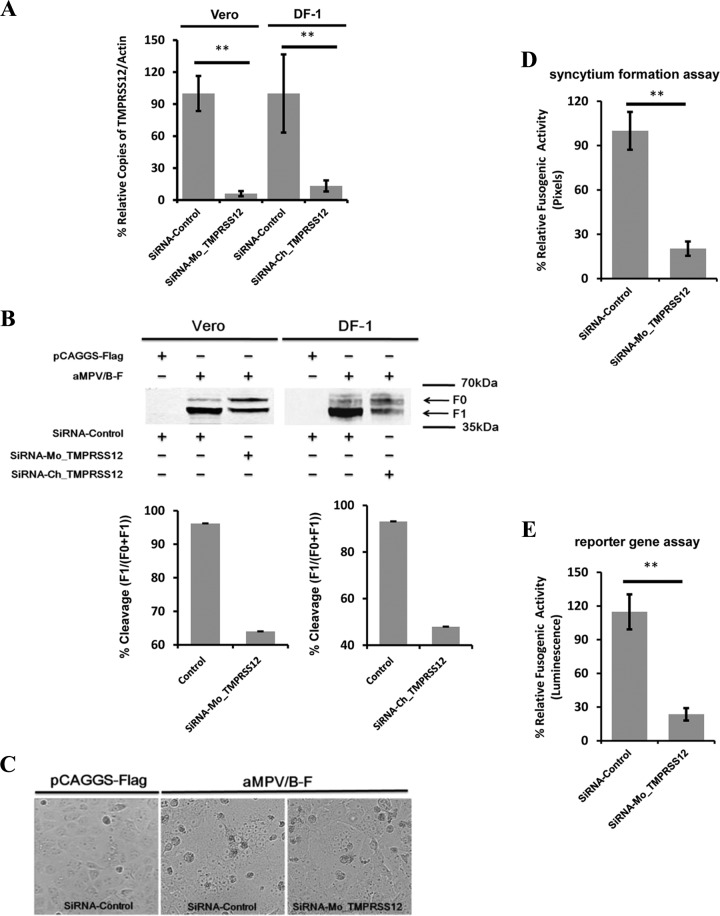
The results described above showed that TMPRSS12 and hepsin enhanced aMPV/B F protein cleavage and fusogenicity and aMPV/B replication. Because hepsin was not detected in DF-1 cells and chicken organs, our subsequent experiments focused on TMPRSS12. To determine whether downregulation of TMPRSS12 diminished aMPV/B F protein cleavage and fusogenic activity, we transfected Vero cells with siRNA targeting Mo_TMPRSS12 and DF-1 cells with siRNA targeting Ch_TMPRSS12, both of which reduced TMPRSS12 mRNA expression (Fig. 4A). Subsequently, these cells were transfected with a plasmid expressing aMPV/B F protein. As shown in Fig. 4B, transfection with TMPRS12-specific siRNAs reduced aMPV/B F protein cleavage compared to nonspecific siRNA transfection.
FIG 4.
Reduced TMPRSS12 expression decreases the cleavage level and fusogenic activity of aMPV/B F protein. (A) siRNA targeting Mo_TMPRSS12 or Ch_TMPRSS12 was transfected into Vero cells or DF-1 cells, respectively. At 48 h posttransfection, TMPRSS12 mRNA expression was analyzed by qRT-PCR. The average copy numbers of Mo_TMPRSS12 mRNA were reduced from 1.10 × 103 to 2.66 × 101. The average copy numbers of Ch_TMPRSS12 mRNA were reduced from 3.02 × 104 to 3.16 × 103. (B) Effect of diminished TMPRSS12 expression on the extent of aMPV/B F protein cleavage. siRNA targeting TMPRSS12 was transfected into cells before transfection with aMPV/B-F. At 48 h posttransfection, the cleavage of aMPV/B F protein was analyzed by biotinylation and Western blot assay. (C) Syncytium formation in Vero cells downregulated with TMPRSS12 was observed and photographed (magnification, ×100; 8.5 cm by 9.1 cm, as shown). (D and E) The effect of downregulated TMPRSS12 expression on aMPV/B F protein fusogenic activity was analyzed by syncytium formation (D) and reporter gene assays (E).
Furthermore, we analyzed the effect of downregulating TMPRSS12 on aMPV/B F protein fusogenic activity. The results showed that syncytium formation was reduced in Vero cells transfected with siRNA targeting Mo_TMPRSS12 compared to that in cells transfected with nonspecific siRNA (Fig. 4C). Further quantitative analysis revealed that downregulation of Mo_TMPRSS12 diminished aMPV/B F protein fusogenic activity approximately 4- to 6-fold (Fig. 4D and E). Overall, the results of these quantitative assays were consistent with results obtained from the syncytium formation assay described above.
To further determine the role of TMPRSS12 in aMPV/B replication, we knocked down Mo_TMPRSS12 in Vero cells and Ch_TMPRSS12 in DF-1 cells using siRNA and then infected these cells with aMPV/B. siRNA targeting Mo_TMPRSS12 in Vero cells impaired aMPV/B replication relative to that with nonspecific siRNA (Fig. 5A). At 72 h p.i., siRNA targeting Mo_TMPRSS12 diminished aMPV/B titers by approximately 10-fold (Fig. 5B). Additionally, Ch_TMPRS12-specific siRNA in DF-1 cells reduced aMPV/B F replication relative to that with nonspecific siRNA (Fig. 5C). At 72 h p.i., Ch_TMPRS12-specific siRNA decreased aMPV/B titers approximately 9-fold (Fig. 5D). To assess whether TMPRSS12 was required for spread of virus to primary cells of avian origin, we knocked down TMPRSS12 in CEF cells (Fig. 5E). As shown in Fig. 5F, siRNA targeting Ch_TMPRSS12 in CEF cells diminished aMPV/B replication relative to that with nonspecific siRNA. At 72 h p.i., Ch_TMPRS12-specific siRNA reduced aMPV/B titers approximately 8-fold (Fig. 5G). Taken together, knockdown of TMPRSS12 expression diminished the cleavage and fusogenicity of aMPV/B F protein as well as aMPV/B replication.
FIG 5.
Reduced TMPRSS12 expression decreases aMPV/B replication. (A and C) aMPV/B replication kinetics in different cells with reduced TMPRSS12 were tested for up to 96 h p.i. (B and D) At 72 h p.i., the supernatant in infected cells was collected to determine titer. (E) siRNA targeting Ch_TMPRSS12 was transfected into CEF cells. At 48 h posttransfection, TMPRSS12 mRNA expression was analyzed by qRT-PCR. The average copy numbers of Ch_TMPRSS12 mRNA were reduced from 2.04 × 103 to 1.78 × 102. (F) aMPV/B replication kinetics in CEF cells with reduced TMPRSS12 were tested for up to 96 h p.i. (G) At 72 h p.i., the supernatant in infected cells was collected to determine titer. (H and I) Mo_TMPRSS12 or Ch_TMPRSS12 was transfected into TMPRSS12 siRNA-treated cells. At 24 h posttransfection, these cells were infected by aMPV/f. At 48 h p.i., the supernatant in infected cells was collected to determine titer. **, P < 0.05; ***, P < 0.005; #, P > 0.05 (nonsignificant).
To determine whether overexpression of TMPRSS12 restored aMPV susceptibility to TMPRSS12 siRNA-treated cells, we knocked down Mo_TMPRSS12 in Vero cells and Ch_TMPRSS12 in DF-1 cells using siRNA. As shown in Fig. 5H and I, transfection with TMPRSS12 restored aMPV replication to the TMPRSS12 siRNA-treated Vero and DF-1 cells, whereas adding trypsin did not. Overall, these results supported the view that TMPRSS12 facilitated aMPV/B infection.
aMPV/B F protein contains a cleavage motif recognized by TMPRSS12.
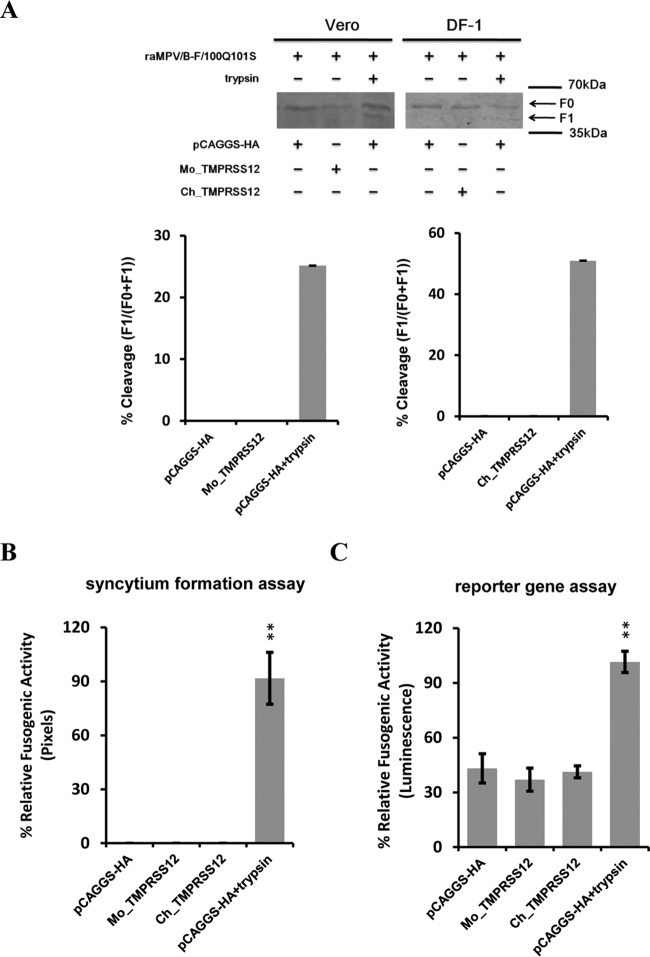
Our previous work showed that wild-type aMPV/B F protein containing 99RKKR102 is cleaved by endogenous proteases, whereas mutant aMPV/B F protein with 99RQSR102 is not (34). Hence, we hypothesized that 99RKKR102 was a cleavage motif recognized by TMPRSS12. To test our hypothesis, we replaced 99RKKR102 in the aMPV/B F protein with 99RQSR102 and analyzed the effect of TMPRSS12 on cleavage and fusogenicity of the mutant aMPV/B F protein with 99RQSR102. As shown in Fig. 6A, trypsin cleaved the mutant aMPV/B F protein, whereas overexpressed TMPRSS12 could not. Moreover, TMPRSS12 did not affect the fusogenic activity of mutant aMPV/B F protein, whereas exogenously added trypsin did (Fig. 6B and C). Collectively, these results suggest that amino acids 100 and 101 of the aMPV/B F protein constitute a cleavage motif recognized by TMPRSS12.
FIG 6.
Cleavage motif of aMPV/B F protein is recognized by TMPRSS12. (A and D) The effect of expression of TMPRSS12 on the extent of cleavage of mutant aMPV/B F protein carrying 100Q101S and hMPV F protein. Trypsin was exogenously added as a positive control. The cleavage of mutant aMPV/B and hMPV F proteins was analyzed by biotinylation and Western blot assay. (B, C, E, and F) The effect of expression of TMPRSS12 on the fusogenicity of mutant aMPV/B F protein with 100Q101S and hMPV F protein was analyzed by syncytium formation (B and E) and reporter gene assays (C and F). **, P < 0.05; ***, P < 0.005; #, P > 0.05 (nonsignificant).
An existing report shows that TMPRSS2 facilitates cleavage of the hMPV F protein (25). Therefore, we investigated whether overexpression of TMPRSS12 increased the extent of hMPV F protein cleavage. As shown in Fig. 6D, overexpression of TMPRSS2 enhanced hMPV F protein cleavage, whereas overexpression of TMPRSS12 did not. Moreover, we found that fusogenicity of hMPV F protein was not promoted by TMPRSS12 (Fig. 6E and F). Overall, these results indicated that TMPRSS12 had no effect on hMPV F protein cleavage and fusogenic activity, which can be explained by the fact that the putative cleavage motif of hMPV F protein was 99RQSR102, which might not be recognized by TMPRSS12.
The HDS triad in TMPRSS12 is essential for promoting cleavage and fusogenicity of aMPV/B F protein and aMPV/B replication.
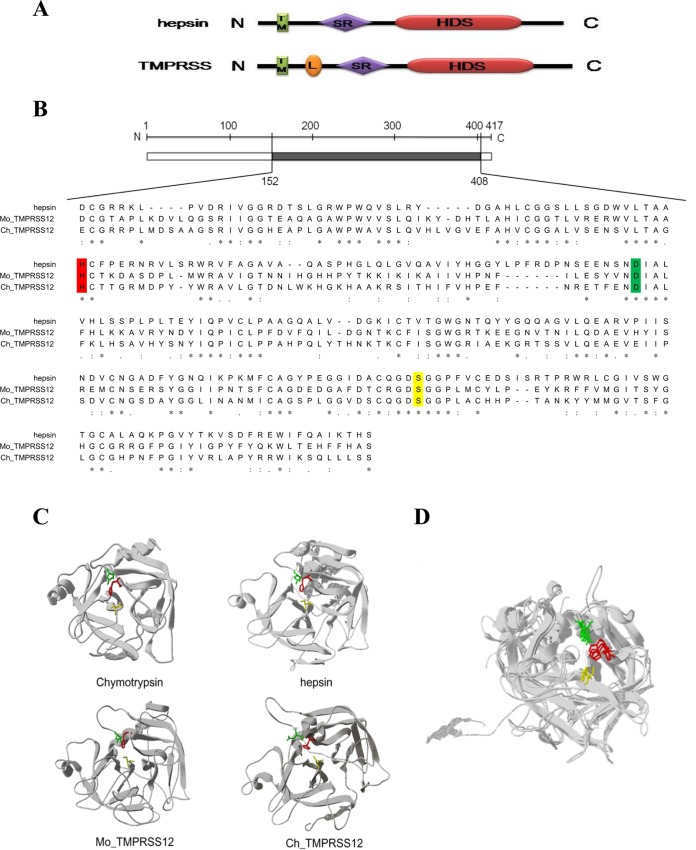
The hallmark of the serine proteases is that they contain a conserved catalytic triad of histidine, aspartic acid, and serine residues (HDS) (Fig. 7A) (31, 44). We predicted the HDS triad in TMPRSS12 by an alignment with a hepsin sequence from previous reports (28, 33). As shown in Fig. 7B, the potential catalytic activity of the HDS triad of TMPRSS12 was marked with a color. Additionally, we constructed a model of the preactive conformation of TMPRSS12 using the SWISS-MODEL server (Fig. 7C). As shown in Fig. 7D, the HDS triad in TMPRSS12 was similar to that of chymotrypsin (PDB entry 2GMT) and hepsin (preactive conformation), suggesting that the HDS triad that we predicted is the active protease site of TMPRSS12.
FIG 7.
HDS triad of TMPRSS12 and hepsin. (A) TM, N-terminal transmembrane signal anchor domain; L, low-density lipoprotein receptor class A domain; SR, group A scavenger receptor domain; HDS, C-terminal serine protease domain. (B) The alignment of hepsin and TMPRSS12 protein sequences was performed using the UniProt alignment facilities (http://www.uniprot.org/align/). The hepsin amino acids are presented as a scale bar, and the amino acid site is presented in Arabic numerals. Dashes represent gaps, and an asterisk indicates complete conservation among the amino acids. Abbreviations for amino acid residues: histidine, H (red); aspartic acid, D (green); serine, S (yellow). (C) The model represents the conformation of the hepsin and TMPRSS12 proteins using the SWISS-MODEL server facilities (http://swissmodel.expasy.org/). The classical structure of chymotrypsin (PDB entry 2GMT) is presented here to show the essential HDS triad. His (H), red; Asp (D), green; Ser (S), yellow. (D) A superposition of the HDS triad of hepsin, TMPRSS12, and chymotrypsin. The HDS triad is marked in color.
To test whether the HDS triad was critical for TMPRSS12 cleavage of aMPV/B F protein, we substituted alanine (A) for each amino acid residue in the HDS triad and compared the effect of the mutant TMPRSS12 with that of the wild type on aMPV/B F protein cleavage. As shown in Fig. 8A, compared to wild-type TMPRSS12, mutant TMPRSS12 showed an impaired capacity to cleave the aMPV/B F protein. The extent of aMPV/B F protein cleavage by TMPRSS12 containing the mutant HDS triad was comparable to that following transfection with an empty vector. Furthermore, we performed biotinylation and Western blot assays and excluded the possibility that HDS triad mutation downregulated cell surface expression of TMPRSS12 (Fig. 8B).
FIG 8.
HDS triad of TMPRSS12 is critical for proteolysis of aMPV/B F protein. (A) The effect of expression of mutant TMPRSS12 on the extent of aMPV/B F protein cleavage. The cleavage of aMPV/B F protein was analyzed by biotinylation and Western blot assay. (B) The expression of wild-type and mutant TMPRSS12 on cellular surfaces was analyzed by biotinylation and Western blot assay. (C to F) The effect of expression of mutant TMPRSS12 on aMPV/B F protein fusogenic activity analyzed by syncytium formation (C and E) and reporter gene assays (D and F). (G and H) Wild-type and mutant TMPRSS12 were transfected into cells before infection with aMPV/B. At 72 h p.i., the supernatant in infected cells was collected to determine titers. **, P < 0.05; ***, P < 0.005; #, P > 0.05 (nonsignificant).
Simultaneously, we tested the effect of TMPRSS12 with the mutant HDS on the fusogenicity of aMPV/B F protein. We analyzed the effect of mutant TMPRSS12 overexpression on the fusogenic activity of aMPV/B F protein using quantitative assays. As shown in Fig. 8C, D, E, and F, transfection of Mo_TMPRSS12 and Ch_TMPRSS12 with mutant HDS did not increase aMPV/B F protein fusogenicity compared to that of an empty vector carrying an HA tag.
Subsequently, we investigated the effect of transient expression of TMPRSS12 with a mutant HDS on aMPV/B replication. We transfected Vero cells with mutant Mo_TMPRSS12 and DF-1 cells with mutant Ch_TMPRSS12 before infection with aMPV/B, and then the supernatant of infected cells was collected to determine viral titers. As shown in Fig. 8G and H, overexpression of TMPRSS12 with a mutant HDS did not increase aMPV/B replication in Vero cells and DF-1 cells compared to transfection with the empty vector pCAGGS. Taken together, these results suggested that the HDS triad of TMPRSS12 was essential for promoting aMPV/B F protein cleavage and fusogenicity as well as aMPV/B replication.
The distribution of TMPRSS12 in chickens.
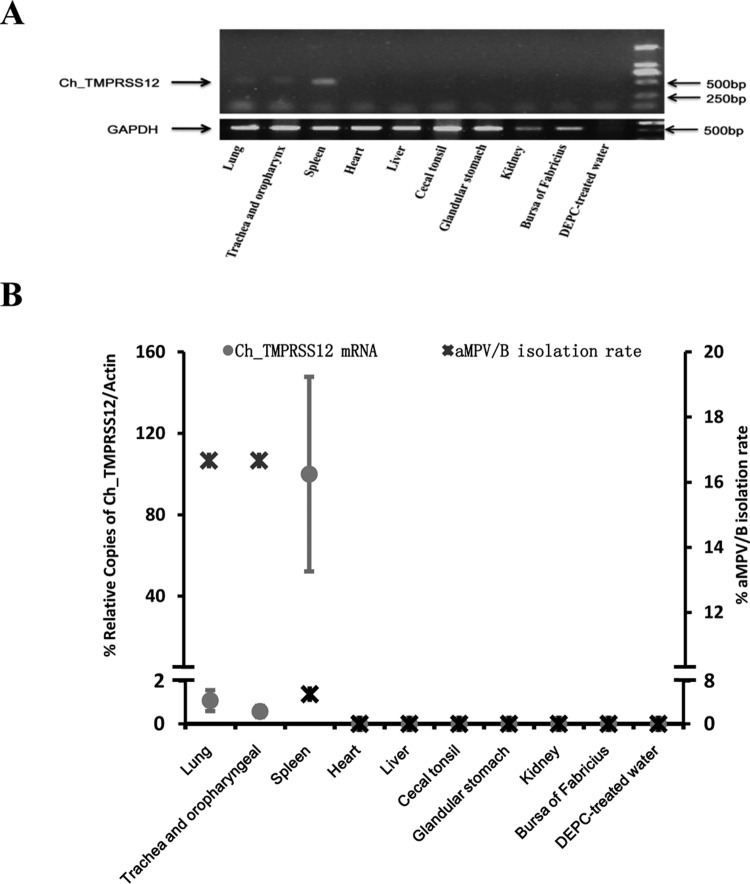
The distribution of a host protease can influence the virulence, infectivity, and pathogenicity of paramyxoviruses (24, 45, 46). Given that Ch_TMPRSS12 promoted aMPV/B replication by enhancing aMPV/B F protein cleavage and fusogenic activity, aMPV/B infection could be affected by the distribution of Ch_TMPRSS12 in chickens. Therefore, we investigated the distribution of Ch_TMPRSS12 in tissues of chickens with RT-PCR and qRT-PCR assays. The results of the RT-PCR assay revealed that spleen, lung, trachea, and oropharynx were positive for Ch_TMPRSS12 mRNA expression (Fig. 9A), and as determined by qRT-PCR assay, the levels of Ch_TMPRSS12 mRNA expressed in the spleen were increased compared to those of the lung, trachea, and oropharynx (Fig. 9B). As Fig. 9B and a previous report indicate (47), the lung, trachea, oropharynx, and spleen of chickens are permissive for aMPV/B infection. These results suggested that these specific tissues facilitated aMPV/B infection via TMPRSS12 cleavage of the F protein.
FIG 9.
TMPRSS12 mRNA is expressed in chicken tissues. (A) Analysis of TMPRSS12 mRNA expression by RT-PCR. Total RNA isolated from chicken tissues was used for the amplification of TMPRSS12 mRNA by RT-PCR. The results analyzed by 2% agarose gel are shown. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.(B) Analysis of TMPRSS12 mRNA expression by qRT-PCR and the rate of aMPV/B isolation. RNA was prepared as described for TMPRSS12 and β-actin and then amplified by qRT-PCR.
DISCUSSION
Proteolysis of aMPV F protein by intracellular proteases is critical for F protein-mediated virus-cell membrane fusion and viral infectivity. In this study, we obtained the first evidence that aMPV/B F protein was proteolytically processed by the endogenous protease TMPRSS12. Moreover, we confirmed that amino acid sites 100 and 101 of aMPV/B F protein were a cleavage motif recognized by TMPRSS12. We also demonstrated that the HDS triad was part of a protease activation site responsible for aMPV/B F protein proteolysis. Furthermore, we detected TMPRSS12 mRNA expression in aMPV/B target tissues in chicken, indicating that TMPRSS12 promotes aMPV/B propagation in this host.
Both aMPV and hMPV are members of the Metapneumovirus genus, and both aMPV/B and hMPV are commonly propagated in Vero cells (24, 48–50). The majority of hMPV strains cultured in Vero cells require the addition of trypsin (24); however, our data indicated that Vero cells without trypsin treatment can be used for the propagation of aMPV/B. Results of this study suggested that aMPV/B F protein cleavage was not dependent on trypsin. aMPV/B growth without trypsin treatment can facilitate the isolation of clinical strains and generation of attenuated aMPV vaccine candidates. Furthermore, TMPRSS12 expression facilitated the replication of aMPV/B. Thus, researchers can establish Vero cell lines that constitutively express TMPRSS12 to harvest high titers of aMPV/B for vaccine manufacture.
Transmembrane serine proteases typically act as activators for the proteolysis of glycoproteins (25, 27, 41–43). In this study, we initially tested whether hepsin, TMPRSS2, and TMPRSS12 facilitated aMPV/B F protein cleavage. We then studied the role of TMPRSS12 in the proteolysis of aMPV/B F protein, because only TMPRSS12 was present in both Vero cells and DF-1 cells. A combination of analyses, using overexpression, knockdown, and mutation approaches, demonstrated that TMPRSS12 was a protease that cleaved aMPV/B F protein. To our knowledge, this is the first report on TMPRSS12 as a protease for viral envelope glycoproteins.
Paramyxovirus F proteins are synthesized as inactive precursors (F0). F0 is cleaved into the disulfide-linked subunits F1 and F2 by host endoprotease(s). Cleavage of F0 then mediates the fusion of virus and host cell membranes (23, 51). Our prior work demonstrated that amino acid sites 100 and 101 are determinants for aMPV/B F protein cleavage by endogenous proteases (34). In this study, we demonstrated that wild-type aMPV/B F protein with 99RKKR102 was cleaved by TMPRSS12, whereas mutant aMPV/B F protein with 99RQSR102 was not. These results support the conclusion that TMPRSS12 cleaved aMPV/B F protein by recognizing amino acid sites 100 and 101. In addition, aMPV/B F protein with the multibasic residues 99RKKR102 was more easily cleaved by intracellular proteases than the protein with the monobasic residues 99RQSR102, which was consistent with the published rule that paramyxovirus F proteins containing basic residues (Arg or Lys) at the putative cleavage motif are easily cleaved by intracellular transmembrane serine proteases (24, 46, 52). The phenomenon of recognition of paramyxovirus F proteins containing multibasic residues by intracellular serine proteases can be explained by the fact that the basic side chain of paramyxovirus F proteins is stabilized by an acidic residue near the underside of the serine protease binding pocket (reviewed in references 28, 29, and 31).
Published reports show that hMPV F protein is largely uncleaved in Vero cells and that TMPRSS2 promotes cleavage of the protein (24, 25). Unlike hMPV F protein, aMPV/B F protein is mostly (80%) cleaved by endogenous proteases (Fig. 2A). However, overexpression of TMPRSS12 increased aMPV/B F protein cleavage to nearly 100% (Fig. 2A), and knockdown of TMPRSS12 decreased aMPV/B F protein cleavage to 60% (Fig. 4B). These results suggest that TMPRSS12 plays an important role in the cleavage of aMPV/B F protein. Furthermore, aMPV/B F protein was partly cleaved when TMPRSS12 was knocked down (Fig. 4B), suggesting that there are relevant activating proteases other than TMPRSS12 in susceptible cells. For example, in mammalian cells, hepsin facilitated aMPV/B F protein cleavage (Fig. 2A).
Serine proteases are involved in a wide variety of cellular as well as extracellular functions, including cell signaling, blood clotting, inflammation, protein digestion, and protein processing (31, 32, 53). A key characteristic of these serine proteases, including TTSPs, is that they contain the classical catalytic HDS triad (28, 31, 33). The HDS triad, which acts like a charge relay system, is crucial for TTSP catalytic activity (31–33). In this study, by mutating each amino acid residue of the HDS triad, we determined that the HDS triad was part of a protease activation site for TMPRSS12 proteolysis of aMPV/B F protein.
Given that cleavage of the F0 precursor is a prerequisite for paramyxovirus F protein mediating virus-cell fusion, the distribution of host protease affects paramyxovirus tropism and pathogenicity (24, 45, 46). For example, in Sendai virus and NDV, the recognition of F proteins from a tissue-specific protease to a nonspecific ubiquitous protease enhances virulence and causes host range changes (24, 45, 46). Here, we found that TMPRSS12, which supports aMPV/B infection, was expressed in only a few chicken tissues, possibly limiting the intrahost spread of aMPV/B.
aMPV causes TRT in turkeys and is associated with SHS in chickens, which are major economic problems for the poultry industry (54, 55). At present, no effective vaccine or drug is available to prevent or treat aMPV in large commercial poultry production (6). A prerequisite for aMPV infection is cleavage of the aMPV F protein, which triggers virus-cell membrane fusion (reviewed in references 12, 37, and 56). Thus, blocking cleavage of the aMPV F protein may be an effective way to control aMPV. Our present study suggests that TMPRSS12 contributes to the proteolysis of aMPV/B F protein in vitro. Therefore, interference with F protein proteolysis by TMPRSS12 could be a therapeutic goal. Further clarification of the factors involved in F protein proteolysis will shed light on the pathogenesis of aMPV and may result in novel therapeutic strategies to control aMPV.
ACKNOWLEDGMENTS
We are grateful to Baoshan Zhang and Changjiang Weng for critical revising of the manuscript.
REFERENCES
- 1.Shin HJ, Cameron KT, Jacobs JA, Turpin EA, Halvorson DA, Goyal SM, Nagaraja KV, Kumar MC, Lauer DC, Seal BS. 2002. Molecular epidemiology of subgroup C avian pneumoviruses isolated in the United States and comparison with subgroup A and B viruses. J Clin Microbiol 40:1687–1693. doi: 10.1128/JCM.40.5.1687-1693.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook J. 2000. Avian pneumovirus infections of turkeys and chickens. Vet J 160:118–125. doi: 10.1053/tvjl.2000.0486. [DOI] [PubMed] [Google Scholar]
- 3.Sun J, Wei Y, Rauf A, Zhang Y, Ma Y, Zhang X, Shilo K, Yu Q, Saif YM, Lu X, Yu L, Li J. 2014. Methyltransferase-defective avian metapneumovirus vaccines provide complete protection against challenge with the homologous Colorado strain and the heterologous Minnesota strain. J Virol 88:12348–12363. doi: 10.1128/JVI.01095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Govindarajan D, Buchholz UJ, Samal SK. 2006. Recovery of avian metapneumovirus subgroup C from cDNA: cross-recognition of avian and human metapneumovirus support proteins. J Virol 80:5790–5797. doi: 10.1128/JVI.00138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei L, Zhu S, Yan X, Wang J, Zhang C, Liu S, She R, Hu F, Quan R, Liu J. 2013. Avian metapneumovirus subgroup C infection in chickens, China. Emerg Infect Dis 19:1092. doi: 10.3201/eid1907.121126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu H, Roth JP, Estevez CN, Zsak L, Liu B, Yu Q. 2011. Generation and evaluation of a recombinant Newcastle disease virus expressing the glycoprotein (G) of avian metapneumovirus subgroup C as a bivalent vaccine in turkeys. Vaccine 29:8624–8633. doi: 10.1016/j.vaccine.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Bäyon-Auboyer M-H, Arnauld C, Toquin D, Eterradossi N. 2000. Nucleotide sequences of the F, L and G protein genes of two non-A/non-B avian pneumoviruses (APV) reveal a novel APV subgroup. J Gen Virol 81:2723–2733. doi: 10.1099/0022-1317-81-11-2723. [DOI] [PubMed] [Google Scholar]
- 8.Catelli E, Cecchinato M, Savage CE, Jones RC, Naylor CJ. 2006. Demonstration of loss of attenuation and extended field persistence of a live avian metapneumovirus vaccine. Vaccine 24:6476–6482. doi: 10.1016/j.vaccine.2006.06.076. [DOI] [PubMed] [Google Scholar]
- 9.Earp L, Delos S, Park H, White J. 2005. The many mechanisms of viral membrane fusion proteins, p 25–66. In Marsh M. (ed), Membrane trafficking in viral replication. Springer, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore J, Jameson BA, Weiss RA, Sattentau Q. 1993. The HIV-cell fusion reaction, p 233–289. In Bentz J. (ed), Viral fusion mechanisms. CRC Press, Boca Raton, FL. [Google Scholar]
- 11.Nussbaum O, Broder CC, Berger EA. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol 68:5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talekar A, DeVito I, Salah Z, Palmer SG, Chattopadhyay A, Rose JK, Xu R, Wilson IA, Moscona A, Porotto M. 2013. Identification of a region in the stalk domain of the Nipah virus receptor binding protein that is critical for fusion activation. J Virol 87:10980–10996. doi: 10.1128/JVI.01646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y, Feng K, Yao X, Cai H, Li J, Mirza AM, Iorio RM, Li J. 2012. Localization of a region in the fusion protein of avian metapneumovirus that modulates cell-cell fusion. J Virol 86:11800–11814. doi: 10.1128/JVI.00232-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Graaf M, Schrauwen EJA, Herfst S, van Amerongen G, Osterhaus ADME, Fouchier RAM. 2009. Fusion protein is the main determinant of metapneumovirus host tropism. J Gen Virol 90:1408–1416. doi: 10.1099/vir.0.009688-0. [DOI] [PubMed] [Google Scholar]
- 15.Yun B, Gao Y, Liu Y, Guan X, Wang Y, Qi X, Gao H, Liu C, Cui H, Zhang Y. 2015. Effect of amino acid sequence variations at position 149 on the fusogenic activity of the subtype B avian metapneumovirus fusion protein. Arch Virol 160:2445–2453. doi: 10.1007/s00705-015-2524-x. [DOI] [PubMed] [Google Scholar]
- 16.Yun B, Guan X, Liu Y, Zhang Y, Wang Y, Qi X, Cui H, Liu C, Zhang Y, Gao H, Gao L, Li K, Gao Y, Wang X. 2016. Integrin αvβ1 modulation affects subtype B avian metapneumovirus fusion protein mediating cell-cell fusion and virus infection. J Biol Chem 291:14815–14825. doi: 10.1074/jbc.M115.711382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naylor CJ, Brown PA, Edworthy N, Ling R, Jones RC, Savage CE, Easton AJ. 2004. Development of a reverse-genetics system for avian pneumovirus demonstrates that the small hydrophobic (SH) and attachment (G) genes are not essential for virus viability. J Gen Virol 85:3219–3227. doi: 10.1099/vir.0.80229-0. [DOI] [PubMed] [Google Scholar]
- 18.Ling R, Sinkovic S, Toquin D, Guionie O, Eterradossi N, Easton AJ. 2008. Deletion of the SH gene from avian metapneumovirus has a greater impact on virus production and immunogenicity in turkeys than deletion of the G gene or M2-2 open reading frame. J Gen Virol 89:525–533. doi: 10.1099/vir.0.83309-0. [DOI] [PubMed] [Google Scholar]
- 19.Lamb RA. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1–11. [DOI] [PubMed] [Google Scholar]
- 20.Bagai S, Lamb RA. 1995. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J Virol 69:6712–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herfst S, Mas V, Ver LS, Wierda RJ, Osterhaus AD, Fouchier RA, Melero JA. 2008. Low-pH-induced membrane fusion mediated by human metapneumovirus F protein is a rare, strain-dependent phenomenon. J Virol 82:8891–8895. doi: 10.1128/JVI.00472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sergel T, McGinnes LW, Peeples ME, Morrison TG. 1993. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology 193:717–726. [DOI] [PubMed] [Google Scholar]
- 23.Morrison TG. 2003. Structure and function of a paramyxovirus fusion protein. Biochim Biophys Acta 1614:73–84. [DOI] [PubMed] [Google Scholar]
- 24.Schickli JH, Kaur J, Ulbrandt N, Spaete RR, Tang RS. 2005. An S101P substitution in the putative cleavage motif of the human metapneumovirus fusion protein is a major determinant for trypsin-independent growth in Vero cells and does not alter tissue tropism in hamsters. J Virol 79:10678–10689. doi: 10.1128/JVI.79.16.10678-10689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirogane Y, Takeda M, Iwasaki M, Ishiguro N, Takeuchi H, Nakatsu Y, Tahara M, Kikuta H, Yanagi Y. 2008. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J Virol 82:8942–8946. doi: 10.1128/JVI.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, Steffen I, Choi S-Y, Park Y, Schneider H, Schughart K, Poehlmann S. 2010. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J Virol 84:10016–10025. doi: 10.1128/JVI.00239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bottcher E, Matrosovich T, Beyerle M, Klenk H-D, Garten W, Matrosovich M. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol 80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barre O, Dufour A, Eckhard U, Kappelhoff R, Beliveau F, Leduc R, Overall CM. 2014. Cleavage specificity analysis of six type II transmembrane serine proteases (TTSPs) using PICS with proteome-derived peptide libraries. PLoS One 9:e105984. doi: 10.1371/journal.pone.0105984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bugge TH, Antalis TM, Wu QY. 2009. Type II transmembrane serine proteases. J Biol Chem 284:23177–23181. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dodson G, Wlodawer A. 1998. Catalytic triads and their relatives. Trends Biochem Sci 23:347–352. doi: 10.1016/S0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 31.Ekici OD, Paetzel M, Dalbey RE. 2008. Unconventional serine proteases: variations on the catalytic Ser/His/Asp triad configuration. Protein Sci 17:2023–2037. doi: 10.1110/ps.035436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polgar L. 2005. The catalytic triad of serine peptidases. Cell Mol Life Sci 62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi N, Okui A, Yamada T, Nakazato H, Mitsui S. 2002. Spinesin/TMPRSS5, a novel transmembrane serine protease, cloned from human spinal cord. J Biol Chem 277:6806–6812. doi: 10.1074/jbc.M103645200. [DOI] [PubMed] [Google Scholar]
- 34.Yun B, Guan X, Liu Y, Gao Y, Wang Y, Qi X, Cui H, Liu C, Zhang Y, Gao L, Li K, Gao H, Gao Y, Wang X. 2015. Trypsin- and low pH-mediated fusogenicity of avian metapneumovirus fusion proteins is determined by residues at positions 100, 101 and 294. Sci Rep 5:15584. doi: 10.1038/srep15584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avitabile E, Lombardi G, Gianni T, Capri M, Campadelli-Fiume G. 2004. Coexpression of UL20p and gK inhibits cell-cell fusion mediated by herpes simplex virus glycoproteins gD, gH-gL, and wild-type gB or an endocytosis-defective gB mutant and downmodulates their cell surface expression. J Virol 78:8015–8025. doi: 10.1128/JVI.78.15.8015-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinzler ER, Compton T. 2005. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J Virol 79:7827–7837. doi: 10.1128/JVI.79.12.7827-7837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirza AM, Aguilar HC, Zhu QY, Mahon PJ, Rota PA, Lee B, Iorio RM. 2011. Triggering of the Newcastle disease virus fusion protein by a chimeric attachment protein that binds to Nipah virus receptors. J Biol Chem 286:17851–17860. doi: 10.1074/jbc.M111.233965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. 2009. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J Virol 83:1511–1522. doi: 10.1128/JVI.01381-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Epidemiol 27:493–497. [Google Scholar]
- 40.Guionie O, Toquin D, Sellal E, Bouley S, Zwingelstein F, Allée C, Bougeard S, Lemière S, Eterradossi N. 2007. Laboratory evaluation of a quantitative real-time reverse transcription PCR assay for the detection and identification of the four subgroups of avian metapneumovirus. J Virol Methods 139:150–158. doi: 10.1016/j.jviromet.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 41.Sakai K, Ami Y, Tahara M, Kubota T, Anraku M, Abe M, Nakajima N, Sekizuka T, Shirato K, Suzaki Y. 2014. The host protease TMPRSS2 plays a major role for in vivo replication of emerging H7N9 and seasonal influenza viruses. J Virol 88:5608–5616. doi: 10.1128/JVI.03677-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abe M, Tahara M, Sakai K, Yamaguchi H, Kanou K, Shirato K, Kawase M, Noda M, Kimura H, Matsuyama S. 2013. TMPRSS2 is an activating protease for respiratory parainfluenza viruses. J Virol 87:11930–11935. doi: 10.1128/JVI.01490-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adeline H, Heike HW, Stefanie G, Thomas L, Olaf J, Stefan PH. 2014. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol 88:1293–1307. doi: 10.1128/JVI.01490-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oyama E, Takahashi H. 2004. Amino acid sequence of a thrombin like enzyme, elegaxobin II, from the venom of Trimeresurus elegans (Sakishima-habu). Toxicon 44:711–721. doi: 10.1016/j.toxicon.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Kido H, Niwa Y, Beppu Y, Towatari T. 1996. Cellular proteases involved in the pathogenicity of enveloped animal viruses, human immunodeficiency virus, influenza virus A and Sendai virus. Adv Enzyme Regul 36:325–347. doi: 10.1016/0065-2571(95)00016-X. [DOI] [PubMed] [Google Scholar]
- 46.Klenk H-D, Garten W. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol 2:39–43. [DOI] [PubMed] [Google Scholar]
- 47.Htut Aung Y, Liman M, Neumann U, Rautenschlein S. 2008. Reproducibility of swollen sinuses in broilers by experimental infection with avian metapneumovirus subtypes A and B of turkey origin and their comparative pathogenesis. Avian Pathol 37:65–74. doi: 10.1080/03079450701802222. [DOI] [PubMed] [Google Scholar]
- 48.Coswig LT, Santos MB, Hafez HM, Ferreira HL, Arns CW. 2010. Propagation of avian metapneumovirus subtypes A and B using chicken embryo related and other cell systems. J Virol Methods 167:1–4. doi: 10.1016/j.jviromet. [DOI] [PubMed] [Google Scholar]
- 49.Sugiyama M, Ito H, Hata Y, Ono E, Ito T. 2010. Complete nucleotide sequences of avian metapneumovirus subtype B genome. Virus Genes 41:389–395. doi: 10.1007/s11262-010-0518-z. [DOI] [PubMed] [Google Scholar]
- 50.Schowalter RM, Smith SE, Dutch RE. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J Virol 80:10931–10941. doi: 10.1128/JVI.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cseke G, Maginnis MS, Cox RG, Tollefson SJ, Podsiad AB, Wright DW, Dermody TS, Williams JV. 2009. Integrin αvβ1 promotes infection by human metapneumovirus. Proc Natl Acad Sci U S A 106:1566–1571. doi: 10.1073/pnas.0801433106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klenk HD, Rott R. 1988. The molecular biology of influenza virus pathogenicity. Adv Virus Res 34:247–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Overall CM, Blobel CP. 2007. In search of partners: linking extracellular proteases to substrates. Nat Rev Mol Cell Biol 8:245–257. doi: 10.1038/nrm2120. [DOI] [PubMed] [Google Scholar]
- 54.Banet-Noach C, Simanov L, Laham-Karam N, Perk S, Bacharach E. 2009. Longitudinal survey of avian metapneumoviruses in poultry in Israel: infiltration of field strains into vaccinated flocks. Avian Dis 53:184–189. doi: 10.1637/8886.1. [DOI] [PubMed] [Google Scholar]
- 55.Olsen RH, Stockholm NM, Permin A, Christensen JP, Christensen H, Bisgaard M. 2011. Multi-locus sequence typing and plasmid profile characterization of avian pathogenic Escherichia coli associated with increased mortality in free-range layer flocks. Avian Pathol 40:437–444. doi: 10.1080/03079457.2011.592822. [DOI] [PubMed] [Google Scholar]
- 56.Bose S, Song AS, Jardetzky TS, Lamb RA. 2014. Fusion activation through attachment protein stalk domains indicates a conserved core mechanism of paramyxovirus entry into cells. J Virol 88:3925–3941. doi: 10.1128/JVI.03741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]