Abstract
Dysbiosis in the periodontal microbiota is associated with the development of periodontal diseases. Little is known about the initiation of dysbiosis. It was hypothesized that some commensal bacteria suppress the outgrowth of pathobionts by H2O2 production. However, serum and blood components released due to inflammation can neutralize this suppressive effect, leading to the initiation of dysbiosis. Agar plate, dual-species and multi-species ecology experiments showed that H2O2 production by commensal bacteria decreases pathobiont growth and colonization. Peroxidase and blood components neutralize this inhibitory effect primarily by an exogenous peroxidase activity without stimulating growth and biofilm formation of pathobionts directly. In multi-species environments, neutralization of H2O2 resulted in 2 to 3 log increases in pathobionts, a hallmark for dysbiosis. Our data show that in oral biofilms, commensal species suppress the amounts of pathobionts by H2O2 production. Inflammation can neutralize this effect and thereby initiates dysbiosis by allowing the outgrowth of pathobionts.
Although it is well known that complex poly-microbial oral biofilms are a necessity to develop tooth decay or periodontal diseases, they are often in balance with the host and consequently do not result in pathology1,2. Not their presence as such but changes in their composition and/or metabolic activity drive pathology. The current etiological model of periodontal disease, termed ‘polymicrobial synergy and dysbiosis’, proposes that changes in the periodontal microbiota or dysbiosis deregulate the host immune response, leading to chronic inflammation3,4. Although dysbiosis is characterized by a proportional increase of pathogenic species and a decrease of commensal species, little is known about the initiation of dysbiosis4.
Most likely, dysbiosis is initiated by a complex interplay between the bacterial community and the host. Commensal bacteria act with the host to prevent colonization or outgrowth of pathobionts that induce inflammation and disrupt the microbial ecology5. Pathobionts are natural members of the human microbiota that under certain perturbations to the host and/or microbiota can cause pathology6,7. Subgingival inter-bacterial correlations and competition between commensal bacteria and pathobionts in relation to disease severity have been shown8. As part of the oral commensal microbiota, highly abundant streptococci produce antimicrobial substances that limit the growth of pathobionts9. It has been shown clinically that in patients with periodontal disease, there is a decline in commensal streptococci such as Streptococcus sanguinis8,10. It was suggested that H2O2 is one of the most important antimicrobials produced by certain streptococci11. For instance, S. sanguinis and Streptococcus gordonii can inhibit the growth of Streptococcus mutans and Aggregatibacter actinomycetemcomitans by H2O2 production12,13. Recently, Rodriguez Herrero and coworkers showed that S. oralis, S. gordonii, S. cristatus, S. parasanguinis, S. mitis and S. sanguinis are good H2O2 producers, even under anaerobic conditions11. Although, it has been suggested that H2O2 has an important role in the formation and the composition of the oral biofilms14, the effect of H2O2 production on oral multi-species biofilm composition has not been investigated yet.
Gingival crevicular fluid (GFC) is an inflammatory exudate of serum that that covers oral biofilms15. It can modulate the composition of oral biofilms by inhibiting or enhancing the growth of certain oral species or by promoting biofilm formation and inhibiting the adhesion of certain species16. Additionally, serum and blood components can have peroxidase activity17. This is of significance since these could interact with the H2O2 produced by commensal bacteria to suppress the outgrowth of pathobionts. Clinically it is known that absence of bleeding on probing is a good marker for periodontal stability18. Recently, it is shown that crevicular myeloperoxidase concentrations are highly correlated with periodontal disease severity19.
Therefore, it can be hypothesized that commensal bacteria suppress the overgrowth of pathobionts by H2O2 but some serum and blood components released during inflammation can neutralize this suppressive effect, leading to the initiation of dysbiosis.
The objective of this study is to determine the neutralizing effect of serum, hemoglobin and hemin on the inhibitory effect of the commensal bacteria towards pathobionts.
Results
Decrease of the inhibitory effect of commensals on agar plates
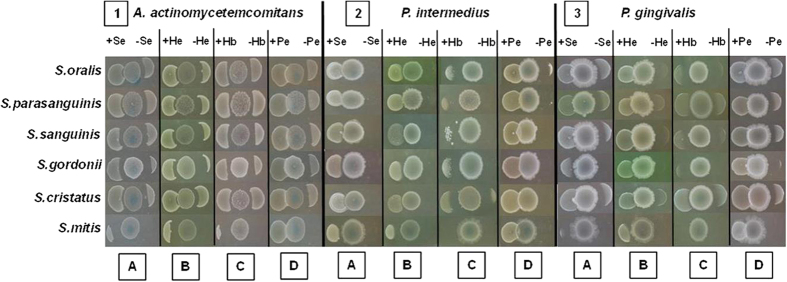
In order to verify the inhibitory effect of some commensal species on pathobionts and the influence of hemin, serum, hemoglobin and peroxidase, a qualitative agar-plate method was used. On agar plates, hemin, serum, hemoglobin and peroxidase significantly lowered the inhibitory effect of commensal bacteria on P. intermedia, P. gingivalis and A. actinomycetemcomitans (Fig. 1). This effect was observed for all commensal species, all substrates on all pathobionts with exception for the effect of hemoglobin on the inhibition of P. intermedia and P. gingivalis by S. mitis (Table 1). Only peroxidase was able to completely neutralize the inhibitory effect of all the commensals. In most cases, hemin lowered the inhibition of the commensals more than serum and hemoglobin.
Figure 1.
Neutralization effect of serum, hemin, hemoglobin and peroxidase on the inhibitory effect of commensal species towards A. actinomycetemcomitans. P. intermedia and P. gingivalis. The commensals were spotted 24 hours before the pathogens in the center of the pictures. The pathogens were spotted at both sides of the commensals, at the left side plus the blood compound (serum (+Se), hemin (+He), hemoglobin (+Hb) and peroxidase (+Pe)) and at the right side without any blood compound.
Table 1. Effect of the addition of serum, hemoglobin, hemin and peroxidase to BHI-2 agar on the antagonistic activity of commensal species.
| Serum | Hemoglobin | Hemin | Peroxidase | ||||||
|---|---|---|---|---|---|---|---|---|---|
| − | + | − | + | − | + | − | + | ||
| S. oralis | Aa | 2,04 ± 0,07 | 0,62 ± 0,17* | 2,08 ± 0,27 | 1,33 ± 0,21* | 2,35 ± 0,23 | 0,67 ± 0,03* | 2,31 ± 0,38 | 0,00 ± 0,00* |
| Pi | TI | 1,43 ± 0,08* | TI | 2,71 ± 0,25* | 3,63 ± 0,81 | 0,46 ± 0,20* | TI | 0,00 ± 0,00* | |
| Pg | TI | 1,37 ± 0,11* | 4,36 ± 0,37 | 2,58 ± 0,24* | 2,23 ± 0,02 | 0,00 ± 0,00* | 2,67 ± 0,16 | 0,00 ± 0,00* | |
| S. sanguinis | Aa | 3,04 ± 0,04 | 1,45 ± 0,17* | 2,67 ± 0,42 | 1,94 ± 0,21* | 2,70 ± 0,25 | 1,14 ± 0,03* | 3,08 ± 0,57 | 0,00 ± 0,00* |
| Pi | TI | 1,29 ± 0,07* | 3,37 ± 0,14 | 1,41 ± 0,28* | 2,75 ± 0,32 | 0,25 ± 0,19* | TI | 0,00 ± 0,00* | |
| Pg | 4,58 ± 0,11 | 0,78 ± 0,15* | 3,04 ± 0,12 | 2,17 ± 0,05* | 2,63 ± 0,39 | 0,00 ± 0,00* | 3,65 ± ± 0,35 | 0,00 ± 0,00* | |
| S. parasanguinis | Aa | 1,99 ± 0,04 | 0,75 ± 0,03* | 1,41 ± 0,27 | 0,47 ± 0,11* | 2,52 ± 0,14 | 0,80 ± 0,02* | 1,80 ± 0,24 | 0,00 ± 0,00* |
| Pi | TI | 0,00 ± 0,00* | TI | 3,05 ± 0,05* | TI | 3,64 ± 0,10* | TI | 0,00 ± 0,00* | |
| Pg | 2,63 ± 0,24 | 0,00 ± 0,00* | 1,14 ± 0,07 | 0,71 ± 0,10 | 1,62 ± 0,12 | 0,00 ± 0,00* | 1,87 ± 0,27 | 0,00 ± 0,00* | |
| S. mitis | Aa | TI | 2,83 ± 0,09* | 5,26 ± 0,85 | 2,56 ± 0,23* | TI | 2,37 ± 0,05* | TI | 0,00 ± 0,00* |
| Pi | TI | 1,76 ± 0,05* | TI | TI | TI | 1,36 ± 0,29* | TI | 0,00 ± 0,00* | |
| Pg | TI | 2,58 ± 0,16* | TI | TI | TI | 1,29 ± 0,13* | TI | 0,00 ± 0,00* | |
| S. gordonii | Aa | 2,68 ± 0,04 | 1,05 ± 0,11* | 3,07 ± 0,21 | 2,41 ± 0,26* | 3,75 ± 0,07 | 1,25 ± 0,02* | 2,11 ± 0,29 | 0,00 ± 0,00* |
| Pi | TI | 1,20 ± 0,13* | TI | 2,78 ± 0,08* | 4,42 ± 0,52 | 0,78 ± 0,31* | TI | 0,00 ± 0,00* | |
| Pg | TI | 0,24 ± 0,17* | TI | 3,40 ± 0,15* | 3,55 ± 1,02 | 0,31 ± 0,08* | 3,21 ± 0,44 | 0,00 ± 0,00* | |
| S. cristatus | Aa | 2,25 ± 0,15 | 0,94 ± 0,09* | 2,00 ± 0,32 | 1,16 ± 0,18* | 1,57 ± 0,06 | 0,45 ± 0,08* | 2,02 ± 0,33 | 0,00 ± 0,00* |
| Pi | TI | 0,82 ± 0,15* | TI | 2,49 ± 0,44* | TI | 1,33 ± 0,21* | TI | 0,00 ± 0,00* | |
| Pg | 2,96 ± 0,26 | 0,00 ± 0,00* | 3,05 ± 0,22 | 1,98 ± 0,24* | 1,61 ± 0,15 | 0,00 ± 0,00* | 2,98 ± 0,41 | 0,00 ± 0,00* | |
Data represent the magnitude of the zone of inhibition expressed in mm (mean ± standard deviation, n = 3) against A. actinomycetemcomitans (Aa), P. intermedia (Pi) and P. gingivalis (Pg). *Statistically significant decrease of the inhibitory effect from commensal bacteria by serum, hemoglobin, hemin and peroxidase (p < 0.05). TI = Total Inhibition. − = without addition; + = with addition, 0,00 = no inhibition.
Decrease of the inhibitory effect of commensals in dual-species planktonic cultures and biofilms
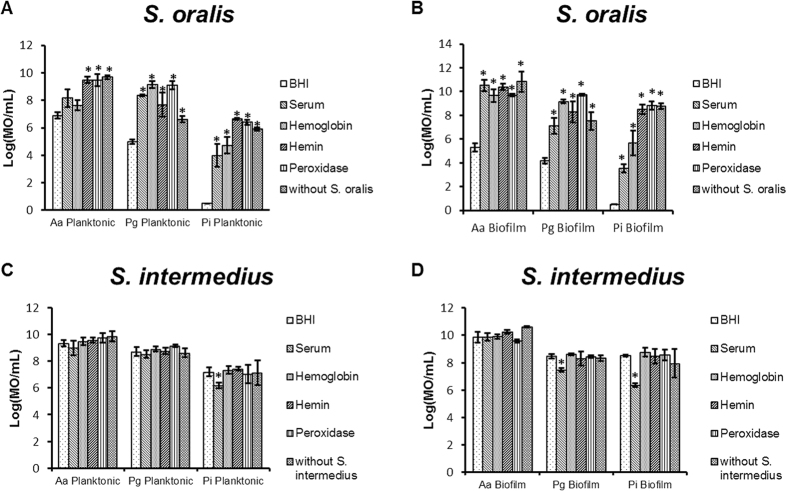
Since bacteria within the oral cavity primarily live as biofilms, which can change their behavior, the decreased inhibitory effect induced by hemin, serum, hemoglobin and peroxidase in agar-plate experiments was verified in dual-species planktonic and biofilm cultures containing a commensal species and a pathobiont. In dual-species planktonic cultures and biofilms, the inhibitory effect of S. oralis on P. intermedia, P. gingivalis and A. actinomycetemcomitans was decreased (p < 0.05) by serum, hemin, hemoglobin and peroxidase (Fig. 2A and B). The effect was not significant for serum and hemoglobin on planktonic A. actinomycetemcomitans. For most of the dual-species experiments, adding serum, hemin, hemoglobin and peroxidase completely abolished the inhibitory effect of S. oralis resulting in numbers of pathobionts similar to the negative control in which S. oralis was not present.
Figure 2. Neutralizing effect of serum and blood compounds in dual species interactions analyzed by PMA-qPCR (mean ± standard deviation, n = 3).
(A,B) Represent the competition between S. oralis and A. actinomycetemcomitans (Aa), P. gingivalis (Pg) and P. intermedia (Pi) in presence of BHI, serum, hemoglobin, hemin and peroxidase in planktonic and biofilm conditions. (C,D) represents the competition between S. intermedius and A. actinomycetemcomitans (Aa), P. gingivalis (Pg) and P. intermedia (Pi) in presence of BHI, serum, hemoglobin, hemin, peroxidase in planktonic and biofilm conditions. Date are expressed as number of micro-organisms (MO)/mL. *Designates a statistically significant increase of the bacterial concentration in respect to BHI (p < 0.05).
S. intermedius, a non-H2O2 producing species, did not show an inhibitory effect on the pathobionts (Fig. 2C and D). Consequently serum, hemin, hemoglobin and peroxidase could not decrease an inhibitory effect on the pathobionts but they also did not increase planktonic growth and biofilm formation in dual-species experiments. However, in these experiments, serum decreased the planktonic growth of P. intermedia and dual species biofilm formation of P. intermedia and P. gingivalis (p < 0.05).
Effect of serum, hemoglobin, hemin and peroxidase on single species cultures
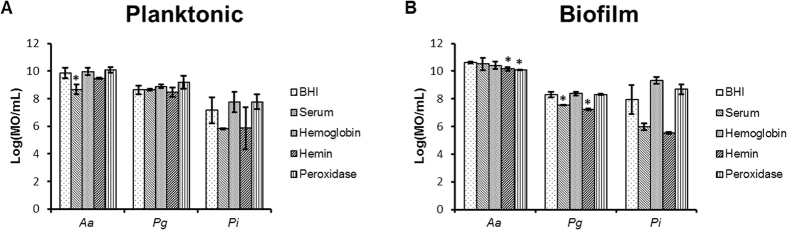
In order to verify if the outgrowth of the pathobionts in the dual-species experiments was due to a decreased inhibitory effect of the commensal species and not due to an increased growth or biofilm formation of the pathobionts by the presence of hemin, serum, hemoglobin or peroxidase. The effect of these substrates on pathobiont growth and biofilm formation was evaluated. Serum, hemin, hemoglobin and peroxidase did not increase growth or biofilm formation of P. intermedia, P. gingivalis and A. actinomycetemcomitans (Fig. 3). Moreover, serum and hemin induced a small, but significant reduction on P. gingivalis biofilm formation (p < 0.05). Additionally, A. actinomycetemcomitans growth was decreased by serum and its biofilm formation by hemin and peroxidase (p < 0.05).
Figure 3.
Effect of serum, hemoglobin, hemin and peroxidase on the growth (A) and biofilm formation (B) of A. actinomycetemcomitans (Aa), P. gingivalis (Pg) and P. intermedia (Pi) analyzed by PMA-qPCR (mean ± standard deviation, n = 3). Date are expressed as MO/mL. *Designates a statistically significant difference of the bacterial concentration in respect to BHI (p < 0.05).
Since it was technically impossible to directly measure the H2O2 concentration in cultures in the presence of hemin, hemoglobin or serum with the Amplex® Red Hydrogen Peroxide kit, the peroxidase activity of hemin, serum, hemoglobin and peroxidase was determined. Although all compounds showed peroxidase activity, Peroxidase and hemin showed a higher peroxidase activity when compared to hemoglobin and serum (Fig. 4). The peroxidase activity was only observed in the highest concentrations of serum and hemoglobin whereas a peroxidase activity was also detected in diluted concentrations of hemin and peroxidase.
Figure 4. Peroxidase activity of serum and blood compounds (mean ± standard deviation, n = 3).
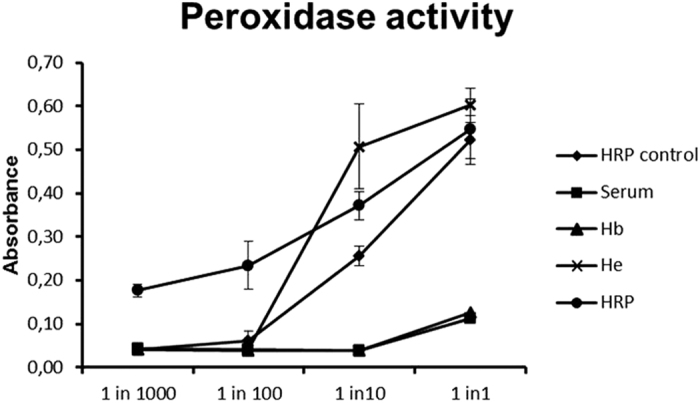
Peroxidase activity of different dilutions of serum and blood compounds was measured via the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit. Data are expressed as absorbance measured at 560 nm.
Reduction of the inhibitory effect of commensals in multi-species ecologies
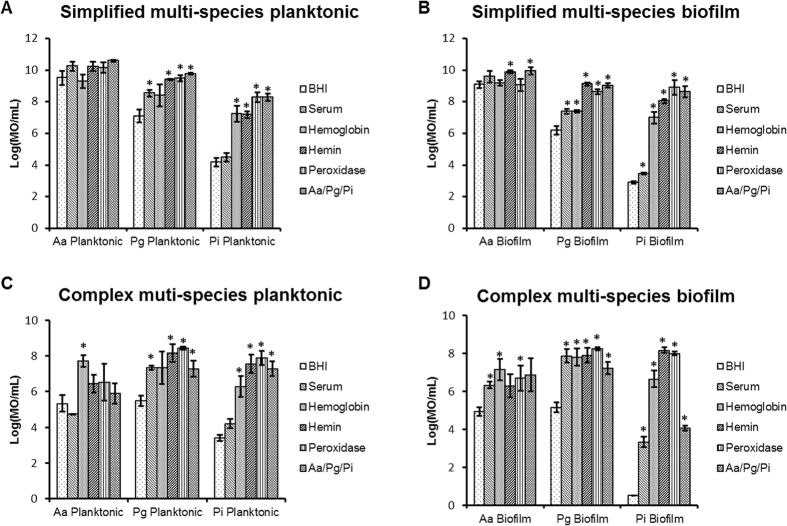
Bacteria within the oral cavity are part of complex microbial ecologies. Since the observed effects in dual-species experiments might be different in more complex ecologies, the effect of hemin, hemoglobin, serum and peroxidase on simplified and complex multi-species ecologies was examined. In both biofilm models, commensal biofilms containing S. oralis, S. gordonii, S. cristatus, S. parasanguinis, S. mitis and S. sanguinis were challenged with either only 3 pathobionts (simplified ecology) or with a complex 14 species ecology (complex ecology). The commensal biofilm significantly inhibited the planktonic and biofilm concentrations of P. gingivalis and P. intermedia both in simplified (Fig. 5A and B) and complex multi-species ecologies (Fig. 5C and D). Its effect on A. actinomycetemcomitans concentrations was limited in simplified multi-species ecologies (Fig. 5A and B). Although it was more pronounced in complex multi-species ecologies, it did not reach statistical significance (Fig. 5C and D). In general, serum, hemin, hemoglobin and peroxidase decreased the inhibitory effect of the commensal biofilm on planktonic and biofilm concentrations of the pathobionts. This inhibition resulted in an outgrowth of A. actinomycetemcomitans, P. gingivalis and P. intermedia respectively of up to 2.37 (±0.98), 4.48 (±0.62) and 2.94 (±0.38) log10 CFU/ml in complex planktonic multi-species ecologies and of up to 2.21 (±0.46), 8.17 (±0.14) and 3.12 (±0.18) log10 CFU/ml in complex multi-species biofilms. The neutralizing effect of serum and hemoglobin was less than that of hemin and peroxidase.
Figure 5. Neutralization effect of serum, hemoglobin, hemin and peroxidase on simple (3 pathobionts) and complex biofilms (14 species community) analyzed by PMA-qPCR (mean ± standard deviation, n = 3).
(A,B) Represent the concentration of A. actinomycetemcomitans (Aa), P. gingivalis (Pg) and P. intermedia (Pi) in presence of BHI, serum, hemoglobin, hemin and peroxidase in planktonic and biofilm conditions. (C,D) represent the concentration of A. actinomycetemcomitans (Aa), P. gingivalis (Pg) and P. intermedia (Pi) in presence of BHI, serum, hemoglobin, hemin and peroxidase in planktonic and biofilm conditions. Date are expressed as MO/mL. *Designates a statistically significant difference of the bacterial concentration in respect to BHI (p < 0.05).
The presence of the commensal biofilm did not only result in decreased planktonic concentrations of P. gingivalis and P. intermedia (Table 2). In the planktonic ecology, also a decreased concentration of S. salivarius and increased concentrations of S. sanguinis and S. oralis were observed when the commensal biofilm was present (p < 0.05). On the other hand, the biofilm concentrations of S. mutans, S. sobrinus and S. salivarius were decreased and the biofilm concentrations of F. nucleatum and S. sanguinis were increased (p < 0.05).
Table 2. Log (MO/ml) of the different species (mean ± standard deviation, n = 3) that form part of the 14 species community exposed to serum, hemoglobin, hemin and peroxidase.
| Planktonic | Biofilm | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BHI | Serum | Hemoglobin | Hemin | Peroxidase | Control | BHI | Serum | Hemoglobin | Hemin | Peroxidase | Control | |
| Aa | 5.34 ± 0.46 | 4.73 ± 0.03 | 7.72 ± 0.34* | 6.45 ± 0.53 | 6.55 ± 1.03 | 5.90 ± 0.55 | 4.93 ± 0.22 | 6.34 ± 0.19* | 7.14 ± 0.56* | 6.29 ± 0.60 | 6.69 ± 0.67* | 6.88 ± 0.88 |
| Pi | 3.41 ± 0.17 | 4.21 ± 0.26 | 6.30 ± 0.57* | 7.57 ± 0.42* | 7.89 ± 0.38* | 7.29 ± 0.42* | 0.00 ± 0.00 | 3.33 ± 0.27* | 6.65 ± 0.43* | 8.17 ± 0.14* | 7.99 ± 0.09* | 4.05 ± 0.14* |
| Pg | 5.49 ± 0.30 | 7.34 ± 0.12* | 7.34 ± 0.90 | 8.17 ± 0.19* | 8.44 ± 0.07* | 7.30 ± 0.45* | 5.15 ± 0.25* | 7.87 ± 0.36* | 7.81 ± 0.44* | 7.91 ± 0.39* | 8.25 ± 0.08* | 7.23 ± 0.32* |
| Fn | 9.23 ± 0.42 | 6.51 ± 0.21# | 9.40 ± 0.47 | 9.36 ± 0.39 | 9.74 ± 0.35 | 9.49 ± 0.26 | 10.42 ± 0.13 | 6.68 ± 0.59# | 10.64 ± 0.09 | 10.30 ± 0.04 | 10.04 ± 0.23 | 9.55 ± 0.10# |
| A. naeslundii | 2.97 ± 0.05 | 2.99 ± 0.19 | 3.46 ± 0.64 | 2.87 ± 0.11 | 3.51 ± 0.82 | 4.52 ± 0.94 | 7.07 ± 0.26 | 5.01 ± 0.40# | 6.87 ± 0.09 | 3.62 ± 1.54# | 5.35 ± 0.98 | 7.32 ± 0.09 |
| A. viscosus | 6.87 ± 0,41 | 3.95 ± 0.52# | 6.98 ± 0.21 | 3.89 ± 0.34# | 6.74 ± 0.44 | 6.62 ± 0.13 | 8.03 ± 0.46 | 6.45 ± 0.74# | 8.25 ± 0.42* | 4.79 ± 1.15# | 8.09 ± 0.27 | 7.81 ± 0.36 |
| S. mutans | 6.83 ± 0.05 | 5.66 ± 0.78 | 6.68 ± 0.11 | 5.75 ± 0.28# | 5.90 ± 0.75 | 7.28 ± 0.18 | 7.40 ± 0.21 | 7.53 ± 0.45 | 7.58 ± 0.11 | 6.97 ± 0.10 | 7.27 ± 0.01 | 8.33 ± 0.09* |
| S. sobrinus | 6.51 ± 0.26 | 5.44 ± 0.51# | 6.34 ± 0.19 | 6.10 ± 0.28 | 5.71 ± 0.30# | 7.08 ± 0.14 | 7.35 ± 0.27 | 7.17 ± 0.45 | 7.57 ± 0.17 | 7.53 ± 0.32 | 6.92 ± 0.05 | 9.01 ± 0.10* |
| S. sanguinis | 6.15 ± 0.30 | 5.85 ± 0.06 | 6.10 ± 0.29 | 5.42 ± 0.33 | 6.09 ± 0.17 | 5.06 ± 0.18# | 7.63 ± 0.09 | 7.69 ± 0.10 | 7.79 ± 0.10* | 6.91 ± 0.26# | 7.71 ± 0.05 | 6.31 ± 0.10# |
| S. gordonii | 8.57 ± 0.30 | 7.43 ± 0.02# | 8.46 ± 0.31 | 6.92 ± 0.20# | 8.48 ± 0.25 | 9.17 ± 0.29 | 9.32 ± 0.24 | 9.35 ± 0.13 | 9.41 ± 0.19 | 7.76 ± 0.35# | 9.32 ± 0.03 | 9.56 ± 0.08 |
| S. oralis | 7.68 ± 0.24 | 5.90 ± 0.11# | 7.54 ± 0.38 | 7.75 ± 0.48 | 7.81 ± 0.24 | 6.81 ± 0.17# | 8.43 ± 0,07 | 8.86 ± 0.10* | 8,64 ± 0.06 | 8.09 ± 0.19 | 8.79 ± 0.16 | 8.04 ± 0.21 |
| S. salivarius | 3.44 ± 0.27 | 3.99 ± 0.40 | 3.41 ± 0.20 | 3.01 ± 0.06 | 3.73 ± 0.29 | 6.23 ± 0.17* | 4.27 ± 0.28 | 4.42 ± 0.27 | 4.39 ± 0.41 | 4.23 ± 0.52 | 4.14 ± 0.68 | 6.41 ± 0.13* |
| V. parvula | 7.60 ± 0.19 | 6.66 ± 0.89 | 7.39 ± 0.14 | 7.74 ± 0.18 | 7.08 ± 0.19 | 7.57 ± 0.12 | 10.06 ± 0.38 | 9.67 ± 0.65 | 10.62 ± 0.13 | 10.36 ± 0.03 | 10.56 ± 0.19 | 10.27 ± 0.12 |
| S. mitis | 4.09 ± 0.14 | 3.48 ± 0.19 | 3.69 ± 0.11 | 2.74 ± 0.06# | 3.51 ± 0.08 | 2.93 ± 0.09 | 4.15 ± 0.22 | 4.34 ± 0.31 | 4.20 ± 0.23 | 3.24 ± 0.49 | 4.27 ± 0.20 | 2.65 ± 0.20 |
Control condition refers to a 14 species community without the addition of the 6 commensal bacteria. *Designates a statistically significant increase of the bacterial concentration in respect to BHI (p < 0.05). #Designates a statistically significant decrease of the bacterial concentration in respect to BHI (p < 0.05).
Moreover, in the complex multi-species ecologies (Table 2), the presence of serum increased the concentration of planktonic P. gingivalis (p < 0.05) and the concentrations of P. intermedia, P. gingivalis, A. actinomycetemcomitans and S. oralis in the biofilm (p < 0.05). Additionally, it decreased the planktonic concentrations of F. nucleatum, A. viscosus, S. oralis, S. sobrinus and S. gordonii (p < 0.05) and the concentrations of F. nucleatum, A. viscosus and A. naeslundii in the biofilm (p < 0.05).
In contrast, the planktonic concentrations of A. actinomycetemcomitans and P. intermedia as well as the biofilm concentrations of A. actinomycetemcomitans, P. gingivalis, P. intermedia, A. viscosus and S. sanguinis were increased in the presence of hemoglobin (p < 0.05).
The addition of hemin increased the planktonic concentrations of P. gingivalis and P. intermedia but decreased the concentrations of A. viscosus, S. mutans, S. gordonii and S. mitis (p < 0.05). In the biofilms, hemin increased the concentrations of P. gingivalis and P. intermedia and decreased the concentrations of A. viscosus, A. naeslundii, S. sanguinis and S. gordonii (p < 0.05).
Peroxidase increased the planktonic concentrations of P. gingivalis and P. intermedia (p < 0.05) and the biofilm concentrations of P. intermedia, P. gingivalis, A. actinomycetemcomitans (p < 0.05).
Discussion
Dysbiosis in oral bacterial communities is characterized by a microbial shift, which is translated in an increase of pathobionts and a decrease of commensal species8,10,20. It has been shown that some commensal species can suppress the growth of pathobionts by H2O2 under specific environmental conditions. More specifically, the sequence of colonization and the presence of oxygen are major influencing factors11. Additionally, dysbiotic biofilms are enriched in virulence factors that stimulate the host inflammatory response21. Although it can be deduced that both absence of commensal species and the presence inflammation are key factors, the factors driving dysbiosis are unclear4. Moreover, as far as the authors know, dysbiosis has never been induced in in vitro multi-species biofilms. In this study, it was hypothesized that commensal bacteria suppress the overgrowth of pathobionts by H2O2 but some serum and blood components released during inflammation can neutralize this suppressive effect, leading dysbiosis. It was shown on agar plates and in dual-species biofilms that H2O2 production by commensal bacteria decreases pathobiont growth and colonization. Although it was not directly tested in the current study, Herrero et al. showed that the amount of inhibition on pathobiont growth is determined by oxygen availability11. Commensal bacteria could produce H2O2 and inhibit pathobiont growth under anaerobic condition. However, H2O2 production and pathobiont inhibition was significantly higher under aerobic conditions. Therefore, oxygen availability must play an important role into the transition from homeostasis to dysbiosis11. If a non-H2O2 producing species was used in the current study, no inhibition was observed. Peroxidase and blood components neutralize the inhibitory effect of H2O2 primarily by a peroxidase activity since they did not stimulate the growth and biofilm formation of the pathobionts directly. In multi-species environments, neutralization of H2O2 by peroxidase or blood components resulted in 2 to 3 log increases in pathobionts which can be considered as a hallmark for dysbiosis.
The agar plate experiments showed an inhibition of the pathobionts when grown in the vicinity of the commensals. This inhibition was completely neutralized in the presence of peroxidase. These results were in concordance with previous studies11,12,22 that also identified H2O2 as the main inhibitory substance. It was observed that the magnitudes of inhibition described by Van Essche et al. were markedly smaller than the ones reported in the current study although the same bacterial strains were used23. This was attributed to the blood agar medium which was used in the Van Essche study and pointed towards an interference of certain blood compounds on the inhibition effect of these streptococcus species.
Gingival inflammation is characterized by an increase in GCF production, which is similar to serum, and bleeding tendency. Among many other components, blood contains serum, hemin and hemoglobin24 but the hemolytic capacity of pathobionts can also increase their concentration25. It is already described that hemoglobin and hemin have a peroxidase activity17,26,27. P. intermedia and P. gingivalis also generate layers of haems with catalytic activity to degrade the H2O2 using hemin and hemoglobin28. Additionally, serum and GCF contains catalytic molecules such as myeloperoxidase that neutralizes the antimicrobial effect of H2O219. Recently, it is shown that crevicular myeloperoxidase concentrations are highly correlated with periodontal disease severity. All these studies substantiate the observation that hemin, hemoglobin and serum decrease the inhibitory effect of the commensal species on agar plates by H2O2 reduction. The effects of hemin, hemoglobin, serum and the role of H2O2 were further demonstrated in dual- and multi-species biofilms. Their addition resulted in increases in pathobionts of up to 8 log values both in planktonic and in biofilm cultures. Although, it has been suggested that hemin and hemoglobin can increase the growth and biofilm formation of some pathogens, this effect was not seen29,30,31,32 (Fig. 3). This can be explained by the qPCR method used to quantify growth and biofilm formation and the growth medium used in this study which might provide sufficient iron sources to the pathobionts so that addition of hemin and hemoglobin did not affected their growth or biofilm formation anymore. The effect of serum on pathobiont growth and biofilm formation is even less reported in literature. Biyikoĝlu et al. reported no effects of serum on A. odontolyticus, V. parvula, F. nucleatum and P. gingivalis over time but a decrease in F. nucleatum biovolume when 4 species biofilms were grown in the presence of serum16. These data are in accordance with the present data.
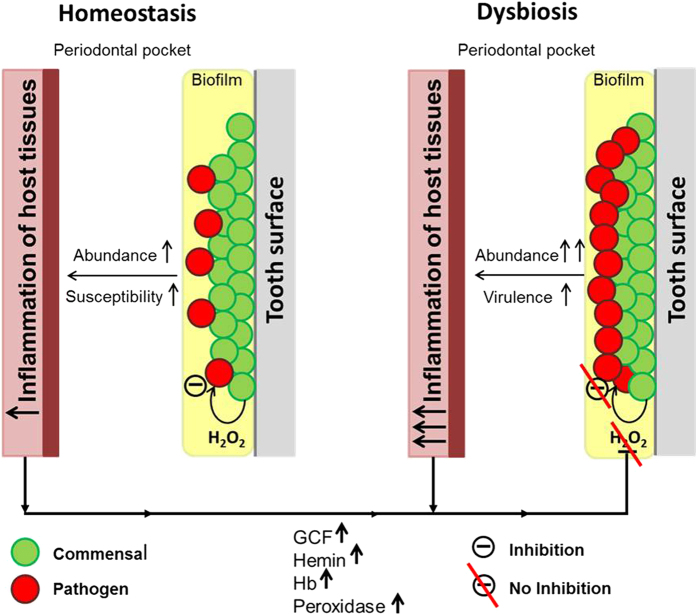
Overall, the study suggests that serum and blood compounds play an important role in the initiation of dysbiosis of oral biofilms by disrupting the inhibitory defensive barrier provided by the commensal bacteria. The data provide a hypothesis for the initiation of dysbiosis in oral biofilms (Fig. 6). In oral biofilms some commensal species can suppress the amounts of pathobionts by H2O2 generation. However, when these biofilms persist over longer periods of time or become more abundant or when the susceptibility of the host changes, the resulting inflammatory reaction neutralizes the effects of H2O2, thereby allowing the outgrowth of pathobionts. Additionally, changes in the oxygen availability within the biofilm might also lower the H2O2 production and subsequently contribute to the outgrowth of pathobionts.
Figure 6. Hypothetical model on the H2O2 neutralizing effect of hemin, hemoglobin and peroxidase on the inhibitory effect of commensal species towards periodontal pathobionts during inflammation.
The neutralization of H2O2 results in the outgrowth of periodontal pathobionts, leading to the initiation of dysbiosis in oral biofilms.
Methods
Bacterial strains and media
All used bacterial species (Streptococcus sanguinis LM14657, Streptococcus cristatus ATCC 49999, Streptococcus gordonii ATCC 49818, Streptococcus parasanguinis DSM 6778, Streptoccocus mitis DSM 12643, Streptoccocus oralis DSM 20627, Streptococcus salivarius TOVE-R, Streptococcus intermedius DSM 20573, Streptococcus mutans ATCC 20523, Streptococcus sobrinus ATCC 20742, Actinomyces viscosus DSM 43327, Actinomyces naeslundii ATCC 51655, Prevotella intermedia ATCC 25611, Porphyromonas gingivalis ATCC 33277, Fusobacterium nucleatum ATCC 20482, Aggregatibacter actinomycetemcomitans ATCC 43718 and Veillonella parvula DSM 2008) were maintained on blood agar (Oxoid, Basingstoke, UK) supplemented with 5 mg/mL hemin (Sigma, St. Louis, USA), 1 mg/mL menadione (Calbiochem-Novabiochem, La Jolla, USA) and 5% sterile horse blood (E&O Laboratories, Bonnybridge, Scotland). Overnight liquid cultures were prepared in Brain Hearth Infusion (BHI) broth (Difco, Detroit, USA). Competitive inhibition experiments were performed in Brain Hearth Infusion 2 (BHI-2) broth or agar containing Brain Heart infusion (Difco, Detroit, USA) supplemented with 2.5 g/L mucin (Sigma-Aldrich, St. Louis, USA), 1.0 g/L yeast extract (Oxoid, Basingstoke, UK), 0.1 g/L cysteine (Calbiochem, San Diego, USA), 2.0 g/L sodium bicarbonate and 0.25% (v/v) glutamic acid (Sigma-Aldrich, St Louis, USA). The bacteria were cultured under aerobic (5% CO2) or anaerobic (80% N2, 10% H2 and 10% CO2) conditions. Optical densities were measured and adjusted using spectrophotometry (OD600, GeneQuant Spectrophotometer, Buckinghamshire, UK).
Serum and blood components
Hemin, human hemoglobin and horseradish peroxidase (Sigma-Aldrich, St. Louis, USA) were dissolved in BHI-2 at concentrations of 5 mg/mL hemin, 0.44 mg/mL hemoglobin33 and 16 μg/mL peroxidase. Human serum was obtained by venipuncture of a single, systemically healthy, male volunteer with no oral disease and who had not taken any antibiotics for 1 year. Peripheral venous blood was immediately centrifuged at 264 × g for 30 min at room temperature. The serum was removed and frozen at −20 °C after aliquotation.
Ethics Statement
The use of human serum was approved by the ethical committee of the KU Leuven and registered with identifier B322201628215. The procedures were executed according to the Helsinki Declaration and the regulations of the University Hospital, which are approved by the ethical committee. The adult subject provided a written and oral consent after having explained to him the purpose of the study. The subject is aware that the results will be used in a scientific study. An informed consent was obtained from all subjects.
Antagonistic experiments on agar plates
The spotting technique was used to quantify the inhibitory effect of 6 commensal species on 3 pathobionts and to identify neutralization effects by serum and blood components11. An overnight culture of a commensal species was adjusted to a concentration of 109 CFU/mL. This solution was spotted on an agar plate and incubated under aerobic conditions. After 24 hours, an overnight culture of the pathobiont (109 CFU/mL) was spotted next to the commensal spot. After 48 hours of anaerobic incubation, a calibrated photograph was taken from the agar plate and the magnitude of inhibition was measured from the edge of the commensal colony to the border of the inhibited pathobiont colony using ImageJ (http://rsb.info.nih.gov/ij/download.html).
Dual-species planktonic and biofilm experiments
S. oralis was selected as the model commensal species with a strong inhibitory effect on pathobionts by H2O2 production. S. intermedius was used as a not-inhibiting commensal species (negative control)11. 10 mL of an overnight culture of these commensals was centrifuged (1438 × g, 10 minutes). The supernatant was discarded and the pellet was re-suspended in 10 mL BHI-2 broth. The density was adjusted to 1 × 108 CFU/ml. 6 ml of this solution was transferred to 6 wells (1 ml/well) of a 24 well-plate (Greiner, Frickenhausen, Germany) and incubated under aerobic conditions. After 24 hours, 500 μL of BHI-2 broth, serum, hemoglobin, hemin or peroxidase was added to the cultures. Additionally, an overnight culture of a pathobiont (A. actinomycetemcomitans, P. gingivalis or P. intermedia) was centrifuged (1438 × g, 10 minutes) and re-suspended in BHI-2 (1 × 108 CFU/ml). 1 mL of this bacterial solution was inoculated in each well with S. oralis or S. intermedius and in an additional well containing 1.5 mL BHI-2 (negative control). After 24 hours of anaerobic incubation, 1 mL was taken from each well, centrifuged (1438 × g, 10 min), re-suspended in PBS and analyzed via vitality q-PCR. Afterwards, the remaining supernatant was removed and the biofilms at the bottom of the wells were washed with phosphate buffered saline (PBS). The biofilms were detached with 500 μL 0,05% Trypsin-EDTA (Gibco, Paisley, UK) for 15 minutes at 37 °C, transferred to Eppendorf tubes, centrifuged (6010 × g, 10 minutes) and after discarding the trypsin, the biofilm pellets were re-suspended in 1 mL of PBS and analyzed by vitality q-PCR.
Effects on the growth and biofilm formation of pathobionts
An overnight culture of a pathobiont (A. actinomycetemcomitans, P. gingivalis or P. intermedia) was centrifuged (1438 × g, 10 minutes) and re-suspended in BHI-2 (1 × 108 CFU/mL). 1 mL of this bacterial solution was inoculated in each well plus 500 μl of BHI-2 broth, serum, hemoglobin, hemin or peroxidase. The wells were incubated for 24 hours under anaerobic conditions where after planktonic bacteria and the biofilms analyzed as described above.
Peroxidase activity of blood compounds and peroxidase on H2O2
50 μl of a 100 μM H2O2 solution was mixed with either 10 μl horseradish peroxidase (10 U/ml) (control) or 10 μl serum or 10 μl hemoglobin (0.44 mg/ml) or 10 μl hemin (5 mg/ml) or 10 μl peroxidase (16 μg/ml). Additionally, 50 μl of a 100 μM H2O2 solution was mixed with 10 fold serial dilutions of peroxidase, serum, hemoglobin, hemin and horseradish peroxidase in PBS (up to 1 in 1000). To 50 μl of these solutions, 50 μl of Amplex Red (50 μM) was added (Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit, Life technologies). After 20 minutes, the absorbance was measured at 560 nm (Powerwave XS Microplate Spectrophotometer, BioTek Instruments, Winooski, USA) according to the manufacturer’s instructions in order to determine the peroxidase activity of serum, hemin, hemoglobin and peroxidase.
Simplified multi-species planktonic and biofilm experiments
Similar to the dual-species experiments, overnight cultures of six commensal bacteria (S. oralis, S. gordonii, S. cristatus, S. parasanguinis, S. mitis and S. sanguinis), with inhibitory effects by producing H2O2, were centrifuged, re-suspended in BHI-2 broth (1 × 108 CFU/mL). Equal volumes of these solutions were mixed and inoculated in 6 wells of a 24 well-plate and incubated under aerobic conditions. After 24 hours, BHI-2, serum, hemoglobin, hemin and peroxidase were added to the wells as described above. Additionally, 1 mL of an overnight co-culture of 3 pathobionts (A. actinomycetemcomitans, P. gingivalis and P. intermedia) was centrifuged (1438 × g, 10 minutes), re-suspended in BHI-2 (1 × 108 CFU/ml) and added to the wells. The latter co-culture was obtained from overnight cultures of the pathobionts (simplified ecology) which were centrifuged (1438 × g, 10 minutes) and re-suspended in 10 ml of BHI-2. 1 mL of each pathobiont culture was added to 7 mL of BHI-2 and incubated for 24 hours under anaerobic conditions to obtain the co-culture. The wells were incubated for 24 hours under anaerobic conditions where after planktonic bacteria and the biofilms analyzed as described above.
Complex multi-species planktonic and biofilm experiments
The experimental set-up was identical to the set-up used for the simplified multi-species experiments with the exception that instead of using an overnight co-culture of 3 pathobionts, a bioreactor derived complex multi-species co-culture of 14 species (complex ecology), as described below, was used. The wells were incubated for 24 hours under anaerobic conditions where after planktonic bacteria and the biofilms analyzed as described above.
Bioreactor derived multi-species community
A multi-species community was established in a BIOSTAT B TWIN (Sartorius, Germany) bioreactor. 750 mL of BHI-2 broth was added to the vessel together with 5.0 mg/mL hemin, 1.0 mg/mL menadione and 200 μl/L Antifoam Y-30 (Sigma, St. Louis, USA). The medium was pre-reduced over 24 hours at 37 °C by bubbling 100% N2 and 5% CO2 in the medium under continuous stirring at 300 rpm. pH was set at 6.7 +/−0.1. After 24 hours, overnight cultures of S. sanguinis, S. gordonii, S. salivarius, S. mitis, S. oralis, S. mutans, S. sobrinus, A. viscosus, A. naeslundii, P. intermedia, P. gingivalis, F. nucleatum, A. actinomycetemcomitans and V. parvula were adjusted to an OD of 1.4 and added to the bioreactor. During the first 48 hours, the medium was not replaced. After that, the medium was replaced at a rate of 200 mL/24 hours.
Vitality q-PCR
DNA extraction and vitality q-PCR using propidium monoazide was previously described34. Table 3 shows primer and probe sequences used in this study.
Table 3. Primers and probes used for the detection and quantification by vitality qPCR.
| STRAIN | Primer/Probe (5′-3′) | Fragment length | |
|---|---|---|---|
| Aggregatibacter actinomycetemcomitans | Forward | GAA CCT TAC CTA CTC TTG ACA TCC GAA | 80 bp |
| Reverse | TGC AGC ACC TGT CTC AAA GC | ||
| Probe | AGA ACT CAG AGA TGG GTT TGT GCC TTA GGG | ||
| Fusobacterium nucleatum | Forward | GGA TTT ATT GGG CGT AAA GC | 162 bp |
| Reverse | GGC ATT CCT ACA AAT ATC TAC GAA | ||
| Probe | CTC TAC ACT TGT AGT TCC G | ||
| Porphyromonas gingivalis | Forward | GCG CTC AAC GTT CAG CC | 68 bp |
| Reverse | CAC GAA TTC CGC CTG C | ||
| Probe | CAC TGA ACT CAA GCC CGG CAG TTT CAA | ||
| Prevotella intermedia | Forward | CGG TCT GTT AAG CGT GTT GTG | 99 bp |
| Reverse | CAC CAT GAA TTC CGC ATA CG | ||
| Probe | TGG CGG ACT TGA GTG CAC GC | ||
| Streptococcus mutans | Forward | GCC TAC AGC TCA GAG ATG CTA TTC T | 114 bp |
| Reverse | GCC ATA CAC CAC TCA TGA ATT GA | ||
| Probe | TGG AAA TGA CGG TCG CCG TTA TGA A | ||
| Streptococcus sobrinus | Forward | TTC AAA GCC AAG ACC AAG CTA GT | 88 bp |
| Reverse | CCA GCC TGA GAT TCA GCT TGT | ||
| Probe | CCT GCT CCA GCG ACA AAG GCA GC | ||
| Actinomyces naeslundii | Forward | TCG AAA CTC AGC AAG TAG CCG | 96 bp |
| Reverse | AGA GGA GGG CCA CAA AAG AAA | ||
| Probe | GGG TAC TCT AGT CCA AAC TGG CGG ATA GCG | ||
| Streptococcus gordonii | Forward | CGG ATG ATG CTA ATC AAG TGA CC | 177 bp |
| Reverse | GTT AGC TGT TGG ATT GGT TGC C | ||
| Probe | AGA ACA GTC CGC TGT TCA GAG CAA | ||
| Actinomyces viscosus | Forward | GTG AAG GAG CCA GCT TGC TGG TTC TG | 155 bp |
| Reverse | CGG AAC AAA CCT TTC CCA GGC | ||
| Probe | ATG AGT GGC GAA CGG GTG AGT AAC | ||
| Streptococcus salivarius | Forward | AAC GTT GAC CTT ACG CTA GC | 192 bp |
| Reverse | ACC GTA ACG TGG GAA AAC TG | ||
| Probe | GTA GCG TCA GAG TGG TTG AC | ||
| Streptococcus oralis | Forward | ACC AGC AGA TAC GAA AGA AGC AT | 229 bp |
| Reverse | AGG TTC GGG CAA GCG ATC TTT CT | ||
| Probe | AAG GCT GCT GTT GCT GAA GAA GT | ||
| Streptococcus mitis | Forward | GGC TCG TAG TCT GGA GAT GG | 133 bp |
| Reverse | TAG GTC GTC GTC CCA AGG AA | ||
| Probe | CGA AGA GCA CCA ATA GCA CCT CCC | ||
| Streptococcus sanguinis | Forward | CAA AAT TGT TGC AAA TCC AAA GG | 75 bp |
| Reverse | GCT ATC GCT CCC TGT CTT TGA | ||
| Probe | AAA GAA AGA TCG CTT GCC AGA ACC GG | ||
| Veillonella parvula | Forward | GAC GAA AGT CTG ACG GAG CA | 171 bp |
| Reverse | TGC CAC CTA CGT ATT ACC GC | ||
| Probe | AGC TCT GTT AAT CGG GAC GAA AGG C |
Statistical analysis
All experiments were repeated on 3 different days. To account for the censored character of the inhibition data from agar plates experiments, differences between treatments (with serum, hemoglobin, hemin or peroxidase) and control (without addition of blood compound or peroxidase) for the inhibition data were analyzed by means of a survival regression model for gaussian data. Comparisons between treatments and control were made for each combination of substance (blood compounds or peroxidase) and bacteria and corrected for simultaneous hypothesis testing according to Sidak. For planktonic and biofilm data, a linear mixed model was fit to model the log-transformed CFU counts using substrate (blood compounds or peroxidase) and sample type (planktonic or biofilm) as fixed factors and run as random factor. Since a residual analysis showed that the model was heteroscedastic, weights, proportional to the inverse of the predicted value, were applied. Comparisons with BHI and control were made separately by calculating the appropriate contrasts and a correction for simultaneous hypothesis testing according to Dunnett was applied. Data were analyzed using S-plus 8.0 for Linux (Tibco, Palo Alto, CA, USA).
Additional Information
How to cite this article: Herrero, E. R. et al. Dysbiosis by neutralizing commensal mediated inhibition of pathobionts. Sci. Rep. 6, 38179; doi: 10.1038/srep38179 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This study was supported by grants of the KU Leuven (OT/12/101) and the Fund for Scientific Research Belgium (FWO G.0584.13).
Footnotes
Author Contributions Conceptualization, E.R.H., K.B., N.B. and W.T.; Methodology, E.R.H., E.H.S. N.B. and W.T.; Analysis, E.R.H. and W.T.; Investigation, E.R.H. and V.S.; Writing—Original Draft, E.R.H.; Writing—Review & Editing V.S., K.B., N.B. and W.T.; Funding Acquisition, W.T. and M.Q.; Resources, W.T., M.Q. and N.B.; Supervision, W.T., N.B. and K.B.
References
- Filoche S., Wong L. & Sissons C. H. Oral biofilms: emerging concepts in microbial ecology. J Dent Res. 89, 8–18 (2010). [DOI] [PubMed] [Google Scholar]
- Kuramitsu H. K., He X., Lux R., Anderson M. H. & Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 71, 653–670 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. & Lamont R. J. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 27, 409–419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. P. Defining functional signatures of dysbiosis in periodontitis progression. Genome Med. 7, 40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abt M. C. & Pamer E. G. Commensal bacteria mediated defenses against pathogens. Curr Opin Immunol. 29, 16–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugini C., Klepac-Ceraj V., Rackaityte E., Riggs J. E. & Davey M. E. Porphyromonas gingivalis: keeping the pathos out of the biont. J Oral Microbiol. 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J., Tang H. & Mazmanian S. K. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 23, 473–480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loozen G. et al. Inter-bacterial correlations in subgingival biofilms: a large-scale survey. J Clin Periodontol. 41, 1–10 (2014). [DOI] [PubMed] [Google Scholar]
- Burton J. P., Wescombe P. A., Cadieux P. A. & Tagg J. R. Beneficial microbes for the oral cavity: time to harness the oral streptococci? Benef Microbes. 2, 93–101 (2011). [DOI] [PubMed] [Google Scholar]
- Stingu C. S., Eschrich K., Rodloff A. C., Schaumann R. & Jentsch H. Periodontitis is associated with a loss of colonization by Streptococcus sanguinis. J Med Microbiol. 57, 495–499 (2008). [DOI] [PubMed] [Google Scholar]
- Herrero E. R. et al. Antimicrobial effects of commensal oral species are regulated by environmental factors. J Dent. 47, 23–33 (2016). [DOI] [PubMed] [Google Scholar]
- Kreth J., Zhang Y. & Herzberg M. C. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 190, 4632–4640 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliepen I., Hofkens J., Van Essche M., Quirynen M. & Teughels W. Aggregatibacter actinomycetemcomitans adhesion inhibited in a flow cell. Oral Microbiol Immunol. 23, 520–524 (2008). [DOI] [PubMed] [Google Scholar]
- Zhu L. & Kreth J. The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid Med Cell Longev. 2012, 717843 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G. S. Formation, collection and significance of gingival crevice fluid. Periodontol 2000. 31, 32–42 (2003). [DOI] [PubMed] [Google Scholar]
- Biyikoglu B., Ricker A. & Diaz P. I. Strain-specific colonization patterns and serum modulation of multi-species oral biofilm development. Anaerobe. 18, 459–470 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov A. et al. Peroxidase activity of hemoglobin-haptoglobin complexes: covalent aggregation and oxidative stress in plasma and macrophages. J Biol Chem. 284, 30395–30407 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang N. P., Adler R., Joss A. & Nyman S. Absence of bleeding on probing. An indicator of periodontal stability. J Clin Periodontol. 17, 714–721 (1990). [DOI] [PubMed] [Google Scholar]
- Leppilahti J. M. et al. Matrix metalloproteinases and myeloperoxidase in gingival crevicular fluid provide site-specific diagnostic value for chronic periodontitis. J Clin Periodontol. 41, 348–356 (2014). [DOI] [PubMed] [Google Scholar]
- Darveau R. P. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 8, 481–490 (2010). [DOI] [PubMed] [Google Scholar]
- Abusleme L. et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J., Merritt J., Shi W. & Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. 187, 7193–7203 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Essche M. et al. Bacterial antagonism against periodontopathogens. J Periodontol. 84, 801–811 (2013). [DOI] [PubMed] [Google Scholar]
- Carneiro L. G., Venuleo C., Oppenheim F. G. & Salih E. Proteome data set of human gingival crevicular fluid from healthy periodontium sites by multidimensional protein separation and mass spectrometry. J Periodontal Res. 47, 248–262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L., Bramanti T. E., Ebersole J. L. & Holt S. C. Hemolytic activity in the periodontopathogen Porphyromonas gingivalis: kinetics of enzyme release and localization. Infect Immun. 59, 1932–1940 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baureder M., Reimann R. & Hederstedt L. Contribution of catalase to hydrogen peroxide resistance in Enterococcus faecalis. FEMS Microbiol Lett. 331, 160–164 (2012). [DOI] [PubMed] [Google Scholar]
- Gregory E. M. & Fanning D. D. Effect of heme on Bacteroides distasonis catalase and aerotolerance. J Bacteriol. 156, 1012–1018 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley J. W., Birss A. J. & Silver J. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the mu-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol Lett. 183, 159–164 (2000). [DOI] [PubMed] [Google Scholar]
- Butler C. A. et al. The Porphyromonas gingivalis ferric uptake regulator orthologue binds hemin and regulates hemin-responsive biofilm development. PloS One. 9, e111168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan S. M., Nagata H., Shizukuishi S. & Wu J. Z. Degradation of human hemoglobin by Prevotella intermedia. Anaerobe. 12, 279–282 (2006). [DOI] [PubMed] [Google Scholar]
- Leung K. P., Subramaniam P. S., Okamoto M., Fukushima H. & Lai C. H. The binding and utilization of hemoglobin by Prevotella intermedia. FEMS Microbiol Lett. 162, 227–233 (1998). [DOI] [PubMed] [Google Scholar]
- Rhodes E. R. et al. Iron acquisition in the dental pathogen Actinobacillus actinomycetemcomitans: what does it use as a source and how does it get this essential metal? Biometals. 20, 365–377 (2007). [DOI] [PubMed] [Google Scholar]
- Smith M. J. & Beck W. S. Peroxidase activity of hemoglobin and its subunits: effects thereupon of haptoglobin. Biochim Biophys Acta. 147, 324–333 (1967). [DOI] [PubMed] [Google Scholar]
- Loozen G., Boon N., Pauwels M., Quirynen M. & Teughels W. Live/dead real-time polymerase chain reaction to assess new therapies against dental plaque-related pathologies. Mol Oral Microbiol. 26, 253–261 (2011). [DOI] [PubMed] [Google Scholar]