Abstract
Short chain fatty acids (SCFAs) produced by intestinal microbes mediate anti-inflammatory effects, but whether they impact on antimicrobial host defenses remains largely unknown. This is of particular concern in light of the attractiveness of developing SCFA-mediated therapies and considering that SCFAs work as inhibitors of histone deacetylases which are known to interfere with host defenses. Here we show that propionate, one of the main SCFAs, dampens the response of innate immune cells to microbial stimulation, inhibiting cytokine and NO production by mouse or human monocytes/macrophages, splenocytes, whole blood and, less efficiently, dendritic cells. In proof of concept studies, propionate neither improved nor worsened morbidity and mortality parameters in models of endotoxemia and infections induced by gram-negative bacteria (Escherichia coli, Klebsiella pneumoniae), gram-positive bacteria (Staphylococcus aureus, Streptococcus pneumoniae) and Candida albicans. Moreover, propionate did not impair the efficacy of passive immunization and natural immunization. Therefore, propionate has no significant impact on host susceptibility to infections and the establishment of protective anti-bacterial responses. These data support the safety of propionate-based therapies, either via direct supplementation or via the diet/microbiota, to treat non-infectious inflammation-related disorders, without increasing the risk of infection.
Host defenses against infection rely on innate immune cells that sense microbial derived products through pattern recognition receptors (PRRs) such as toll-like receptors (TLRs), c-type lectins, NOD-like receptors, RIG-I-like receptors and cytosolic DNA sensors. The interaction of microbial ligands with PRRs activates immune cells to produce immunomodulatory molecules like cytokines and co-stimulatory molecules1,2,3. Pro-inflammatory cytokines play an essential role in coordinating the development of the innate and adaptive immune responses aimed at the eradication or containment of invading pathogens. Yet, inflammation has to be timely and tightly regulated since it may become life-threatening both by default and by excess3,4,5,6.
Short chain fatty acids (SCFAs) are end products of the fermentation of resistant starches and dietary fibers by intestinal bacteria, with the most abundant metabolites produced being acetate, propionate and butyrate7. SCFAs reach elevated concentrations in the gut lumen (50–100 mM) and are absorbed into the portal circulation, acting as a source of SCFAs in the bloodstream (0.1–1 mM)8,9,10,11. SCFAs, primarily butyrate, not only serve as a source of energy, but also stimulate neural and hormonal signals regulating energy homeostasis12. Beside their trophic effects, SCFAs possess antioxidative, anticarcinogenic and anti-inflammatory properties and play an essential role in maintaining gastrointestinal and immune homeostasis7,10,11,13.
Both extracellular and intracellular SCFAs exert immunosuppressive effects. Extracellular SCFAs act through metabolite sensing G-protein coupled receptors (GPCRs) such as GPR41, GPR43 and GPR109A7,14. Although conflicting results have been reported13,15, GPCRs were recently proposed to mediate the anti-inflammatory effects of SCFAs and protect form colitis, rheumatoid arthritis and airway hyper-responsiveness16,17,18,19. GPCRs propagate anti-inflammatory effects at least through a β2-arrestin-dependent stabilization of IκBα and inhibition of NF-κB-dependent transcription, and by promoting the generation of T regulatory cells (Tregs)7,10.
Upon diffusion into cells, intracellular SCFAs inhibit zinc-dependent histone deacetylases (i.e. HDAC1-11)20. HDACs are major epigenetic erasers catalyzing the deacetylation of histones, leading to chromatin compaction and transcriptional repression21. HDACs also target signaling molecules and transcription factors. Inhibitors of HDAC (HDACi) directly or indirectly impair NF-κB and Foxp3 activity, mediating anticancer, anti-neurodegenerative and anti-inflammatory activities, in part by inducing Treg generation13,21,22,23. Numerous HDACi are tested in clinical trials and several have reached the clinic. Besides valproate that is used since decades as a mood stabilizer and anti-epileptic, vorinostat, romidepsin and belinostat are used for the treatment of cutaneous and/or peripheral T-cell lymphoma, and panobinostat is used to treat patients with multiple myeloma who experienced two prior therapies23. HDACi are also viewed as promising latency-reversing agents to purge the HIV reservoir24. In agreement with their powerful anti-inflammatory properties, several HDACi interfere with the development of innate immune responses, protect against lethal sepsis, and increase susceptibility to infection25,26,27,28,29,30,31,32.
The development of SCFA-mediated therapies, either through direct supplementation with SCFAs or diet-induced modifications of the microbiota and production of endogenous SCFAs is an active area of research16,17,18,19,33. Considering that SCFAs carry anti-inflammatory activity and that HDACi were shown to increase susceptibility to infections in preclinical models and in patients enrolled in oncologic clinical studies29,34,35,36,37,38, an important question is whether SCFA-mediated therapies are safe. Here we focused on propionate as a representative SCFA reported to modulate adaptive immune responses in vivo39,40,41,42,43. We analyzed the response of macrophages, dendritic cells (DCs), splenocytes and whole blood to microbial compounds. Additionally, we performed proof of concept studies using a large panel of preclinical mouse models of endotoxemia, gram-positive and gram-negative bacterial and fungal infection of diverse severity. The results show that propionate to some extent inhibits innate immune responses in vitro, but does not alter susceptibility to infection in vivo nor inhibit passive or natural immunization. These data support the safety of therapies using propionate for treating non-infectious inflammation-related disorders.
Results
Impact of propionate on the response of immune cells to microbial stimulation
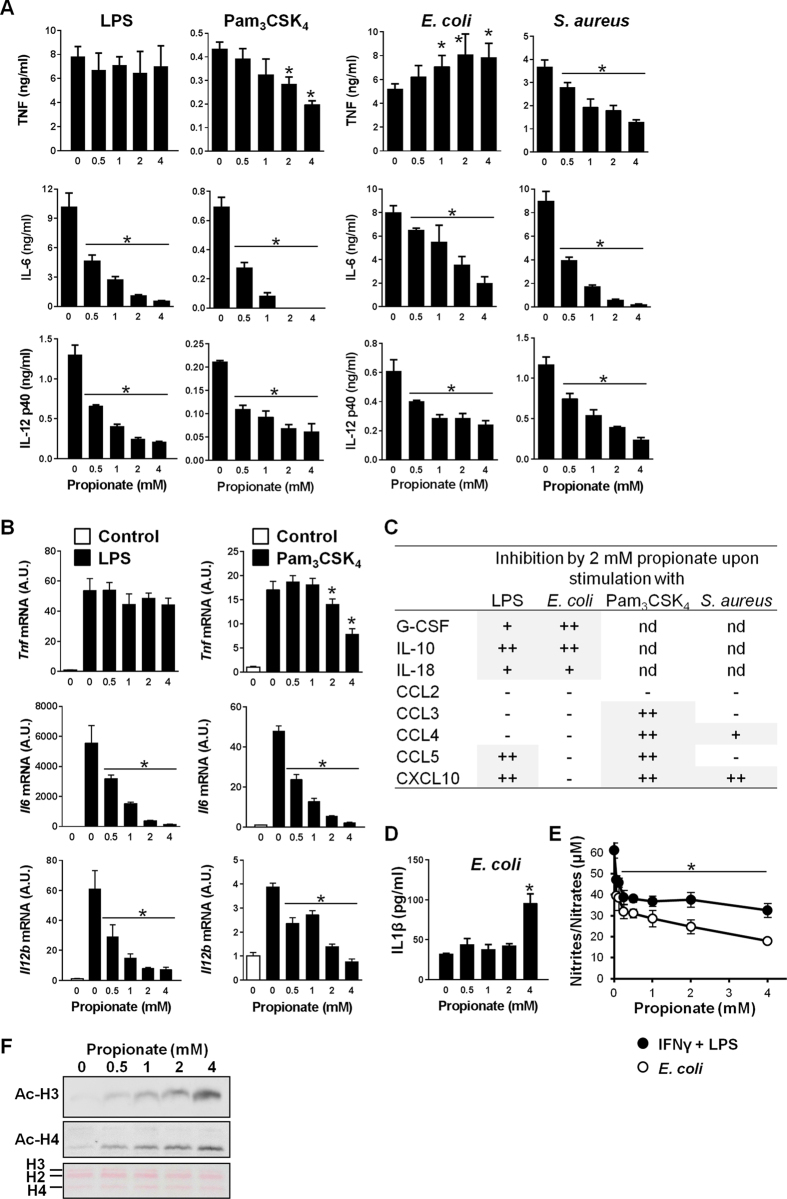
To address the effects of propionate on the response of immune cells to microbial stimulation, bone marrow-derived macrophages (BMDMs) were exposed for 8 h to LPS (a TLR4 agonist), Pam3CSK4 (a lipopeptide triggering cells through TLR1/TLR2) and Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus), used as representative gram-negative and gram-positive bacteria. The levels of TNF, IL-6 and IL-12p40 produced by BMDMs were quantified by ELISA (Fig. 1A). Propionate (0.5–4 mM) dose-dependently inhibited TNF production induced by Pam3CSK4 and S. aureus, and IL-6 and IL-12p40 production induced by LPS, Pam3CSK4, E. coli and S. aureus. Similar to other HDACi29,44,45, propionate did not inhibit TNF production induced by LPS and E. coli, and slightly amplified TNF response to E. coli. Accordingly, propionate powerfully inhibited LPS and Pam3CSK4-induced Il6 and Il12b mRNA, to a lesser extent Pam3CSK4-induced Tnf mRNA, but not LPS-induced Tnf mRNA expression (Fig. 1B).
Figure 1. Impact of propionate on the response of macrophages to microbial stimulation.
BMDMs were pre-incubated for 1 h with increasing concentrations (0, 0.06, 0.12, 0.25, 0.5, 1, 2 and 4 mM) of propionate before exposure for 4, 8 or 24 h to LPS (10 ng/ml), Pam3CSK4 (10 ng/ml), E. coli (106 CFU/ml), S. aureus (107 CFU/ml) or a combination of IFNγ (100 U/ml) plus LPS (10 ng/ml). (A,B) TNF, IL-6 and IL-12p40 concentrations in cell culture supernatants and Tnf, Il6, Il12b mRNA levels were quantified by ELISA (A, t = 8 h) and real time-PCR (B, t = 4 h). No cytokine was detected in the supernatants of unstimulated cells (P < 0.001 vs stimulus alone). Tnf, Il6 and Il12b mRNA levels were normalized to Hprt mRNA levels. Data are means ± SD of triplicate samples from one experiment performed with 4 mice and representative of 2 experiments. *P < 0.05 vs stimulus without propionate. A.U.: arbitrary units. (C) The production of G-CSF, IL-10, IL-18, CCL2, CCL3, CCL4, CCL5 and CXCL10 was assessed by the Luminex technology (t = 8 h). Data summarize the impact of 2 mM propionate on mediators produced in response to LPS, E. coli, Pam3CSK4 and S. aureus: −, no inhibition; +, 1.5-2-fold inhibition; ++, >2-fold inhibition. Quantification is from one experiment performed with 4 mice. (D) IL-1β in cell culture supernatants. Data are means ± SD of triplicate samples from one experiment performed with 2 mice. *P < 0.05 vs no propionate. (E) Nitrites/nitrates were quantified using the Griess reagent (t = 24 h). Data are means ± SD of quadruplicate samples from one experiment performed with 4 mice. *P < 0.05 when comparing propionate at all concentrations vs no propionate. (F) Western blot analysis of acetylated histone 3 (Ac-H3) and Ac-H4 in BMDMs treated for 18 h with propionate. Ponceau staining of the membrane shows equal loading of total histones. Full-length blots are presented in Supplementary Figure S1.
The anti-inflammatory activity of propionate was compared to that of butyrate and valproate by defining the IC50 of each of the SCFAs for LPS-induced IL-6 and IL-12p40 production. Similar IC50s were obtained for IL-6 and IL-12p40: 0.01–0.05 mM for butyrate, 0.2–0.4 mM for valproate and 0.2–0.3 mM for propionate. Thus, propionate is as potent as valproate at inhibiting IL-6 and IL-12p40 but 8–20 fold less efficient than butyrate. The concentrations of G-CSF, IL-10, IL-18, CCL2/MCP-1, CCL3/MIP-1α, CCL4/MIP-1β, CCL5/RANTES and CXCL10/IP10 released by BMDMs exposed to LPS, E. coli, Pam3CSK4 and S. aureus were measured by Luminex (Fig. 1C). Whereas LPS and E. coli induced the secretion of all mediators, Pam3CSK4 and S. aureus did not induce the production of G-CSF, IL-10 and IL-18. Propionate inhibited G-CSF, IL-10 and IL-18 induced by LPS and E. coli, and CCL5 and CXCL10 induced by LPS. Propionate also inhibited CCL3, CCL4, CCL5 and CXCL10 induced by Pam3CSK4 and CCL4 and CXCL10 induced by S. aureus. Overall, propionate impaired more powerfully cytokine/chemokine secretion induced by Pam3CSK4 than LPS, like structurally unrelated HDACi29,44,45, and more efficiently cytokine production induced by pure microbial ligands than whole bacteria triggering similar PRRs (i.e. LPS vs E. coli, and Pam3CSK4 vs S. aureus). Notably, and in line with recent reports46,47, propionate at 4 mM increased E. coli-induced IL-1β secretion by BMDMs (Fig. 1D). Thus propionate impacts on inflammation in a cytokine dependent manner. Propionate also inhibited the production of nitric oxide (NO) induced by E. coli or IFNγ/LPS in BMDMs (50% inhibition using 0.6 mM and 4 mM propionate, respectively (Fig. 1E)).
To answer the question whether propionate acted through HDAC inhibition or via GPCRs, we first quantified mRNA levels of Hdac1-11 and free fatty acid receptor 2 (Ffar2) and Ffar3 encoding for GPR43 and GPR41. Ffar2 and Ffar3 mRNAs were not detected in BMDMs, in line with a previous report41. Incubation of BMDMs with propionate (0–4 mM for 4 or 18 hours) slightly modulated Hdac1-11 expression (range: 1.2–2.5 fold increase or decrease). Yet, propionate strongly increased histone 3 (H3) and H4 acetylation in a dose-dependent manner (Fig. 1F), indicating that propionate inhibits histone deacetylase activity in BMDMs.
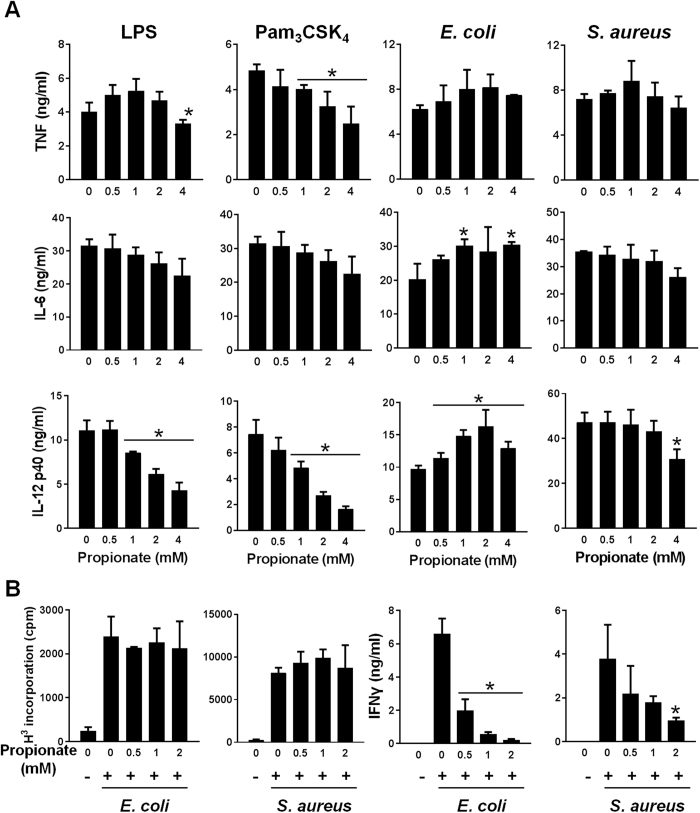
Bone marrow-derived dendritic cells (BMDCs) were less sensitive than BMDMs to the anti-inflammatory effects of propionate. In BMDCs, propionate only significantly inhibited Pam3CSK4-induced TNF and IL-12p40 production in response to LPS, Pam3CSK4 or S. aureus (Fig. 2A). Of note, propionate slightly increased E. coli-induced IL-6 and IL-12p40 production by BMDCs. The viability of BMDMs and BMDCs incubated for 18 h with up to 8 mM propionate was greater than 98%, suggesting that propionate’s effects were not related to cytotoxicity. Along with a good tolerability of immune cells to propionate, propionate barely affected the proliferation of splenocytes exposed to E. coli and S. aureus whereas it efficiently inhibited IFNγ production (Fig. 2B).
Figure 2. Impact of propionate on the response of dendritic cells and splenocytes.
(A) BMDCs were pre-incubated for 1 h with increasing concentrations (0, 0.5, 1, 2 and 4 mM) of propionate before exposure for 8 h to LPS (10 ng/ml), Pam3CSK4 (10 ng/ml), E. coli (106 CFU/ml) and S. aureus (107 CFU/ml). TNF, IL-6 and IL-12p40 concentrations in cell culture supernatants were quantified by ELISA. Data are means ± SD of triplicate samples from one experiment performed with 4 mice and representative of 2 experiments. No cytokine was detected in the supernatants of unstimulated cells (P < 0.001 vs stimulus alone). (B) Mouse splenocytes were incubated for 48 h with or without propionate and E. coli or S. aureus (106 CFU/ml). Proliferation was measured by 3H-thymidine incorporation. IFNγ concentrations in cell culture supernatants were quantified by ELISA. Data are means ± SD of triplicate samples from one experiment performed with 4 mice. *P < 0.05 vs stimulus without propionate.
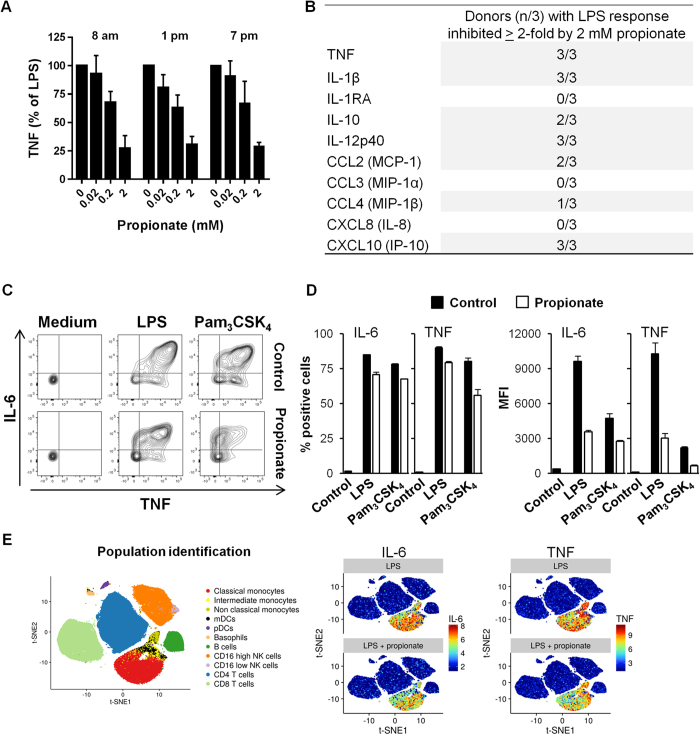
The impact of propionate was tested on human cells. Propionate dose-dependently inhibited TNF production by whole blood exposed to LPS (Fig. 3A), albeit less efficiently than butyrate (TNF: 61 ± 6% vs 96 ± 4% and IL-6: 41 ± 7% vs 70 ± 10% inhibition using propionate vs butyrate at 2 mM, n = 3 donors collected at 8 am; P < 0.05). The extent of TNF inhibition by propionate was similar using blood collected at various times of the day (8 am, 1 pm and 7 pm), excluding a circadian rhythm-dependent effect. A Luminex quantification of 10 mediators produced by whole blood exposed to LPS extended to IL-1β, IL-10, IL-12p40, CCL2 and CXCL10 the spectrum of cytokines and chemokines whose expression was significantly inhibited (≥2-fold) by 2 mM propionate in at least 2 out of the 3 donors tested (Fig. 3B). In parallel experiments, butyrate inhibited more powerfully than propionate the secretion of IL-10 (in 3/3 vs 2/3 donors), CCL2 (3/3 vs 2/3) and CCL4 (2/3 vs 1/3). Butyrate also impaired the release of IL-1RA (3/3 donors) and CXCL8 (1/3). In a confirmation approach, flow cytometry analyses of intracellular cytokine expression in purified human CD14+ monocytes exposed to LPS and Pam3CSK4 revealed that propionate reduced the percentage (11–24% reduction) and the mean fluorescence intensity (1.7–3.4 fold reduction) of TNF and IL-6 positive cells (Fig. 3C and D). Additionally, mass cytometry (CyTOF48) analyses on human whole blood demonstrated that propionate inhibited IL-6 and TNF production by both classical and non-classical monocytes (Fig. 3E). Altogether, propionate inhibited, in a cell and stimulus-specific manner, the response of mouse and human immune cells in vitro. We next investigated the impact of propionate in vivo.
Figure 3. Impact of propionate on the response of human whole blood and monocytes.
Whole blood from 3 healthy subjects was incubated for 18 h with propionate and LPS (100 ng/ml). (A) TNF released by whole blood collected at 8 am, 1 pm and 7 pm was quantified by ELISA. Data are expressed as the percentage of maximal (LPS without propionate) TNF release. No TNF was detected in the absence of LPS stimulation (not shown). Data are means ± SD from 3 healthy subjects. P < 0.005 when comparing 0.2 and 2 mM propionate with 0 mM propionate. (B) TNF, IL-1β, IL-1RA, IL-10, IL-12p40, CCL2, CCL3, CCL4, CXCL8 and CXCL10 were quantified by Luminex. Results summarize the number of donors in whom propionate inhibited significantly (P < 0.05) and by at least 2-fold cytokine release. (C,D) PBMCs were incubated for 1 h with 2 mM propionate and stimulated for 4 h with LPS (100 ng/ml) and Pam3CSK4 (1 μg/ml). TNF and IL-6 expression in CD14+ monocytes was analyzed by flow cytometry to calculate the percentage of positive cells (C) and mean fluorescence intensity (MFI) (D). Data are means ± SD from one experiment performed with 2 donors. (E) Whole blood incubated for 4 h with 2 mM propionate and 100 ng/ml LPS was fixed with Smart Tube stabilizer, and processed by CyTOF as described in Materials and Methods. Left: t-SNE scatter plot of non-granulocyte events. Right: t-SNE plot with arcsinh transformed signal intensity of IL-6 and TNF. Data are representative of results obtained with 3 donors.
Propionate does not protect from lethal endotoxemia and severe sepsis
Following common procedures used to study the impact of SCFAs in vivo17,18,39,41,42,43, mice were fed with propionate at 200 mM in the drinking water for 3 weeks, unless otherwise specified, before being used in preclinical models of toxic shock and infection. Attesting of the effectiveness of the treatment, 3 weeks of propionate regimen increased the number of splenic Foxp3+ Tregs (116% when compared to control mice; n = 8–11 animals per group; P = 0.0007) and of acetylated H4 in stomach, blood and bone marrow (3.5, 3.6 and 1.8 fold increased versus control mice, n = 2).
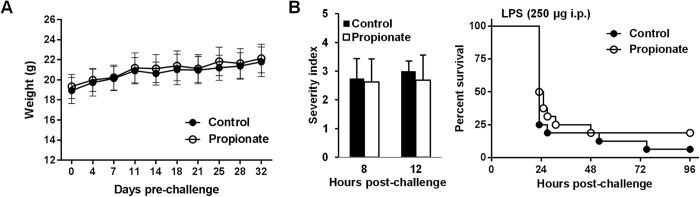
Overwhelming inflammatory responses are deleterious for the host, and inhibition of the release of pro-inflammatory mediators confers protection in preclinical models of sepsis3,4,5. Moreover, several HDACi were shown to protect from toxic shock49. Therefore, we first tested propionate in a mouse model of acute endotoxemia. One month of propionate treatment had no impact on animal weight (Fig. 4A). In mice challenged with a lethal dose of LPS, severity scores and survival rates were similar whether or not animals were treated with propionate (P > 0.5 and P = 0.3; Fig. 4B).
Figure 4. Propionate does not protect from lethal endotoxemia.
BALB/c mice (n = 16 per group) were treated with or without 200 mM propionate in drinking water for 1 month. (A) Weight of animals under propionate treatment. (B) Severity scores (P > 0.1) and survival (P = 0.3) of mice challenged with LPS (250 μg i.p.).
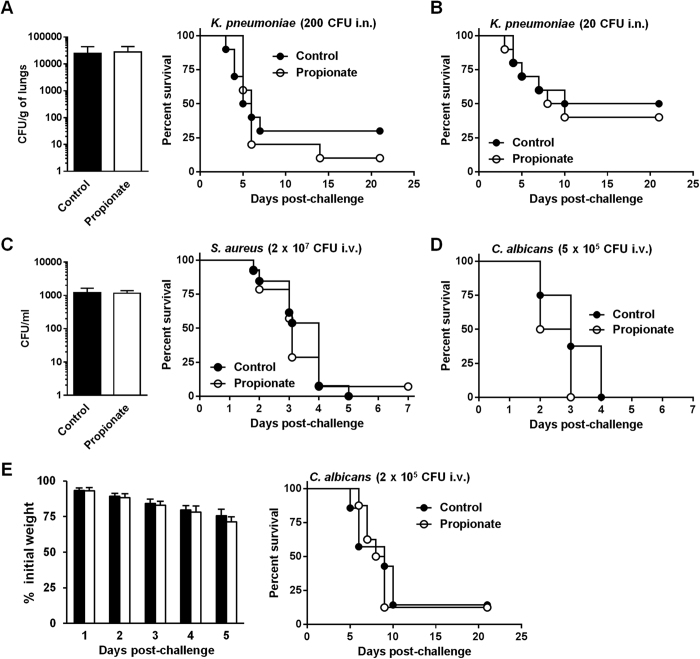
The class of innate immune responses challenged by propionate was extended using models of severe sepsis induced by gram-negative (Klebsiella pneumoniae, K. pneumoniae), gram-positive (S. aureus) and fungal (Candida albicans, C. albicans) pathogens administrated either intranasally (i.n., K. pneumoniae) or intravenously (i.v., S. aureus and C. albicans). In mice challenged with 200 CFU K. pneumoniae, bacterial loads in lungs (P = 0.4) and mortality (70% vs 90% in control vs propionate groups; P = 0.8) were not affected by propionate (Fig. 5A). Mortality was also similar in control and propionate-treated mice infected with 20 CFU of K. pneumoniae (50% vs 60% in control vs propionate group; P = 0.7; Fig. 5B). In the severe model of systemic infection with S. aureus, bacterial counts in blood (P = 0.9) and mortality (100% vs 93% in control vs propionate groups; P = 0.6) were comparable with or without propionate treatment (Fig. 5C). In the acute model of candidiasis all mice died within 4 days, irrespective of the treatment applied (P = 0.1; Fig. 5D). The inoculum of C. albicans was then adjusted to produce a milder form of candidiasis during which mortality occurs 5 to 10 days after infection. Weight loss (P > 0.1) monitored during the first 5 days and survival (14.3% and 12.5%; P = 0.8) were comparable in untreated and propionate-treated mice (Fig. 5E).
Figure 5. Propionate does not protect from lethal sepsis.
BALB/c mice were treated with or without 200 mM propionate in drinking water for 3 weeks. (A,B) Bacterial counts in lungs 48 h post-infection and survival of mice (n = 10 per group) challenged i.n. with 200 CFU (A) or 20 CFU (B) of K. pneumoniae. P = 0.4, 0.8 and 0.7, respectively. (C) Bacterial counts in blood 24 h post-infection and survival of mice (n = 15 per group) challenged with S. aureus (2 × 107 CFU i.v.). P = 0.9 and P = 0.6. (D,E) Survival and body weight of mice (n = 8 per group) challenged with C. albicans (5 × 105 CFU i.v. in D and 2 × 105 CFU i.v. in E). P = 0.1, P > 0.1 and P = 0.8, respectively.
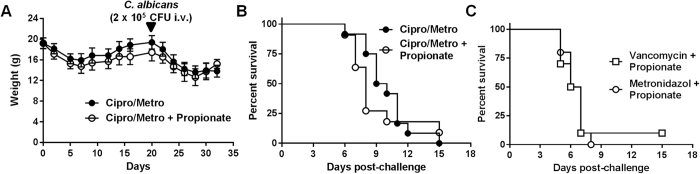
Even though propionate was shown to impact on immune parameters of mice with a normal microbiota17,18,39,43,45,50, propionate produced by gut bacteria may attenuate the impact of propionate supplementation in models of infection. To address this issue, mice were treated with a combination of ciprofloxacin and metronidazole (CM) to deplete the gut flora and decrease endogenous SCFAs levels39,51. CM-treated mice lost 17% weight during the first week of treatment and recovered initial weight after 3 weeks. CM-treated mice were more sensitive to candidiasis (median survival time: 9.5 days for CM vs 11.5 days for controls mice run in parallel; n = 10 mice/group; P = 0.05). Co-treatment with CM plus propionate slightly increased weight loss and impaired weight rebound of uninfected mice (Fig. 6A). CM-treated mice died in between days 6 and 15 after C. albicans challenge, and propionate supplementation did not protect CM-treated mice from candidiasis (P = 0.4; Fig. 6B). To delineate the impact of gram-positive and gram-negative bacteria, mice were treated, together with propionate, with either metronidazole to target anaerobic gram-negative bacteria or vancomycin to target gram-positive bacteria. The two treatments resulted in identical survival profiles (Fig. 6C). Overall, propionate did not interfere with acute, lethal, bacterial and fungal infections.
Figure 6. Propionate does not protect from candidiasis mice depleted of gut microbiota.
BALB/c mice (n = 10 per group) were treated with ciprofloxacin (0.2 mg/ml) and metronidazole (1 mg/ml) or metronidazole or vancomycin (1 mg/ml) with or without 200 mM propionate in drinking water for 3 weeks and challenged with C. albicans (2 × 105 CFU i.v.). (A) Body weight. (B,C) Survival of mice. P > 0.05.
Propionate does not sensitize to mild infection
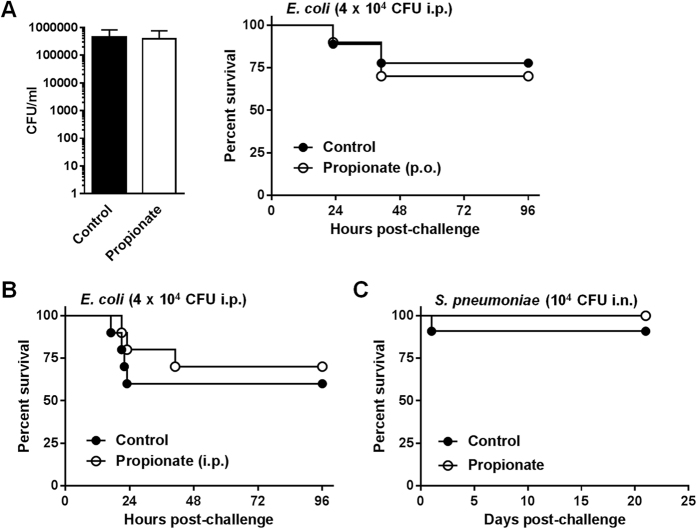
Compromising innate immune responses may increase susceptibility to infection. To analyze the impact of propionate on mild infection, and to test another route of administration of propionate, propionate was given either per os or intraperitoneally (p.o.: 200 mM in water; i.p.: 1 g/kg i.p. every other day43) to mice subsequently challenged with E. coli titrated to cause a mild infection. Bacterial counts (P = 0.9) and survival rates (77% vs 70% and 60% vs 70% in control vs propionate groups upon p.o. and i.p. treatments; P = 0.7 and P = 0.6) were similar in all groups of treatment (Fig. 7A and B). Confirming that propionate does not sensitize mice to infection, 90% (9/10) of control mice and 100% (9/9) of propionate-treated mice infected i.n. with 104 CFU Streptococcus pneumoniae (S. pneumoniae) survived infection (P = 0.4; Fig. 7C). Hence, propionate did not increase susceptibility to E. coli peritonitis and pneumococcal pneumonia.
Figure 7. Propionate does not sensitize to mild infection by E. coli and S. pneumoniae.
BALB/c mice were treated with or without 200 mM propionate in drinking water (A,C) or 1 g/kg propionate given i.p. every other day (B) for 3 weeks and challenged with E. coli (4 × 104 CFU i.p.; n = 10 per group; (A,B) or S. pneumoniae (104 CFU i.p.; n = 9–10; C). (A) Bacterial counts in blood 24 h post-infection and survival of mice. P = 0.9 and 0.7. (B,C) Survival of mice. P = 0.6 and 0.4.
Propionate does not impair passive and natural immunization
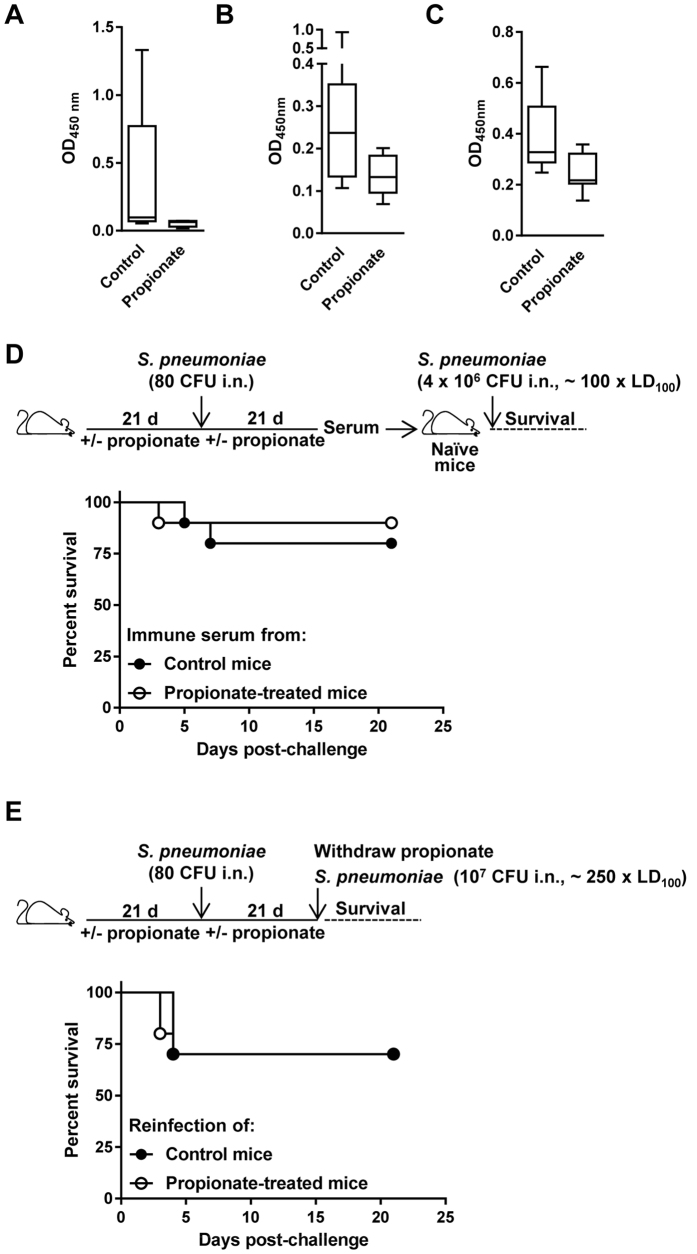
We measured anti-K. pneumoniae and anti-S. pneumoniae IgG titers in mice surviving infection with 20 CFU K. pneumoniae (4 controls and 5 propionate-treated mice; Fig. 5B) and 104 CFU S. pneumoniae (9 controls and 9 propionate-treated mice; Fig. 7C). Anti-bacterial IgG titers were reduced in propionate-treated mice (P = 0.1 and 0.01 for anti-K. pneumoniae and S. pneumoniae IgG titers, respectively; Fig. 8A and B). To confirm this observation, we measured IgG titers in mice infected 3 weeks earlier with a non-lethal inoculum of C. albicans (2 × 104 CFU i.v.). Anti-C. albicans IgG titers were reduced in propionate fed mice (P = 0.02; Fig. 8C). In addition, splenic Foxp3+ Tregs were increased in propionate treated and C. albicans infected mice (113% when compared to control mice; n = 10 mice per group; P = 0.006). Therefore, although propionate did not interfere with morbidity and mortality in the models of infection presented above, it impacted to some extent on anti-microbial host responses. Two approaches were used to assess the relevance of this observation. Mice treated with or without propionate for 3 weeks were challenged with a non-lethal inoculum of S. pneumoniae (80 CFU). In a first setting, 3 weeks after infection, sera were collected and transferred into naive, non-treated, mice that were infected 24 h later with S. pneumoniae used at around 100 x LD100 (Fig. 8D). In a second setting, 3 weeks after infection, propionate treatment was withdrawn and mice were re-challenged with S. pneumoniae used at around 250 x LD100 (Fig. 8E). Overall, propionate treatment during the primary infection had no impact on outcome, and both transfer of immune serum and pre-exposure to a low S. pneumoniae inoculum protected mice from a lethal S. pneumoniae inoculum.
Figure 8. Propionate does not impair passive immunization and protection to secondary infection.
Anti-K. pneumoniae (A), anti-S. pneumoniae (B) and anti-C. albicans (C) IgG titers in BALB/c mice surviving infection with 20 CFU K. pneumoniae (n = 4 control and 5 propionate-treated mice; Fig. 5B), 104 CFU S. pneumoniae (n = 9 control and 10 propionate-treated mice; Fig. 7C) and 4 × 104 CFU C. albicans (n = 9 control and 9 propionate-treated mice, serum was collected 3 weeks post-infection). Box and min-to-max whisker plots represent the OD450 nm using plasma (diluted 1/200) collected on day 21 after infection. P = 0.1, 0.01 and 0.02, respectively. No signal was detected using plasma from uninfected mice. (D,E) BALB/c mice (n = 18–21 per group) were treated with or without 200 mM propionate in drinking water for 3 weeks, challenged i.n. with 80 CFU S. pneumoniae, and used for subsequent experimentation 3 weeks later. (D) Sera collected from 8 water and 11 propionate-treated mice were pooled and transferred (120 μl i.p.) into naive mice (n = 10 per group) infected 24 h later with 4 × 106 CFU S. pneumoniae (~100 x LD100). Survival was monitored for 21 days. P = 0.6. (E) Propionate treatment was withdrawn. Mice (n = 10 per group) were infected with 107 CFU S. pneumoniae (~250 x LD100). Survival was monitored for 21 days. P = 0.8.
Discussion
The gut microbiota and its metabolites exert strong influences on human health. Among bacterial metabolites, SCFAs have attracted much attention because of their beneficial influence on the development of inflammation-related pathologies in combination with the fact that their production can be influenced by the diet7,10,52,53. Here we show that propionate has powerful yet selective anti-inflammatory activity in vitro, and that it does not have a major impact on host susceptibility to infection in vivo. This observation is particularly relevant in light of the development of diet or microbiota targeting strategies to treat immune related diseases.
Propionate impaired cytokine production by innate immune cells, albeit differently according to the cell type, the microbial trigger and the cytokine analyzed. Similar disparities have been observed with other SCFAs29,40,45,46,54. BMDCs were more resistant to propionate than BMDMs, human monocytes and whole blood. In human monocyte-derived DCs (moDCs) and BMDCs, propionate modestly affected IL-6 but efficiently limited IL-12p40 production induced by LPS (ref. 40 and Fig. 2). Furthermore, propionate did not inhibit MHC-II and CD86 expression but impaired CD83 expression by moDCs40. Disparate cell responses to propionate may reflect, at least in part, differential expression of GPCRs. In depth analyses of the pattern and the expression levels of cell-surface GPCRs by immune and non-immune cells is still missing, and could give clues about the contradictory findings reported concerning the inflammatory phenotype of GPR41 and GPR43 knockout mice15. Moreover, SCFA specificity of GPCRs and how redundant behave GPCRs in vitro and in vivo are largely unresolved issues. For example, mice deficient in either GPR43 or GPR109A were susceptible to gut inflammation and developed exacerbated colitis, and mice deficient in either GPR41 or GPR43 were susceptible to allergic airway inflammation17,18,19,43,55. At least in the gut, expression of both GPR43 and GPR109A by non-hematopoietic cells contributed to the protective effects of high-fiber regimen against colitis19.
Besides acting through GPCRs, SCFAs act as inhibitors of class I and II HDACs (HDACi). HDACi impair innate and adaptive immune responses at multiple levels, including TLR and IFN signaling, cytokine production, bacterial phagocytosis and killing, leukocyte adhesion and migration, antigen presentation by DCs, cell proliferation and apoptosis, and Treg development and function21,49,56. In T cells, acetate, propionate and butyrate suppressed HDAC activity independently of GPR41 and GPR4341. Whether SCFAs mediate HDAC inhibition through GPCRs is debatable13, but it is worth mentioning that GPCR signaling modulates kinase, redox and acetylation pathways that, in turn, influence the cellular distribution and activity of histone acetyl transferases and HDACs57.
Like other structurally unrelated HDACi (trichostatin A and suberanilohydroxamic acid, i.e. vorinostat), acetate, propionate and butyrate increased acetylation of FOXP3 and potentiated the generation of peripheral Treg cells39,42,58. Moreover, SCFAs were recently reported to promote the generation of Th1 and Th17 cells during Citrobacter rodentium infection, suggesting a complex, context-dependent impact of SCFAs on immune responses41. Butyrate is a more potent HDACi than propionate, which is more potent than acetate. This ranking parallels the effectiveness of the anti-inflammatory activity of SCFAs39,59,60. Propionate failed to inhibit TNF but not IL-6 and IL-12p40 induced by LPS in BMDMs, which mirrored previous observations obtained with trichostatin A and suberanilohydroxamic acid29,44,61,62,63. Propionate strongly increased H3 and H4 acetylation in BMDMs which, like BMDCs, barely expressed Ffar2 and Ffar341. Thus, the effect of propionate on BMDMs is at least in part mediated by inhibition of HDACs. The fact that propionate slightly increased cytokine production under certain conditions is reminiscent of the paradoxical impact of HDACi on TNF secretion in human macrophages46. Further work will be required to unravel how propionate differentially affects cytokine expression induced by pure microbial ligands versus whole bacteria, for example by analyzing chromatin structure, modifications and activation of transcriptional regulators, and signaling pathways.
Acetate, propionate and butyrate are found at molar ratios of 60/20/20 in the intestinal tract and 90-55/35-5/10-4 in blood, depending on portal, hepatic and peripheral origins, where they altogether reach 50–150 mM and 0.1–1 mM, respectively8,43. The high plasma concentrations of propionate compared to butyrate may counterbalance its weaker anti-inflammatory activity. Further work will be required to analyze the effects of combinational treatments with SCFAs on innate immune cells. Propionate is produced primarily by Bacteroidetes via the succinate pathway and some Firmicutes through the lactate and succinate pathways, acetate by enteric bacteria and butyrate by Firmicutes10. SCFAs themselves modify the composition of the gut microbiota. Propionate stimulates the growth of Bifidobacterium64. Proportions of Bacteroidaceae and Bifidobacteriaceae increased in the gut of mice fed with a high-fiber diet, elevating acetate and propionate levels but decreasing butyrate concentrations in cecal content and blood43. Thus, changing microbiome composition affects SCFA levels both locally and systemically. Moreover, the gut microbiota protects from pneumococcal pneumonia65. In the perspective of targeting the diet or the microbiota for treating inflammatory conditions7,10,52,53, it was of prime interest to analyze the impact of propionate in preclinical models of infection.
Unlike powerful broad-spectrum HDACi27,29,66,67, propionate had no obvious impact on morbidity and mortality parameters in models of endotoxemia and infections. This contrasts with the effectiveness of SCFAs at ameliorating the clinical outcome in chronic inflammatory diseases like rheumatoid arthritis, colitis and airway allergy17,18,19,33,39,43,45,50. The propionate regimen itself was unlikely responsible of the failure to protect septic animals since it increased H4 acetylation in organs and identical or shorter treatments had an immune impact39,43. Moreover, as expected39,43,50, propionate increased the frequency of peripheral Tregs and reduced anti-microbial IgG responses, indicating that propionate influenced immune parameters during the course of infections. Multiple mechanisms may account for the reduced humoral response. Besides increasing the frequency of Tregs, SCFAs and HDACi have been shown to inhibit development and migration of DCs. Moreover, they decreased expression of costimulatory and MHC molecules, reduced production of T-cell polarizing factors and increased production of indoleamine 2,3-dioxygenase (a negative regulator of T-cell activation) by DCs. In mouse models, HDACi inhibited antigen presentation and allogeneic and syngeneic responses and decreased antibody generation29,40,44,56.
Albeit surprising at first glance, propionate did not increase the mortality of mice subjected to sub-lethal/mild infections. Indeed, one of the possible collateral damages of administrating immunomodulatory compounds is an increased risk of infections. A well-known example is anti-TNF therapies that are associated with reactivation of latent tuberculosis and viral infections as well as an increased risk of opportunistic infections68. Moreover, episodes of severe infection have been reported in patients treated with HDACi34,35,36,37,38. In the present study, we tested models of systemic and local infections using the most common etiologic agents of bacterial sepsis in humans (E. coli, S. aureus and K. pneumoniae) to investigate the safety of propionate supplementation for clinical purposes.
The production of propionate by intestinal bacteria has been proposed to represent a mechanism through which the host response to commensals is kept under control and avoid local inflammation and tissue damage. It is however now well established that SCFAs have much broader effects on human health. Using several preclinical mouse models, we report that administration of propionate neither protects from lethal sepsis nor increases susceptibility to mild infections. These results are encouraging in the perspective of developing propionate-based therapies, e.g. direct supplementation or via the diet/microbiota, without putting patients at risk of developing infections.
Materials and Methods
Ethics statement
Animal experimentations were approved by the Service de la Consommation et des Affaires vétérinaires (SCAV) du Canton de Vaud (Epalinges, Switzerland) under authorizations n° 876.7, 876.8, 877.7 and 877.8, and performed according to our institutional guidelines and ARRIVE guidelines (http://www.nc3rs.org.uk/arrive-guidelines). Experiments on human whole blood samples were carried out in accordance with guidelines and regulations of the Swiss Ethics Committees on research involving humans. The procedure, using anonymized human whole blood samples from healthy subjects without possibility to trace back subject identity, did not require prior ethics committee authorization. Written informed consent was obtained from blood donors at the Infectious Diseases Service, CHUV, Lausanne.
Mice, cells and reagents
Female BALB/cByJ mice (8–10 week-old; Charles River Laboratories, Saint-Germain-sur-l’Arbresle, France) were housed under specific pathogen-free conditions. Bone marrow cells were cultured for 7 days in IMDM containing 50 μM 2-ME and 30% L929 supernatant as a source of M-CSF to generate bone marrow-derived macrophages (BMDMs), or GM-CSF to generate bone marrow-derived dendritic cells (BMDCs)69. Splenocytes were cultured in RPMI 1640 medium containing 2 mM glutamine and 50 μM 2-ME70. Culture media (Invitrogen, San Diego, CA) were supplemented with 10% (v/v) heat-inactivated FCS (Sigma-Aldrich St. Louis, MO), 100 IU/ml penicillin and 100 μg/ml streptomycin (Invitrogen). Propionate and butyrate were from Sigma-Aldrich, valproate from Desitin (Hamburg, Germany), Salmonella minnesota ultra pure lipopolysaccharide (LPS) from List Biologicals Laboratories (Campbell, CA) and Pam3CSK4 from EMC microcollections (Tübingen, Germany). E. coli O18:K1:H7 (E. coli), K. pneumoniae caroli (K. pneumoniae), S. aureus AW7 (S. aureus), S. pneumoniae 6303 (S. pneumoniae), and C. albicans were isolated from septic patients71,72. E. coli, K. pneumoniae, S. aureus and S. pneumoniae were grown in brain heart infusion broth, C. albicans in yeast extract-peptone-dextrose (BD Biosciences, Erembodegem, Belgium). For in vitro experiments, microorganisms were heat-inactivated for 2 h at 56 °C before usage.
Cell viability assay
Cell viability was assessed using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) Cell Proliferation and Viability Assay and a Synergy H1 microplate reader (BioTek, Winooski, VT)73. On each 96-well cell culture plate, serial quantities of cells (0.3 × 104–5 × 105) were seeded to establish a standard curve.
Whole blood assay
Heparinized whole blood (50 μl) obtained from healthy subjects was diluted 5-fold in RPMI 1640 medium and incubated with or without propionate and microbial products in 96-wells plates. Reaction mixtures were incubated for 24 h at 37 °C in the presence of 5% CO2. Cell-free supernatants were stored at −80 °C until cytokine measurement.
Cytokine and NO measurements and flow cytometry analyses
Cell culture supernatants and plasma were used to quantify the concentrations of TNF, IL-1β, IL-6, IL-12p40 and IFN-γ by DuoSet ELISA kits (R&D Systems, Abingdon, UK), cytokines/chemokines using mouse (G-CSF, IL-1β, IL-10, IL-12p70, IL-18, CCL2/MCP1, CCL3/MIP-1α, CCL4/MIP-1β, CCL5/RANTES, CXCL10/IP10) and human (TNF, IL-1β, IL-1ra, IL-10, IL-12p40, CCL2, CCL3, CCL4, CXCL8/IL-8, CXCL10) Luminex assays (Affimetrix eBioscience, Vienna, Austria), and NO using the Griess reagent71,74. Intracellular cytokine staining was performed essentially as described75. Half a million peripheral blood mononuclear cells (PBMCs) were incubated for 1 h with propionate and then for 4 h with 1 μg/ml brefeldin A (BioLegend, San Diego, CA) with or without LPS (100 ng/ml) or Pam3CSK4 (1 μg/ml). PBMCs were stained with the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Molecular Probes, ThermoFisher Scientific, Zug, Switzerland), washed with Cell Stain Medium (CSM: PBS, 0.5% BSA, 0.02% NaN3 and 2 mM EDTA), incubated with Human TruStain FcX™ (BioLegend) to block Fc receptors and stained with anti-CD3-PerCP/Cy5.5 (clone UCHT1) and anti-CD14-Pacific Blue (clone M5E2). Cells were washed with CSM, fixed using 2.4% (w/v) formaldehyde in PBS, washed with CSM-S (CSM containing 0.3% saponin, Sigma-Aldrich) and stained with anti-IL-6-allophycocyanin and anti-TNF-phycoerythrin/Dazzle™ 594 antibodies (clones MQ2-13A5 and MAB11). Single cell suspensions of splenocytes were stained with anti-CD4-APC/Cy7 (clone Gk1.5) and anti-CD25-APC (clone 3C7). Staining steps were performed at 20 °C for 20 min. All antibodies were from BioLegend. Foxp3 staining (anti-Foxp3-PE, clone FJK-16S,) was performed with the Foxp3/Transcription Factor Staining Buffer Set from eBioscience according to the manufacturer’s instructions. Unstained and single stained samples were used to calculate compensation for PBMCs while UltraComp eBeads (eBioscience) were used for splenocytes. Acquisition was performed on a LSR II flow cytometer (BD Biosciences). Foxp3 and CD25 gates were determined using FMO controls. Data was analyzed using FlowJo vX (FlowJo LCC, Ashland, OR).
RNA analyses by real-time PCR
Total RNA was isolated, reverse transcribed and used for real-time PCR analyses using a QuantStudio™ 12 K Flex system (Life Technologies, Carlsbad, CA). Reactions consisted of 1.25 μl cDNA, 1.25 μl H2O, 0.62 μl primers and 3.12 μl Fast SYBR® Green Master Mix (Life Technologies). Primer pairs for amplifying Tnf, Il6, Il12b and Hprt (hypoxanthine guanine phosphoribosyl transferase) cDNA were as published69. Samples were tested in triplicates. Gene specific expression was normalized to Hprt expression and expressed relative to that of untreated cells.
Western blot analyses
Histones were extracted from cells or organs (disrupted using a gentleMACSTM Octo Dissociator, Miltenyi Biotec; Bergisch Gladbach, Germany), run through SDS-PAGE and detected by Western blotting29 using antibodies (diluted 1:1000) directed against acetylated histone 3 (H3) and H4 (06-599, 06-860) (EMD Millipore, Billerica, MA). Blots were revealed with the enhanced chemiluminescence Western blotting system (GE Healthcare, Little Chalfont, Great Britain). Images were recorded using a Fusion Fx system (Viber Lourmat, Collégien, France).
Proliferation assay
The proliferation of splenocytes (1.5 × 105 cells) cultured for 48 h in 96-well plates was assessed by measuring 3H-thymidine incorporation over 18 h using a β-counter (Packard Instrument Inc, Meriden, CT).
Mass cytometry (CyTOF) analysis
Blood was collected from healthy donors in heparin tubes and stimulated with LPS (100 ng/ml) with or without propionate (2 mM) and brefeldin A. After 4 h, EDTA (2 mM) was added and cells incubated for 15 min at room temperature (RT). Cells were fixed using a proteomic stabilizer (Smart Tube Inc., San Carlos, CA) and stored at −80 °C. Thawing and erythrocyte lysis was performed according to instructions from Smart Tube Inc. Fc receptors were blocked with Human TruStain FcX Receptor Blocker. Cells were stained using metal-conjugated antibodies according to the CyTOF manufacturer’s instructions (Fluidigm, South San Franciso, CA). Briefly, individual samples were stained with unique combinations of CD45 antibodies for 30 min at RT, followed by 3 washes with CSM. Samples were pooled and stained using a cocktail of antibodies for cell surface markers, washed with CSM and PBS, fixed with 2.4% formaldehyde, washed with CSM-S, and stained for intracellular targets. Antibodies directed against CD1c (L161), CD3 (UCHT1), CD4 (RPA-T4), CD7 (CD7-6B7), CD8 (SK1), CD11c (Bu15), CD14 (M5E2), CD16 (3G8), CD20 (2H7), CD45 (HI30), CD66b (G10F5), CD123 (6H6), HLA-DR (L243), IL-6 (MQ2-13A5) and TNF (Mab11) were from BioLegend, Slan (DD1) from Miltenyi Biotec and CD56 (R19-760) and CD141 (1A4) from BD. After intracellular staining, cells were resuspended in DNA-intercalation solution (PBS, 1 μM Ir-Intercalator, 1% formaldehyde, 0.3% saponin) and stored at 4 °C until analysis. For analysis, cells were washed 3 times with MilliQ water and resuspended at 0.5 × 106 cells/ml in 0.1% EQ™ Four Element Calibration Beads solution (Fluidigm). Samples were acquired on an upgraded CyTOF 1 using a syringe pump at 45 μl/min. FCS files were concatenated and normalized using the cytobank concatenation tool and matlab normalizer respectively76. Data was processed and analyzed with cytobank and R using the OpenCyto and cytofkit packages77,78. Dimensionality reduction with t-SNE was performed on a merged dataset, consisting of a random selection of 10’000 non granulocyte events from each sample.
In vivo models
Mice (n = 8–16/group) treated or not with propionate (200 mM in drinking water or 1 g/kg i.p. every other day) were challenged with LPS (250 μg i.p.), E. coli (4 × 104 CFU i.p.), K. pneumoniae (20 or 200 CFU i.n.), S. pneumoniae (80, 104, 4 × 106 or 107 CFU i.n.), S. aureus (2 × 107 CFU i.v.) or C. albicans (4 × 104, 2 × 105 or 5 × 105 conidia i.v.)29,70,71,79,80,81. Unless specified, propionate treatment was continued after microbial challenge. To analyze the impact of propionate on acquired immunity to bacterial infection, BALB/c mice with or without propionate treatment were infected with 80 CFU S. pneumoniae. Three weeks later, propionate treatment was stopped. Mice were either re-infected with 107 CFU S. pneumoniae or sacrificed to collect sera. Sera from water or propionate-treated mice were pooled and transferred (120 μl i.p.) into naïve mice that were infected 24 h later with 4 × 106 CFU S. pneumoniae. In selected experiments, mice were treated with ciprofloxacin (0.2 mg/ml; Fresenius, Brézins, France), metronidazole (1 mg/ml; Sintetica S.A., Couver, Switzerland) and vancomycin (TEVA, North Wales, PA) in drinking water to deplete the gut microbiota17,18,39,43,82. Body weight, severity scores and survival were registered at least once daily as described previously71.
Detection of anti-bacterial and anti-Candida IgG by ELISA
Briefly, 96-well plates (Maxisorp, Affimetrix eBioscience) were coated with 5 × 106 heat-killed K. pneumoniae or S. pneumoniae or 100 μg/ml C. albicans in bicarbonate/carbonate buffer (100 mM, pH 9.6), blocked with PBS containing 3% BSA (PBS-BSA) and incubated with mouse serum diluted 1/200 in PBS-BSA. IgGs were detected with peroxidase-goat anti-mouse IgG (H+L) and then 3,3′,5,5′-tetramethylbenzidine (TMB) Substrate Solution (ThermoFisher Scientific). Reactions were stopped using 0.16 M sulfuric acid and absorbance measured at 450 nm using a VersaMax ELISA microplate reader (Molecular devices, Sunnyvale, CA). All washing steps were performed using PBS containing 0.05% (v/v) Tween-20.
Statistical analyses
Comparisons between the different groups were performed by analysis of variance followed by two-tailed unpaired Student’s t-test or Mann-Whitney test when appropriate. The Kaplan-Meier method was used for building survival curves and differences were analyzed by the log-rank sum test. All analyses were performed using PRISM (GraphPad Software). P values are two-sided, and values < 0.05 were considered to indicate statistical significance.
Additional Information
How to cite this article: Ciarlo, E. et al. Impact of the microbial derived short chain fatty acid propionate on host susceptibility to bacterial and fungal infections in vivo. Sci. Rep. 6, 37944; doi: 10.1038/srep37944 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank François Carlen, Xiaoyue Zhang and Niklaus Hurlimann for technical help. TR is supported by grants from the Swiss National Science Foundation (SNF 145014, 149511 and 147662) and an interdisciplinary grant from the Faculty of Biology and Medicine of the University of Lausanne (Switzerland). XZ was supported by the Summer Undergraduate Research (SUR) program of the University of Lausanne.
Footnotes
Author Contributions E.C., T.H. and M.M. performed in vitro experiments, J.H. performed flow cytometry experiments, J.H. and C.F. designed and performed CyTOF study, E.C., T.H. and D.L.R. performed in vivo experiments. T.R. conceived the project, designed the experiments and wrote the paper. All authors reviewed the manuscript.
References
- Broz P. & Monack D. M. Newly described pattern recognition receptors team up against intracellular pathogens. Nature reviews. Immunology 13, 551–565, doi: 10.1038/nri3479 (2013). [DOI] [PubMed] [Google Scholar]
- Chu H. & Mazmanian S. K. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nature immunology 14, 668–675, doi: 10.1038/ni.2635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savva A. & Roger T. Targeting toll-like receptors: promising therapeutic strategies for the management of sepsis-associated pathology and infectious diseases. Frontiers in immunology 4, 387, doi: 10.3389/fimmu.2013.00387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. et al. Sepsis: a roadmap for future research. The Lancet infectious diseases 15, 581–614, doi: 10.1016/S1473-3099(15)70112-X (2015). [DOI] [PubMed] [Google Scholar]
- Rittirsch D., Flierl M. A. & Ward P. A. Harmful molecular mechanisms in sepsis. Nature reviews. Immunology 8, 776–787, doi: 10.1038/nri2402 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcais A., Abel L. & Casanova J. L. Human genetics of infectious diseases: between proof of principle and paradigm. The Journal of clinical investigation 119, 2506–2514, doi: 10.1172/JCI38111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn A. N., Macia L. & Mackay C. R. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 40, 833–842, doi: 10.1016/j.immuni.2014.05.014 (2014). [DOI] [PubMed] [Google Scholar]
- Cummings J. H., Pomare E. W., Branch W. J., Naylor C. P. & Macfarlane G. T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28, 1221–1227 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman E. N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiological reviews 70, 567–590 (1990). [DOI] [PubMed] [Google Scholar]
- Louis P., Hold G. L. & Flint H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nature reviews. Microbiology 12, 661–672, doi: 10.1038/nrmicro3344 (2014). [DOI] [PubMed] [Google Scholar]
- Natarajan N. & Pluznick J. L. From microbe to man: the role of microbial short chain fatty acid metabolites in host cell biology. American journal of physiology. Cell physiology 307, C979–985, doi: 10.1152/ajpcell.00228.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara A. Contributions of colonic short-chain Fatty Acid receptors in energy homeostasis. Frontiers in endocrinology 5, 144, doi: 10.3389/fendo.2014.00144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Oliveira R., Fachi J. L., Vieira A., Sato F. T. & Vinolo M. A. Regulation of immune cell function by short-chain fatty acids. Clinical & translational immunology 5, e73, doi: 10.1038/cti.2016.17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloes D. A., Kretschmer D. & Peschel A. Enemy attraction: bacterial agonists for leukocyte chemotaxis receptors. Nature reviews. Microbiology 13, 95–104, doi: 10.1038/nrmicro3390 (2015). [DOI] [PubMed] [Google Scholar]
- Ang Z. & Ding J. L. GPR41 and GPR43 in Obesity and Inflammation - Protective or Causative? Frontiers in immunology 7, 28, doi: 10.3389/fimmu.2016.00028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. H., Kang S. G., Park J. H., Yanagisawa M. & Kim C. H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145, 396–406 e391-310, doi: 10.1053/j.gastro.2013.04.056 (2013). [DOI] [PubMed] [Google Scholar]
- Maslowski K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286, doi: 10.1038/nature08530 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui R. et al. G protein-coupled receptor 43 moderates gut inflammation through cytokine regulation from mononuclear cells. Inflammatory bowel diseases 19, 2848–2856, doi: 10.1097/01.MIB.0000435444.14860.ea (2013). [DOI] [PubMed] [Google Scholar]
- Macia L. et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nature communications 6, 6734, doi: 10.1038/ncomms7734 (2015). [DOI] [PubMed] [Google Scholar]
- Segain J. P. et al. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut 47, 397–403 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg K. J. & Johnstone R. W. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nature reviews. Drug discovery 13, 673–691, doi: 10.1038/nrd4360 (2014). [DOI] [PubMed] [Google Scholar]
- Carafa V., Miceli M., Altucci L. & Nebbioso A. Histone deacetylase inhibitors: a patent review (2009–2011). Expert opinion on therapeutic patents 23, 1–17, doi: 10.1517/13543776.2013.736493 (2013). [DOI] [PubMed] [Google Scholar]
- Guha M. HDAC inhibitors still need a home run, despite recent approval. Nature reviews. Drug discovery 14, 225–226, doi: 10.1038/nrd4583 (2015). [DOI] [PubMed] [Google Scholar]
- Darcis G., Van Driessche B. & Van Lint C. Preclinical shock strategies to reactivate latent HIV-1: an update. Current opinion in HIV and AIDS 11, 388–393, doi: 10.1097/COH.0000000000000288 (2016). [DOI] [PubMed] [Google Scholar]
- Leoni F. et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proceedings of the National Academy of Sciences of the United States of America 99, 2995–3000, doi: 10.1073/pnas.052702999 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni F. et al. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Molecular medicine 11, 1–15, doi: 10.2119/2006-00005.Dinarello (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. T. et al. Sodium butyrate prevents lethality of severe sepsis in rats. Shock 27, 672–677, doi: 10.1097/SHK.0b013e31802e3f4c (2007). [DOI] [PubMed] [Google Scholar]
- Mombelli M. et al. Histone deacetylase inhibitors impair antibacterial defenses of macrophages. The Journal of infectious diseases 204, 1367–1374, doi: 10.1093/infdis/jir553 (2011). [DOI] [PubMed] [Google Scholar]
- Roger T. et al. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood 117, 1205–1217, doi: 10.1182/blood-2010-05-284711 (2011). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Protective effect of suberoylanilide hydroxamic acid against LPS-induced septic shock in rodents. Shock 32, 517–523, doi: 10.1097/SHK.0b013e3181a44c79 (2009). [DOI] [PubMed] [Google Scholar]
- Jones R. B. et al. Histone deacetylase inhibitors impair the elimination of HIV-infected cells by cytotoxic T-lymphocytes. PLoS pathogens 10, e1004287, doi: 10.1371/journal.ppat.1004287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugrin J. et al. Histone deacetylase inhibitors repress macrophage migration inhibitory factor (MIF) expression by targeting MIF gene transcription through a local chromatin deacetylation. Biochimica et biophysica acta 1793, 1749–1758, doi: 10.1016/j.bbamcr.2009.09.007 (2009). [DOI] [PubMed] [Google Scholar]
- Thorburn A. N. et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nature communications 6, 7320, doi: 10.1038/ncomms8320 (2015). [DOI] [PubMed] [Google Scholar]
- Gojo I. et al. Phase 1 and pharmacologic study of MS-275, a histone deacetylase inhibitor, in adults with refractory and relapsed acute leukemias. Blood 109, 2781–2790, doi: 10.1182/blood-2006-05-021873 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane S. et al. Valproic acid combined with cytosine arabinoside in elderly patients with acute myeloid leukemia has in vitro but limited clinical activity. Leukemia & lymphoma 53, 1077–1083, doi: 10.3109/10428194.2011.642302 (2012). [DOI] [PubMed] [Google Scholar]
- Moskowitz C. H. et al. Brentuximab vedotin as consolidation therapy after autologous stem-cell transplantation in patients with Hodgkin’s lymphoma at risk of relapse or progression (AETHERA): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 385, 1853–1862, doi: 10.1016/S0140-6736(15)60165-9 (2015). [DOI] [PubMed] [Google Scholar]
- Witta S. E. et al. Randomized phase II trial of erlotinib with and without entinostat in patients with advanced non-small-cell lung cancer who progressed on prior chemotherapy. Journal of clinical oncology 30, 2248–2255, doi: 10.1200/JCO.2011.38.9411 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes A. et al. Mocetinostat for relapsed classical Hodgkin’s lymphoma: an open-label, single-arm, phase 2 trial. The Lancet. Oncology 12, 1222–1228, doi: 10.1016/S1470-2045(11)70265-0 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N. et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455, doi: 10.1038/nature12726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nastasi C. et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Scientific reports 5, 16148, doi: 10.1038/srep16148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal immunology 8, 80–93, doi: 10.1038/mi.2014.44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. M. et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573, doi: 10.1126/science.1241165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompette A. et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature medicine 20, 159–166, doi: 10.1038/nm.3444 (2014). [DOI] [PubMed] [Google Scholar]
- Brogdon J. L. et al. Histone deacetylase activities are required for innate immune cell control of Th1 but not Th2 effector cell function. Blood 109, 1123–1130, doi: 10.1182/blood-2006-04-019711 (2007). [DOI] [PubMed] [Google Scholar]
- Chang P. V., Hao L., Offermanns S. & Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proceedings of the National Academy of Sciences of the United States of America 111, 2247–2252, doi: 10.1073/pnas.1322269111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman R. J. et al. Differential Anti-inflammatory Activity of HDAC Inhibitors in Human Macrophages and Rat Arthritis. The Journal of pharmacology and experimental therapeutics 356, 387–396, doi: 10.1124/jpet.115.229328 (2016). [DOI] [PubMed] [Google Scholar]
- Stammler D. et al. Inhibition of Histone Deacetylases Permits Lipopolysaccharide-Mediated Secretion of Bioactive IL-1beta via a Caspase-1-Independent Mechanism. Journal of immunology 195, 5421–5431, doi: 10.4049/jimmunol.1501195 (2015). [DOI] [PubMed] [Google Scholar]
- Herderschee J., Fenwick C., Pantaleo G., Roger T. & Calandra T. Emerging single-cell technologies in immunology. Journal of leukocyte biology 98, 23–32, doi: 10.1189/jlb.6RU0115-020R (2015). [DOI] [PubMed] [Google Scholar]
- Ciarlo E., Savva A. & Roger T. Epigenetics in sepsis: targeting histone deacetylases. International journal of antimicrobial agents 42 Suppl, S8–12, doi: 10.1016/j.ijantimicag.2013.04.004 (2013). [DOI] [PubMed] [Google Scholar]
- Furusawa Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450, doi: 10.1038/nature12721 (2013). [DOI] [PubMed] [Google Scholar]
- Kelly C. J. et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell host & microbe 17, 662–671, doi: 10.1016/j.chom.2015.03.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrestein P. C., Mazmanian S. K. & Knight R. Finding the missing links among metabolites, microbes, and the host. Immunity 40, 824–832, doi: 10.1016/j.immuni.2014.05.015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G. et al. Specialized metabolites from the microbiome in health and disease. Cell metabolism 20, 719–730, doi: 10.1016/j.cmet.2014.10.016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt B. E. et al. Butyrate increases IL-23 production by stimulated dendritic cells. American journal of physiology. Gastrointestinal and liver physiology 303, G1384–1392, doi: 10.1152/ajpgi.00540.2011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139, doi: 10.1016/j.immuni.2013.12.007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakespear M. R., Halili M. A., Irvine K. M., Fairlie D. P. & Sweet M. J. Histone deacetylases as regulators of inflammation and immunity. Trends in immunology 32, 335–343, doi: 10.1016/j.it.2011.04.001 (2011). [DOI] [PubMed] [Google Scholar]
- Spiegelberg B. D. G protein coupled-receptor signaling and reversible lysine acetylation. Journal of receptor and signal transduction research 33, 261–266, doi: 10.3109/10799893.2013.822889 (2013). [DOI] [PubMed] [Google Scholar]
- Tao R. et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nature medicine 13, 1299–1307, doi: 10.1038/nm1652 (2007). [DOI] [PubMed] [Google Scholar]
- Iraporda C. et al. Lactate and short chain fatty acids produced by microbial fermentation downregulate proinflammatory responses in intestinal epithelial cells and myeloid cells. Immunobiology 220, 1161–1169, doi: 10.1016/j.imbio.2015.06.004 (2015). [DOI] [PubMed] [Google Scholar]
- Tedelind S., Westberg F., Kjerrulf M. & Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World journal of gastroenterology 13, 2826–2832 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode K. A. et al. Histone deacetylase inhibitors decrease Toll-like receptor-mediated activation of proinflammatory gene expression by impairing transcription factor recruitment. Immunology 122, 596–606, doi: 10.1111/j.1365-2567.2007.02678.x (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S. L., Hargreaves D. C. & Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978, doi: 10.1038/nature05836 (2007). [DOI] [PubMed] [Google Scholar]
- Serrat N. et al. The response of secondary genes to lipopolysaccharides in macrophages depends on histone deacetylase and phosphorylation of C/EBPbeta. Journal of immunology 192, 418–426, doi: 10.4049/jimmunol.1203500 (2014). [DOI] [PubMed] [Google Scholar]
- Kaneko T., Mori H., Iwata M. & Meguro S. Growth stimulator for bifidobacteria produced by Propionibacterium freudenreichii and several intestinal bacteria. Journal of dairy science 77, 393–404, doi: 10.3168/jds.S0022-0302(94)76965-4 (1994). [DOI] [PubMed] [Google Scholar]
- Schuijt T. J. et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 65, 575–583, doi: 10.1136/gutjnl-2015-309728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jin S., Wang C., Jiang R. & Wan J. Histone deacetylase inhibitors attenuate acute lung injury during cecal ligation and puncture-induced polymicrobial sepsis. World journal of surgery 34, 1676–1683, doi: 10.1007/s00268-010-0493-5 (2010). [DOI] [PubMed] [Google Scholar]
- Ji M. H. et al. Valproic acid attenuates lipopolysaccharide-induced acute lung injury in mice. Inflammation 36, 1453–1459, doi: 10.1007/s10753-013-9686-z (2013). [DOI] [PubMed] [Google Scholar]
- Murdaca G. et al. Infection risk associated with anti-TNF-alpha agents: a review. Expert opinion on drug safety 14, 571–582, doi: 10.1517/14740338.2015.1009036 (2015). [DOI] [PubMed] [Google Scholar]
- Ciarlo E. & Roger T. Screening the Impact of Sirtuin Inhibitors on Inflammatory and Innate Immune Responses of Macrophages and in a Mouse Model of Endotoxic Shock. Methods Mol Biol 1436, 313–334, doi: 10.1007/978-1-4939-3667-0_21 (2016). [DOI] [PubMed] [Google Scholar]
- Roger T. et al. Protection from lethal gram-negative bacterial sepsis by targeting Toll-like receptor 4. Proceedings of the National Academy of Sciences of the United States of America 106, 2348–2352, doi: 10.1073/pnas.0808146106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger T. et al. Macrophage migration inhibitory factor deficiency is associated with impaired killing of gram-negative bacteria by macrophages and increased susceptibility to Klebsiella pneumoniae sepsis. The Journal of infectious diseases 207, 331–339, doi: 10.1093/infdis/jis673 (2013). [DOI] [PubMed] [Google Scholar]
- Savva A. et al. Functional polymorphisms of macrophage migration inhibitory factor as predictors of morbidity and mortality of pneumococcal meningitis. Proceedings of the National Academy of Sciences of the United States of America 113, 3597–3602, doi: 10.1073/pnas.1520727113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawadros T. et al. Release of macrophage migration inhibitory factor by neuroendocrine-differentiated LNCaP cells sustains the proliferation and survival of prostate cancer cells. Endocrine-related cancer 20, 137–149, doi: 10.1530/ERC-12-0286 (2013). [DOI] [PubMed] [Google Scholar]
- Vacher G. et al. Innate Immune Sensing of Fusarium culmorum by Mouse Dendritic Cells. Journal of toxicology and environmental health. Part A 78, 871–885, doi: 10.1080/15287394.2015.1051201 (2015). [DOI] [PubMed] [Google Scholar]
- Perreau M. et al. Exhaustion of bacteria-specific CD4 T cells and microbial translocation in common variable immunodeficiency disorders. The Journal of experimental medicine 211, 2033–2045, doi: 10.1084/jem.20140039 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R. et al. Normalization of mass cytometry data with bead standards. Cytometry 83A, 483–494, doi: 10.1002/cyto.a.22271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. et al. Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline. PLoS computational biology 12, e1005112, doi: 10.1371/journal.pcbi.1005112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G. et al. OpenCyto: an open source infrastructure for scalable, robust, reproducible, and automated, end-to-end flow cytometry data analysis. PLoS computational biology 10, e1003806, doi: 10.1371/journal.pcbi.1003806 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugrin J. et al. The sirtuin inhibitor cambinol impairs MAPK signaling, inhibits inflammatory and innate immune responses and protects from septic shock. Biochimica et biophysica acta 1833, 1498–1510, doi: 10.1016/j.bbamcr.2013.03.004 (2013). [DOI] [PubMed] [Google Scholar]
- Meller S. et al. T(H)17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nature immunology 16, 970–979, doi: 10.1038/ni.3211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger T. et al. High expression levels of macrophage migration inhibitory factor sustain the innate immune responses of neonates. Proceedings of the National Academy of Sciences of the United States of America 113, E997–E1005, doi: 10.1073/pnas.1514018113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkara L. T., Jiang W. & Zhang G. Modulation of antimicrobial host defense peptide gene expression by free fatty acids. PloS one 7, e49558, doi: 10.1371/journal.pone.0049558 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.