Abstract
Salmonella Typhi (S. Typhi) causes typhoid fever which is a disease characterised by high mortality and morbidity worldwide. In order to curtail the transmission of this highly infectious disease, identification of new markers that can detect the pathogen is needed for development of sensitive and specific diagnostic tests. In this study, genomic comparison of S. Typhi with other enteric pathogens was performed, and 6 S. Typhi genes, that is, STY0201, STY0307, STY0322, STY0326, STY2020, and STY2021, were found to be specific in silico. Six PCR assays each targeting a unique gene were developed to test the specificity of these genes in vitro. The diagnostic sensitivities and specificities of each assay were determined using 39 S. Typhi, 62 non-Typhi Salmonella, and 10 non-Salmonella clinical isolates. The results showed that 5 of these genes, that is, STY0307, STY0322, STY0326, STY2020, and STY2021, demonstrated 100% sensitivity (39/39) and 100% specificity (0/72). The detection limit of the 5 PCR assays was 32 pg for STY0322, 6.4 pg for STY0326, STY2020, and STY2021, and 1.28 pg for STY0307. In conclusion, 5 PCR assays using STY0307, STY0322, STY0326, STY2020, and STY2021 were developed and found to be highly specific at single-gene target resolution for diagnosis of typhoid fever.
1. Introduction
To date, there are more than 2,500 serotypes identified within the Salmonella enterica species [1]. Most are harmless to humans but one serotype, Salmonella enterica subspecies enterica serovar Typhi (S. Typhi), causes typhoid fever, a severe and life-threatening systemic infection in humans. Worldwide, typhoid fever causes 269,000 deaths from 26.9 million new cases each year [2]. Travellers, children, the elderly, and immune-compromised individuals are especially at risk [3, 4]. The clinical manifestations of typhoid fever are similar to other febrile illnesses. Therefore, diagnosis based on clinical signs and symptoms alone is difficult [5]. The emergence of multidrug-resistant S. Typhi strains and development of the typhoid carrier state have further complicated the management of typhoid fever [6, 7]. Delay in diagnosis and initiation of antibiotic treatment can cause serious clinical complications and fatality [8]. Thus, early and correct laboratory diagnosis of typhoid fever is critical to reduce the morbidity and mortality, as well as curtail transmission of the disease.
DNA-based detection methods, such as polymerase chain reaction (PCR), have proven to be sensitive, specific, and rapid compared to conventional culture-based methods for the diagnosis of many infectious diseases [9–11]. Several target genes have been used for S. Typhi identification using PCR, such as the O antigen somatic genes (tyv and prt) [12], H antigen flagellar gene (fliC-d) [13], and Vi capsular antigen gene (viaB) [14]. However, these genes cannot stand alone as single S. Typhi-specific diagnostic marker since they are not specific to S. Typhi and are also found in other Salmonella serotypes. Thus, these markers provide provisional rather than differential diagnosis of typhoid fever. For example, the fliC-d gene of S. Typhi shares the same nucleic acid sequence as S. Muenchen [15]; the prt gene is present in S. Typhi, S. Paratyphi A, and S. Enteritidis [12]; and the viaB gene is found not only in S. Typhi but also in S. Dublin, a few strains of S. Paratyphi C [16] and Citrobacter freundii [17]. Due to the lack of specificity of these target genes, a combination of different pairs of primers using multiplex PCR [18] or nested PCR [19] are needed to increase the sensitivity and specificity of the PCR diagnostic test. This, however, will increase the cost, time, and complexity of the laboratory diagnosis.
Diagnostic markers which can detect pathogens at single-gene target resolution could lead to a simpler, cost-effective, and more functional DNA-based detection method since less primers are needed for target detection. Many approaches, such as subtractive hybridization [20], next generation sequencing [21], and microarray [22] techniques, have been used to identify genes that are specific or unique to a pathogen. However, these high-end technologies are cumbersome and expensive and sometimes yield false negative or false positive results [23]. Since bacterial genome databases have expanded tremendously over the past decade and advancement in computing technologies has made nucleic acid sequence alignment services readily accessible at NCBI, in silico comparative hybridization approach coupled with in vitro PCR (wet-lab) validation is sufficient to facilitate the translation of genomic data into diagnostic marker discoveries. In this study, a low-cost and simple attempt was made to identify new DNA diagnostic markers specific for S. Typhi by utilizing genome data (stored in NCBI databases) and nucleic acid sequence alignment tools (BLASTn) that are readily available in the public domain. The diagnostic sensitivities and specificities of the primers designed for amplifying whole gene sequences can be validated using a panel of confirmed bacteria isolates selected from S. Typhi, non-Typhi Salmonella, and non-Salmonella clinical isolates. To serve as a control for the PCR reaction, 16S rRNA gene, that is ubiquitous among bacteria species, can be used as a PCR amplification control [24].
2. Materials and Methods
2.1. Bacterial Strains
A total of 111 bacteria isolates including 39 S. Typhi, 62 non-Typhi Salmonella serotypes, and 10 non-Salmonella strains were used in this study. S. Typhi strains consisted of 1 S. Typhi reference strains (ATCC 7251) and 38 different pulsed-field types (PFTs) representing all strains in the state of Kelantan in Malaysia. These 38 PFTs were the result of screening 279 S. Typhi clinical isolates using pulsed-field gel electrophoresis (PFGE) [25]. Non-Typhi Salmonella serotypes were closely related Salmonella species made up of 26 different serotypes (Table 2) and 10 ATCC strains including S. Paratyphi A (ATCC 9150), S. Paratyphi B (ATCC BAA 1250), S. Paratyphi C (ATCC 9068), S. Enteritidis (ATCC 13076), S. Typhimurium (ATCC 14028), S. Weltevreden (NCTC 6534), S. Agona (ATCC 51957), S. Heidelberg (ATCC 8326), S. Poona (ATCC 04840), and S. Braenderup (ATCC BAA-664). In addition, 10 other non-Salmonella strains such as Shigella dysenteriae, Shigella flexneri, Shigella boydii, Shigella sonnei, Vibrio cholera, Enterohemorrhagic E. coli, Enteropathogenic E. coli, Aeromonas hydrophila, Yersinia enterocolitica, and Klebsiella pneumonia were also included. All clinical strains were procured from the Department of Clinical Microbiology and Parasitology, Hospital Universiti Sains Malaysia (HUSM), Kelantan, Malaysia, and the Biobank of the Institute for Research in Molecular Medicine (INFORMM), Kelantan, Malaysia. All bacteria strains were stored in glycerol stocks at −80°C until being ready for use. Ethical clearance for this project was obtained from the Human Research Ethics Committee, Universiti Sains Malaysia (reference number USMKK/PPP/JEPeM [235.3.(16)]).
Table 2.
Evaluation of the specificities of the 6 target genes for identification of S. Typhi using PCR (total of 111 clinical isolates).
| Test bacteria strains | Positive PCR amplification for each target gene | |||||
|---|---|---|---|---|---|---|
| STY0201 | STY0307 | STY0322 | STY0326 | STY2020 | STY2021 | |
| S. Typhi (n = 39) | 39/39 | 39/39 | 39/39 | 39/39 | 39/39 | 39/39 |
| S. Paratyphi A (n = 10) | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| S. Paratyphi B (n = 10) | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| S. Paratyphi C (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Enteritidis (n = 10) | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| S. Typhimurium (n = 10) | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| S. Weltevreden (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Agona (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Hadar (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Heidelberg (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Poona (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Braenderup (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Albany (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Oslo (n = 1) | 1/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Kibi (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Newport (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Tshiongwe (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Uppsala (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Richmond (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Bardo (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Emek (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Kissi (n = 1) | 1/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Virchow (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Bordeaux (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Regent (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Java (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| S. Farsta (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Shigella dysenteriae (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Shigella flexneri (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Shigella sonnei (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Shigella boydii (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Vibrio cholerae (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Enterohemorrhagic E. coli (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Enteropathogenic E. coli (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Aeromonas hydrophila (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Yersinia enterocolitica (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Klebsiella pneumoniae (n = 1) | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
2.2. Culture Conditions and Confirmation Tests
All bacteria isolates used in this study were confirmed by traditional culture, biochemical, and serotyping methods as described in ISO6579 with some modifications. Bacteria isolates were revived from frozen glycerol stocks by pipetting 100 μL thawed cells into 10 mL nutrient broth and incubated at 37°C for 18 hours in an orbital shaker at 200 rpm. The bacteria were streaked on Xylose Lysine Deoxycholate (XLD) selective agar and incubated at 37°C for 18 hours. Colonies grown on the agar were tested with a panel of biochemical tests, including Triple Sugar Iron (TSI), urease, Methyl Red Voges Proskauer (MRVP), citrate, and indole tests. Suspected Salmonella isolates were then sent to the Salmonella Reference Centre, Institute for Medical Research (IMR), Malaysia, to confirm their serotypes using specific antisera and latex agglutination method.
2.3. Identification of S. Typhi-Specific Genes Using Bioinformatics (In Silico)
Full genome sequence of S. Typhi CT18 (GenBank accession number AL513382) was downloaded from the National Center for Biotechnology Information database (NCBI) and used as the reference genome. The 2 plasmids, namely, pHCM1 and pHCM2, which resided in S. Typhi CT18 were excluded since plasmids are genetically unstable. The 6 complete S. Typhi whole-genome sequences available in NCBI were used for data mining. They comprised CT18 (Genbank accession number AL513382) [27], Ty2 (Genbank accession number AE014613) [28], P-stx-12 (Genbank accession number CP003278) [29], Ty21a (Genbank accession number CP002099) [30], B/SF/13/03/195 (Genbank accession number CP012151) [31], and PM016/13 (Genbank accession number CP012091) [32]. In order to ascertain whether the genomic regions were conserved and specific to S. Typhi, the nucleotide Basic Local Alignment Search Tool (BLASTn), a free online software for nucleic acid analysis, was used to compare the whole-genome sequence of S. Typhi CT18 with the other 5 complete S. Typhi genomes and other bacteria genomes in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Genes found in unique regions which have no nucleotide similarity with other enteric organisms were identified and retrieved from the Genebank of NCBI. These genes were further screened individually using similarity searches against the NCBI non-redundant nucleotide (nr/nt) database to reconfirm their specificities. The program was set for “somewhat similar sequences search,” which allowed nucleotide sequence matching down to 7 bases (the smaller the nucleotide size, the more sensitive the result). Realizing the high genome similarity among the enteric pathogens and the possibility that different geographical areas may result in different bacterial genotypes, only genes which have 100% sequence conservation (an E-value threshold = 0.0) in all 6 complete S. Typhi genomes and had little or no similarity (E-value threshold ≥ 1e −41) to other bacterial sequences in the NCBI database were considered as potential targets and were subjected to wet-lab analysis. The experimental pipeline is as shown in Figure 1.
Figure 1.
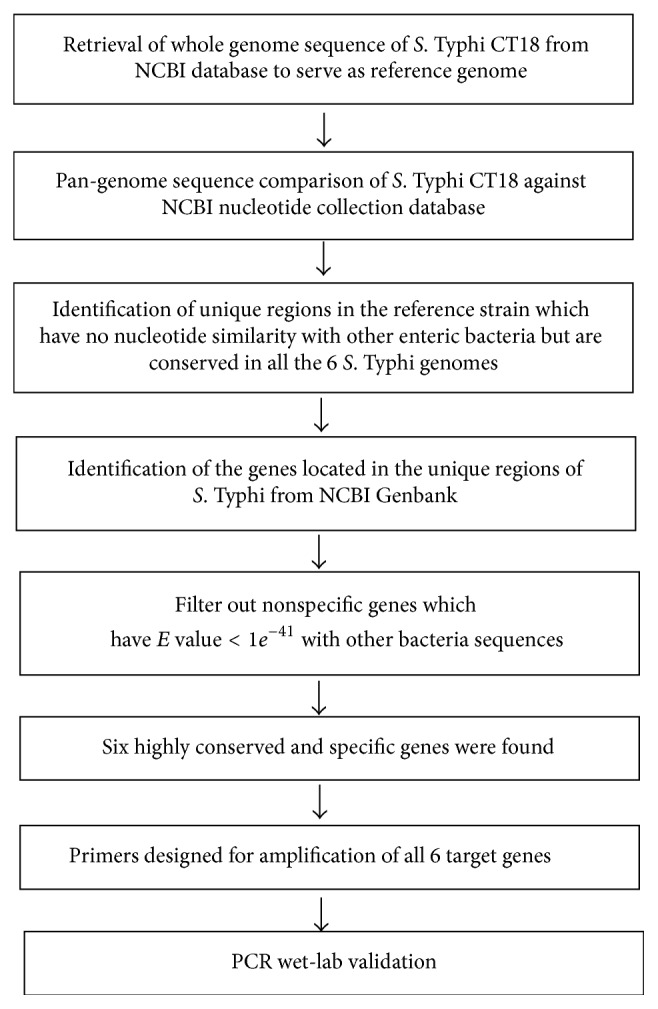
Experimental workflow describing the comparative genomic and wet-lab approaches used to identify and validate S. Typhi-specific DNA diagnostic markers.
2.4. Design of Oligonucleotide Primers for PCR Amplification
Primers were designed manually to amplify the S. Typhi-specific genes identified previously, including the start and the stop codons. A pair of primers specific for 16S rRNA gene amplification as described by Marchesi and colleagues [24] were also incorporated into each PCR assay to serve as an internal amplification control (IAC). This is a universal gene target which is highly conserved in bacteria [24]. All primers were synthesized by Integrated DNA Technologies (IDT) Pte. Ltd., Malaysia.
2.5. Template DNA Extraction
DNA from all bacteria isolates were extracted using DNeasy Blood & Tissue kit® (Qiagen, USA) according to the manufacturer's instructions. The purity and concentration of the extracted DNA were determined using Nanodrop Spectrophotometer ND-1000 (Thermo Fisher Scientific, USA). DNA concentration was measured from the absorbance at 260 nm. Ratio of the absorbance at 260 and 280 nm (A 260/280) and ratio of the absorbance at 230 and 260 nm (A 230/260) were used to evaluate the DNA quality. The extracted DNAs were diluted to a final stock concentration of 50 ng/μL using ultrapure water and stored at −20°C until ready for PCR amplification.
2.6. Optimization of PCR
Each PCR assay was optimized using a modified Taguchi method as described by Cobb and Clarkson [33]. The effects and interactions of the 4 main PCR components (IAC primers, S. Typhi-specific gene primers, MgCl2, and annealing temperatures) each at 3 different levels (IAC primers: 0.05, 0.10, and 0.15 μM; S. Typhi primers: 1.00, 1.50, and 2.00 μM; MgCl2: 2.00, 2.50, and 3.00 mM, and annealing temperatures: 50, 55, and 60°C) were investigated in a balanced orthogonal array of 9 experimental combinations. The PCR amplifications were carried out in a total reaction volume of 20 μL, and the PCR products were analysed on a 1.2% (w/v) agarose gel containing SYBR® Safe DNA Gel Stain (Invitrogen, USA), visualized using a blue-light transilluminator (Syngene, UK).
2.7. Analytical Specificities of Genes Unique to S. Typhi
Analytical specificities of the PCR assays were assessed by running each PCR assay on a panel of bacteria strains consisting of 39 S. Typhi, 62 non-Typhi Salmonella, and 10 non-Salmonella clinical isolates.
2.8. Detection Limit of the PCR Assays
Detection limit of the PCR assays was defined as the minimum amount of S. Typhi DNA (ng/μL) that yielded positive PCR amplicons. The assay sensitivities were determined by amplification of a 5-fold serial dilution of S. Typhi ATCC 7251 DNA, ranging from 50 ng to 25.6 fg. Two microliters of the DNA was subjected to PCR amplification. The analytical sensitivity was indicated by the presence of visible PCR product bands on the agarose gel using the transilluminator as described above.
2.9. DNA Sequencing
To confirm the PCR products were indeed derived from the S. Typhi strains, PCR amplicons from all assays produced using Phusion® High-Fidelity DNA Polymerase (New England Biolabs, USA) were purified and sent to First BASE Laboratories Pte. Ltd., Malaysia, for sequencing. The resultant nucleotide sequences were compared with the reference S. Typhi CT18 gene sequences in NCBI using BioEdit software.
3. Results
Using the bioinformatic method for whole-genome comparison (Figure 1), 6 potential diagnostic markers with NCBI locus tags, STY0201, STY0307, STY0322, STY0326, STY2020, and STY2021, were found. They exhibit 100% query coverage and identity (E-value = 0) with all 6 S. Typhi gene sequences but had low or no significant similarity (E-value ≥ 1e −41) with other enteric bacteria nucleotide sequences as of 11 March 2016. These genes were found to be (bioinformatically) highly conserved and specific and thus were selected for further wet-lab validation using PCR method. The primers designed to amplify these selected genes are shown in Table 1.
Table 1.
List of primers targeting S. Typhi-specific genes for the development of 6 PCR assays.
| Target genes | Primer labels | Primer sequences (5′-3′) | Target lengths (bp) |
|---|---|---|---|
| STY0201 | 0201F | ATGCTTTTAAAAAACACAACATGG | 1176 |
| 0201R | TTACGGATAGGTGATTGAAAATTG | ||
| STY0307 | 0307F | ATGAAACCTTTATTCTCAGTGC | 495 |
| 0307R | TTAGCGTAATTCCCAGAACC | ||
| STY0322 | 0322F | ATGAAATATAAAAAAATAAGAG | 678 |
| 0322R | CTATGGATTCATTTCCATTTC | ||
| STY0326 | 0326F | ATGAATACGAATAATTCACC | 261 |
| 0326R | TTACCCTCCCCATGTCAC | ||
| STY2020 | 2020F | ATGCCTGTTATGCATAATTG | 429 |
| 2020R | TTATGCTGTTAACGAGTCGTC | ||
| STY2021 | 2021F | ATGAGTTTAGCGCAGCCTAAATCC | 732 |
| 2021R | TTAGAAGTCTCCTGCCTGGAAAC | ||
| 16S rRNAa | 16SF | CAGGCCTAACACATGCAAGTC | 1362 |
| 16SR | GGGCGGTGTGTACAAGGC |
a16S ribosomal RNA gene served as internal amplification control (IAC) [24].
F represents forward primer.
R represents reverse primer.
The results showed that all 6 designed primer pairs successfully amplified their target genes with amplicon sizes of 1176, 495, 678, 261, 429, and 732 bps, respectively. DNA sequencing results of the amplicons showed 100% identity with their corresponding S. Typhi genes, confirming the fidelity and sensitivity of the primers.
The 6 single-gene target PCR assays were then optimized using Taguchi method with the incorporation of IAC which targeted the 16S rRNA gene. The optimized master mix for the PCR assays targeting STY0201, STY0307, and STY2020 genes consisted of 1x Green GoTaq Flexi Buffer, 2.0 mM MgCl2, 0.2 mM dNTPs, 1.5 μM S. Typhi-specific gene primers, 0.10 μM IAC primers, 0.75 U GoTaq Flexi DNA Polymerase (Promega, USA), and 5% glycerol in a total volume of 20 μL. Two microliters of test DNA (50 ng/μL) was added to the master mix and amplified using the following optimized thermal-cycling parameters: initial denaturation at 95°C for 1 min, followed by 30 cycles elongation at 95°C for 30 s, 55°C for 30 s, 72°C for 1 min and a final extension at 72°C for 5 min. Similar PCR conditions were used for amplification of STY0322, STY0326, and STY2021 genes except for the concentration of MgCl2 and IAC primers which were set at 3.0 mM and 0.15 μM, respectively. The optimal annealing temperature was set at 50°C. Under these conditions, the IAC primer pair produced an amplicon of 1,362 bp for all bacteria isolates tested (111/111).
The optimized PCR assays for STY0307, STY0322, STY0326, STY2020, and STY2021 correctly identified all S. Typhi (39/39) isolates, whereas none of the non-Typhi Salmonella (0/62) and none of the non-Salmonella (0/10) isolates were detected. This showed a 100% sensitivity and 100% specificity for the PCR assays (Table 2) and indicate that the 5 genes were unique to S. Typhi (Figures 2, 3, and 4).
Figure 2.
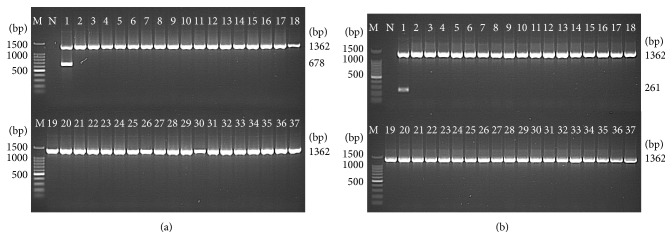
Analytical specificity of the PCR assay for detection of (a) STY0201 and (b) STY0307 genes (representative figures). Lane: M = 100 bp DNA ladder (Promega); N = negative control; 1 = S. Typhi; 2 = S. Paratyphi A; 3 = S. Paratyphi B; 4 = S. Paratyphi C; 5 = S. Enteritidis; 6 = S. Typhimurium; 7 = S. Weltevreden; 8 = S. Agona; 9 = S. Heidelberg; 10 = S. Poona; 11 = S. Hadar; 12 = S. Braenderup; 13 = S. Albany; 14 = S. Oslo; 15 = S. Kibi; 16 = S. Newport; 17 = S. Tshiongwe; 18 = S. Uppsala; 19 = S. Richmond; 20 = S. Bardo; 21 = S. Emek; 22 = S. Kissi; 23 = S. Virchow; 24 = S. Bordeaux; 25 = S. Regent; 26 = S. Java; 27 = S. Farsta; 28 = Shigella dysenteriae; 29 = Shigella flexneri; 30 = Shigella sonnei; 31 = Shigella boydii; 32 = Vibrio cholerae; 33 = Enterohemorrhagic Escherichia coli; 34 = Enteropathogenic Escherichia coli; 35 = Aeromonas hydrophila; 36 = Yersinia enterocolitica; and 37 = Klebsiella pneumonia. The PCR amplicon sizes for genes 16S rRNA, STY0201, and STY0307 were 1326 bp, 1176 bp, and 732 bp, respectively.
Figure 3.
Analytical specificity of the PCR assay for detection of (a) STY0322 and (b) STY0326 genes, respectively (representative gels). Lane: M = 100 bp DNA ladder (Promega); N = negative control; 1 = S. Typhi; 2 = S. Paratyphi A; 3 = S. Paratyphi B; 4 = S. Paratyphi C; 5 = S. Enteritidis; 6 = S. Typhimurium; 7 = S. Weltevreden; 8 = S. Agona; 9 = S. Heidelberg; 10 = S. Poona; 11 = S. Hadar; 12 = S. Braenderup; 13 = S. Albany; 14 = S. Oslo; 15 = S. Kibi; 16 = S. Newport; 17 = S. Tshiongwe; 18 = S. Uppsala; 19 = S. Richmond; 20 = S. Bardo; 21 = S. Emek; 22 = S. Kissi; 23 = S. Virchow; 24 = S. Bordeaux; 25 = S. Regent; 26 = S. Java; 27 = S. Farsta; 28 = Shigella dysenteriae; 29 = Shigella flexneri; 30 = Shigella sonnei; 31 = Shigella boydii; 32 = Vibrio cholerae; 33 = Enterohemorrhagic Escherichia coli; 34 = Enteropathogenic Escherichia coli; 35 = Aeromonas hydrophila; 36 = Yersinia enterocolitica; and 37 = Klebsiella pneumonia. The PCR amplicon sizes for genes 16S rRNA, STY0322, and STY0326 were 1,326 bp, 678 bp, and 261 bp, respectively.
Figure 4.
Analytical specificity of the PCR assay for detection of (a) STY2020 and (b) STY2021 genes, respectively (representative figures). Lane: M = 100 bp DNA ladder (Promega); N = negative control; 1 = S. Typhi; 2 = S. Paratyphi A; 3 = S. Paratyphi B; 4 = S. Paratyphi C; 5 = S. Enteritidis; 6 = S. Typhimurium; 7 = S. Weltevreden; 8 = S. Agona; 9 = S. Heidelberg; 10 = S. Poona; 11 = S. Hadar; 12 = S. Braenderup; 13 = S. Albany; 14 = S. Oslo; 15 = S. Kibi; 16 = S. Newport; 17 = S. Tshiongwe; 18 = S. Uppsala; 19 = S. Richmond; 20 = S. Bardo; 21 = S. Emek; 22 = S. Kissi; 23 = S. Virchow; 24 = S. Bordeaux; 25 = S. Regent; 26 = S. Java; 27 = S. Farsta; 28 = Shigella dysenteriae; 29 = Shigella flexneri; 30 = Shigella sonnei; 31 = Shigella boydii; 32 = Vibrio cholerae; 33 = Enterohemorrhagic Escherichia coli; 34 = Enteropathogenic Escherichia coli; 35 = Aeromonas hydrophila; 36 = Yersinia enterocolitica; and 37 = Klebsiella pneumonia. The PCR amplicon sizes for genes 16S rRNA, STY2020, and STY2021 were 1,326 bp, 429 bp, and 732 bp, respectively.
The results of serial dilution of S. Typhi genomic DNA showed that the detection limit of the optimized PCR assays was 32 pg for gene STY0322, 6.4 pg for genes STY0326, STY2020, and STY2021, and 1.28 pg for gene STY0307.
Although gene STY0201 exhibited 100% sensitivity (detection of 39/39 S. Typhi isolates), it showed cross-reactivity with S. Oslo and S. Kissi (Table 2), resulting in a specificity of only 97.2% (detection of 2/72 of non-Typhi isolates). Sequencing of their PCR products showed a substitution of nucleotide C → T at position 89 and T → C at positions 354 and 1,026 for both S. Kissi and S. Oslo. The sequence variation between S. Kissi and S. Oslo with the S. Typhi CT18 reference genome was very small (only 3 nucleotide differences), indicating that the false positive results were due to sequence similarity among themselves.
4. Discussion
The diagnosis of typhoid fever based on clinical signs and symptoms is often ambiguous, while phenotypic detection of S. Typhi bacteria based on biochemical and serotyping methods is laborious and time-consuming. Thus, rapid molecular detection methods, such as nucleic acid-based amplification, such as PCR assay, is critically needed to help diagnose this contagious disease. Development of this test requires diagnostic markers that are sensitive and specific.
This is the first report on the use of genes STY0307, STY0322, STY0326, STY2020, and STY2021 as S. Typhi-specific diagnostic markers. Unlike other S. Typhi PCR targets that were selected based on immunological properties, these genes are individually highly specific for S. Typhi and therefore can be used as single-gene target PCR assays without the need for nested or multiplex PCR. Also, these targets are whole gene sequences (from start to stop codon for the purpose of whole gene amplification) unlike other diagnostic markers which are only partial gene sequences. The idea of using this strategy is that if the whole gene sequence is specific to the bacteria then primers can be designed at any location of the gene. Thus, these gene sequences not only serve as specific targets for PCR assay, but also are suitable for more advance diagnostic tests that require multiple DNA sites, such as loop-mediated isothermal amplification (LAMP) and strand displacement amplification (SDA) which requires multiple primer annealing sites [34]. These genes could be utilized for the development of innovative Point-of-Care (POC) diagnostics to address the need for low-cost, simple, rapid, and accurate diagnostics for low resource settings.
The gene STY0201 has been used as a PCR target, and the PCR assays that were developed based on this gene were reported to be 100% sensitivity and specificity [35, 36]. However, this study found that this gene was only 97.2% specific and cross-reacted with S. Oslo and S. Kissi. The incorrect bioinformatic prediction of the specificity of gene STY0201 may be due to the incomplete genome sequence available for the 2 bacteria in the NCBI database that limit the matching accuracy of the BLASTn search. This is a limitation of the alignment-based marker identification method, as it relies on the availability of a complete genome sequence. Thus, whenever new sequence data becomes available for the target organism, the bioinformatic analysis should be repeated to align the current diagnostic markers with the new gene sequence to ensure the specificity.
The other 5 genes identified in this study showed no sequence homology to proteins of known function using protein BLAST (BLASTp) programs. Genes STY0307, STY0322, and STY0326 encode for hypothetical proteins, while genes STY2020 and STY2021 encode for putative bacteriophage proteins. Interestingly, genes STY0307, STY0322, and STY0326 are located in the Salmonella Pathogenicity Island 6 (SPI-6). Yet, their role in bacteria virulence and pathogenicity remains unknown. More importantly, antigenicity prediction scores using SCRATCH protein prediction software [26] showed that genes STY0201, STY0207, STY0307, STY0326, and STY2020 were highly antigenic and may have potential to serve as antigens for serodiagnosis of typhoid fever (Table 3). When compared with the deduced amino acid sequence of S. Paratyphi A, which is the closest relative of S. Typhi [37], the putative proteins showed weak or no similarity to S. Typhi (Table 3). These findings provide an opportunity for gene cloning and protein expression to investigate their serodiagnostic value for development of low-cost antibody-based diagnostic tests or vaccines for typhoid fever.
Table 3.
Details of the 5 target genes and their description, antigenicity prediction, protein coverage, and identity with S. Paratyphi A.
| Number | Target genes (NCBI locus tag) |
Gene description | GC content (%) | Antigenicity prediction∗ | Protein coverage with S. Paratyphi A (%) | Protein identity with S. Paratyphi A (%) |
|---|---|---|---|---|---|---|
| 1 | STY0307 | Hypothetical protein | 43 | 0.66 | 0 | 0 |
| 2 | STY0322 | Hypothetical protein | 29 | 0.37 | 21 | 33 |
| 3 | STY0326 | Conserved hypothetical protein | 37 | 0.79 | 0 | 0 |
| 4 | STY2020 | Putative bacteriophage protein | 42 | 0.66 | 0 | 0 |
| 5 | STY2021 | Putative bacteriophage protein | 42 | 0.27 | 0 | 0 |
∗Antigenicity of the S. Typhiproteins predicted using SCRATCH Protein Prediction software [26].
In conclusion, 5 S. Typhi-specific genes, namely, STY0307, STY0322, STY0326, STY2020, and STY2021, were found to be highly conserved among S. Typhi strains. Wet-lab experiments found no false positive reaction with non-Typhi serotypes or non-Salmonella enteric pathogens. These genes could serve as useful diagnostic markers for development of DNA-based diagnostics for sensitive and specific detection of typhoid fever.
Acknowledgments
This work was supported by Universiti Sains Malaysia (USM) Research University Individual (RUI) Grant (1001/CIPPM/812096) and the Ministry of Higher Education Prototype Research Grant Scheme (PRGS) (203/CIPPM/6740030). Yuan Xin Goay was a recipient of the National Science Fellowship (NSF) (M/0071/03/2010/S&T) and USM Fellowship [P-NFD0004/12(R)]. The authors would like to thank the Department of Clinical Microbiology and Parasitology, Hospital Universiti Sains Malaysia (HUSM), and INFORMM Biobank, Kelantan, Malaysia, for providing the bacterial strains.
Competing Interests
The authors declare there is no conflict of interests regarding the publication of this paper.
References
- 1.Akiba M., Kusumoto M., Iwata T. Rapid identification of Salmonella enterica serovars, typhimurium, choleraesuis, infantis, hadar, enteritidis, dublin and gallinarum, by multiplex PCR. Journal of Microbiological Methods. 2011;85(1):9–15. doi: 10.1016/j.mimet.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Buckle G. C., Walker C. L., Black R. E. Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010. Journal of Global Health. 2012;2, article 10401 doi: 10.7189/jogh.01.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darton T. C., Blohmke C. J., Pollard A. J. Typhoid epidemiology, diagnostics and the human challenge model. Current Opinion in Gastroenterology. 2014;30(1):7–17. doi: 10.1097/mog.0000000000000021. [DOI] [PubMed] [Google Scholar]
- 4.Ja'afar J. N., Goay Y. X., Mohammed Zaidi N. F., et al. Epidemiological analysis of typhoid fever in Kelantan from a retrieved registry. Malaysian Journal of Microbiology. 2013;9(2):147–151. [Google Scholar]
- 5.Nga T. V. T., Karkey A., Dongol S., et al. The sensitivity of real-time PCR amplification targeting invasive Salmonella serovars in biological specimens. BMC Infectious Diseases. 2010;10, article 125 doi: 10.1186/1471-2334-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganjali M. D., Abdeshahian P., Sudesh K., Phua K. K. Optimization of Salmonella Typhi biofilm assay on polypropylene microtiter plates using response surface methodology. Biofouling. 2016;32(4):477–487. doi: 10.1080/08927014.2015.1135328. [DOI] [PubMed] [Google Scholar]
- 7.Zaki S. A., Karande S. Multidrug-resistant typhoid fever: a review. Journal of Infection in Developing Countries. 2011;5(5):324–337. doi: 10.3855/jidc.1405. [DOI] [PubMed] [Google Scholar]
- 8.Ugochukwu A. I., Amu O. C., Nzegwu M. A. Ileal perforation due to typhoid fever—review of operative management and outcome in an urban centre in Nigeria. International Journal of Surgery. 2013;11(3):218–222. doi: 10.1016/j.ijsu.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi M., Natori T., Kubota-Hayashi S., et al. A new protocol to detect multiple foodborne pathogens with PCR dipstick DNA chromatography after a six-hour enrichment culture in a broad-range food pathogen enrichment broth. BioMed Research International. 2013;2013:10. doi: 10.1155/2013/295050.295050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar G., Pratap C. B., Mishra O. P., Kumar K., Nath G. Use of urine with nested PCR targeting the flagellin gene (fliC) for diagnosis of typhoid fever. Journal of Clinical Microbiology. 2012;50(6):1964–1967. doi: 10.1128/jcm.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Xu Z. Q., Niu P. H., et al. A two-tube multiplex reverse transcription PCR assay for simultaneous detection of viral and bacterial pathogens of infectious diarrhea. BioMed Research International. 2014;2014:9. doi: 10.1155/2014/648520.648520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirose K., Itoh K.-I., Nakajima H., et al. Selective amplification of tyv (rfbE), prt (rfbS), viaB, and fliC genes by multiplex PCR for identification of Salmonella enterica serovars Typhi and Paratyphi A. Journal of Clinical Microbiology. 2002;40(2):633–636. doi: 10.1128/jcm.40.02.633-636.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song J.-H., Cho H., Park M. Y., Na D. S., Moon H. B., Pai C. H. Detection of Salmonella typhi in the blood of patients with typhoid fever by polymerase chain reaction. Journal of Clinical Microbiology. 1993;31(6):1439–1443. doi: 10.1128/jcm.31.6.1439-1443.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolyva S., Waxin H., Popoff M. Y. The Vi antigen of Salmonella typhi: molecular analysis of the viaB locus. Journal of General Microbiology. 1992;138(2):297–304. doi: 10.1099/00221287-138-2-297. [DOI] [PubMed] [Google Scholar]
- 15.Frankel G., Newton S. M., Schoolnik G. K., Stocker B. A. Intragenic recombination in a flagellin gene: characterization of the H1-j gene of Salmonella typhi . The EMBO Journal. 1989;8:3149–3152. doi: 10.1002/j.1460-2075.1989.tb08468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seth-Smith H. M. SPI-7: salmonella's Vi-encoding Pathogenicity Island. Journal of Infection in Developing Countries. 2008;2(4):267–271. doi: 10.3855/jidc.220. [DOI] [PubMed] [Google Scholar]
- 17.Daniels E. M., Schneerson R., Egan W. M., Szu S. C., Robbins J. B. Characterization of the Salmonella paratyphi C Vi polysaccharide. Infection and Immunity. 1989;57:3159–3164. doi: 10.1128/iai.57.10.3159-3164.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A., Balachandran Y., Gupta S., Khare S., Suman Quick PCR based diagnosis of typhoid using specific genetic markers. Biotechnology Letters. 2010;32(5):707–712. doi: 10.1007/s10529-010-0211-2. [DOI] [PubMed] [Google Scholar]
- 19.Khan S., Harish B. N., Menezes G. A., Acharya N. S., Parija S. C. Early diagnosis of typhoid fever by nested PCR for flagellin gene of salmonella enterica serotype typhi. Indian Journal of Medical Research. 2012;136(5):850–854. [PMC free article] [PubMed] [Google Scholar]
- 20.Agron P. G., Walker R. L., Kinde H., et al. Identification by subtractive hybridization of sequences specific for salmonella enterica serovar enteritidis. Applied and Environmental Microbiology. 2001;67(3–12):4984–4991. doi: 10.1128/aem.67.11.4984-4991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leekitcharoenphon P., Nielsen E. M., Kaas R. S., Lund O., Aarestrup F. M. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica . PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0087991.e87991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porwollik S., Wong R. M.-Y., McClelland M. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(13):8956–8961. doi: 10.1073/pnas.122153699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poly F., Threadgill D., Stintzi A. Identification of Campylobacter jejuni ATCC 43431-specific genes by whole microbial genome comparisons. Journal of Bacteriology. 2004;186(14):4781–4795. doi: 10.1128/jb.186.14.4781-4795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchesi J. R., Sato T., Weightman A. J., et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Applied and Environmental Microbiology. 1998;64(2):795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamaruzzaman N. F., Jaafar N. J., Hani M. H., et al. Pulsed-field gel electrophoresis analysis of Salmonella enterica serovar typhi isolates in the north-east region of Peninsular Malaysia between 2002 and 2009. Journal of Applied Life Sciences International. 2016;5(2):2394–1103. doi: 10.9734/jalsi/2016/25486. [DOI] [Google Scholar]
- 26.Magnan C. N., Zeller M., Kayala M. A., et al. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010;26(23):2936–2943. doi: 10.1093/bioinformatics/btq551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkhill J., Dougan G., James K. D., et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413(6858):848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 28.Deng W., Liou S.-R., Plunkett G., III, et al. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. Journal of Bacteriology. 2003;185(7):2330–2337. doi: 10.1128/jb.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ong S. Y., Pratap C. B., Wan X., et al. Complete genome sequence of Salmonella enterica subsp. Enterica serovar typhi P-stx-12. Journal of Bacteriology. 2012;194(8):2115–2116. doi: 10.1128/JB.00121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu D., Cisar J. O., Poly F., et al. Genome sequence of Salmonella enterica serovar Typhi oral vaccine strain Ty21a. Genome Announcements. 2013;1(4) doi: 10.1128/genomeA.00650-13.e00650-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harish S. M., Sim K. S., Mohd Nor F., et al. Complete genome sequence of Salmonella enterica subsp. enterica serovar Typhi isolate B/SF/13/03/195 associated with a Typhoid carrier in pasir mas, Kelantan, Malaysia. Genome Announcements. 2015;3(6) doi: 10.1128/genomeA.01285-15.e01285-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhamad Harish S., Sim K.-S., Najimudin N., Aziah I. Genome sequence of Salmonella enterica subsp. enterica serovar Typhi isolate PM016/13 from untreated well water associated with a typhoid outbreak in pasir mas, Kelantan, Malaysia. Genome Announcements. 2015;3(6) doi: 10.1128/genomeA.01261-15.e01261-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cobb B. D., Clarkson J. M. A simple procedure for optimising the polymerase chain reaction (PCR) using modified Taguchi methods. Nucleic Acids Research. 1994;22(18):3801–3805. doi: 10.1093/nar/22.18.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang C.-C., Chen C.-C., Wei S.-C., Lu H.-H., Liang Y.-H., Lin C.-W. Diagnostic devices for isothermal nucleic acid amplification. Sensors (Switzerland) 2012;12(6):8319–8337. doi: 10.3390/s120608319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pratap C. B., Kumar G., Patel S. K., et al. Targeting of putative fimbrial gene for detection of S. typhi in typhoid fever and chronic typhoid carriers by nested PCR. Journal of Infection in Developing Countries. 2013;7(7):520–527. doi: 10.3855/jidc.2561. [DOI] [PubMed] [Google Scholar]
- 36.Yin Ngan G. J., Ng L. M., Lin R. T. P., Teo J. W. P. Development of a novel multiplex PCR for the detection and differentiation of Salmonella enterica serovars Typhi and Paratyphi A. Research in Microbiology. 2010;161(4):243–248. doi: 10.1016/j.resmic.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Holt K. E., Thomson N. R., Wain J., et al. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics. 2009;10, article 36 doi: 10.1186/1471-2164-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]