Abstract
Integrated approaches provide better understanding of HIV/AIDS epidemics. We optimised a multiassay algorithm (MAA) and assessed HIV incidence, correlates of recent infections, viral diversity, plus transmission clusters among participants screened for Kisumu Incidence Cohort Study (KICoS1) (2007–2009). We performed BED-CEIA, Limiting antigen (LAg) avidity, Biorad avidity, and viral load (VL) tests on HIV-positive samples. Genotypic analyses focused on HIV-1 pol gene. Correlates of testing recent by MAA were assessed using logistic regression model. Overall, 133 (12%, 95% CI: 10.2–14.1) participants were HIV-positive, of whom 11 tested recent by MAA (BED-CEIA OD-n < 0.8 + LAg avidity OD-n < 1.5 + VL > 1000 copies/mL), giving an incidence of 1.46% (95% CI: 0.58–2.35) per year. This MAA-based incidence was similar to longitudinal KICoS1 incidence. Correlates of testing recent included sexually transmitted infection (STI) treatment history (OR = 3.94, 95% CI: 1.03–15.07) and syphilis seropositivity (OR = 10.15, 95% CI: 1.51–68.22). Overall, HIV-1 subtype A (63%), D (15%), C (3%), G (1%) and recombinants (18%), two monophyletic dyads and intrinsic viral mutations (V81I, V81I/V, V108I/V and K101Q) were observed. Viral diversity mirrored known patterns in this region, while resistance mutations reflected likely non-exposure to antiretroviral drugs. Management of STIs may help address ongoing HIV transmission in this region.
Sub-Saharan Africa bears the greatest burden of the HIV/AIDS epidemic. In 2014, the number of people living with HIV in this region was approximately 25.8 million, and about 1.4 million were newly infected, accounting for two thirds of new daily infections worldwide1. In Kenya, the overall prevalence was 5.6% by 2012, with Nyanza region of western Kenya recording the highest prevalence (15.1%)2.
Recently, Kenyan leaders launched a visionary program employing geographic and populations targeted measures to achieve zero new HIV infections by 20303. While prevalence is a basic epidemiological tool in countries with >1% HIV prevalence, it is a poor measurement of finer changes in an epidemic4, and would not sufficiently support policies like the Kenyan ‘prevention-revolution’.
Characterising new HIV infections in a population provides better insights about the epidemic, including the impact of the disease on the public’s health and related prevention efforts. It also allows for identification of factors responsible for onward transmission of the disease5,6, an important prerequisite for implementing robust prevention programs. Owing to the cost-prohibitive nature of longitudinal HIV incidence studies7, several serological assays have been developed for cross-sectional estimation of HIV incidence. However, these assays falsely classify some established infections as recent, thereby overestimating population incidence8,9,10. A multi-assay algorithm (MAAs), is a hierarchical recent infection testing algorithm which employs one or more serological incidence assays, combined with viral load and other available non-serological biomarkers such as metabolites for antiretroviral drug (ARV) drugs, to reduce misclassification of chronic HIV infections as recent on the serological incidence assays11,12,13,14,15,16,17,18,19. Serological incidence assays target a biomarker that is indicative of early phase HIV infection5,20. However, changes in such biomarkers over time often vary per person21, hence the use of incidence assays at population rather than individual level.
Apart from incidence testing, techniques like genetic sequencing provide insights on vital viral properties that could boost understanding of HIV epidemics. HIV-1 exhibits high diversity with nine distinct subtypes: A–D, F–H, J and K and several circulating recombinant forms (CRFs) which show distinct geographical distribution22. In Kenya, subtype A is most predominant, however, subtypes C, D, G and CRFs are also common23,24,25. Viral diversity remains a challenge to HIV diagnosis, treatment, and other biomedical interventions26. Hence knowledge and continuous monitoring of circulating viral strains is an indispensible requisite for effective HIV prevention.
Current technological advances have made multi-pronged assessment integrating serological and molecular viral data, together with host’s clinical, demographic and socio-behavioral indices, easily attainable. This could provide highly useful and timely information on which to base formulation and review of transmission prevention initiatives. In this paper, we aimed to optimise an MAA strategy and utilise viral and host data to characterise MAA-identified recent and established HIV infections among participants screened for Kisumu HIV Incidence Cohort Study (KICoS)27.
Results
General characteristics of study participants
Of the 1106 participants, the median age was 21 years (range 16–34 years), and HIV-1 infection prevalence was 12% (n = 133) (95% CI: 10.2–14.1). Among the 133 HIV positives, 98 (73.7%) were females and 35 (26.3%) males, with an overall median age of 23 years (range 16–34 years) (Table 1). Briefly, fewer participants (286/1053, 27.2%) had acquired post secondary education and a majority (833/1099, 75.8%) had never been married. While most participants reported having had no previous treatment for sexually transmitted infections (STIs), a majority (66.2%) of the HIV infected individuals had HSV-2 co-infection. We noticed a reduced rate of reporting, especially pertaining to sexual behaviour questions, but a majority (168/235, 71.5%) of those who responded reported having received money in exchange for sex, while condom use during sex in the previous three months was low, especially among HIV positive individuals (43/110, 39.1) (Table 1).
Table 1. General characteristics of HIV infected and uninfected study participants, Kisumu Incidence Cohort Study (KICoS): 2007–2009.
| Characteristic | Total, N = 1106 | HIV positive, n = 133 | HIV negative, n = 973 |
|---|---|---|---|
| Gender | |||
| Male | 534 (48.3) | 35 (26.3) | 499 (51.3) |
| Female | 572 (51.7) | 98 (73.7) | 474 (48.7) |
| Age | |||
| 16–17 | 260 (23.5) | 10 (7.5) | 250 (25.7) |
| 18–24 | 635 (57.4) | 71 (53.4) | 564 (58.0) |
| 25–29 | 149 (13.5) | 31 (23.3) | 118 (12.1) |
| 30–34 | 62 (5.6) | 21 (15.8) | 41 (4.2) |
| Ever attended school | |||
| Yes | 625/671 (93.1) | 102/116 (87.9) | 523/555 (94.2) |
| Highest level of education | |||
| Primary | 386/1053 (36.7) | 60/118 (50.8) | 326/935 (34.9) |
| Secondary | 381/1053 (36.2) | 38/118 (32.2) | 343/935 (36.7) |
| Technical | 65/1053 (6.2) | 5/118 (4.2) | 60/935 (6.4) |
| College | 201/1053 (19.1) | 15/118 (12.7) | 186/935 (19.9) |
| University | 20/1053 (1.9) | 0 | 20/935 (2.1) |
| Marital status | |||
| Single/never married | 747/1099 (68.0) | 57/131 (43.5) | 690/968 (71.3) |
| Single/stable partner | 86/1099 (7.8) | 10/131 (7.6) | 76/968 (7.9) |
| Married | 219/1099 (19.9) | 46/131 (35.1) | 173/968 (17.9) |
| Separated/divorce | 32/1099 (2.9) | 9/131 (6.9) | 23/968 (2.4) |
| Widowed | 15/1099 (1.4) | 9/131 (6.9) | 6/968 (0.6) |
| Sex for gifts | |||
| Yes | 203/1024 (19.8) | 19/122 (15.6) | 184/902 (20.4) |
| Sex for money | |||
| Yes | 168/235 (71.5) | 21/30 (70.0) | 147/205 (71.7) |
| Any partners last 3 months | |||
| Yes | 137/197 (69.5) | 17/28 (60.7) | 120/169 (71.0) |
| Ever treated for STI last 3 months | |||
| Yes | 141/1096 (12.7) | 33/132 (25.0) | 108/964 (11.2) |
| Syphilis test result | |||
| Positive | 17 (1.5) | 7 (5.3) | 10 (1.0) |
| Negative | 1089 (98.5) | 126 (94.7) | 963 (99.0) |
| HSV-2 | |||
| Positive | 272 (24.6) | 88 (66.2) | 184 (18.9) |
| Negative | 752 (68.0) | 29 (21.8) | 723 (74.3) |
| Indeterminate | 82 (7.4) | 16 (12.0) | 66 (6.8) |
| Condom use | |||
| Yes | 435/967 (45.0) | 43/110 (39.1) | 392/857 (45.7) |
Note: Denominators indicated for variables with missing values.
Recent and established HIV-1 infections
Of the HIV-positive samples (n = 133), a total of 8 samples were excluded from the MAAs evaluation due to missing Biorad and BED-CEIA results (5 of which also lacked LAg avidity results) occasioned by sample depletion in preceding tests, resulting into 125 samples with complete incidence assays and VL results.
Considering single assays, Biorad avidity had the highest proportion of recent infections compared to the other two assays i.e. 20.0% versus 15.2% and 13.2% for BED-CEIA and LAg avidity respectively. Similarly, there was a trend where MAAs with single incidence assay plus VL generally had higher estimates of recent infections compared to MAAs with more than one incidence assay (Table 2). Narrowing our focus on the MAAs in the last category reported above, we reviewed the incidence estimates against the KICoS1 incidence (1.4%) estimated prospectively and selected the MAA in which recent infections were defined by BED-CEIA OD-n < 0.8 + LAg OD-n < 1.5 + VL > 1000 copies/mL, giving 11 (8.8%, 95% CI: 5.0–15.1) recent infections and incidence of 1.46% (95% CI: 0.58–2.35) per year.
Table 2. Performance evaluation of different single incidence assays and multiassay algorithms (MAAs), Kisumu Incidence Cohort Study (KICoS): 2007–2009.
| Assay/MAA | Recent, n (%) | w (95%CI) | Incidence-% (95% CI) |
|---|---|---|---|
| BED, n = 125 | |||
| BED only | 19 (15.2) | 300 (270–329) | 2.53 (1.35–3.71) |
| BED + VL | 16 (12.8) | 300 (270–329) | 2.13 (1.05–3.20) |
| BED + LAg + VL* | 11 (8.8) | 300 (270–329) | 1.46 (0.58–2.35) |
| LAg, n = 128 | |||
| LAg only | 17 (13.2) | 184 (161–208) | 3.60 (1.81–5.39) |
| LAg + VL | 16 (12.5) | 184 (161–208) | 3.39 (1.65–5.12) |
| Biorad, n = 125 | |||
| Biorad only | 25 (20.0) | 293 (263–323) | 3.40 (2.00–4.81) |
| Biorad + VL | 23 (18.4) | 293 (263–323) | 3.13 (1.79–4.48) |
Note: Number of persons in the survey = 1106, persons HIV negative = 973, persons HIV positive = 133. BED, BED-CEIA OD-n < 0.8; LAg, LAg avidity OD-n < 1.5; Biorad, Biorad avidity AI < 30%; VL, viral load > 1000 copies/mL; w, mean duration of recency in days; CI, confidence intervals. *selected MAA.
Of the 125 samples used in evaluating the MAAs, 114 (91.2%) (12 recent and 102 established) were concordantly classified by BED-CEIA and LAg avidity assays (Kappa score of 0.635 (P = 0.001, 95% CI: 0.429–0.841) and Pearson’s phi coefficient (ϕ) of 0.638 (P = 0.001)) (Supplementary Table S1). One of these 12 recent infections, with VL of 761 copies/mL, was reclassified as established based on the MAA’s criteria. We show the general overview of the classification of all the 125 samples by all the five parameters and by the selected MAA (Supplementary Fig. S1).
We further analysed the performance of the three incidence assays on 144 samples that were classified as established by the MAA. Biorad avidity had the highest rate of misclassification of samples as recent, followed by BED-CEIA (Table 3). In addition, VL mean values were lower among samples classified as recent infections than the established infections, for all the three incidence assays. However the differences were not statistically significant (Table 3). Although A, D and AD were the subtypes misclassified as recent, statistical analysis showed that overall misclassification by the three incidence assays were not significantly linked with viral diversity, Chi-square p-values > 0.05 (Supplementary Fig. S2).
Table 3. HIV viral load characteristics of samples classified as established by the MAA (N = 114) categorised by three single incidence assays, Kisumu Incidence Cohort Study (KICoS): 2007–2009.
| Recent |
Established |
|||
|---|---|---|---|---|
| n (%) | Viral load | n (%) | Viral load | |
| Biorad < 30 | 15 (13.2) | 81838.27 | 99 (86.8) | 161325.55 |
| BED < 0.8 | 8 (7.0) | 40014.38 | 106 (93.0) | 159232.91 |
| LAg < 1.5 | 5 (4.4) | 98989.40 | 109 (95.6) | 153246.39 |
Note: There were no differences in HIV viral load between recent and established infections (t-test).
Characteristics of individuals with multiassay algorithm-determined recent infections
Among individuals tested as recently infected on the BED + Lag + VL MAA, 7/11 (63.6%) were females, a similar number were <25 years old, and a majority (8/11, 72.7%) had never married. The most prevalent HIV subtype was A (6/11, 54.5%), followed by C and AD (both with 2/11, 18.2%), and lastly D (1/11, 9.1%) (Supplementary Table S2). In the regression model for HIV recent infection versus HIV negatives, after controlling for age, gender, sex for gifts, history of STI treatment, syphilis and HSV-2, only history of STI treatment and syphilis sero-positivity explained the recent infections in this population (Table 4). Persons reporting past treatment for STI were nearly 4 times more likely to be recently infected than those never treated for STI (OR = 3.94; 95% CI: 1.03–15.07), while those who tested positive for syphilis were ten times more likely to be recently infected compared to the syphilis negative (OR = 10.15; 95% CI: 1.51–68.22). Only history of STI treatment was linked to recent infection in the second model comparing recent against established infections (OR = 9.91; 95% CI: 1.55–63.46) (Supplementary Table S3).
Table 4. Factors potentially associated with testing recent on MAA as compared to HIV negative persons, Kisumu Incidence Cohort Study (KICoS): 2007–2009.
| Characteristic | Bivariate |
Multivariate |
||
|---|---|---|---|---|
| OR* (95% CI) | p-value | OR (95% CI) | p-value | |
| Gender | ||||
| Male | 1 | 1 | ||
| Female | 1.84 (0.54–6.33) | 0.332 | 1.42 (0.35–5.77) | 0.625 |
| Age | ||||
| 24–34 | 1 | 1 | ||
| 16–23 | 0.35 (0.10–1.14) | 0.082 | 0.43 (0.11–1.64) | 0.218 |
| Sex for gifts | ||||
| No | 1 | 1 | ||
| Yes | 0.39 (0.05–3.06) | 0.371 | 0.68 (0.08–5.75) | 0.723 |
| Ever treated for STI | ||||
| No | 1 | 1 | ||
| Yes | 6.60 (1.98–22.01) | 0.002 | 3.94 (1.03–15.07) | 0.045 |
| Syphilis test result | ||||
| Negative | 1 | 1 | ||
| Positive | 0.046 (0.01–0.24) | 0.001 | 10.15 (1.51–68.22) | 0.017 |
| HSV–2 | ||||
| Negative | 1 | 1 | ||
| Positive | 4.91 (1.31–18.47) | 0.019 | 2.64 (0.60–11.55) | 0.199 |
| Indeterminate | 5.47 (0.98–30.47) | 0.052 | 4.11 (0.68–25.00) | 0.125 |
¥Odds ratios with 95% confidence intervals (CI).
Viral subtyping analysis
Of the 133 HIV-positive samples, a total of 100 (75.2%) samples were successfully genotyped (GenBank accession numbers KX306376-KX306475). The remaining 33 samples included: 14 with insufficient plasma quantities, 5 with viral load <1000 copies/mL (the amplification sensitivity threshold for sequencing), 9 with undetectable viral load, and 5 that failed amplification.
Overall, the predominant HIV-1 subtype was A (63%), followed by D (15%), C (3%) and G (1%). Recombinant variants constituted 18% of all the samples with AD dominating at 15%, followed by AC (2%) and AG (1%) (Fig. 1a). Subtypes G, AC and AG, were only present in established infections (Fig. 1b). There was no significant difference in subtype distribution between recent and established infections.
Figure 1. HIV subtype distribution among study participants (n = 100).
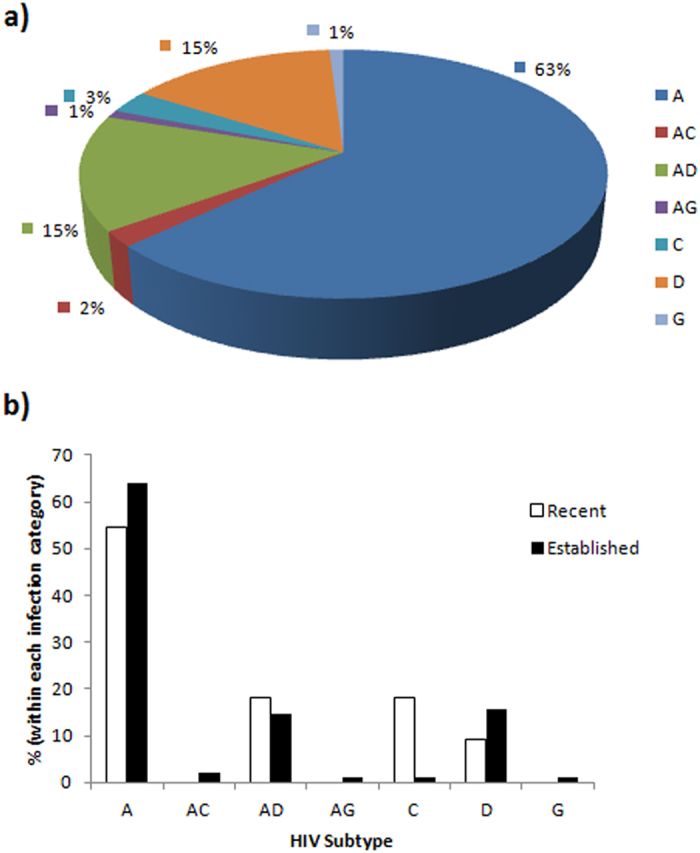
(A) overall distribution. (B) Categorised by recent (open bars, n = 11) and established (closed bars, n = 89) HIV infections, Kisumu Incidence Cohort Study (KICoS): 2007–2009.
HIV transmission network analysis
Evaluation of evolutionary relatedness among 98 samples (after removing two sequences with sequence quality issues) revealed two monophyletic dyads (Fig. 2) of HIV-1 subtype A and AD. Each dyad had an older male and a younger female. One dyad had single individuals while individuals in the other dyad were both in married relationships. All the individuals had MAA-classified established infections characterised by higher VL in males compared to females. Discordant variable outcomes within dyads, for instance condom use, imply the possibility of transmission networks being larger than captured by this study. Additionally, the small size of the dyads could be due to limited sample size and not an indication of transmission pairs (Supplementary Table S4).
Figure 2. Molecular phylogenetic analysis for HIV transmission clustering.
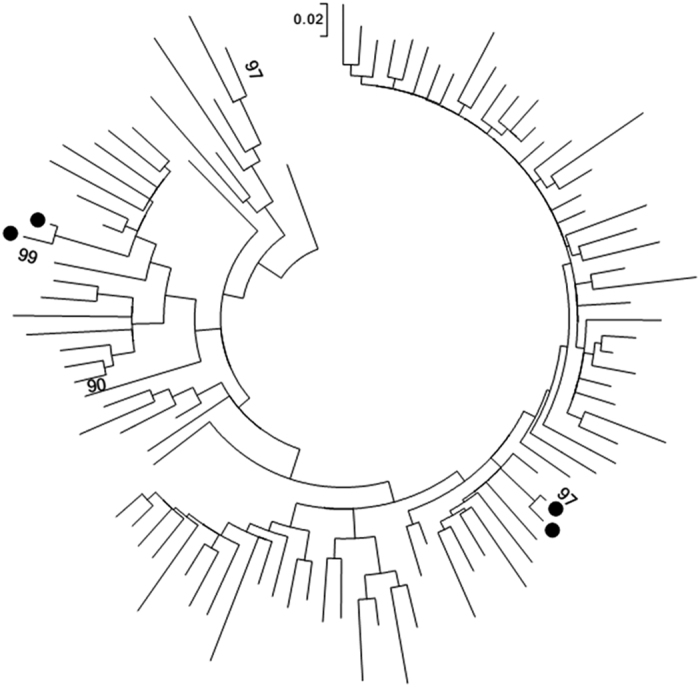
Evolutionary history was inferred using Maximum Likelihood method based on the Tamura-Nei model in Mega v.7.0. The tree with the highest log likelihood is shown. Nodes with high bootstrap values indicated. Black dots represent individuals in transmission clusters.
Viral resistance mutations analysis
Observed viral mutations included V81I and V81I/V in the protease gene, and V108I/V and K101Q in the reverse transcriptase gene, all of which were intrinsic mutations not associated with ARV exposure. Four of the five individuals bearing the mutations were females, while two individuals (one male) had recent infections (Table 5).
Table 5. Characteristics of individuals with HIV resistance mutations, Kisumu Incidence Cohort Study (KICoS): 2007–2009.
| Individual | Resistance mutation |
HIV-1 subtype | Duration of infection | Gender | |
|---|---|---|---|---|---|
| PR | RT | ||||
| 1 | V82I/V | — | C | Recent | Female |
| 2 | V82I | V108I/V | AD | Recent | Male |
| 3 | V82I | — | G | Established | Female |
| 4 | — | K101Q | AD | Established | Female |
| 5 | — | K101Q | D | Established | Female |
Abbreviations: PR = protease gene, RT = reverse transcriptase gene.
Discussion
This is one of the few applications of a multifactorial strategy and the recent approaches and recommendations for cross-sectional HIV incidence testing algorithms in Africa16,17. We optimised an MAA that identified potential recent HIV infections in the study population. We then analysed host factors and viral molecular properties characterising this population.
From our analysis, single incidence assays or MAAs with single incidence assay plus one or two non-serological biomarkers seemed to give poor outcomes compared to MAAs integrating at least two incidence assays and one or more non-serological biomarkers. This pattern generally corroborates the findings from previous evaluations of single incidence assays and MAAs in different setups8,12,17,28,29. Single incidence assays in our study gave more than twofold higher incidence estimates compared to the longitudinal KICoS1 incidence. This emphasises the inappropriateness of using the current incidence assays singly in cross-sectional incidence studies9,13. The selected MAA comprised of BED-CEIA, a relatively inexpressive and widely used assay28, plus LAg avidity, an assay with commendable performance in different epidemics, and VL, a test that is currently recommended for incidence MAAs9. The concordance between BED-CEIA and LAg avidity assays was statistically significant, with modest Pearson and Kappa statistical values. With expansion of capacity for dry blood spot specimens and point of care VL testing, such MAA will even be more attainable in resource limited settings.
In our study, HIV prevalence was approximately 12% (95% CI: 10.2–14.1), about threefold higher than the national prevalence of 4.04% (95%CI: 3.6–4.53) for persons aged 15–34 years in Kenya in 201230. Coupled with the high prevalence of 14.9% (KAIS 2007)31 and 15.1% (KAIS 2012)30 reported for Nyanza, these statistics collectively emphasise the large HIV burden in this region, hence the need for well designed prevention strategies. As commonly observed in other parts of sub-Saharan Africa, HIV was more prevalent in females in this study than males32,33, potentially due to disproportionate social and biological factors influencing vulnerability as previously reported34, and possibly an imbalance in health seeking behaviours.
According to Kenya’s census projections from 2000 to 2020, the total population of persons aged 15–34 years in Kisumu in 2007 was 233,57035. Assuming that the 2007 HIV incidence rate among persons aged 15–34 years was 1.46% in Kisumu based on the MAA, approximately 3,400 persons in Kisumu aged 15–34 years had recent HIV infection. This is approximately 3% of the estimated number of recent HIV cases that occurred in Kenya among persons aged 15–49 years which ranged from 120,000 to 140,000 in 200736. While this may not be an exact estimate due to design differences, it illustrates the case of ongoing HIV transmission in this area despite HIV prevention efforts. While HIV risk patterns appear similar between established and recent infections, symptomatic and diagnosed STI infections could be among key factors driving new HIV transmission in this younger population. This is consistent with HIV risk factor analysis reported previously34. Two recently published studies also identified history of STI as a risk factor for recent HIV infections in both rural western Kenya and the country as a whole16,25. Prompt diagnosis and treatment of STIs, accompanied with risk reduction counselling remain vital to the success of HIV prevention initiatives in this setting. Additional efforts are needed to share risk knowledge with younger adults who are sexually active and promote early HIV testing.
The overall distribution of HIV-1 subtypes in our study was synonymous to patterns earlier reported for this region24,25. Although slight changes were observed between recent and established infections, where some subtypes were apparently lower (A and D) or not found (AC, AG and G), while AD and C apparently increased among recent infections, the variations were not statistically significant, implying a mature epidemic. Additionally, Kenya is bordered by five countries with variable distribution of HIV-1 subtypes. Subtypes C and AC dominate in Somalia and Ethiopia, C and D in Sudan, while A and D are the most common subtypes in Uganda. Tanzania has subtypes A, C, D, AC, AD and CRF_CD in varying proportions37. Owing to the ongoing regional integration among East African states, frequent transfer of different viral subtypes between states is highly likely, hence continuous monitoring of HIV strains remains an important consideration when carrying out biological investigations in this region.
Although only polymorphic drug-resistance mutations, which have a low effect on HIV therapy, were observed in this study, such polymorphisms could lead to rapid treatment failure and development of drug resistant HIV-1 variants following initiation of therapy38. For instance, V82I polymorphism in subtype G contributes to emergence of I82M/T/S resistance after protease inhibitor based treatment failure39. The pattern of resistance mutations in this study could be a reflection of lack of prior ARV exposure. Nevertheless, with the increase in ARV use and consequent primary drug resistance mutations in Kenya as well as southern and eastern Africa regions, the importance of frequent drug resistance surveillance cannot be overstated40,41.
Finally, the small sample size of recent infections, and the convenience sampling method employed to screen participants for KICoS1, may have affected the statistical power of various variables. We also lacked professional panels to generate local incidence assays window periods. Although we utilised longitudinal KICoS1 incidence to validate the derived cross-sectional incidences, the possibility of misclassification by the MAA cannot be completely ruled out. These factors may reduce the representativeness of our findings.
Conclusion
In summary, our study presents an MAA that estimated cross-sectional HIV incidence with perfect concordance to longitudinal incidence, with a mean recency of infection below one year. This offers important insights on the performance of MAAs in local African epidemics. This MAA allowed us to demonstrate the possibility of comprehensive evaluations covering key groups in the HIV epidemic, i.e. the HIV negative, recent and established infections. This study showed that current/past STI infections could be possible independent factors for new HIV infection in this population. We observed limited viral resistance mutations, four pure HIV-1 subtypes (A, C, D, and G) plus a number of recombinant viruses, and existence of transmission clusters, consistent with previous molecular surveys in this region. Application of our strategy in larger cross-sectional studies will enable a more in-depth assessment with definite outcomes that will support progressive approaches for tackling the spread of HIV.
Methodology
Study population
As previously described27, KICoS was an observational prospective cohort study that sought to longitudinally determine HIV incidence among sexually active 16–34 year olds in Kisumu, western Kenya. A total of 1106 individuals were screened for eligibility for recruitment from 2007 to 2009. Through convenience sampling, the study enrolled male or female residents of Kisumu aged 16–34 years, sexually active in the past three months, not pregnant, and HIV uninfected27. Demographic and behavioural information was collected at screening via Audio Computer Assisted Self Interview (ACASI), followed by medical examination and testing for common sexually transmitted infections (STIs). This was termed KICoS1, followed by later design modifications to target at-risk individuals (KICoS2 and 3)42. Procedures in this study were conducted in accordance with, and under the oversight of the approving ethical bodies. In the current study, we focus on the baseline data and samples collected from persons screened for participation in KICoS1.
Sample collection
Whole blood samples were collected in vacutainer tubes with EDTA (Becton Dickinson, San Jose, CA, USA). Plasma was separated and stored frozen until use.
Viral load measurements
HIV-1 plasma RNA VL was quantified using CobasAmpliprep/CobasTaqman HIV-1 test v.2.0 according to manufacturer’s instructions (Roche Diagnostic System Branchburg, NL, USA).
Cross-sectional HIV incidence assays
HIV positive samples were tested by three incidence assays: BED-Capture enzyme immunoassay (BED-CEIA) (Calypte, USA; Sedia BioSciences, USA), Limiting antigen (LAg) avidity (Sedia BioSciences, USA), and Biorad avidity assay, modified from the Genetic Systems 1/2 + O ELISA (Bio-Rad Laboratories, Redmond, WA). The BED-CEIA and LAg avidity were performed according to manufacturer’s instructions as previously described5,20,43. Normalised optical density (OD-n) <0.8 and <1.5 represented recent infections on BED-CEIA and LAg avidity respectively, while values above the cut-offs were considered established. Same was done for avidity index (AI) <30% for Biorad avidity assay.
HIV genetic sequencing
Protease (1–99 amino acids) and part of reverse transcriptase (1–250 amino acids) regions of HIV-1 were sequenced by a broadly sensitive in-house assay as previously described44. Briefly, HIV-1 RNA was extracted using QiaAmp Viral RNA mini kit following manufacturer’s instruction (QiagenInc, Chatsworth, CA). Using primers spanning the target pol region, RT-PCR and nested PCR were conducted sequentially followed by Big Dye Terminator sequencing and resolution using an ABI 3100 Genetic Analyser (Applied Biosystems, Foster City, CA, USA). The sequences were assembled with Sequencher v.3.1 (Genecodes, Ann Arbor, MI) and quality checks done using sequence quality assessment tool (SQUAT).
To assess genetic diversity, sequences were analysed by REGA HIV-1 subtyping tool v.3.0, and further compared with NCBI-BLAST and MEGA v.7.0. Sequences showing ambiguous subtyping were selected for recombination analysis using SimPlot software v3.5.5 in a 400 base pair (bp) sliding window with 20 bp increments.
We also investigated potential existence of transmission clusters by evaluating evolutionary relatedness between the sequences in MEGA v.7. We used pair-wise Tamura Nei 93 (TN93) model, assuming gamma distribution (shape parameter = 0.3305). Potential transmission clusters were defined as ≥2 sequences with ≤1.5% genetic distance and high bootstrap values (>95%) from 1000 re-samplings45,46. The trees were rooted using subtype K reference sequence (Los Alamos Database accession number AJ249239_CM_K).
HIV viral resistance mutations were assessed by the algorithm in the Stanford University HIV Drug Resistance Database and categorised according to the International AIDS Society–USA Drug Resistance Mutations Group December 2010 updates47.
Statistical methods
From the incidence assay results, we evaluated the performance of each assay singly and in various MAAs with and without VL (cut-off of >1000 copies/mL)48. Samples with missing values by any of the four parameters (three incidence assays or VL) were excluded from this analysis. We derived mean duration of recency (w) and 95% confidence intervals (95% CI) for the three assays from a previous publication49, and estimated percent incidence by individual assays and MAAs as previously described50, assuming missing at random to adjust for the samples missing incidence test data. We transformed the five parameters’ data into binomial values according to their respective cut-offs and generated heat maps using PermutMatrix-1.9.3 to evaluate the classification of samples by the five parameters. We considered published incidence of 1.4%, derived from the longitudinal phase of KICoS142, as a guide to select suitable MAA for subsequent analyses. Kappa coefficient and Pearson’s phi coefficient (ϕ), with their respective 95% CIs and p-values, were calculated to measure agreement between incidence assays. We further sought to characterise misclassification by the three incidence assays based on the optimised MAA.
To assess factors potentially associated with the MAA-identified recent infections, we fitted two models using logistic regression for both bivariate and multivariate analyses. One model assessed recent infection versus HIV negatives and another model assessed recent infections versus established infections. All variables with bivariate p-value ≤ 0.2 or set a priori were included in the final multivariable models. In the multivariable models, covariates were added one by one to assess their individual effect on the outcome, while controlling for other covariates as potential confounders. We used likelihood ratio statistical test to select the best models. Chi-square and t-test were used to compare proportions and means respectively, considering statistical significance as p-value ≤ 0.05. Statistical analyses were performed using STATA version 13.0 (STATA Corporation, College Station, Texas, USA).
Ethical Approval
Ethical approval was obtained from the Ethical Review Committee of the Kenya Medical Research Institute (KEMRI) as well as the Institutional Review Board of the U.S. Centers for Disease Control and Prevention (CDC), Atlanta, GA, USA. Written informed consent (with assent obtained in addition to parental consent for minors) was obtained from participants.
Additional Information
Accession codes: KX306376-KX306475.
How to cite this article: Otecko, N. et al. Viral and Host Characteristics of Recent and Established HIV-1 Infections in Kisumu based on a Multiassay Approach. Sci. Rep. 6, 37964; doi: 10.1038/srep37964 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors are grateful to the study participants, the Kisumu cohort study (KICoS) team, and the Kenya Medical Research Institute and Kenya (KEMRI)/Centers for disease control and prevention (CDC) HIV Research branch for making the study possible. KEMRI/CDC staff participated in the design, data collection, analysis, and interpretation of the data; writing the report; and the decision to submit the paper for publication. This paper is published with the permission of the Director of KEMRI. The findings and conclusions in this work are those of the authors and do not necessarily represent the views of CDC. Use of trade names is for identification purposes only and does not constitute endorsement by the CDC or the Department of Health and Human Services. This work was supported by Interagency Agreement Y1-AI-7278-01/ST07015 between CDC and the National Institutes of Health (NIH).
Footnotes
Author Contributions N.O., C.Z., A.A.K. designed the study; N.O., F.O., V.O., K.W., K.N., A.M. collected data and conducted experiments; N.O., A.A.K., G.O., C.O. and S.I. analysed the data; N.O. and C.Z. wrote the paper; all authors reviewed and approved the final manuscript.
References
- UNAIDS. Core Epidemiology Slides., (Geneva. http://www.unaids.org/sites/default/files/media_asset/20150714_epi_core_en.ppt. Accessed on, 20th March, 2016, 2015).
- Kimanga D. O. et al. Prevalence and incidence of HIV infection, trends, and risk factors among persons aged 15–64 years in Kenya: results from a nationally representative study. J Acquir Immune Defic Syndr 66 Suppl 1, S13–26, doi: 10.1097/QAI.0000000000000124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- NACC. Kenya HIV prevention revolution road map: countdown to 2030. Report No. ISBN 978 99 6 603802 9, (Nairobi, Kenya. http://hivhealthclearinghouse.unesco.org/sites/default/files/resources/kenya_hiv_prevention_revolution_road_map.pdf. Accessed on 3rd March, 2016, 2014).
- Kim A. A. et al. Evaluating the BED capture enzyme immunoassay to estimate HIV incidence among adults in three countries in sub-Saharan Africa. AIDS Res Hum Retroviruses 26, 1051–1061 (2010). [DOI] [PubMed] [Google Scholar]
- Duong Y. T. et al. Detection of recent HIV-1 infection using a new limiting-antigen avidity assay: potential for HIV-1 incidence estimates and avidity maturation studies. PLoS One 7, e33328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett T. B. et al. Estimating incidence from prevalence in generalised HIV epidemics: methods and validation. PLoS Med 5, e80 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal J. S. et al. Comparison of HIV type 1 incidence observed during longitudinal follow-up with incidence estimated by cross-sectional analysis using the BED capture enzyme immunoassay. AIDS Res Hum Retroviruses 22, 945–952 (2006). [DOI] [PubMed] [Google Scholar]
- Konikoff J. et al. Performance of a limiting-antigen avidity enzyme immunoassay for cross-sectional estimation of HIV incidence in the United States. PLoS One 8, e82772, doi: 10.1371/journal.pone.0082772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS/WHO. Technical Update on HIV Incidence Assays for Surveillance and Epidemic Monitoring., (Geneva, Switzerland. http://www.aidsdatahub.org/sites/default/files/publication/HIV_incidence_assays_surveillance_monitoring_2015.pdf. Accessed on 3rd March, 2016, 2015).
- Guy R. et al. Accuracy of serological assays for detection of recent infection with HIV and estimation of population incidence: a systematic review. Lancet Infect Dis 9, 747–759 (2009). [DOI] [PubMed] [Google Scholar]
- Braunstein S. L. et al. High burden of prevalent and recently acquired HIV among female sex workers and female HIV voluntary testing center clients in Kigali, Rwanda. PLoS One 6, e24321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein S. L. et al. Dual testing algorithm of BED-CEIA and AxSYM Avidity Index assays performs best in identifying recent HIV infection in a sample of Rwandan sex workers. PLoS One 6, e18402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookmeyer R., Konikoff J., Laeyendecker O. & Eshleman S. H. Estimation of HIV incidence using multiple biomarkers. American journal of epidemiology 177, 264–272, doi: 10.1093/aje/kws436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookmeyer R., Laeyendecker O., Donnell D. & Eshleman S. H. Cross-sectional HIV incidence estimation in HIV prevention research. J Acquir Immune Defic Syndr 63 Suppl 2, S233–239, doi: 10.1097/QAI.0b013e3182986fdf (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins M. M. et al. A comparison of two measures of HIV diversity in multi-assay algorithms for HIV incidence estimation. PLoS One 9, e101043, doi: 10.1371/journal.pone.0101043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. A. et al. Identifying Risk Factors for Recent HIV Infection in Kenya Using a Recent Infection Testing Algorithm: Results from a Nationally Representative Population-Based Survey. PLoS One 11, e0155498, doi: 10.1371/journal.pone.0155498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeyendecker O. et al. Development of methods for cross-sectional HIV incidence estimation in a large, community randomized trial. PLoS One 8, e78818, doi: 10.1371/journal.pone.0078818 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeyendecker O. et al. Improved Precision of Cross-sectional HIV Incidence Testing Using a Multi-assay Algorithm that Includes BED and an Avidity Assay with Modified Assay Cut-offs. 17th Conference on Retroviruses and Opportunistic Infections. 16–19 February 2010; San Francisco, California, United States of America (2010).
- Murphy G. & Parry J. V. Assays for the detection of recent infections with human immunodeficiency virus type 1. Euro Surveill 13 (2008). [PubMed] [Google Scholar]
- Dobbs T., Kennedy S., Pau C. P., McDougal J. S. & Parekh B. S. Performance characteristics of the immunoglobulin G-capture BED-enzyme immunoassay, an assay to detect recent human immunodeficiency virus type 1 seroconversion. J Clin Microbiol 42, 2623–2628 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiamma A. et al. Can HIV incidence testing be used for evaluating HIV intervention programs? A reanalysis of the Orange Farm male circumcision trial (ANRS-1265). BMC Infect Dis 10, 137 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J., Gouws E., Ghys P. D. & Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. Aids 20, W13–23 (2006). [DOI] [PubMed] [Google Scholar]
- Lihana R. W. et al. The changing trend of HIV type 1 subtypes in Nairobi. AIDS Res Hum Retroviruses 25, 337–342 (2009). [DOI] [PubMed] [Google Scholar]
- Yang C. et al. Genetic diversity and high proportion of intersubtype recombinants among HIV type 1-infected pregnant women in Kisumu, western Kenya. AIDS Res Hum Retroviruses 20, 565–574 (2004). [DOI] [PubMed] [Google Scholar]
- Zeh C. et al. Molecular Epidemiology and Transmission Dynamics of Recent and Long-Term HIV-1 Infections in Rural Western Kenya. PLoS One 11, e0147436, doi: 10.1371/journal.pone.0147436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. S., Sobieszczyk M. E., McCutchan F. E. & Hammer S. M. The challenge of HIV-1 subtype diversity. N Engl J Med 358, 1590–1602 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbe A. et al. Correlates of prevalent HIV infection among adults and adolescents in the Kisumu incidence cohort study, Kisumu, Kenya. Int J STD AIDS 26, 929–940, doi: 10.1177/0956462414563625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeyendecker O. et al. HIV incidence determination in the United States: a multiassay approach. J Infect Dis 207, 232–239, doi: 10.1093/infdis/jis659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longosz A. F. et al. Incorrect identification of recent HIV infection in adults in the United States using a limiting-antigen avidity assay. AIDS 28, 1227–1232, doi: 10.1097/QAD.0000000000000221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- NASCOP. Kenya AIDS Indicator Survey 2012: Final Report., (Nairobi, Kenya. http://www.nacc.or.ke/index.php?option=com_content&view=article&id=403:kais-2012-final-report&catid=157:publications Accessed on 20th March, 2015, 2012).
- NASCOP. Kenya AIDS Indicator Survey 2007: Preliminary Report., (Nairobi, Kenya. http://www.wofak.or.ke/Publications/kais__preliminary_report_july_29.pdf. Accessed on 27th November, 2011, 2008).
- Buve A. et al. Multicentre study on factors determining differences in rate of spread of HIV in sub-Saharan Africa: methods and prevalence of HIV infection. Aids 15 Suppl 4, S5–14 (2001). [DOI] [PubMed] [Google Scholar]
- Joesoef M. R. et al. Differential of HIV prevalence in women and men who attended sexually transmitted disease clinics at HIV sentinel surveillance sites in Kenya, 1990-2001. Int J STD AIDS 14, 193–196 (2003). [DOI] [PubMed] [Google Scholar]
- Amornkul P. N. et al. HIV prevalence and associated risk factors among individuals aged 13-34 years in Rural Western Kenya. PLoS One 4, e6470 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Central Bureau of Statistics (CBS) [Kenya], M. o. P. a. N. D. Population Projections for Kenya 2000-2020 (Revised) (2006).
- Kim A. A. et al. Estimating HIV incidence among adults in Kenya and Uganda: a systematic comparison of multiple methods. PLoS One 6, e17535, doi: 10.1371/journal.pone.0017535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyaro M. et al. Molecular characterization of HIV type 1 among HIV-infected respondents in a cohort being prepared for HIV Phase III vaccine clinical trials, Western Kenya. AIDS Res Hum Retroviruses 27, 257–264 (2011). [DOI] [PubMed] [Google Scholar]
- Handema R. et al. Emergence of new HIV-1 subtypes other than Subtype C among antenatal women in Lusaka, Zambia. AIDS Res Hum Retroviruses 17, 759–763 (2001). [DOI] [PubMed] [Google Scholar]
- Dumans A. T. et al. Synonymous genetic polymorphisms within Brazilian human immunodeficiency virus Type 1 subtypes may influence mutational routes to drug resistance. J Infect Dis 189, 1232–1238 (2004). [DOI] [PubMed] [Google Scholar]
- Price M. A. et al. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses 27, 5–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigaloff K. C. et al. Short communication: High prevalence of transmitted antiretroviral drug resistance among newly HIV type 1 diagnosed adults in Mombasa, Kenya. AIDS Res Hum Retroviruses 28, 1033–1037 (2012). [DOI] [PubMed] [Google Scholar]
- Mdodo R. et al. Investigation of HIV Incidence Rates in a High-Risk, High-Prevalence Kenyan Population: Potential Lessons for Intervention Trials and Programmatic Strategies. Journal of the International Association of Providers of AIDS Care 15, 42–50, doi: 10.1177/2325957413511667 (2016). [DOI] [PubMed] [Google Scholar]
- Duong Y. T. et al. Recalibration of the limiting antigen avidity EIA to determine mean duration of recent infection in divergent HIV-1 subtypes. PLoS One 10, e0114947, doi: 10.1371/journal.pone.0114947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. et al. Development and application of a broadly sensitive dried-blood-spot-based genotyping assay for global surveillance of HIV-1 drug resistance. J Clin Microbiol 48, 3158–3164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G. & Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol (accepted) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. & Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular biology and evolution 10, 512–526 (1993). [DOI] [PubMed] [Google Scholar]
- Johnson V. A. et al. 2011 update of the drug resistance mutations in HIV-1. Top Antivir Med 19, 156–164 (2011). [PMC free article] [PubMed] [Google Scholar]
- Braunstein S. L. et al. Risk Factor Detection as a Metric of STARHS Performance for HIV Incidence Surveillance Among Female Sex Workers in Kigali, Rwanda. Open AIDS J 6, 112–121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassanjee R. et al. Viral load criteria and threshold optimization to improve HIV incidence assay characteristics. AIDS 30, 2361–2371, doi: 10.1097/QAD.0000000000001209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte A., McWalter T. A. & Barnighausen T. A Simplified Formula for Inferring HIV Incidence from Cross-Sectional Surveys Using a Test for Recent Infection. AIDS Res Hum Retroviruses 25, 125–126, doi: 10.1089/aid.2008.0150 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


