Abstract
Breast cancer is a complex disease, characterized by gene deregulation. There is less systematic investigation of the capacity of long intergenic non-coding RNAs (lincRNAs) as biomarkers associated with breast cancer pathogenesis or several clinicopathological variables including receptor status and patient survival. We designed a two-stage study, including 1,000 breast tumor RNA-seq data from The Cancer Genome Atlas (TCGA) as the discovery stage, and RNA-seq data of matched tumor and adjacent normal tissue from 50 breast cancer patients as well as 23 normal breast tissue from healthy women as the replication stage. We identified 83 lincRNAs showing the significant expression changes in breast tumors with a false discovery rate (FDR) < 1% in the discovery dataset. Thirty-seven out of the 83 were validated in the replication dataset. Integrative genomic analyses suggested that the aberrant expression of these 37 lincRNAs was probably related with the expression alteration of several transcription factors (TFs). We observed a differential co-expression pattern between lincRNAs and their neighboring genes. We found that the expression levels of one lincRNA (RP5-1198O20 with Ensembl ID ENSG00000230615) were associated with breast cancer survival with P < 0.05. Our study identifies a set of aberrantly expressed lincRNAs in breast cancer.
The advances in high-throughput sequencing technologies provide a systematic landscape in the complexity of transcriptome, especially the discovery of a wealth of long non-coding transcripts1,2. This phenomenon is referred to as pervasive transcription3, albeit some transcripts are probably the byproduct of the transcription noise4. Characterized as transcription from the intergenic regions, lincRNAs are specific types of long non-coding RNAs (lncRNAs), ranging in length from 200 nt to >100 kb5. Biochemically, many lincRNAs harbor mRNA-like features, such as the poly-adenosine (polyA+) tail and splicing6, enabling them to be detectable by polyA+ RNA-sequencing (RNA-seq) method. Functionally, there is mounting evidence that lincRNAs may act as modular scaffolds to recruit other active or repressive regulators7, resulting in widespread roles in imprinting control8, chromosome remodeling9, pluripotency and differentiation10, immune responses11 and cancer progression12. Many lincRNAs are expressed in high tissue or cell specificity10, serving lincRNA molecules as potential disease biomarkers.
Recently, the Cancer Genome Atlas (TCGA) has revealed that the aberrant expression of hundreds of protein-coding genes and tens of microRNAs are associated with breast cancer development, progression, metastasis13,14,15,16 and subtypes17,18. One large-scale study also reported hundreds of lincRNAs showing expression changes in breast cancer19, but this study has a lack of replication and comprehensive analyses. To further use lincRNAs as robust biomarkers, we performed an integrative analysis on RNA-seq data from approximately 1,000 breast tumors from the TCGA and an independent RNA-seq dataset of 50 pairs of matched tumors and adjacent normal tissues from breast cancer patients as well as 23 normal breast tissues from healthy women.
Results
Data summary
In the discovery stage, RNA-seq data for 85 pairs of breast tumors and adjacent normal tissues were collected, along with 830 primary breast tumors available from TCGA. In the replication stage, RNA-seq data from paired tumor and adjacent normal tissues from 50 breast cancer patients and normal breast tissue from 23 healthy women were included. Overall, the study subjects had similar clinical characteristics between the two stages (Table 1).
Table 1. Clinical characteristic of study subjects in this study.
| Discovery stage |
Replication stage |
|||
|---|---|---|---|---|
| Paired tumor-normal | Only tumor tissuesb | Paired tumor-normal tissues | Healthy Women | |
| No. of samples | 85 | 830 | 50 | 23 |
| Age (yr ± s.d.) | 57.2 ± 15.4 | 58.4 ± 12.9 | 51.7 ± 10.9 | 52.0 ± 11.3 |
| Ethnicity | ||||
| White | 79 (92.9%) | 588 (70.8%) | 39 (78%) | 18 (78%) |
| Asian | 1 (1.2%) | 56 (6.8%) | 0 | 0 |
| Black | 5 (5.9%) | 79 (9.5%) | 11 (22%) | 5 (22%) |
| Unknown | 0 | 107 (12.9%) | 0 | 0 |
| ER status | ||||
| ER+ | 67 (78.9%) | 597 (71.9%) | 40 (80%) | NA |
| ER− | 15 (17.6%) | 181 (21.8%) | 10 (20%) | NA |
| Unknown | 3 (3.5%) | 52 (6.3%) | 0 | NA |
| Stage | ||||
| 0-I | 16 (18.8%) | 135 (16.2%) | 2 (4%) | NA |
| II | 46 (54.1%) | 469 (56.5%) | 14 (28%) | NA |
| III | 20 (23.5%) | 179 (21.6%) | 6 (12%) | NA |
| IV | 2 (2.4%) | 13 (1.6%) | 1 (2%) | NA |
| Unknown | 1 (1.2%) | 34 (4.1%) | 27 (54%) | NA |
aBRCA (breast invasive carcinoma) with matched adjacent normal tissues.
bBRCA without matched adjacent normal tissues.
Also included in the present study were Genome-wide protein-DNA interaction data using chromatin immunoprecipitation followed by sequencing (ChIP-seq) for 26 transcription factors (TFs) in MCF-7 cell lines from the ENCODE project, and two independent ChIP-seq datasets for transcription factor ERα each with replicated experiments in the MCF-7 cells20,21, as well as RNA-seq data across 14 breast cancer cell lines (Table S1).
Global changes of the lincRNA species in breast cancer
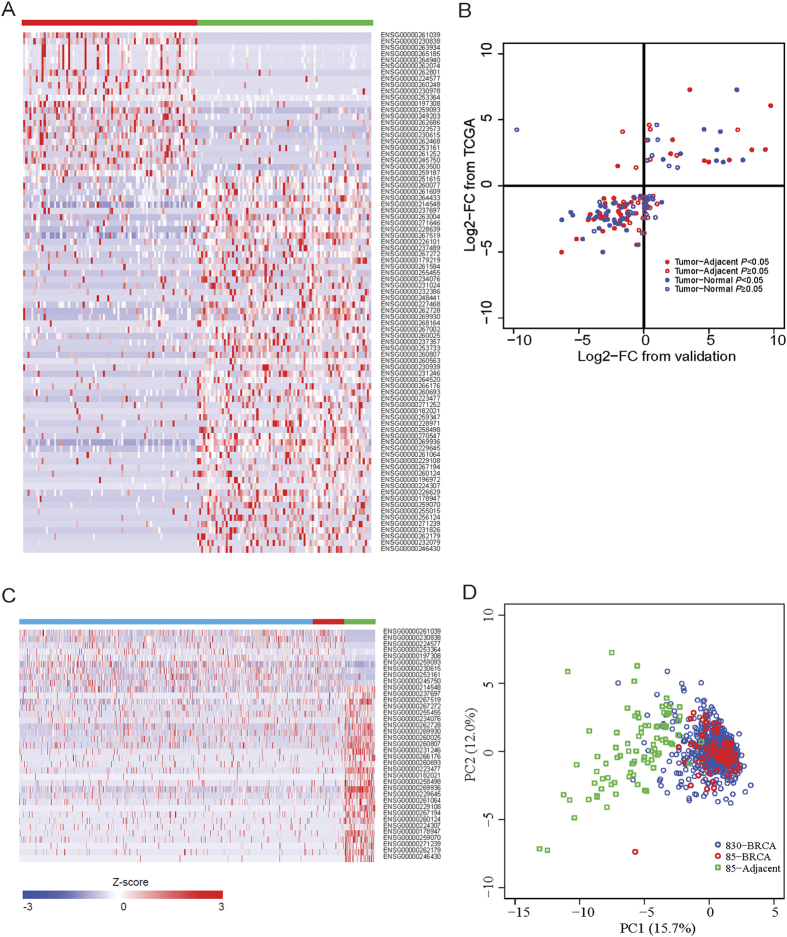
Our transcriptomic analyses focused on the lincRNA species, a specific type of lncRNAs. First, we investigated the lincRNA expression profile in 85 paired samples of primary breast tumor tissue and adjacent normal tissue from TCGA. After stringent filtering (see Methods), we totally identified 2,171 lincRNAs expressed in at least one sample. As expected, most lincRNAs expressed at low levels and sparsely expressed in <10% of all samples (Figure S1). Similar observations were also reported elsewhere22,23,24. For subsequent analysis, we focused on 584 lincRNAs that expressed in at least 20% of samples. Compared with the matched adjacent normal tissues, we observed that these lincRNAs showed a global decreased expression profile in breast tumors (Figure S2). The similar trend has been reported for small non-coding RNAs in multiple cancer types25,26. Specifically, among these 584 lincRNAs, 83 were significantly differential expression (DE) with a permutation-based FDR < 1% (Fig. 1A). Among them, 60 lincRNAs displayed down-regulation in breast tumor tissues.
Figure 1. Expression differences of lincRNAs between human breast cancer and normal tissues.
(A) Heatmap of 83 differentially expressed lincRNAs in 85 pair-matched breast cancer samples. Red and green bars denote 85 breast cancer specimens and adjacent normal tissues from TCGA, respectively. Colors ranging from blue to red represent the relative expression levels (Z-score) of lincRNAs. (B) Dot plot of the Log2-transformed FC for 83 lincRNAs between 50 paired breast tumor and adjacent normal tissues (Tumor−Adjacent), and between 50 breast tumors and 23 normal tissues from healthy women (Tumor−Normal) as validation (X-axis) versus that from TCGA breast cancer patients (Y-axis). (C) Heatmap of 37 differentially expressed and replicated lincRNAs in 830 unmatched tumors (blue bar) in combined with 85 pairs of tumors (red bar) and adjacent normal tissues (green bar). Values range in color from blue to red, meaning the relative expression levels. (D) PCA of 85 paired breast cancer and adjacent normal samples (85 BRCA and 85 Adjacent) and 830 unpaired breast cancer samples (830 BRCA) based on 37 differentially expressed lincRNAs.
Validation of lincRNA expression in an independent study
To validate the DE lincRNAs identified in TCGA, we generated an independent RNA-seq dataset from 50 pairs of matched tumor and adjacent normal samples from breast cancer patients and 23 normal breast tissues from healthy women in the replication study. Among the 83 DE lincRNAs, 66 lincRNAs (80%) showed consistent expression change direction (Fig. 1B). Specifically, 37 lincRNAs (45%) were replicated with absolute log2-FC ≥ 1 and P < 0.05 in the 50 pairs of matched tumor and adjacent normal breast tissues in replication stage (Table 2). Over 80% of them also showed significant expression difference in the tumors compared to normal breast tissues in the validation stage (Table 2). When we relaxed the cutoff to absolute log2-FC ≥ 0.585 and P < 0.05 in the replication dataset (Table S2), 47 DE lincRNAs (57%) could be replicated in our independent study, which is consistent with the previous report19.
Table 2. Expression alteration of 37 lincRNAs in discovery and replication stages.
| Ensembl ID | Symbol | Discovery stagea |
Replication Stageb |
Replication Stagec |
|||
|---|---|---|---|---|---|---|---|
| Log2-FCd | P | Log2-FCd | P | Log2-FCd | P | ||
| ENSG00000261039 | RP11-417E7 | 7.04 | 1.47E-08 | 3.49 | 2.42E-03 | 7.11 | 6.45E-05 |
| ENSG00000230838 | AC093850 | 5.84 | 6.87E-08 | 9.72 | 1.19E-10 | 15.03 | 1.42E-11 |
| ENSG00000224577 | AC017048 | 3.26 | 4.31E-05 | 2.17 | 2.69E-02 | 1.03 | 4.56E-01 |
| ENSG00000253364 | RP11-731F5 | 2.52 | 4.88E-03 | 9.31 | 1.78E-09 | 13.17 | 1.16E-08 |
| ENSG00000197308 | GATA3-AS1 | 2.49 | 2.08E-05 | 8.27 | 6.36E-07 | 5.58 | 1.43E-02 |
| ENSG00000259093 | RP11-1112J20 | 2.20 | 6.87E-08 | 2.02 | 8.65E-07 | 2.78 | 4.02E-06 |
| ENSG00000230615 | RP5-1198O20 | 1.72 | 2.80E-04 | 6.53 | 1.75E-13 | 7.58 | 3.50E-09 |
| ENSG00000253161 | RP11-150O12 | 1.68 | 1.00E-03 | 4.61 | 4.12E-05 | 5.54 | 9.67E-04 |
| ENSG00000245750 | RP11-279F6 | 1.57 | 1.17E-03 | 4.96 | 1.63E-04 | 6.07 | 1.47E-03 |
| ENSG00000214548 | MEG3 | −1.17 | 2.47E-07 | −2.35 | 1.15E-07 | −0.75 | 2.18E-01 |
| ENSG00000237697 | LINC00312 | −1.18 | 9.13E-03 | −3.10 | 1.24E-04 | 0.41 | 7.27E-01 |
| ENSG00000267519 | CTD-3252C9 | −1.48 | 6.89E-11 | −1.01 | 4.12E-05 | 1.33 | 2.00E-05 |
| ENSG00000267272 | RP5-1052I5 | −1.56 | 2.64E-05 | −3.02 | 2.65E-04 | −2.37 | 3.67E-02 |
| ENSG00000255455 | RP11-890B15 | −1.62 | 1.28E-04 | −1.59 | 3.60E-04 | −1.35 | 3.52E-02 |
| ENSG00000234076 | TPRG1-AS1 | −1.74 | 3.52E-03 | −1.93 | 1.12E-03 | −3.37 | 2.06E-05 |
| ENSG00000262728 | AC123768 | −1.82 | 4.90E-08 | −1.58 | 1.27E-02 | −3.01 | 2.80E-04 |
| ENSG00000269930 | RP11-932O9 | −1.85 | 9.95E-07 | −1.82 | 8.56E-05 | −1.74 | 7.45E-03 |
| ENSG00000260025 | RP11-490M8 | −2.15 | 1.10E-06 | −1.99 | 1.18E-02 | −2.78 | 6.05E-03 |
| ENSG00000260807 | RP11-161M6 | −2.24 | 1.64E-06 | −1.96 | 4.49E-02 | −5.65 | 1.34E-05 |
| ENSG00000231246 | RP5-965F6 | −2.32 | 4.87E-07 | −3.42 | 5.22E-03 | −2.94 | 7.71E-02 |
| ENSG00000266176 | RP11-855A2 | −2.39 | 9.44E-04 | −4.16 | 9.90E-05 | 0.16 | 9.21E-01 |
| ENSG00000260693 | AC026150 | −2.44 | 1.08E-06 | −1.81 | 3.04E-04 | −1.19 | 8.99E-02 |
| ENSG00000223477 | LINC00842 | −2.46 | 8.86E-08 | −2.93 | 7.21E-04 | −5.50 | 1.07E-06 |
| ENSG00000182021 | RP11-381O7 | −2.57 | 2.33E-05 | −3.26 | 1.17E-06 | −3.67 | 8.14E-05 |
| ENSG00000258498 | DIO3OS | −2.68 | 1.01E-04 | −3.34 | 2.49E-05 | −3.76 | 9.62E-05 |
| ENSG00000269936 | MIR145 | −2.70 | 9.87E-13 | −1.34 | 5.52E-04 | −1.61 | 1.80E-03 |
| ENSG00000229645 | LINC00341 | −2.77 | 6.89E-11 | −3.89 | 7.12E-06 | −2.82 | 1.50E-02 |
| ENSG00000261064 | RP11-1000B6 | −2.79 | 9.95E-07 | −1.33 | 7.23E-03 | −1.77 | 6.81E-03 |
| ENSG00000229108 | AC005550 | −2.81 | 2.28E-05 | −4.31 | 4.11E-04 | −6.34 | 7.85E-05 |
| ENSG00000267194 | RP1-193H18 | −2.81 | 2.97E-06 | −2.32 | 1.43E-03 | −3.05 | 7.26E-04 |
| ENSG00000260124 | RP4-791K14 | −2.84 | 8.18E-06 | −1.84 | 8.56E-06 | −2.32 | 4.72E-05 |
| ENSG00000224307 | RP11-344B5 | −3.10 | 7.24E-04 | −2.07 | 2.87E-03 | −1.88 | 3.44E-02 |
| ENSG00000178947 | LINC00086 | −3.23 | 1.94E-09 | −1.59 | 2.60E-02 | −2.66 | 5.14E-03 |
| ENSG00000259070 | LINC00639 | −3.45 | 2.88E-07 | −2.30 | 8.87E-04 | −3.48 | 1.40E-04 |
| ENSG00000271239 | RP11-238F2 | −3.67 | 3.37E-06 | −3.19 | 3.08E-02 | −3.86 | 5.24E-02 |
| ENSG00000262179 | RP1-302G2 | −4.26 | 6.95E-07 | −5.21 | 3.39E-06 | −4.74 | 1.56E-03 |
| ENSG00000246430 | RP11-16M8 | −5.25 | 2.61E-06 | −6.36 | 4.27E-09 | −3.19 | 3.97E-02 |
aThe discovery stage from TCGA.
b,cThe replication stage from an independent study; bcomparison between paired tumor and adjacent normal breast tissue from cancer patients; ccomparison between tumor and normal breast tissue from healthy controls.
dLog2-FC denotes log2-transformed fold change between breast tumor and non-tumor samples.
We further examined the expression profiling of the 37 replicated lincRNAs in the remaining 830 breast tumor tissues samples without matched normal tissues from the TCGA dataset. The expression patterns of these 37 lincRNAs in the 830 tumor samples were very similar to those in 85 normal-paired breast tumor samples, but very different from those in adjacent normal breast tissue samples in the discovery stage (Fig. 1C). Principal component analysis (PCA) based on these 37 lincRNAs also showed that all breast tumor samples are clustered together, but separated from the adjacent normal tissues (with a few exceptions) (Fig. 1D). In addition, we also checked the expression levels of these 37 lincRNAs across 14 breast cancer cell lines that are commonly used in the lab (Table S3). Most of these 37 lincRNAs were expressed in at least one common breast cancer cell line, making these cell lines as possible in vitro model to study lincRNAs. Altogether, these results suggested that these 37 lincRNAs showed aberrant expression in breast tumors.
Association of LincRNAs with transcription factors
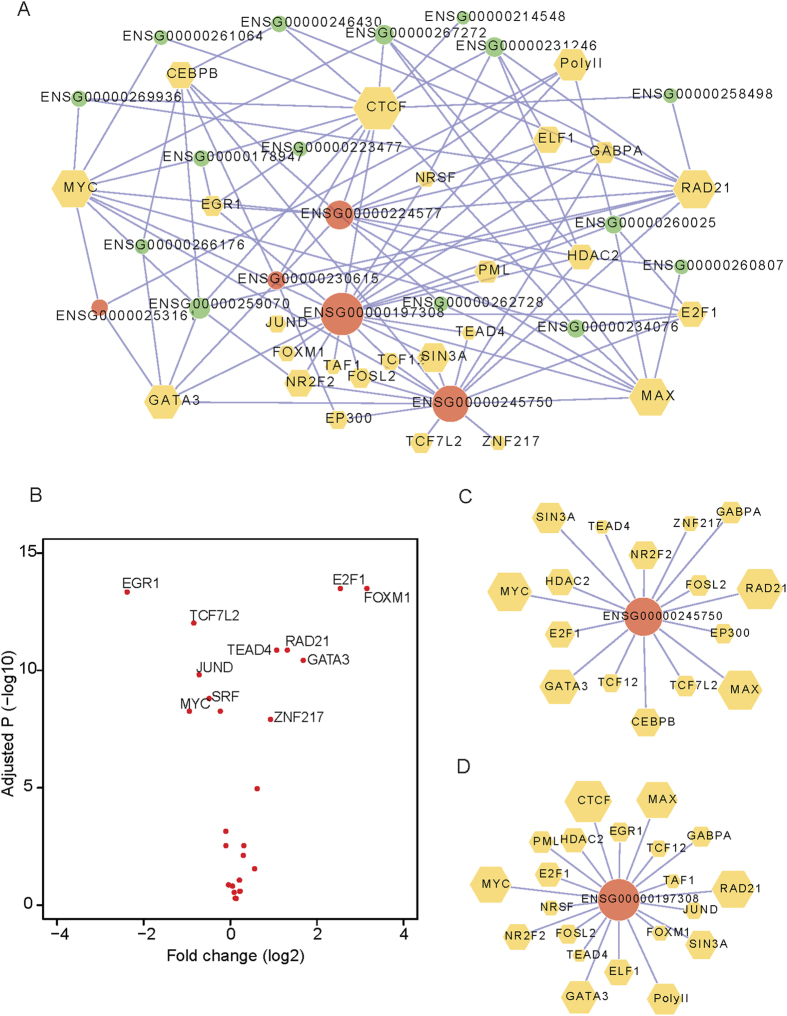
We then integrated 26 known TFs ChIP-seq data in MCF-7 cells, an ER+ breast cancer cell line from the ENCODE project (Table S4), to investigate whether and to what extent the 37 differentially expressed lincRNAs are functionally related to TFs. We constructed a lincRNA-TF bipartite network (see Methods), which consisted of 45 nodes and 101 edges (Fig. 2A). We found that 20 (54.1%) lincRNAs were bound by at least one TF, and 25 of the 26 TFs regulated at least one lincRNA. Especially, each of the eight key TFs, including GATA3, RAD21, MYC, CTCF, MAX, E2F1, CEBPB and RNA polymerase II, binds ≥ 5 lincRNAs. Among them, 11 TFs exhibited a significant differential expression (absolute log2-FC ≥ 0.585, BH-adjusted P < 0.01) in breast cancer tissues compared with adjacent normal tissues (Fig. 2B). We also found that several over-expressed lincRNAs may be related to these key TFs. For example, two lincRNA genes (RP11-279F6 with Ensemble ID ENSG00000245750 and GATA3-AS1 with Ensemble ID ENSG00000197308) had DNA binding peaks by 16 and 20 TFs, respectively. Specifically, all three over-expressed key regulators in breast tumors, GATA3, E2F1 and RAD21, showed connection with these two lincRNAs (Fig. 2C,D). These results suggest that the expression alteration of lincRNAs in breast cancer is probably functionally associated by one or multiple TFs in a complex way.
Figure 2. LincRNAs link to the regulation by transcription factors.
(A) LincRNA-transcription factors (TFs) bipartite network. Each node with size scaled by its degree (the number of directly linked nodes) denotes one TF (yellow hexagon) or lincRNA (circle). Each edge represents the regulation of lincRNAs by TFs. Red and green circles represent the over-expressed and down-expressed lincRNAs in breast cancer samples, respectively. (B) Volcano plot of the log2-transformed fold change between breast cancer and adjacent normal tissues (X-axis, n = 85 pairs) versus –log10-transformed BH adjusted P value (Y-axis) for TFs (n = 26). The marked TFs show significantly differential expression in breast cancers relative to adjacent normal tissues. (C,D) Examples of two lincRNA genes (RP11-279F6 with Ensemble ID ENSG00000245750 and GATA3-AS1 with Ensemble ID ENSG00000197308) regulated by multiple TFs.
Co-expression of lincRNAs with nearby protein-coding genes
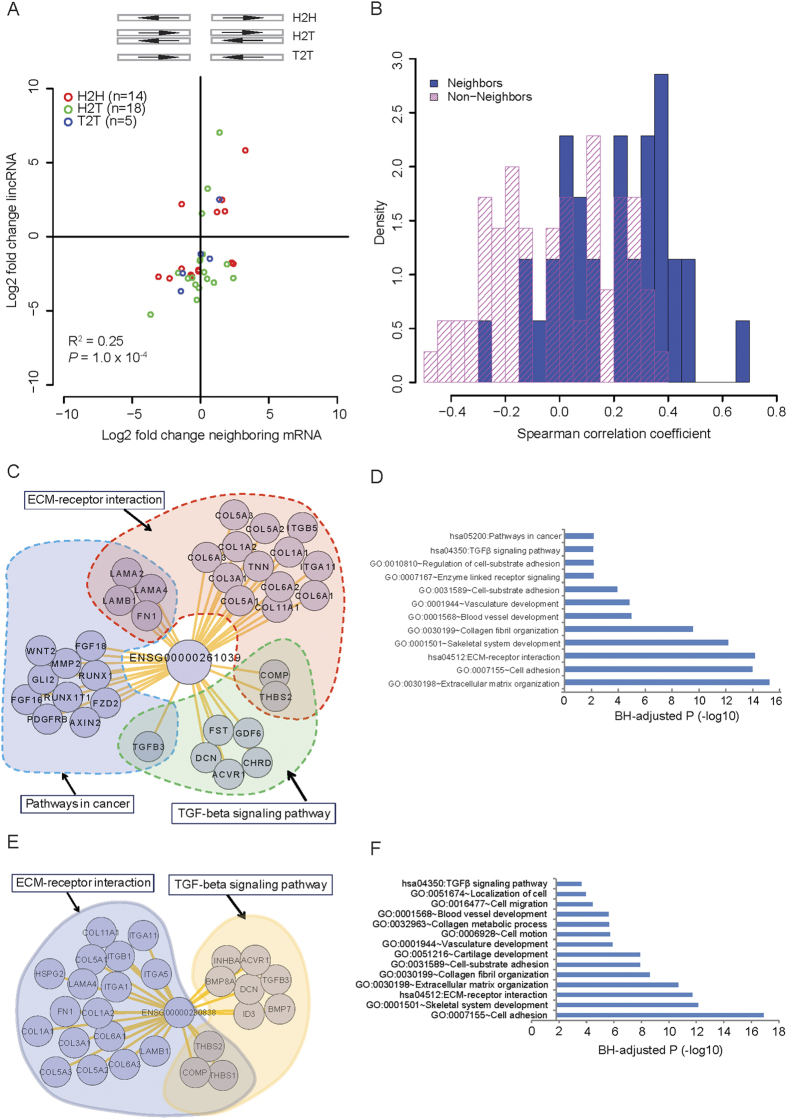
Several studies have reported that lincRNAs show an expression correlation with their neighboring protein-coding genes in normal tissues/cells5,27. We examined whether 37 DE lincRNAs could also exhibit the differential co-expression profile with the adjacent genes in breast cancer development. We classified these 37 lincRNA/mRNA pairs into three types: head-head (H2H), head-tail (H2T) and tail-tail (T2T) based on the paired transcriptional orientation (Fig. 3A). We observed a coordinated expression change (r2 = 0.25, P = 1.0 × 10−4) between lincRNAs and their neighbor mRNAs via a comparison of 85 paired breast tumor and adjacent normal breast tissues (Fig. 3A and B). We detected 14 (37.8%) lincRNA/mRNA pairs with the H2H type, which shows marginally significant over-representation (p = 0.06, one-sided Binomial test) relative to the global distribution. Similar results were observed when we analyzed 83 initially detected DE lincRNAs (Figure S3). Taking the H2H pair of GATA3-AS1 (Ensembl ID ENSG00000197308) and its adjacent (aparting from 1220 bp) protein-coding gene GATA3 as an example (Figure S4), the correlation coefficient between these two genes was 0.54 with P = 6.7 × 10−5. These observations are consistent with the previous reports that both GATA3-AS1 and GATA3 are co-expressed in mouse and human TH2 cells28,29. Altogether, the lincRNAs divergently transcribed with protein-coding genes are more likely to show the differentially co-expression profiles.
Figure 3. Co-expression differences between lincRNAs and neighboring mRNAs.
(A) Plot of log2-transformed FC between 85 paired breast cancers for 37 lincRNAs (Y-axis) versus nearby mRNAs (X-axis). H2H, H2T and T2T represent the head-to-head, head-to-tail and tail-to-tail orientation between lincRNA and neighboring mRNAs, respectively (top panel). (B) Distribution of Spearman correlation coefficient from 85 adjacent normal tissues for lincRNA-neighboring pairs (blue) and lincRNA-non-neighboring pairs (pink). Functional enrichment and remarkable correlated protein-coding genes associated with lincRNA ENSG00000261039 (C,D) and ENSG00000230838 (E,F).
To further determine the uniqueness of the differential co-expression observation of lincRNA-mRNA adjacent pairs, we characterized the co-expression patterns of non-neighboring lincRNA-mRNA pairs and randomly shuffled lincRNA-mRNA pairs using the same 85 pairs of breast cancer/adjacent normal tissue in discovery stage (see Methods). As expected, there were no co-expression changes for the random pairs (Figure S5). In addition, the neighboring lincRNA-mRNA pairs showed a higher Spearman’s rank correlation coefficient than those of non-neighboring lincRNA-mRNA pairs (Fig. 3B).
Functional Prediction of lincRNAs
To date, thousands of lincRNAs have been annotated, while the biological functions are unclear for most of them. Based on the Refseq genes showing strong co-expression relationship with lincRNAs, a method commonly used for functional prediction of unknown genes30,31, we predicted the biological functions for these 37 lincRNAs (see Methods). Enrichment analysis of GO terms and KEGG pathways showed that these down-regulated lincRNAs might be associated with transcriptional regulation, RNA processing and translational elongation processes, etc. In contrast, the up-regulated lincRNAs probably participate in cancer cell migration and proliferation, including cell adhesion, regulation of epithelial cell proliferation and regulation of cell cycle (Table S5). For example, two over-expressed lincRNAs (RP11-417E7 with Ensemble ID ENSG00000261039 and AC093850 with Ensemble ID ENSG00000230838) showed over-expression in both the discovery and the replication stage. These two lincRNAs may be involved in the ECM-receptor interaction and cell adhesion, TGFβ signaling pathway and others (Fig. 3C–F). The TGFβ signaling pathway is well documented with a promoter of tumor progression and invasion32. Together, these two lincRNAs probably participate in the pathogenesis of breast cancer.
LincRNAs associated with breast cancer subtypes
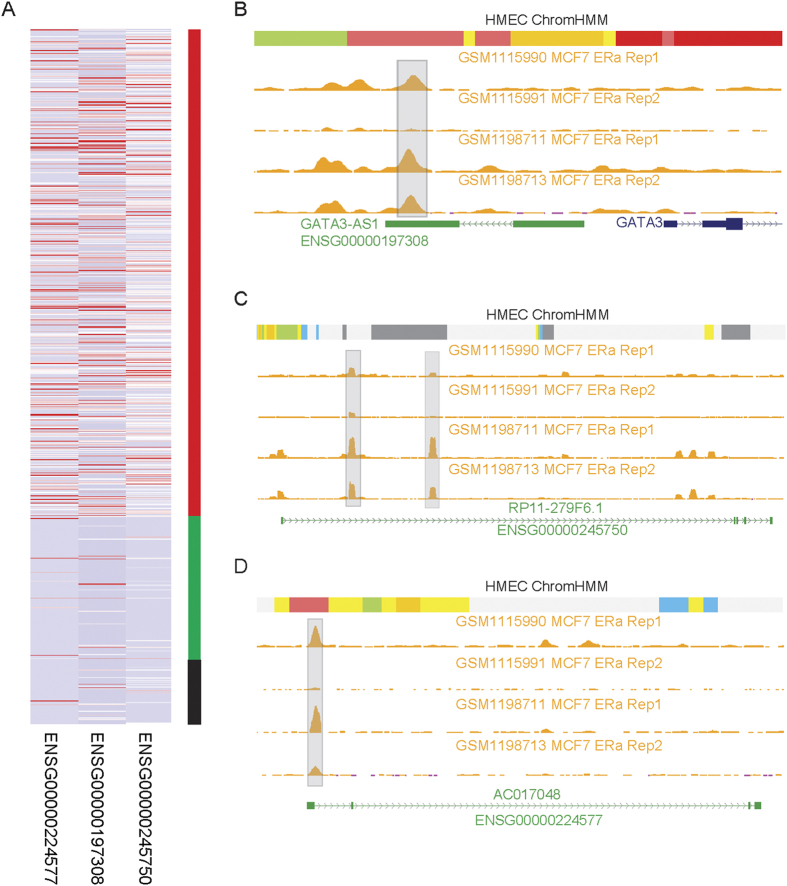
We also investigated whether these 37 DE lincRNAs exhibited expression difference across different breast cancer subtypes. We found three lincRNAs: GATA3-AS1 (ENSG00000197308), RP11-279F6 (ENSG00000245750) and AC017048 (ENSG00000224577), showed specifically high expression levels in ER-positive (ER+), compared to ER-negative (ER-) cancers and normal breast tissue samples (Fig. 4A). The specific expression alteration of these three lincRNAs in ER+ subtype was also validated in the replication stage. We also used the data of differentially expressed lincRNAs across four breast cancer subtypes (Luminal A, Lumnal B, Her2, and Basal-like) reported in Su et al. study30 to evaluate the subtype associations for these 37 lincRNAs. We found that the 22 (59.5%) lincRNAs (8 up-regulated lincRNAs and 14 down-regulated lincRNAs) were differentially expressed in at least one breast cancer subtype (Table S6). Three above reported lincRNAs over-expressed in the ER+ subtype (Fig. 4A) showed much higher expression levels in Luminal A and Lumnal B subtypes (enriched for ER+), relative to Her2, and Basal-like subtypes.
Figure 4. Specific expression of lincRNAs in breast cancer subtypes.
(A) Heatmap of three lincRNAs specifically over-expressed in ER+ breast cancer. Red and green represent 664 ER+ and 196 ER- cancer samples from TCGA, respectively. Black bar denotes 85 adjacent normal tissues. Distribution of DNA binding by ERα in three lincRNA genes, (B) GATA3-AS1 (Ensemble ID ENSG00000197308), (C) RP11-279F6 (Ensemble ID ENSG00000245750) and (D) AC017048 (Ensemble ID ENSG00000224577). The gray bars represent the DNA binding enrichment for the ERα in the MCF-7 cells. The track in the top for each lincRNA is the chromatin states from the chromHMM algorithm in the HMEC cell line. Chromatin states with bright red and light red, orange and yellow, blue, green and grey represent active promoter and weak promoter, strong enhancer and weak/poised enhancer, insulator, transcriptional region and heterochromatin/low signal, respectively.
Further integrating two independent estrogen receptor alpha (ERα) ChIP-seq dataset (each with two replicates) in MCF-7 cancer cells, we observed the ERα binding sites in all three lincRNA loci (Fig. 4B–D). For example, an ERα DNA binding site was found near the transcriptional terminal region of the GATA3-AS1 based on the analysis of ERα ChIP-seq data (Fig. 4B). This binding region was further annotated as the active enhancer region using chromatin states in human mammary epithelial cells (HMECs). Similarly, this enhancer state was also observed in the other two lincRNAs (Fig. 4C,D). These results indicated that these three ER status associated lincRNAs may be regulated by ERα.
LincRNAs associated with breast cancer survival
We assessed the association of those 37 lincRNAs expression levels with breast cancer survival and found one candidate (RP5-1198O20 with Ensembl ID ENSG00000230615, Fig. 5). Patients with expression levels of lincRNA ENSG00000230615 higher than the median value (median FPKM value is 2.8) had worse survival rates than those women with expression levels less than the median value (median FPKM value is 0.5) (Fig. 5).
Figure 5. Association of lincRNA expression levels with breast cancer survival.
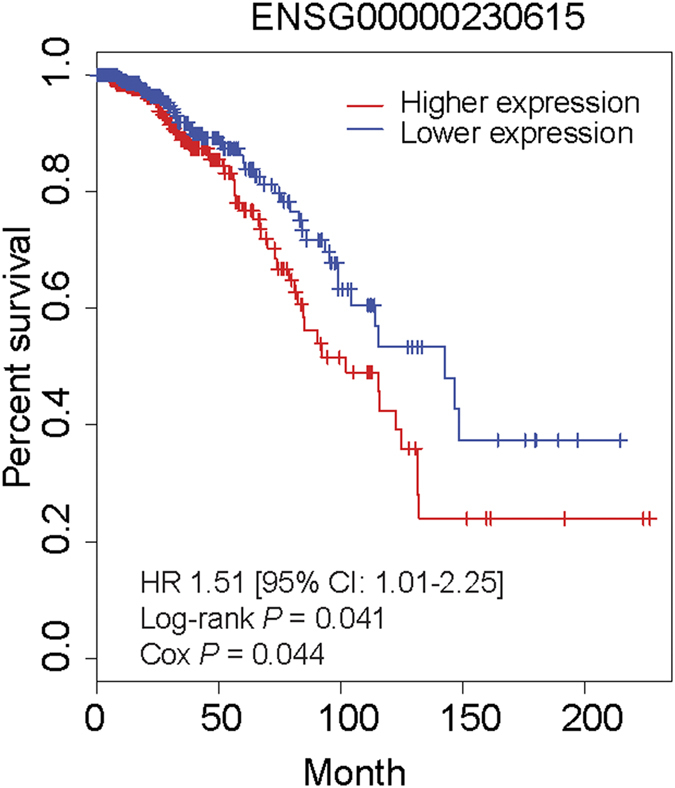
Kaplan-Meier survival plots for the lincRNA RP5-1198O20 (Ensemble ID ENSG00000230615). Two groups (higher and lower expression) are separated on the basis of the median expression level of lincRNAs across breast cancer patients.
Discussion
In this study, we analyzed lincRNA transcriptome in over 1000 breast tissue samples. Two independent sequencing datasets consistently identified a set of lincRNAs deregulated in breast carcinogenesis. Importantly, the protein-coding genes neighboring these deregulated lincRNA loci also showed expression alternation in breast cancer tissues, implying the transcriptional concordance between lincRNAs loci and neighboring genomic regions in cancer development. The expression aberration of lincRNAs in breast cancer may be associated with the expression alteration of multiple TFs. Our work substantially extends the biological understanding of the lincRNA repertoire in the pathogenesis of breast cancer.
It should be noted that the aberrant expression of several lincRNAs previously identified in multiple cancers showed deregulation in the present study. For example, in both discovery and validation stages, expression levels of the MEG3 decreased in breast tumor samples, which is consistent with the proposed tumor suppressor role for MEG312,30. Another un-regulated lincRNA, GATA3-AS1 (Ensembl ID ENSG00000197308) was also identified in Ding et al. study33. Our in silico functional prediction (Table S5) indicated the GATA3-AS1 probably performs an immune response associated role in breast cancer progression. This result was supported by previous two studies29,34 showing that the GATA3-AS1 is highly expressed in T helper subsets. Spurlock et al. also speculated that the GATA3-AS1 might play a role in allergic or asthmatic responses34. In addition, this lincRNA was also reported to have a decreased expression level in brain, bladder and prostate cancers22. Another breast cancer survival associated lincRNA RP5-1198O20 (Ensembl ID ENSG00000230615) showed the up-regulation in both stages of this study. Re-examining Gibb et al. integrated SAGE-seq findings22, we confirmed that this lincRNA also showed an increased expression in breast cancers. Another study about transcriptome analysis of aging identified the down-regulation of this lincRNA35. Further functional investigation of the lincRNA RP5-1198O20 in either carcinogenesis or aging would be interesting. In another example, our data showed the down-regulation of the miRNA-145 host gene (MIR145 with Ensembl ID ENSG00000269936) in breast cancer, consistent with previous reports in breast cancer36 and other cancer types37,38. However, several well-characterized lincRNAs12,22,23,39,40, including HOTAIR, H19, GAS5, PCA3, PVT1, were not investigated in this study. Those lncRNAs are not transcribed from intergenic regions, belonging to either anti-sense or other types of long non-coding transcripts, according to the GENCODE (version17) annotation. That means these lncRNAs are difficult to distinguish from their host protein-coding genes using the typical RNA-seq technology. There are several lncRNAs with no polyA tails41,42 which are unmeasured in the current study as well. Therefore, more sophisticated methods, such as strand-specific and non-poly(A) tail RNA-seq technology, are required to distinguish anti-sense transcripts from protein-coding genes, and to comprehensively capture the lncRNA transcriptome.
Several lincRNAs show expression alteration in other cancers, including NEAT1 (Ensembl ID ENSG00000245532) down-regulated in retinoblastoma22, MALAT1 (Ensembl ID ENSG00000251562) up-regulated in lung and colorectal cancer43,44. However, these lincRNAs do not exhibit expression aberration in breast cancer. Likewise, the PCAT1 (Ensembl ID ENSG00000253438), a prostate-specific lincRNA regulating cell proliferation, shows over-expression in a subset of prostate cancers45. In our study, we almost did not detect the expression level of the PCAT1 in either breast normal or breast tumor samples. These results partially support the observation that many lincRNAs are expressed in a tissue- and cancer-type restricted manner, making them useful as prognostic markers12.
However, some limitations remain in this study. Firstly, no functional validations on these DE lincRNAs prevent us draw the further conclusion about how the aberrant expression of lincRNAs contribute to tumorigenesis. Secondly, the relative small sample size and the unavailability of breast cancer subtype data in the replication stage also prevent us replicate the subtype-specific lincRNAs. Finally, due to the tumor heterogeneity and the cell mixture in tumor tissues, global comparison of gene expression profiles among breast tumor, adjacent and normal tissues is insufficient, particularly for lincRNAs whose expression patterns broadly show the cell- or tissue-type specificity. Thus, single-cell or sorted cell population based transcriptomic analysis46,47 will be favorable to determine these lincRNAs as robust biomarkers.
Conclusions
We identified a signature of lincRNA expression profile for breast cancer. Further functional surveys of these lincRNAs will be warranted to discover the biological mechanisms of lincRNA in breast cancer development and progression.
Materials and Methods
Discovery dataset
After approval, raw RNA-seq data (Level 1) of breast tumor tissues (N = 915) and adjacent normal breast tissues (N = 85) were acquired from the TCGA. Clinical data (Biotab format) for these 915 breast invasive carcinoma (BRCA) were also acquired from the TCGA.
Replication dataset
Our replication study consisted of 50 breast cancer cases and 23 healthy controls. The patients were pathologically confirmed for primary breast cancer diagnosed at one of three hospitals in Indianapolis, Indiana, between 1998 and 2009: Indiana University Hospital, Wishard Hospital (now Eskenazi Hospital), and the Indiana University Simon Cancer Center (IUSCC). Controls were randomly selected from a pool of healthy women who donated normal breast tissues to the Susan G. Komen Tissue Bank between 2005 and 2009, and were free of breast cancer at the time of donation. The participants completed a questionnaire on medical histories and health-related exposures at the time of donation. Controls were frequency matched to cases based on self-reported ancestry and age (within 5 years).
Breast tissue (untreated tumor or normal) biospecimens were collected from each case and control under standard operating procedures at IUSCC. All breast tissue samples were snap-frozen immediately after removal and stored in liquid nitrogen until processed, and were determined to be of high quality through histological and molecular quality control tests. Tumor samples were pathologically verified for high tumor content. Information concerning demographics, clinical data, and personal risk factors, including age and race, were either extracted from medical records (for cases) or obtained through the questionnaires completed by the participants (for controls).
Total RNA was extracted from freshly frozen breast tissue samples using the Qiagen® miRNeasy Mini Kit. Construction of cDNA libraries and subsequent RNA sequencing of paired-end reads (2 × 50 nt reads) were performed according to the standard Illumina protocol using the HiSeq2000 sequencing systems. The raw sequencing output was 25–35 million reads per sample.
Other dataset
The annotation data for known lincRNAs (n = 6,020) were extracted from the gencode.v17.long_noncoding_RNAs.gtf.gz file from the GENCODE database. Other data used in this study include: human Refseq genes obtained from the NCBI database; protein-DNA interactions from ChIP-seq data in MCF-7 breast cancer cell lines downloaded from ENCODE project48; DNA binding by ERα using ChIP-seq in MCF-7 cells, and RNA-seq data across 14 breast cancer cell lines49,50 downloaded from the Gene Expression Omnibus (GEO) database51.
RNA-seq data processing and lincRNA annotation
In the discovery stage, for the 85 pair of matched breast tumor and normal tissues, mapped reads in BAM format (Level 1) were assembled with Cufflinks (v2.1.1)52. The lincRNA annotation was conducted by the following procedures: 1) we retained assembled transcripts whose genomic loci are overlapped or imbedded with known annotated lincRNAs; 2) we removed transcripts in length of <200 nt; 3) we eliminated transcripts showing coding potential (score <0.5) predicted with iSeeRNA program (version 1.2)53; 4) if two or more transcripts (isoforms) mapped to a lincRNA locus, we assigned the mean coverage and expression value to that lincRNA. We applied the same procedures for the remaining RNA-seq data, except for publically accessible RNA-seq data that were processed as described elsewhere54. To reduce the noise caused by the lincRNA expression variability between samples, we plotted the calling rate (occurrence of lincRNA transcription based on Cufflinks FPKM ≥ 0.3) versus lincRNAs ranked by missing rate across 170 samples. As Ramskold et al. proposed55, this cutoff (FPKM ≥ 0.3) is an optimized threshold for detectable expression above background. Then, we set the calling rate ≥ 20% as a threshold to ensure high-confidence lincRNAs for further analyses.
For RNA-seq data in the replication stage, quality control (QC) filtering was first performed on raw RNA-seq data to remove adapter sequences and poor quality bases using the FastqMCF clipping algorithm. RNA-seq reads were then mapped by Bowtie v1.0.0, to GENCODE lncRNA reference (release 17) for lncRNA (including lincRNA) annotations. Transcript abundances were quantified using NGSUtils. Samples were further filtered based on percentage of genes detected (less than 50%) and percentage of reads mapped to the reference (less than 25%). Extreme outliers were further identified and filtered from the dataset using principal component analysis (PCA). After these steps, a total of 7,450 lncRNAs retained and were used in further analyses.
ChIP-seq data processing
For ChIP-seq data from the ENCODE project, we directly downloaded files with called peaks for subsequent analyses. For two independent ERα ChIP-seq data whose processed peaks were unavailable, we downloaded the raw data in FASTQ format and conducted the peak calling as follows. Reads were mapped to the human reference genome (hg19) using Bowtie2 program56 in the default parameters. Aligned data were processed and converted into BAM files using SAMtools program57. Then, we used the MACS14 (version 1.4.2) program58 to call peaks in 20 bp resolution. We visualized the results in the UCSC Genome Browser.
Differential expression of lincRNAs
The fragments per kilobase of exon per million fragments mapped (FPKM) values were calculated from the Cufflinks program to represent lincRNA expression levels. The differential expression of each lincRNA between breast tumor and adjacent normal tissues was defined as: fold change (FC) ≥ 2 with Benjamini-Hochberg (BH) adjusted P < 0.01 based on non-parametric Wilcoxon rank sum paired test.
Following the permutation-based method developed by Xie et al.59, we estimated the FDR in identifying lincRNAs with differential expression. Briefly, FDR is estimated as 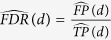 , where d is a fixed threshold,
, where d is a fixed threshold, 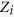 is a test statistic and B = 1000 permutations.
is a test statistic and B = 1000 permutations.  . The 1000 permutations were achieved by shuffling the tumor/normal samples in the lincRNA-sample matrix (584 lincRNAs and 170 samples).
. The 1000 permutations were achieved by shuffling the tumor/normal samples in the lincRNA-sample matrix (584 lincRNAs and 170 samples).
For RNA-seq data in the replications stage, differential expression (DE) analyses were performed using edgeR v2.6.12, implemented in the Bioconductor package to identify differentially expressed lncRNAs between tumor and adjacent normal, as well as tumor and normal breast tissue. The trimmed mean of M-values (TMM) method was used to normalize the raw read counts. Biological coefficients of variation between the samples were estimated using an empirical Bayes approach under the assumption that the data follows a negative binomial distribution. Differential expression between tumor and adjacent normal or normal breast tissue was analyzed using a generalized linear model to regress lncRNA (expression on tissue type, adjusting for age, race, and sequencing batch). Statistical significance was defined as FDR p-value < 0.05 and a two-fold change (FC) of expression level between comparison of tumor and adjacent normal or normal breast tissue.
Construction of transcription factor and lincRNA network
We downloaded ChIP-seq peak files of 26 transcription factors (TFs) in the MCF-7 breast cancer cells from the ENCODE project. A lincRNA bound by a TF was determined, if the TF binding peak is located within the region of 2 kb upstream of its transcription start site (TSS) to transcription end site (TES) of the lincRNA locus. Then, a lincRNA-TF bipartite network was constructed using the Cytoscape (v2.8.3) software60, where each node denotes a lincRNA or TF and each edge represents a lincRNA-TF interaction. The degree of each node in this bipartite network was calculated. We regarded the nodes representing TFs having ≥ 5 degrees as key TFs.
Co-expression analysis
We used similar methods for lincRNA annotation to annotate Refseq protein-coding genes and calculate their FPKM values. The neighboring protein-coding genes were defined as the ones with the closest physical distance to known lincRNAs. We coupled lincRNA-encoding locus and neighboring protein-coding genes, and referred to it as a lincRNAs-mRNA pair. We further classified lincRNAs-mRNA pairs into three types, according to their transcriptional orientation: head-to-head (H2H), head-to-tail (H2T) and tail-to-tail (T2T). The linear relationship of log2-transformed fold change in a comparison of 85 paired samples for lincRNA-mRNA pairs was used to evaluate the coordinated changes.
We defined the non-neighboring genes as the ones over a 1 Mb physical distance from lincRNAs on both strands. We coupled them as non-neighboring lincRNAs-mRNA pairs. Similarly, we determined the coordinated changes for non-neighboring lincRNA/mRNA pairs. We repeated the analysis using randomly selected 1,000 lincRNA-mRNA pairs (regardless of the distance) from the entire transcriptome data.
Functional prediction of lincRNAs
To predict biological functions for these DE lincRNAs, we calculated the correlation coefficients between DE lincRNAs and all Refseq protein-coding genes using the Spearman rank correlation analysis. We regarded the Spearman rank correlation coefficients calculated from randomly shuffling lincRNA/mRNA pairs (1000 times) as null distribution. Compared with the null distribution, we set a threshold for the Spearman rank correlation coefficient ≥ 0.4 (or ≤ −0.4) (Figure S6) to reflect the strong co-expression between lincRNAs and Refseq genes in high confidence. On this basis, a set of Refseq genes passing this threshold were regarded as the functional association to lincRNAs, and used for functional enrichment analysis using DAVID annotation61. GO terms with BH-adjusted P ≤ 0.05 served as functional enrichment for lincRNAs.
Survival analyses
Excluding participants with unknown survival information (n = 16), the remaining 899 subjects were retained in survival analyses. We split patients into two groups (higher and lower expressions of lincRNA) based on the median level of lincRNA expression. The Kaplan-Meier curve and hazard ratio (HR) of higher versus lower expressed groups were generated in R (versions 2.15.0) using the survival package.
Ethical consent
Utilization of data was conducted in accordance with TCGA data access policies. Signed informed consent was obtained from each subject in the replication study. The study was approved by Indiana University institutional review board.
Additional Information
How to cite this article: Zhang, Y. et al. Long intergenic non-coding RNA expression signature in human breast cancer. Sci. Rep. 6, 37821; doi: 10.1038/srep37821 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Jing He and Yan Guo for data processing and analyses, and Marshal S Younger for clerical support in the preparation of this manuscript. The results here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/). Data analyses were conducted using the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University. We thank the technical staff at Q Squared Solutions Expression Analysis LLC for their collaboration and scientific support in performing the RNA-sequencing for the replication study. We acknowledge Indiana CTSI Specimen Storage Facility (SSF), which is supported by NCRR Clinical and Translational Sciences Award (U54-RR025761) and NCRR construction award (C06-RR020128-01). We also thank the Indiana University Simon Cancer Center at Indiana University School of Medicine for the use of the Tissue Procurement & Distribution Core, which provided breast tumor specimens. Samples from the Susan G. Komen Tissue Bank at the Indiana University Simon Cancer Center were used in this study. We thank contributors, including Indiana University who collected samples used in this study, as well as donors and their families, whose help and participation made this work possible. This work was supported in part by US National Institutes of Health grants R01CA137013 to JL and NIH R01 grant R01CA194030 and Susan G. Komen Career Catalyst Research grant CCR15333233 to CH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions Y.Z., J.L. and C.H. conceived and designed the study; Y.Z., E.W., X.G., I.M. carried out data analysis; Y.Z., E.W., J.L., C.H. wrote the manuscript. C.H., Q.C. and W.Z. directed the data management. All authors read and approved the final manuscript.
References
- Core L. J., Waterfall J. J. & Lis J. T. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science 322, 1845–1848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H., Nislow C., Blencowe B. J. & Hughes T. R. Most Dark Matter Transcripts Are Associated With Known Genes. PLoS Biol 8, e1000371 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F. F. Non-coding RNAs: Meet thy masters. BioEssays 32, 599–608 (2010). [DOI] [PubMed] [Google Scholar]
- Ponjavic J., Ponting C. P. & Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Research 17, 556–565 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I. & Bartel D.-P. lincRNAs: Genomics, Evolution, and Mechanisms. Cell 154, 26–46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927, doi: 10.1101/gad.17446611 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M. & Rinn J. L. Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan C. I., Dees E. C., Ingram R. S. & Tilghman S. M. The product of the H19 gene may function as an RNA. Molecular and Cellular Biology 10, 28–36 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106, 11667–11672, doi: 10.1073/pnas.0904715106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova A. A. et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci USA 110, 2876–2881, doi: 10.1073/pnas.1221904110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S. et al. A Long Noncoding RNA Mediates Both Activation and Repression of Immune Response Genes. Science 341, 789–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham S. W., Gruhl F., Mattick J. S. & Dinger M. E. Long noncoding RNAs and the genetics of cancer. Br J Cancer 108, 2419–2425, doi: Minireview (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorfer C. A., Necela B. M., Thompson E. A. & Perez E. A. MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer. Trends in Molecular Medicine 17, 313–319 (2011). [DOI] [PubMed] [Google Scholar]
- Cianfrocca M. & Gradishar W. New Molecular Classifications of Breast Cancer. CA: A Cancer Journal for Clinicians 59, 303–313 (2009). [DOI] [PubMed] [Google Scholar]
- Volinia S. et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci USA 109, 3024–3029, doi: 10.1073/pnas.1200010109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B., Peterse J. L. & van’t Veer L. J. Breast cancer metastasis: markers and models. Nat Rev Cancer 5, 591–602, doi: 10.1038/nrc1670 (2005). [DOI] [PubMed] [Google Scholar]
- Curtis C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352, doi: 10.1038/nature10983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752, doi: 10.1038/35021093 (2000). [DOI] [PubMed] [Google Scholar]
- Iyer M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47, 199–208, doi: 10.1038/ng.3192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. F. et al. Co-regulated gene expression by oestrogen receptor a and liver receptor homolog-1 is a feature of the oestrogen response in breast cancer cells. Nucleic Acids Research 41, 10228–10240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb E. A. et al. Human Cancer Long Non-Coding RNA Transcriptomes. PLoS ONE 6, e25915, doi: doi: 10.1371/journal.pone.0025915 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 32, 1616–1625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A. & Ponting C. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biology 10, R124, doi: 10.1186/gb-2009-10-11-r124 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838, doi: 10.1038/nature03702 (2005). [DOI] [PubMed] [Google Scholar]
- Martello G. et al. A MicroRNA Targeting Dicer for Metastasis Control. Cell 141, 1195–1207 (2010). [DOI] [PubMed] [Google Scholar]
- Wei W., Pelechano V., Jarvelin A. I. & Steinmetz L. M. Functional consequences of bidirectional promoters. Trends in Genetics 27, 267–276 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G. et al. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol 14, 1190–1198, doi: 10.1038/ni.2712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. Profiling of human CD4+ T-cell subsets identifies the TH2-specific noncoding RNA <em>GATA3-AS1</em>. Journal of Allergy and Clinical Immunology 132, 1005–1008, doi: 10.1016/j.jaci.2013.05.033 (2013). [DOI] [PubMed] [Google Scholar]
- Su X. et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmire L. X. et al. A global clustering algorithm to identify long intergenic non-coding RNA–with applications in mouse macrophages. PLoS One 6, e24051, doi: 10.1371/journal.pone.0024051 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield L. M. & Roberts A. B. TGF-beta signaling: positive and negative effects on tumorigenesis. Current Opinion in Genetics & Development 12, 22–29 (2002). [DOI] [PubMed] [Google Scholar]
- Ding X. et al. Long Intergenic Non-Coding RNAs (LincRNAs) Identified by RNA-Seq in Breast Cancer. PLoS ONE 9, e103270, doi: 10.1371/journal.pone.0103270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurlock Iii C. F. et al. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat Commun 6, doi: 10.1038/ncomms7932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peffers M. J. et al. Transcriptome analysis of ageing in uninjured human Achilles tendon. Arthritis Research & Therapy 17, 1–18, doi: 10.1186/s13075-015-0544-2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M. V. et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Research 65, 7065–7070 (2005). [DOI] [PubMed] [Google Scholar]
- Bandres E. et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Molecular Cancer 5, 29, doi: 10.1186/1476-4598-5-29 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198 (2006). [DOI] [PubMed] [Google Scholar]
- Du Z. et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol 20, 908–913, doi: Resource (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076, doi: 10.1038/nature08975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q. et al. Poly A- Transcripts Expressed in HeLa Cells. PLoS ONE 3, e2803, doi: doi: 10.1371/journal.pone.0002803 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Duff M., Graveley B., Carmichael G. & Chen L. L. Genomewide characterization of non-polyadenylated RNAs. Genome Biology 12, R16, doi: 10.1186/gb-2011-12-2-r16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P. et al. MALAT-1, a novel noncoding RNA, and thymosin [beta]4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003). [DOI] [PubMed] [Google Scholar]
- Shi X., Sun M., Liu H., Yao Y. & Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Letters 339, 159–166 (2013). [DOI] [PubMed] [Google Scholar]
- Prensner J. R. et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotech 29, 742–749, doi: 10.1038/nbt.1914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk Aleksandra A., Kim J. K., Svensson V., Marioni, John C. & Teichmann, Sarah A. The Technology and Biology of Single-Cell RNA Sequencing. Molecular Cell 58, 610–620, doi: 10.1016/j.molcel.2015.04.005 (2015). [DOI] [PubMed] [Google Scholar]
- Grun D. & van Oudenaarden A. Design and Analysis of Single-Cell Sequencing Experiments. Cell 163, 799–810, doi: 10.1016/j.cell.2015.10.039 (2015). [DOI] [PubMed] [Google Scholar]
- Consortium T. E. P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, doi: 10.1038/nature11247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. et al. Integrated Analysis of Gene Expression, CpG Island Methylation, and Gene Copy Number in Breast Cancer Cells by Deep Sequencing. PLoS ONE 6, e17490, doi: doi: 10.1371/journal.pone.0017490 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner A. L. et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biology 13, 1–13, doi: 10.1186/gb-2012-13-8-r75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T. et al. NCBI GEO: archive for functional genomics data sets-10 years on. Nucleic Acids Research 39, D1005–D1010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Trapnell C., Donaghey J., Rinn J. & Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biology 12, R22, doi: 10.1186/gb-2011-12-3-r22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K. et al. iSeeRNA: identification of long intergenic non-coding RNA transcripts from transcriptome sequencing data. BMC Genomics 14, S7, doi: 10.1186/1471-2164-14-S2-S7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Delahanty R., Guo X., Zheng W. & Long J. Integrative genomic analysis reveals functional diversification of APOBEC gene family in breast cancer. Human Genomics 9, 1–12, doi: 10.1186/s40246-015-0056-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D., Wang E. T., Burge C. B. & Sandberg R. An Abundance of Ubiquitously Expressed Genes Revealed by Tissue Transcriptome Sequence Data. PLoS Comput Biol 5, e1000598, doi: 10.1371/journal.pcbi.1000598 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. & Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nat Meth 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079, doi: 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biology 9, R137, doi: 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Pan W. & Khodursky A. B. A note on using permutation-based false discovery rate estimates to compare different analysis methods for microarray data. Bioinformatics 21, 4280–4288 (2005). [DOI] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat.Protocols 4, 44–57, doi: 10.1038/nprot.2008.211 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.