Abstract
Tuberculosis (TB) is the deadliest infectious disease worldwide. One obstacle hindering the elimination of TB is our lack of understanding of host-pathogen interactions. Exosomes, naturally loaded with microbial molecules, are circulating markers of TB. Changes in the host protein composition of exosomes from Mycobacterium tuberculosis (Mtb)-infected cells have not been described, can contribute to our understanding of the disease process, and serve as a direct source of biomarkers or as capture targets to enrich for exosomes containing microbial molecules. Here, the protein composition of exosomes from Mtb-infected and uninfected THP-1-derived macrophages was evaluated by tandem-mass-spectrometry and differences in protein abundances were assessed. Our results show that infection with Mtb leads to significant changes in the protein composition of exosomes. Specifically, 41 proteins were significantly more abundant in exosomes from Mtb-infected cells; 63% of these were predicted to be membrane associated. Thus, we used a novel biotinylation strategy to verify protein localization, and confirmed the localization of some of these proteins in the exosomal membrane. Our findings reveal another important scenario where Mtb could be influencing changes in host cells that unveil new features of the host-pathogen interaction and may also be exploited as a source of biomarkers for TB.
Tuberculosis (TB) is the deadliest infectious disease worldwide; despite available treatment, 1.5 million people died from TB during 20141. Additionally, during the last 5 years the global incidence of TB remained above 100 cases/100,000 population per year2. A major obstacle to control TB is our lack of understanding of host-pathogen interaction. Upon infection Mycobacterium tuberculosis (Mtb, the causal agent of TB) invades and grows mainly inside alveolar macrophages3. It is known that Mtb is able to modulate several mechanisms in the host to its favor, such as the blockage of phagosome maturation4, the disruption of the phagosome5 and the immune response6,7. The definitive explanation of these phenomena still needs to be defined. Exosomes are a type of extracellular vesicle involved in cell to cell communication8,9 and may unveil additional information about the mechanisms of interaction between Mtb and host cells.
Another scenario where exosomes are equally relevant is in biomarker discovery and diagnosis of TB. Historically, the identification of patients with Mtb infection is confirmed through the visualization of acid-fast bacilli in infected sputum samples using light microscopy. This test has shown very low sensitivity (50% to 60%), and is even lower in cases of HIV co-infected TB patients which account globally for the 51% of TB cases1. Of equal concern is the reservoir of latently infected individuals, comprising a third of the global population, from which 5% to 15% will develop active disease especially within the first 5 years of infection10. The available strategies to detect latent TB infection (LTBI) include tuberculin skin test (TST) or interferon gamma release assays (IGRA); both rely on sufficient T-cell based immune response against Mtb antigens11. In the case of TST, people that have received BCG-vaccination often have false positive results. This is important considering that roughly 90% of the countries have a policy of universal BCG vaccination12. IGRA is a suitable alternative designed to detect T-cell mediated gamma-interferon release in response to specific Mtb-complex antigens. Both tests show low sensitivity against HIV infected patients and neither can discriminate LTBI from TB or identify LTBI at high risk to develop TB11. As such, additional biomarker tests for LTBI identification and TB diagnosis are urgently needed.
Exosomes have emerged as a promising alternative source of biomarkers for several diseases, since they can be obtained from almost all biological fluids8,9,13. Exosome composition depends on the cell of origin, as well as the physiological status of the host8,9. In the case of infectious diseases, exosomes represent a source of biomarkers at two different levels: first, exosomes can carry pathogen associated molecular patterns13 and second, host proteins in the exosomes change as a result of an infectious process. In the first scenario, several studies have demonstrated that exosomes released from Mtb-infected cells contain mycobacterial products including proteins, RNAs and glycolipid14,15,16. More importantly, our laboratory recently published the discovery of a panel of Mtb peptides that were identified in exosomes isolated from the sera of TB patients and LTBI individuals, confirming the fact that Mtb infection modifies exosome composition and could be a source of biomarkers for TB17.
Regarding the host proteome, little is known about alterations in the exosome content influenced by Mtb infection. Taking into account that Mtb affects the protein composition of several organelles in infected cells (i.e. mitochondria, endoplasmic reticulum and the phagosome)4,18,19, and the intrinsic features of exosomes previously described, it is very likely that Mtb affects the composition of the human proteins loaded in exosomes of infected cells. Here, we aimed to identify significant changes in human proteins of exosomes derived from Mtb infected cells. Considering that protein localization within the exosome can play an important role in function of these vesicles, we implemented a novel biotinylation scheme to differentiate the surface-exposed proteins within the exosome membrane from those protected within the vesicle. This work serves as a proof-of-concept that intracellular Mtb affects the protein composition of exosomes released from host cells. In view of that, the host protein composition of exosomes from individuals infected with Mtb could aid in our understanding of the host-Mtb interaction and further TB biomarker efforts.
Results and Discussion
General characterization of exosomes is stable regardless of infection status
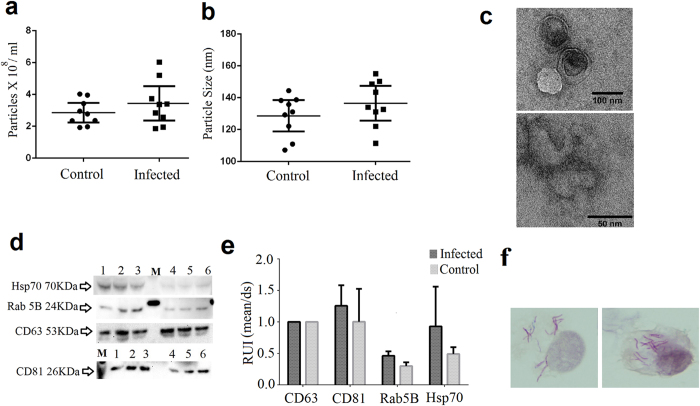
The nanovesicle concentration when normalized by total protein was similar between samples, ranging from 2 × 108 to 6 × 108 particles per ml (t-test, p = 0.299). Exosomes from infected macrophages (MΦ) appeared to be slightly larger; however, the size difference was not significant (t-test, p = 0.236) (Fig. 1a/b). In addition to size and concentration, we detected the presence of CD63, CD81, and Rab-5B, considered exosome hallmark proteins, by western blot in all biological replicates (Fig. 1d/e). Abundance of these proteins was invariant amongst all samples. Finally, exosomes were visualized with transmission electron microscopy (Fig. 1c). These findings confirmed that the nanovesicles in this study were consistent with exosomes.
Figure 1. Characterization and validation of the human macrophages derived exosomes.
Vesicle concentration (a) and size (b) from Mtb-infected and uninfected control cells. Differences between infected and control were not statistically different as determined by t-test, p = 0.299 and p = 0.236, respectively. More detailed results from the light scattering analysis are presented in the Supplementary Fig S1. Values represent three independent experiments and three technical replicates. Exosomes from Mtb-infected cells (c), upper picture) and from control cells (c), lower picture) viewed using transmission electron microscopy. Western blot (d) and densitometry analysis (e) of exosome hallmark proteins from three independent experiments. Exosomes from infected cells, lanes: 1, 2 and 3. M: Molecular weight marker. Exosomes from control cells lanes: 4, 5 and 6. The original images of the western blots can be found in Supplementary Fig. S2. THP-1 macrophages infected with Mtb (f) stained after exosome collection 100X magnification (Modified Kinyoun and hematoxylin staining). RUI: relative units of intensity.
To determine the viability of Mtb-infected cells and controls cells after exosome collection, we did an assay based on the reducing capacity of viable cells using resaruzin (AlamarBlue-Thermo Scientific), a non-toxic permeable substrate, which once in the cytoplasm is reduced to resorufin. In general, the proportion of viable infected cells relative to their corresponding control ranged from 92% to 114% confirming the sufficient viability of the cells.
Finally, we confirmed the extent of bacterial infection at the final point of the experiment microscopically (Fig. 1f) and by bacterial enumeration. The average number of colony forming units (CFUs) was 1.05 × 107 ± 5 × 106 CFU/ml. This average represents the 33.8% resident bacilli from the initial bacterial inoculum. Both results allowed us to confirm that the isolated exosomes were produced from Mtb-infected cells.
Proteome of exosomes released from MΦ infected with Mtb
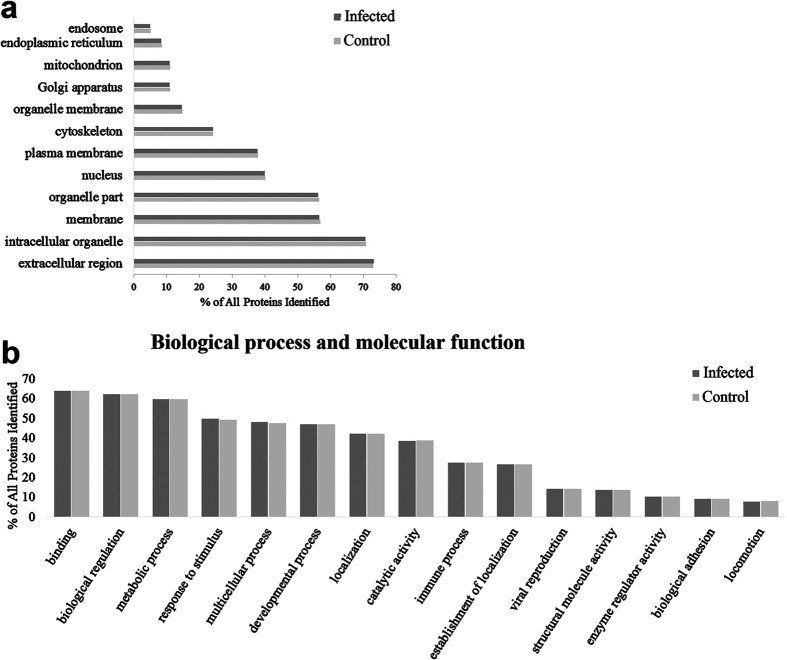
Here we describe for the first time the human proteome of exosomes released from MΦ infected with Mtb. Overall we identified 355 proteins, from which 201 were classified as membrane associated proteins based on the GO term annotations from the NCBI database (Fig. 2a; Supplementary Table S1). Twenty-eight percent of the identified proteins were exclusively present in exosomes from infected MΦ.
Figure 2. Function and localization of the proteins found in exosomes released from Mtb-infected macrophages and uninfected control cells.
(a) Subcellular localization of most of the proteins found in the macrophage-derived exosomes. (b) The top 15 identified molecular functions and biological process associated with the proteins found in exosomal samples. Information obtained directly from the NCBI database.
Most of the proteins found in our exosome samples are involved in binding, immunological and metabolic processes (Fig. 2b). All of these processes have been previously associated with potential roles for exosomes20,21,22.
Comparative proteomic analysis reveals significant differences between exosomes from infected and control cells
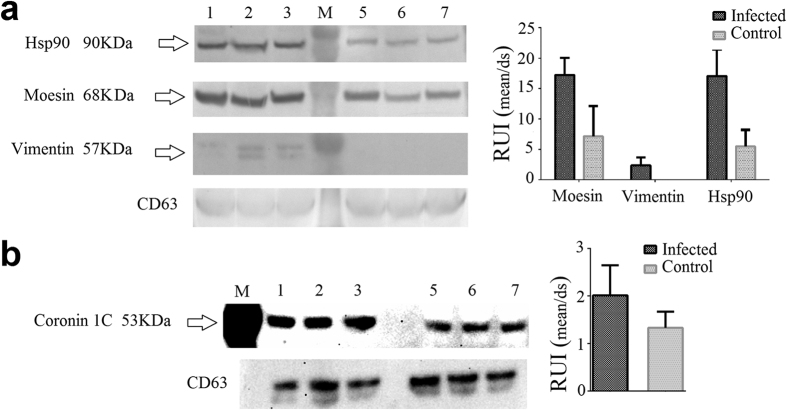
As we predicted, the infection of MΦ with Mtb impacted the protein composition of exosomes. To analyze the difference in protein abundance between exosomes from Mtb-infected and control cells, we used normalized spectral abundance factor (NSAF)23 to accurately compare individual protein abundances among our two samples. Forty-one proteins were significantly more abundant in exosomes in Mtb-infected cells (Table 1), a subset of these were confirmed by western blot (Fig. 3), including: HSP90, vimentin, Coronin 1 C and moesin. Previous studies have shown that some of these proteins play important roles during Mtb infection. Shekhawat et al., showed that human HSP proteins (including HSP90) were increased in sera from individuals with a high risk of being latently infected with Mtb, suggesting HSP proteins as potential biomarkers for LTBI24. We observed that HSP90 was significantly higher in exosomes from infected cells. In this study, we also found that infected MΦ produced exosomes with a higher concentration of vimentin compared with control cells. Vimentin is a ligand for NKp46, a receptor in Natural Killer cells (NK). The interaction between NKp46 and vimentin mediates the lysis of Mtb-infected cells by NK25. Exosomes enriched with vimentin could interfere in the interaction of NKp46 with the cell membrane associated vimentin (from MΦ), thus delaying the killing of Mtb infected cells. In this way, intracellular mycobacteria could be modulating the loading of vimentin in exosomes as a defense mechanism. Further investigations into this phenomenon could give us new insights about the complex host-pathogen interaction in TB. Lastly, L-amino acid oxidase (LAAO) was more abundant in exosomes from Mtb-infected MΦ in our study. This protein plays important roles in the innate immune response acting as an antibacterial enzyme that catabolizes the deamination of L-amino acids producing H2O2 and ammonia26. Our results suggest that Mtb-infected cells could be using exosomes to export endogenous antimicrobial molecules as a defense mechanism. This phenomenon has been previously observed with IFNα that was transported between cells via exosomes20. Collectively, these changes in exosomes as a consequence of Mtb infection could reveal important information regarding the host-pathogen interaction.
Table 1. Proteins significantly different between exosomes from Mtb-infected and control macrophages.
| Identified Proteins | Infected NSAF | Control NSAF | p value NSAF-inf versus NSAF-control | Fold Change NSAF-inf/NSAF-control |
|---|---|---|---|---|
| 60 S acidic ribosomal protein P0 | 0.064 | 0.000 | 0.00012 | INF |
| Coronin-1C | 0.023 | 0.000 | 0.00017 | INF |
| Lupus La protein | 0.023 | 0.000 | 0.00019 | INF |
| Heterogeneous nuclear ribonucleoprotein K | 0.075 | 0.003 | 0.00029 | 28.4 |
| Heat shock 70 kDa protein 4 | 0.013 | 0.000 | 0.00031 | INF |
| Alanine-tRNA ligase, cytoplasmic | 0.006 | 0.000 | 0.00035 | INF |
| Calreticulin | 0.017 | 0.000 | 0.001 | INF |
| Protein disulfide-isomerase A3 | 0.040 | 0.000 | 0.002 | INF |
| L-amino-acid oxidase | 0.018 | 0.000 | 0.003 | INF |
| Moesin | 0.151 | 0.062 | 0.0032 | 2.4 |
| Nucleolin | 0.063 | 0.007 | 0.0032 | 8.6 |
| Vimentin | 0.251 | 0.072 | 0.0034 | 3.5 |
| Protein disulfide-isomerase A6 | 0.046 | 0.003 | 0.0035 | 16.6 |
| Spliceosome RNA helicase DDX39B | 0.027 | 0.000 | 0.0039 | INF |
| Fermitin family homolog 3 | 0.046 | 0.002 | 0.0047 | 19.8 |
| Programmed cell death 6-interacting protein | 0.005 | 0.000 | 0.0047 | INF |
| S-adenosylmethionine synthase isoform type-2 | 0.029 | 0.000 | 0.0048 | INF |
| Glyceraldehyde-3-phosphate dehydrogenase | 0.293 | 0.201 | 0.0059 | 1.5 |
| ATP-dependent RNA helicase A | 0.005 | 0.000 | 0.0068 | INF |
| 60 kDa heat shock protein, mitochondrial | 0.013 | 0.000 | 0.0082 | INF |
| Cytosol aminopeptidase | 0.041 | 0.000 | 0.0084 | INF |
| Ubiquitin-like modifier-activating enzyme 1 | 0.056 | 0.007 | 0.0089 | 8.5 |
| ITIH4 protein | 0.011 | 0.000 | 0.01 | INF |
| Serine/threonine-protein phosphatase 2 A 65 kDa regulatory subunit A alpha isoform | 0.011 | 0.002 | 0.011 | 7.0 |
| Tryptophan-tRNA ligase, cytoplasmic | 0.031 | 0.000 | 0.011 | INF |
| Transketolase | 0.082 | 0.015 | 0.012 | 5.6 |
| Zyxin (Fragment) | 0.007 | 0.000 | 0.012 | INF |
| Heat shock protein HSP 90-beta | 0.361 | 0.221 | 0.014 | 1.6 |
| Tyrosine-tRNA ligase, cytoplasmic | 0.014 | 0.000 | 0.017 | INF |
| 6-phosphogluconate dehydrogenase, decarboxylating | 0.075 | 0.021 | 0.024 | 3.6 |
| X-ray repair cross-complementing protein 6 | 0.061 | 0.004 | 0.026 | 13.8 |
| 78 kDa glucose-regulated protein | 0.109 | 0.047 | 0.028 | 2.3 |
| Eukaryotic initiation factor 4A-I | 0.115 | 0.038 | 0.028 | 3.1 |
| Thrombospondin-4 | 0.011 | 0.002 | 0.028 | 6.2 |
| Bifunctional purine biosynthesis protein PURH | 0.028 | 0.000 | 0.028 | INF |
| Staphylococcal nuclease domain-containing protein 1 | 0.012 | 0.001 | 0.031 | 9.2 |
| Heat shock cognate 71 kDa protein | 0.443 | 0.273 | 0.036 | 1.6 |
| Integrin beta-1 | 0.006 | 0.000 | 0.046 | INF |
| UDP-glucose 6-dehydrogenase | 0.013 | 0.000 | 0.046 | INF |
| Purine nucleoside phosphorylase | 0.039 | 0.000 | 0.048 | INF |
| Lamin-B1 | 0.021 | 0.003 | 0.049 | 6.5 |
| Transforming growth factor-beta-induced protein ig-h3 | 0.061 | 0.128 | 0.0047 | 0.5 |
| Palmitoyl-protein thioesterase 1 | 0.053 | 0.106 | 0.033 | 0.5 |
| Complement C4-A | 0.056 | 0.073 | 0.035 | 0.8 |
INF: When the value in the denominator is zero (the fold change is the normalized spectral abundance factor23 (NSAF)-infected divided by NSAF-control), *p value from t-test comparing the averages of three independent experiments between NSAF-infected versus NSAF-control.
Figure 3. Western blot confirmed proteins significantly more abundant in exosomes from Mtb-infected cells originally detected by LC-MS/MS.
Hsp90, Moesin and Vimentin were detected using a chromogenic substrate (a). The intensity of the bands was evaluated relative to the intensity of CD63. Additionally, Coronin 1 C was detected using a chemiluminescent substrate (b). The intensity of the bands was evaluated relative to the intensity of CD63. Samples from three independent experiments were evaluated. Exosomes from infected cells, lanes: 1, 2 and 3. M: Molecular weight marker. Exosomes from control cells lanes: 5, 6 and 7. The original images of the western blots can be found in Supplementary Fig. S3. RUI: relative units of intensity.
Significant changes of the exosome membrane proteome after infection with Mtb
Exosomes are an important source of biomarkers for TB by virtue of the discovery of mycobacterial molecules packaged within these vesicles14,15,17,27. Here, the effect of the infection on host exosomal protein composition illustrates another potential opportunity to exploit exosomes as biomarkers of TB. In this regard, we sought to explore the differential abundance of membrane associated proteins since they represent a more accessible set of targets for downstream development of a biomarker assay. Using GO-term annotation analysis, we found that 63% (26/41) of the proteins that were significantly more abundant in exosomes from infected cells were also membrane associated. To better characterize this subset of proteins we used transmembrane helix prediction software and found that 31% of the 26 proteins contained at least one probable transmembrane domain or residues that were potentially membrane associated (Table 2).
Table 2. Membrane associated proteins significantly more abundant in exosomes from Mtb-Infected cells and their biotinylation pattern.
| Membrane associated protein‡ | Proteins with AA residues in transmembrane domains** | LC-LC Biotin | Biotin |
|---|---|---|---|
| 60S acidic ribosomal protein P0 | X | ||
| Coronin-1C | X | ||
| Heterogeneous nuclear ribonucleoprotein K | |||
| Alanine-tRNA ligase, cytoplasmic | |||
| Calreticulin | X | ||
| Protein disulfide-isomerase A3 | X | ||
| Moesin | X | ||
| Nucleolin | |||
| Vimentin | X | X | |
| Protein disulfide-isomerase A6 | X | ||
| Fermitin family homolog 3 | |||
| Programmed cell death 6-interacting protein | |||
| Glyceraldehyde-3-phosphate dehydrogenase | X | X | |
| ATP-dependent RNA helicase A | X | ||
| 60 kDa heat shock protein, mitochondrial | |||
| Cytosol aminopeptidase | |||
| Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | |||
| Transketolase | X | ||
| Heat shock protein HSP 90-beta | X | ||
| 78 kDa glucose-regulated protein | |||
| Eukaryotic initiation factor 4A-I | X | X | |
| Bifunctional purine biosynthesis protein PURH | |||
| Staphylococcal nuclease domain-containing protein 1 | |||
| Heat shock cognate 71 kDa protein | X | ||
| Integrin beta-1 | X | ||
| Lamin-B1 |
‡Classification based on the Go-term annotations from the NCBI database.
**Based on TMHMM Server v. 2.0 prediction of transmembrane helices in proteins (AA: Aminoacids).
To further ascertain the localization of the proteins that were significantly more abundant in exosomes from infected cells, we conducted an experiment to differentially label proteins based on their localization within the exosome. First, intact exosomes were exposed to NHS-Sulfo-LC-LC-Biotin, a negatively charged molecule unable to permeate biological membranes, to label proteins that were exclusively facing-out of the exosomal membrane. NHS-Sulfo-LC-LC-Biotin binds amino-terminus and lysine residues, increasing the molecular mass of the labeled peptide by 452 Da. The fact that this molecule has a spacer arm of 30.5 Å improves its accessibility to folded proteins in the membrane of intact exosomes. After cleaning the excess of NHS-Sulfo-LC-LC-Biotin, the biotinylated exosomes were lysed followed by secondary labeling of proteins on the internal-face of the exosomal membrane or within the lumen of the exosome, using an alternate biotin reagent (Sulfo-NHS-Biotin) that increases the molecular weight of the labelled residues by 226 Da. This labeling strategy allowed for the identification and differentiation of protein populations by mass spectrometry analysis, including determination of protein domains exposed to the external side of the exosome.
Our labeling studies demonstrated that 6 of 26 differentially abundant membrane proteins identified by GO-term analysis were differentially labeled with LC-LC biotin, and thus are surface exposed (Table 2). In addition, this technique afforded the identification of one additional surface exposed protein, a nucleoside diphosphate kinase (Ndk) (Table 3). Interestingly, Mycobacterium-derived Ndk has been associated with a greater survival of infected macrophages28. Ndk metabolizes extracellular ATP which plays important roles during the inflammatory response and macrophage activation via P2X7 receptor29. Although human derived-Ndk has been mostly related with intracellular vesicle trafficking29, human-derived Ndk in secreted exosomes could be acting as an ecto-enzyme that modulates macrophage activation and survival in an ATP-dependent manner; favoring mycobacterial persistence. Three of the seven LC-LC biotin labeled proteins (Table 3) were also labeled with the shorter biotin (226 Da label) (Table 2); these are indicative of transmembrane proteins with both external and internal segments. In the same way, the presence of proteins exhibiting only the small biotin implies that the protein is confined to the exosome lumen. Our biotinylation strategy represents an original approach that allows for a more specific characterization of exosomes.
Table 3. List of proteins and the specific peptide(s) labelled with LC-LC biotin.
| Protein name | Peptide sequence |
|---|---|
| Eukaryotic initiation factor 4AI | EVQkLQMEAPHIIVGTPGRVF |
| Glyceraldehyde-3-phosphate dehydrogenase | DNFGIVEGLMTTVHAITATQkTV |
| Heat shock cognate 71 kDa protein | DPVEkALR |
| Heat shock protein HSP 90-beta | ERIMkAQALR |
| Moesin | EEAkEALLQASR |
| Nucleoside diphosphate kinase | ERTFIAIkP |
| Vimentin | DVRQQYESVAAkNLQEA |
k: designates a lysine residue with the LC-LC biotin modification.
Conclusions
Our study demonstrates that the infection with Mtb influences changes in the protein composition of exosomes released from infected cells. Even though our findings do not have an immediate translational application they represent the proof-of-concept that Mtb-infected cells will produce exosomes with a characteristic proteome. The confirmation of these phenomena in a clinically relevant sample set (i.e. TB-patient sera derived exosomes) will advance our knowledge about Mtb-host interactions and will offer a potential source for new TB biomarkers.
Methods
Human monocytes growth and activation
THP-1 human monocytes (American Type Culture Collection, ATCC/TIB-202) were cultured at 37 °C at 5% CO2 in complete RPMI (cRPMI) media. This media contained RPMI 1640 base medium (ATCC) supplemented with 2-mercaptoethanol (Gibco) (final concentration of 0.05 mM) and 10% exosome-depleted fetal bovine serum (EXO-FBS, SBI). Monocytes (2.5 × 105 cells/ml) were activated to macrophages (MΦ) using 200 nM of Phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich) for 72 h. After the activation, unbound cells and remaining media were removed, and the adherent cells were washed three times using phosphate buffer saline (PBS).
Macrophage infection
All procedures involving live Mtb were completed in a biosafety level 3 laboratory at Colorado State University. Infectivity stocks of Mtb strain H37Rv were thawed and spun down at 1,200 × g for 10 minutes. The pellet was suspended in cRPMI and bath-sonicated for 1 minute to disrupt bacteria clumps. THP-1 MΦ were infected with Mtb at a 1:5 ratio (MΦ: Bacteria), for 4 h at 37 °C/5% CO2. After infection, the THP-1 MΦs were washed three times with PBS and then 25 ml of fresh cRPMI was added. The cells were returned to culture conditions for 24 h for the production of exosomes. Identical flasks of THP-1 MΦ were treated in the same way but without Mtb to produce control exosomes. Every experiment with Mtb infected and uninfected THP-1 MΦ was done in triplicate and three independent experiments at different days were done as biological replicates.
Exosome purification
Approximately, 25 ml of supernatant from infected and control THP-1 MΦ were collected after 24 h of infection and filtered through 0.2 μm membrane to remove cellular debris, potential membrane fragments from lysed cells, large vesicles (>500 nm) and whole bacteria. The collected material was filtered using an Amicon centrifugal filter unit with a molecular weight cut-off (MWCO) of 100 KDa (EMD Millipore) to 2 ml concentration to remove most of the soluble proteins. The exosome-rich retentate was diluted to 15 ml with PBS and filtered again through the 100 KDa to elute remaining soluble proteins. After this, the retained sample was centrifuged at 18,000 × g for 30 min to pellet larger vesicles. The resultant supernatant was mixed with 400 μl of ExoQuick-TC (SBI) and incubated at 4 °C overnight to precipitate exosomes. After this, the mix was centrifuged at 2,600 × g for 30 min. The pellet-containing exosomes was suspended in 1 ml of PBS and the total protein concentration was measured using the microbicinchoninic acid assay (microBCA, Thermo Scientific). The exosomes were aliquoted (50 μg/vial) and stored at −20 °C until further analysis.
THP-1 MΦ viability test after Mtb infection
To guarantee the accurate comparison between exosomes from Mtb-infected and control cells, the viability of both types of cells was evaluated after exosome collection. Briefly, after each experiment, the cells were washed with PBS and detached from the flask using 5 ml of 0.25% trypsin-EDTA (Gibco) at 25 °C for 10 min. Five ml of cRPMI were added and the cells were centrifuged at 600 × g per 10 min and washed with cRPMI once. Then, cells were suspended in cRPMI to perform the Alamar Blue (Invitrogen) viability assay. Briefly, 100  l of previously suspended control and infected cells were added to a 96 wells plate in triplicate. Subsequently, 10 μl of Alamar Blue reagent was added to each well and the plate was incubated at 37 °C/5%CO2 for 4 h in the dark. The viability of the infected and control cells was determined following the manufacturer’s recommendations.
l of previously suspended control and infected cells were added to a 96 wells plate in triplicate. Subsequently, 10 μl of Alamar Blue reagent was added to each well and the plate was incubated at 37 °C/5%CO2 for 4 h in the dark. The viability of the infected and control cells was determined following the manufacturer’s recommendations.
Validation of Mtb infection following exosome purification
Ten μl of infected cells were pipetted onto a microscope slide, fixed with paraformaldehyde 4% for 24 h and stained by the Kinyoun acid-fast procedure and microscopically evaluated. Then, the remaining infected THP-1 MΦ were pelleted and lysed using 0.05% SDS for 3 min. The SDS treatment was enough to lyse the THP-1 MΦ but not the intracellular bacteria. The suspension was centrifuged at 1,200 × g for 10 min and the pellet was suspended in Middlebrook 7H9 broth. Seven 10-fold serial dilutions were plated on 7H11 quad-plates and incubated at 37 °C for 3 weeks for colony forming units (CFU) enumeration to verify the number of infecting bacteria after exosome production.
Characterization of exosomes
The concentration and size of the exosomes for all samples were evaluated by nanoparticle tracking analysis (NTA), using the NanoSight NS300 (Malvern Instruments). From each exosome-sample, 5 μg of total protein were diluted in 1 ml of PBS and loaded into the instrument. Each sample was analyzed in triplicate. The presence of hallmark exosome proteins was evaluated by Western Blot.
Western Blot (WB) analysis
Exosome samples (50 ug) from three independent experiments were resolved in a NuPAGE Novex 4–12% Bis-Tris Gel (Life Technologies) and transferred to a nitrocellulose membrane, 0.2 μm (BIO-RAD). Then, the membrane was blocked with bovine serum albumin 2% for 1 h. After this, the primary antibody was added and incubated during 1 h. Subsequently, the conjugated antibody was added and finally the membrane was exposed to the developer reagent. For exosome characterization the primary antibodies: anti-CD63 (SBI), anti-CD81 (SBI) and anti-Rab5B (A-20) (Santa Cruz Biotechnology) were used. For validation of the proteomics results, the primary antibodies: anti-Coronin 1 C (G-R2) (Santa Cruz Biotechnology), anti-Moesin (Life Technologies), anti-Vimentin Antibody (9E7E7) (Santa Cruz Biotechnology) and anti-HSP 90 (F8) (Santa Cruz Biotechnology) were used. Two different HRP-conjugated antibodies were used: Goat Anti-Rabbit F(ab)2 fragment (Thermo Scientific) and Goat anti-Mouse IgG (H + L) (Thermo Scientific) depending on the source of the primary antibody. For the WB detecting moesin and HSP 90, the colorimetric substrate 4-Chloro-1-Naphtol (Sigma) was used. For the analysis of the other proteins the chemiluminescent substrate Super signal West Pico (Thermo Scientific) was used. WB images and band intensities were determined using the Chemi-Doc XRS+ with Image Lab software version 3.0 (BIORAD). For some of the assays, after transferring the protein to the nitrocellulose membrane, a Ponceau S stain was done to verify the efficiency of the transfer (Supplementary Fig. S3). For this, the membrane was soaked in Ponceau S solution (Ponceau S 0.2%, Acetic acid 3% in distilled water) for 5 minutes, washed with distilled water for approximately 5 minutes to remove unspecific staining and a scan of the membrane was saved. After this, the membrane was washed with PBS 1X for 5 minutes, 3 times. Then, the western blot procedure was followed as described above.
Transmission electron microscopy
For electron microscopy analysis, exosome samples from Mtb-infected and control cells were prepared following the protocol described by Théry et al.30. Briefly, exosome samples were mixed with paraformaldehyde 4% (1:1 ratio) and incubated overnight at 4 °C. Then, a formvar/carbon coated grid (Electron Microscopy Science) was placed on top of a drop of 10 ul of exosome preparation for 20 min. Then, the grid was transferred to a drop of 100 ul of PBS, and then, transfer to a 50 ul drop of glutaraldehyde 1% for 5 min. After this, the grid was washed 7 times with 100 ul drops of distilled water. Next, the grid was transferred to a 50 ul drop of uranyl-oxalate for 5 min. Finally, the grid was transferred to a 50 ul drop of methylcellulose/uranyl acetate (9:1 ratio) for 10 min on ice. After this, excess of methylcellulose/uranyl acetate was removed by blotting on Whatman #1. The grids were air dried and observed in the transmission electron microscope JEOL 2100 F at 200 kV.
Biotinylation of exosome proteins
Two different types of biotin reagents were used in this experiment. Sulfo-NHS-Biotin (Thermo Scientific), containing a shorter, 13.5 Å spacer arm biotin, was used to label the proteins in the lumen or in the internal leaflet of the exosomal membrane and Sulfo-NHS-LC-LC-Biotin (Thermo Scientific), was used to label the proteins exposed to the external leaflet of intact exosomes and contains a larger, 30.5 Å spacer arm between the biotin and amine reactive linker—the size of this linker helps to overcome steric hindrance and increases labeling efficiency at the crowded exosome surface. Two hundred micrograms of intact exosomes were mixed with 10 mM Sulfo-NHS-LC-LC-Biotin at room temperature for 30 min. Four conditions were taken into account during this experiment: (a) an excess of Sulfo-NHS-LC-LC-Biotin was used to favor a complete saturation of exposed lysine residues and potential N-terminus, (b) the presence of the sulfonate group in Sulfo-NHS-LC-LC-Biotin blocks the reagent from penetrating the exosomal membrane, (c) Sulfo-NHS-LC-LC-Biotin has an spacer arm of 30.5 angstroms which improves the biotinylation of proteins in their natural conformation, and (d) amino acids labeled with Sulfo-NHS-LC-LC-Biotin will have an increase in mass of 452 Da. After incubation, the excess of Sulfo-NHS-LC-LC-Biotin was removed using a 10 KDa MWCO filtration device. Biotinylated exosomes were washed with 10X volumes of 1X PBS and concentrated to a final volume of 30 μl. Biotinylated exosomes were lysed with 300 ul of RIPA buffer (Thermo Scientific) for 1 h, followed by three freeze and thaw cycles. After lysis, buffer exchange was done to replace RIPA buffer with 1X PBS. RIPA buffer contains primary amines that interfere with the next biotinylation step. Lysed-biotinylated exosomes were exposed to 10 mM Sulfo-NHS-Biotin (Thermo Scientific), at room temperature for 30 min, for labeling of remaining free amines. Excess of biotin was removed as mentioned above.
Protein digestion
Two strategies were applied based on exosome preparation (biotinylated or unlabeled)
Unlabeled samples
50 μg of exosomes were mixed with Laemmli SDS-PAGE buffer which contains: 2-Mercaptoethanol (5%), Bromophenol blue (0.01%), Glycerol (50%), SDS (electrophoresis-grade, 2%), and Tris-HCl, 63 mM (pH 6.8). Then, samples were heated at 100 °C for 5 minutes. After this, the samples were resolved in a NuPAGE Novex 4–12% Bis-Tris Gel (Life Technologies) for 5 min. Then, the gels were stained with Coomassie Blue (Invitrogen) for 1 h to visualize the localization of the proteins and destained briefly in water to clarify protein bands. Afterwards, the entire lane of gel containing the proteins was excised and cut into 1 mm3 pieces that were transferred to an Eppendorf tube and mixed with destaining solution (60% acetonitrile (ACN) in ammonium bicarbonate 0.2 M) for 1 hour at 37 °C, twice. After destaining, the gel pieces were vacuum dried. Dried gel pieces were mixed with trypsin (Roche) in 0.2 M ammonium bicarbonate at a ratio of 50:1 (w/w-sample: trypsin) at 37 °C, overnight. The next day, the tryptic peptides were extracted by adding 100 μl of 60% ACN, 0.1% trifluoroacetic acid in HPLC-grade water and incubated at 37 °C for 1 hour, twice. The extracted peptides were vacuum dried, suspended in Solvent A (3% ACN and 0.1% formic acid in HPLC-grade water), centrifuged at 13,000 × g for 5 min to remove debris, solution transferred into auto-sampler vials (Agilent technologies), and samples stored at −20 °C in until analysis by LC-MS/MS.
Biotinylated samples
50 μg of labeled exosome lysates were subject to gel electrophoresis and staining as described above. The biotin label binds to the free amine group of lysine residues, thus, interfering with one of the cleavage sites for trypsin; as an alternative, the endoproteinase AspN (Roche) was used for protein digestion. After obtaining the dried and destained gel pieces as described above, protein samples were reduced by incubation with 5 mM dithiothreitol (Sigma) for 20 min at 50 °C and alkylated by incubation with 15 mM iodoacetamide (Sigma) at 25 °C for 15 min in the dark, following the AspN manufacturer’s recommendations. Proteins were then digested with AspN in 0.2 M ammonium bicarbonate using a 50:1 ratio (w/w-sample: enzyme). Subsequent steps after enzymatic digestion were similarly performed as described for trypsin digestion.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Approximately 1 μg of digested peptides for each sample was injected using an EASY nanoLC-II system (Thermo Scientific, San Jose, CA). Peptides were purified and concentrated using an on-line enrichment column (EASY-Column, 100 μm ID × 2 cm ReproSil-Pur C18). Subsequent chromatographic separation was performed on a reverse phase nanospray column (EASY-Column, 3 μm, 75 μm ID × 100 mm ReproSil-Pur C18) using a 90 minute linear gradient from 5–45% solvent B (100% ACN, 0.1% formic acid) at a flow rate of 400 nanoliters/min. Peptides were eluted directly into the mass spectrometer (Thermo Scientific Orbitrap Velos). The instrument was operated in Orbitrap-LTQ mode where precursor measurements were acquired in the Orbitrap (60,000 resolution) and MS/MS spectra (top 20) were acquired in the LTQ ion trap with a normalized collision energy of 35%. Mass spectra were collected over a m/z range of 400–2000 Da using a dynamic exclusion limit of 2 MS/MS spectra of a given peptide mass for 30 s (exclusion duration of 90 s). Compound lists of the resulting spectra were generated using Xcalibur 2.2 software (Thermo Scientific) with an S/N threshold of 1.5 and 1 scan/group.
Data Analysis
Tandem mass spectra were extracted, charge state deconvoluted and deisotoped by ProteoWizard (MSConvert version 3.0). Raw data files were converted to mzXML format and submitted to the Sorcerer2 integrated data analysis platform (Sage-N Research, version 5.0.1); subsequent MS/MS analysis was performed using SEQUEST (Sage-N Research, Milpitas, CA, USA; version v. 3.5). SEQUEST was set up to search the Uni-Prot Homo sapiens Reference Proteome (ID: UP000005640, 70076 entries) assuming the enzymatic digestion with trypsin (after Arg or Lys) or AspN (before Asp or Glu) depending on which enzyme was used. SEQUEST was searched with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 50 PPM. Oxidation of methionine (+15.99) and carbamidomethyl of cysteine (+57.02) were specified in SEQUEST as variable modifications for the unlabeled experiment. Biotin (+226) and LC-LC Biotin (+452) in lysine and N-termini were also included as variable modification for the labeling experiment.
Scaffold (version Scaffold_4.5.1, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identification thresholds were set such that a peptide FDR of 1% and a peptide confidence threshold of 95% was achieved based on hits to the reverse database31. Protein identifications were accepted if they could be established at greater than 99.0% probability to achieve an FDR less than 1.0% and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm32. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins were annotated with GO terms from NCBI (downloaded Dec 31, 2015)33. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository34 with the dataset identifier PXD004062 and DOI 10.6019/PXD004062. Membrane associated proteins were also explored using the free online software TMHMM (Version 2.0)35,36.
Differences in protein abundances between exosomes from Mtb-infected MΦ versus control cells were evaluated by t-test, using normalized spectral abundance factor (NSAF)23. P values < 0.05 were accepted as statistically significant. For validation of the proteomics results a subset of proteins that were significantly higher in exosomes from infected cells were evaluated by western blot as described above.
Additional Information
How to cite this article: Diaz, G. et al. Changes in the Membrane-Associated Proteins of Exosomes Released from Human Macrophages after Mycobacterium tuberculosis Infection. Sci. Rep. 6, 37975; doi: 10.1038/srep37975 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We would like to thank Dr. Roy Geiss from the Central Instrument Facility at Colorado State University for his assistance with TEM. This work was supported by the Bill & Melinda Gates Foundation (Fund#: OPP1039688) and for the scholarship for graduate studies abroad Call 529–2011. Administrative Department of Science, Technology and Innovation – COLCIENCIAS. Colombia.
Footnotes
Author Contributions K.M.D., N.K.G. and G.D. conceived the experiments. G.D. conducted the experiments. K.M.D., N.K.G., L.M.W. and G.D. analyzed the results. All authors contributed to writing and reviewing the manuscript.
References
- The Twentieth Global Report on Tuberculosis; World Health Organization: Geneva, Switzerland (2015).
- Sulis G., Roggi A., Matteelli A. & Raviglione M. C. Tuberculosis: epidemiology and control. Mediterr J Hematol Infect Dis 6, e2014070, doi: 10.4084/MJHID.2014.070 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengenbacher M. & Kaufmann S. H. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev 36, 514–532, doi: 10.1111/j.1574-6976.2012.00331.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne I. et al. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci USA 102, 4033–4038, doi: 10.1073/pnas.0409716102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriff M. J., Purdy G. E. & Lewinsohn D. M. Escape from the Phagosome: The Explanation for MHC-I Processing of Mycobacterial Antigens? Front Immunol 3, 40, doi: 10.3389/fimmu.2012.00040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia J. K., Georgieva M. & Rengarajan J. Innate Immune Defenses in Human Tuberculosis: An Overview of the Interactions between Mycobacterium tuberculosis and Innate Immune Cells. J Immunol Res 2015, 747543, doi: 10.1155/2015/747543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme I. M., Robinson R. T. & Cooper A. M. The balance between protective and pathogenic immune responses in the TB-infected lung. Nat Immunol 16, 57–63, doi: 10.1038/ni.3048 (2015). [DOI] [PubMed] [Google Scholar]
- Raposo G. & Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200, 373–383, doi: 10.1083/jcb.201211138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoorvogel W., Kleijmeer M. J., Geuze H. J. & Raposo G. The biogenesis and functions of exosomes. Traffic 3, 321–330 (2002). [DOI] [PubMed] [Google Scholar]
- Getahun H. et al. Management of latent Mycobacterium tuberculosis infection: WHO guidelines for low tuberculosis burden countries. Eur Respir J., doi: 10.1183/13993003.01245-2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai M. et al. Gamma interferon release assays for detection of Mycobacterium tuberculosis infection. Clin Microbiol Rev 27, 3–20, doi: 10.1128/CMR.00034-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerling A. et al. The BCG World Atlas: a database of global BCG vaccination policies and practices. PLoS Med 8, e1001012, doi: 10.1371/journal.pmed.1001012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar S., Shinagawa K., Castellino F. J. & Schorey J. S. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood 110, 3234–3244, doi: 10.1182/blood-2007-03-079152 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri P. K., Kruh N. A., Dobos K. M. & Schorey J. S. Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics 10, 3190–3202, doi: 10.1002/pmic.200900840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. & Schorey J. S. Exosomes carrying mycobacterial antigens can protect mice against Mycobacterium tuberculosis infection. Eur J Immunol 43, 3279–3290, doi: 10.1002/eji.201343727 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey J. S., Cheng Y., Singh P. P. & Smith V. L. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep 16, 24–43, doi: 10.15252/embr.201439363 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruh-Garcia N. A. et al. Detection of Mycobacterium tuberculosis peptides in the exosomes of patients with active and latent M. tuberculosis infection using MRM-MS. PLoS One 9, e103811, doi: 10.1371/journal.pone.0103811 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamwal S. et al. Characterizing virulence-specific perturbations in the mitochondrial function of macrophages infected with Mycobacterium tuberculosis. Sci Rep 3, 1328, doi: 10.1038/srep01328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saquib N. M., Jamwal S., Midha M. K., Verma H. N. & Manivel V. Quantitative Proteomics and Lipidomics Analysis of Endoplasmic Reticulum of Macrophage Infected with Mycobacterium tuberculosis. Int J Proteomics 2015, 270438, doi: 10.1155/2015/270438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat Immunol 14, 793–803, doi: 10.1038/ni.2647 (2013). [DOI] [PubMed] [Google Scholar]
- Théry C., Zitvogel L. & Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2, 569–579, doi: 10.1038/nri855 (2002). [DOI] [PubMed] [Google Scholar]
- Anand P. K. Exosomal membrane molecules are potent immune response modulators. Commun Integr Biol 3, 405–408, doi: 10.4161/cib.3.5.12474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wen Z., Washburn M. P. & Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem 82, 2272–2281, doi: 10.1021/ac9023999 (2010). [DOI] [PubMed] [Google Scholar]
- Shekhawat S. D., Purohit H. J., Taori G. M., Daginawala H. F. & Kashyap R. S. Evaluation of heat shock proteins for discriminating between latent tuberculosis infection and active tuberculosis: A preliminary report. J Infect Public Health, doi: 10.1016/j.jiph.2015.07.003 (2015). [DOI] [PubMed] [Google Scholar]
- Garg A. et al. Vimentin expressed on Mycobacterium tuberculosis-infected human monocytes is involved in binding to the NKp46 receptor. J Immunol 177, 6192–6198 (2006). [DOI] [PubMed] [Google Scholar]
- Puiffe M. L., Lachaise I., Molinier-Frenkel V. & Castellano F. Antibacterial properties of the mammalian L-amino acid oxidase IL4I1. PLoS One 8, e54589, doi: 10.1371/journal.pone.0054589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. et al. Intracellular trafficking in Mycobacterium tuberculosis and Mycobacterium avium-infected macrophages. J Immunol 153, 2568–2578 (1994). [PubMed] [Google Scholar]
- Zaborina O., Li X., Cheng G., Kapatral V. & Chakrabarty A. M. Secretion of ATP-utilizing enzymes, nucleoside diphosphate kinase and ATPase, by Mycobacterium bovis BCG: sequestration of ATP from macrophage P2Z receptors? Mol Microbiol 31, 1333–1343 (1999). [DOI] [PubMed] [Google Scholar]
- Spooner R. & Yilmaz Ö. Nucleoside-diphosphate-kinase: a pleiotropic effector in microbial colonization under interdisciplinary characterization. Microbes Infect 14, 228–237, doi: 10.1016/j.micinf.2011.10.002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Amigorena S., Raposo G. & Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit 3. 22, doi: 10.1002/0471143030.cb0322s30 (2006). [DOI] [PubMed] [Google Scholar]
- Käll L., Storey J. D., MacCoss M. J. & Noble W. S. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res 7, 29–34, doi: 10.1021/pr700600n (2008). [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A. I., Keller A., Kolker E. & Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75, 4646–4658 (2003). [DOI] [PubMed] [Google Scholar]
- Ashburner M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29, doi: 10.1038/75556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A. et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41, D1063–1069, doi: 10.1093/nar/gks1262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G. & Sonnhammer E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580, doi: 10.1006/jmbi.2000.4315 (2001). [DOI] [PubMed] [Google Scholar]
- Sonnhammer E. L., von Heijne G. & Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6, 175–182 (1998). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.