Abstract
Goat fecundity is important for agriculture and varies depending on the genetic background of the goat. Two excellent domestic breeds in China, the Jining Grey and Laiwu Black goats, have different fecundity and prolificacies. To explore the potential miRNAs that regulate the expression of the genes involved in these prolific differences and to potentially discover new miRNAs, we performed a genome-wide analysis of the miRNAs in the ovaries from these two goats using RNA-Seq technology. Thirty miRNAs were differentially expressed between the Jining Grey and Laiwu Black goats. Gene Ontology and KEGG pathway analyses revealed that the target genes of the differentially expressed miRNAs were significantly enriched in several biological processes and pathways. A protein-protein interaction analysis indicated that the miRNAs and their target genes were related to the reproduction complex regulation network. The differential miRNA expression profiles found in the ovaries between the two distinctive breeds of goats studied here provide a unique resource for addressing fecundity differences in goats.
The domestic goat (Capra hircus) is raised all over the world and is a great sector in the consumer market, providing humans with meat, skin, fur, fiber, and so on. The Laiwu Black and Jining Grey goats are two excellent local breeds in Shandong, China. The Jining Grey goat is an excellent local breed in China that possesses the characteristics of high prolificacy and year-round estrus. The fecundity of the Laiwu Black goat is much lower than that of the Jining Grey goat1. Specifically, the mean litter size for the Jining Grey goats is 2.942. In contrast, the fecundity of the Laiwu Black goat is only 1.643. In the goat industry, these reproductive traits are largely important because even a slight increase in litter size can lead to a large profit4.
Previous studies indicate that the bone morphogenetic protein receptor-1B (BMPR1B) gene regulates the fecundity and ovulation rate of sheep5,6,7. The goat genome sequence was recently resolved8,9, and despite the progress made in goat fecundity studies, the genes involved in the regulation of fecundity in goats are largely unknown. Thus, it may not be possible to improve fecundity at this point, and this is mostly due to the fact that reproductive traits are complex quantitative traits involving multiple genes, loci, and interactions. To beter understand goat fecundity, it is necessary to analyze the combined effect of multiple genes or loci on reproductive traits. Therefore, it is neceasary to identify more functional genes, to clarify the molecular mechanisms of actions and to identify regulatory networks that might be involved in goat fecundity4.
miRNAs are endogenous small non-coding RNAs that play an important role in gene regulation in animals and plants by pairing to the mRNAs of protein-coding genes to direct their posttranscriptional repression10,11. Generally, miRNAs are initially transcribed by RNA polymerase II and mature miRNAs form the RNA-induced silencing complex (RISC) with other components, such as dicer. Gene silencing occurs through the paring of complementary mRNA and miRNA, which, thus, leads to mRNA degradation or the prevention of translation11. More and more evidence suggests that hundreds of miRNAs affect a large fraction of the transcriptome and a broad range of biological processes and signaling pathways, such as the regulation of the transforming growth factor-β, Wnt, Notch, and epidermal growth factor signaling pathways12,13.
Recently, the RNA-Seq approach, which is a powerful tool based on next-generation deep-sequencing technologies, was utilized for RNA analysis14. The sequence reads from these analyses are individually mapped to the source genome and counted to obtain the number and density of reads corresponding to the RNA from each known exon, splice event or new candidate gene15. This technique has been successfully applied in multiple organisms for the genome-wide analysis of RNAs, such as yeast16, sheep17,18,19,20,21, and human22. The current study aimed to characterize distinct gene regulation between two domestic goats raised in China using genome-wide miRNA profiling via high-throughput deep RNA sequencing (RNA-Seq) technology.
Results
Mapping the small RNA reads
The small RNA reads obtained from the ovaries of the two goat breeds were first mapped using the goat miRBase database, and the resulting mapping rate was 92.1% (1.86 of 2.02 M) and 93.7% (2.47of 2.61 M) of total clean reads for the Jining Grey goats and the Laiwu Black goats, respectively (Table 1). Additional mapping to other species was attempted on the remaining reads, and a ~0.4% mapping rate was obtained that covered 297 and 278 miRNAs from other species for the Jining Grey and Laiwu Black goats, respectively (Table 2). For reads that did not map in the miRBase databases, the miRNA prediction process was performed to identify pre-mature miRNAs and to discover potential novel miRNAs.
Table 1. Reads mapping statistics.
| Category | Jining Grey | % | Laiwu Black | % |
|---|---|---|---|---|
| Mapped in goat miRBase | 1,861,671 | 92.05 | 2,446,554 | 93.70 |
| Mapped in other species | 7,305 | 0.36 | 7,736 | 0.30 |
| Novel annotated | 10,383 | 0.51 | 11,355 | 0.43 |
| Novel non-annotated | 8,116 | 0.40 | 8,831 | 0.34 |
| Total mapped | 1,887,475 | 93.33 | 2,474,476 | 94.77 |
| Unmapped | 134,885 | 6.67 | 136,694 | 5.23 |
| Total celan reads | 2,022,360 | 100.00 | 2,611,170 | 100.00 |
Table 2. Species of mapped miRNAs.
| Category | Jining Grey | Laiwu Black | |
|---|---|---|---|
| mature | goat | 383 | 373 |
| sheep | 9 | 7 | |
| cow | 81 | 76 | |
| pig | 22 | 17 | |
| rat | 33 | 29 | |
| mouse | 75 | 69 | |
| human | 77 | 80 | |
| predict | non-annotated | 1146 | 1316 |
| sheep | 15 | 17 | |
| cow | 112 | 121 | |
| pig | 33 | 32 | |
| rat | 8 | 7 | |
| mouse | 7 | 10 | |
| human | 14 | 10 | |
| Total mapped | 2015 | 2164 |
Novel miRNA prediction
The novel miRNAs were predicted using Mireap software23, and the predicted miRNAs were all essentially with a hair-pin structure. The predicted miRNAs were first mapped to the goat database and were subsequently mapped to the sheep, cow, pig, rat, mouse, and human miRBase databases for annotations. As shown in Table 1, 56.1% (10,383/18,499) and 56.3% (11,355/20,186) of the predicted miRNA reads in the Jining Grey and Laiwu Black goat samples, respectively, were mapped to the miRBase databases corresponding to these species.
Length distribution and base preference of the miRNAs
The length of the mapped miRNAs in this study was around 22 nt. Previous studies suggest that the miRNAs with a base U at the 5′ terminus are more conserved than miRNAs with a non-U base at the 5′ terminus24. We analyzed the base at the 5′ terminus of the miRNAs identified, and the results showed that U is the most common base in both of the goat breeds.
Differential regulation of the miRNAs between the two breeds of goats
A total of 5254 miRNAs were identified in the two goat samples, of which 603 (314 mature and 289 predicted, Table S1) had at least 10 read counts in the two goat samples. The expression level of these 603 conserved and novel miRNAs was assessed by a differential analysis, a downstream target prediction and biological annotation enrichment analyses. At a false discovery rate (FDR) <0.05 and an absolute value of log2 (fold change) > = 1, 30 miRNAs (22 mature and 8 predicted) were defined as differentially expressed miRNAs between the Laiwu Black and Jining Grey goats (Table 3), of which 10 miRNAs were down-regulated and 20 were up-regulated in the Jining Grey goat as compared with the Laiwu Black goat.
Table 3. 30 differentially expressed miRNAs.
| miRNA Name | Normalized counts - Jining Grey | Normalized counts - Laiwu Black | Log2FC | FDR | Style | Raw counts - Jining Grey | Raw counts - Laiwu Black |
|---|---|---|---|---|---|---|---|
| chr14_11964_mature | 0 | 19.50487369 | −20 | 6.12E-05 | down | 0 | 20 |
| chi-miR-9-3p | 19.48231021 | 152.1380148 | −2.965144 | 1.22E-11 | down | 19 | 156 |
| chi-miR-9-5p | 51.26923741 | 285.7463996 | −2.47857 | 1.29E-11 | down | 50 | 293 |
| chr12_10768_star | 14.35538647 | 78.01949477 | −2.442242 | 3.98E-06 | down | 14 | 80 |
| chr18_14930_mature | 7.177693237 | 30.23255422 | −2.074511 | 0.019925 | down | 7 | 31 |
| chi-miR-183 | 31.78692719 | 102.4005869 | −1.687719 | 0.000535 | down | 31 | 105 |
| chi-miR-449a-5p | 212.2546429 | 578.319505 | −1.446071 | 4.46E-05 | down | 207 | 593 |
| chr4_3193_mature | 25.6346187 | 69.24230161 | −1.43356 | 0.021011 | down | 25 | 71 |
| chr4_3446_mature | 44.09154417 | 112.1530237 | −1.346895 | 0.008989 | down | 43 | 115 |
| chi-miR-874-3p | 48.19308316 | 112.1530237 | −1.218571 | 0.027479 | down | 47 | 115 |
| chi-miR-30f-5p | 252.244648 | 124.8311916 | 1.014845 | 0.04378 | up | 246 | 128 |
| chi-miR-3958-3p | 253.2700328 | 124.8311916 | 1.020698 | 0.041048 | up | 247 | 128 |
| chi-miR-493-3p | 235.8384921 | 115.0787548 | 1.035178 | 0.038424 | up | 230 | 118 |
| chr20_16009_mature@@bta-miR-2478 | 280.955421 | 135.5588722 | 1.051422 | 0.026344 | up | 274 | 139 |
| chi-miR-450-5p | 3051.545011 | 1434.58346 | 1.088908 | 0.007197 | up | 2976 | 1471 |
| chi-miR-202-5p | 147.6554037 | 67.29181424 | 1.133731 | 0.030467 | up | 144 | 69 |
| chi-miR-424-5p | 3025.910392 | 1378.99457 | 1.133752 | 0.00353 | up | 2951 | 1414 |
| chi-miR-136-3p | 1258.147086 | 566.6165808 | 1.150856 | 0.002616 | up | 1227 | 581 |
| chi-miR-494 | 568.0631505 | 251.6128706 | 1.174846 | 0.002685 | up | 554 | 258 |
| chi-miR-655 | 91.25924259 | 38.0345037 | 1.262662 | 0.030556 | up | 89 | 39 |
| chr23_18096_mature | 173.2900224 | 72.16803267 | 1.263757 | 0.005829 | up | 169 | 74 |
| chi-miR-369-3p | 97.41155107 | 39.98499107 | 1.284634 | 0.021533 | up | 95 | 41 |
| chi-miR-145-3p | 1201.750925 | 487.6218423 | 1.301303 | 0.000224 | up | 1172 | 500 |
| chi-miR-542-3p | 1539.102507 | 620.2549835 | 1.311156 | 0.000173 | up | 1501 | 636 |
| bta-miR-132 | 156.8838665 | 61.44035213 | 1.352439 | 0.002894 | up | 153 | 63 |
| chi-miR-450-3p | 141.5030952 | 52.66315897 | 1.425968 | 0.001808 | up | 138 | 54 |
| chi-miR-223-3p | 95.36078158 | 34.13352896 | 1.482206 | 0.004285 | up | 93 | 35 |
| chi-miR-187 | 37.93923568 | 11.70292422 | 1.696822 | 0.037147 | up | 37 | 12 |
| chi-miR-497-3p | 60.49770014 | 10.72768053 | 2.495542 | 2E-05 | up | 59 | 11 |
| chr11_10274_mature | 16.40615597 | 1.950487369 | 3.072331 | 0.022057 | up | 16 | 2 |
Prediction of the miRNA targets
The programs TargetFinder25 and miRanda26 were used to assess the 3′-UTR regions of the potential target genes. A total of 15,848 gene targets without redundancy were prediced from the 30 differential miRNAs (30,172 pairs) with a free enengy (ΔG) of less than or equal to −15.0, with 1–7 miRNAs targeting a single gene or 17–4357 genes targeted by a single miRNA. Of these 15, 848 gene targets, known miRNAs gene targets without redundancy were 13,699, and predicted miRNAs gene targets without redundancy were 6,703 (Table S2).
Gene Ontology and KEGG pathway analysis
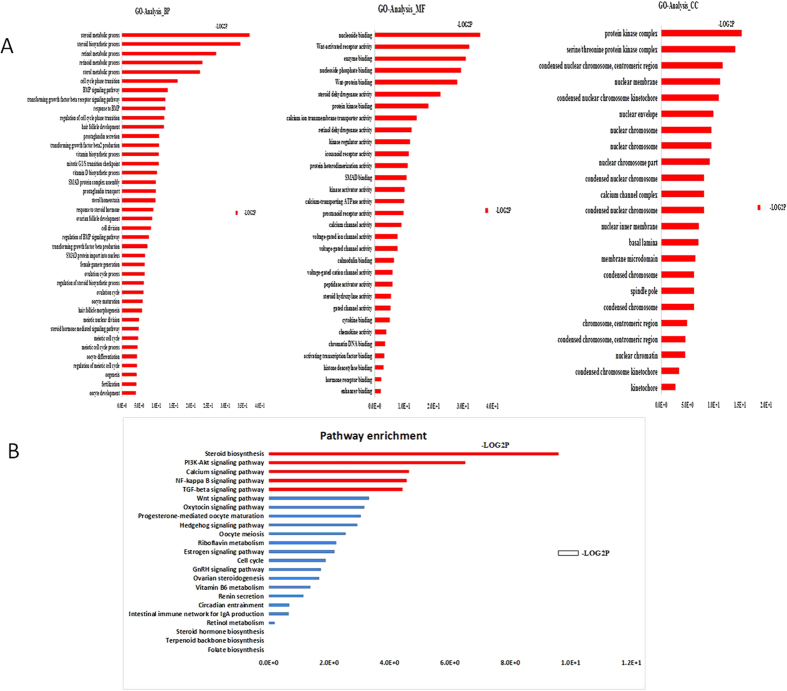
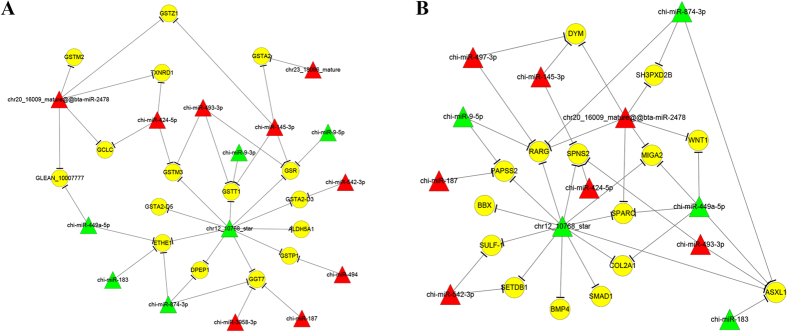
The predicted miRNA-targeted genes were further analyzed by Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The target genes that interact with conserved and predicted miRNAs were analyzed separately, with regard to the enrichment of the GO terms and the KEGG pathway analysis. For the mature miRNA-targeted genes, there were no significantly enriched GO terms and KEGG pathways. In contrast, the predicted miRNA-targeted genes were enriched significantly in 28 biological processes and 2 KEGG pathways (Fig. 1). Among the 28 biological processes, 19 were associated with reproduction, such as steroid biosynthetic process, the BMP signaling pathway, and the transforming growth factor beta receptor signaling pathway (Figs 1A and 2A). Glycosaminoglycans that are localized to the extracellular matrix of bone are thought to play a key role in mediating aspects of bone development. The two enriched KEGG pathways were the TGF-β signaling pathway and steroid biosynthesis (Figs 1B and 2B). Indeed, these functional annotation results demonstrate that this database contains clues that might lead to uncovering potential regulators of fecundity between these 2 breeds of goats.
Figure 1. Summary of the gene ontology and KEGG pathway analyses of the targets of the differentially expressed miRNAs.
(A) Gene ontology and (B) KEGG pathway. The processes and pathways in red are significant.
Figure 2. Two specific miRNA-mRNA regulation networks.
(A) glutathione metabolic process and (B) bone development. The yellow circle indicates mRNA. Red and green triangles represent miRNAs. Red and green represents up and down regulation respectively.
Interaction networks between miRNAs and the target genes related to reproduction
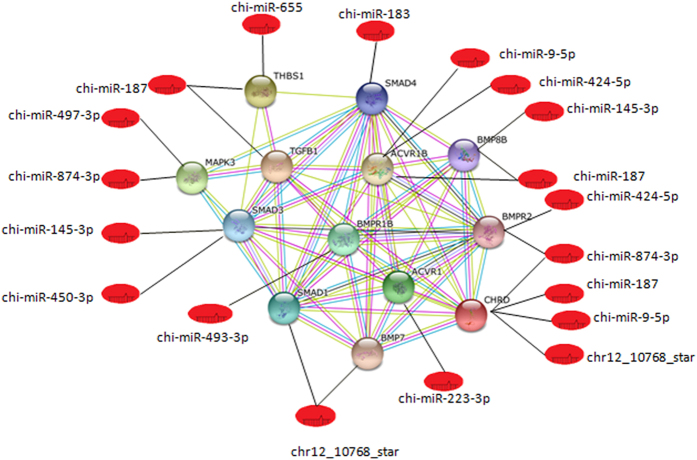
Given the fact that we are interested in exploring the fecundity differences between these 2 breed of goats, the relationships between the differentially expressed miRNAs and the targeted genes related with reproduction are described based on the STRING database analysis. Figure 3 demonstrates that the relationships between the miRNAs and the genes BMPR 1B, SMAD1, BMP7, ACVR1, CHRD, BMPR2, BMP88, ACVR18, SMAD4, THBS1, TGFB1, MAPK3 and SMAD3, which are related to reproduction, are complicated. The BMPR1B gene, which is regulated by chi-miR-493-3P alone, is a very important finding. In addition, chi-miR-187 regulates four genes, including TGFB1, THBS1, ACVR18 and BMP88, and chr12_10768_star regulates three genes, including CHRD, SMAD1 and BMP7. In addition, chi-miR-874-3P regulates three genes, including MAPK3, BMPR2 and CHRD. Therefore, chi-miR-187, chr12_10768_star and chi-miR-874-3P might play an important role in reproductive regulation processes.
Figure 3. Network of the miRNAs with the targeted genes related to reproduction.
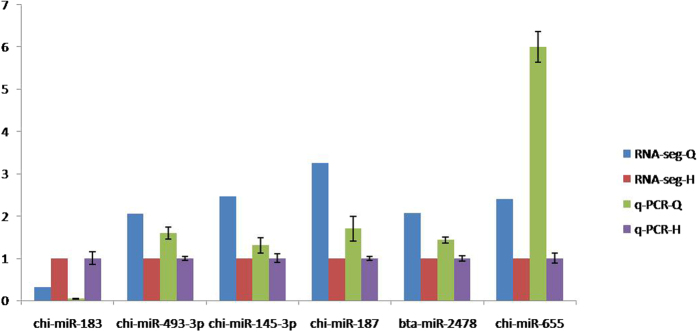
qRT-PCR validation of RNA-Seq data
To confirm the RNA-Seq data, 6 differentially expressed miRNAs were examined by qRT-PCR, and these data were consistent with the above RNA-Seq results (Table 4 and Fig. 4). The genes potentially targeted by the 6 miRNAs are involved in diverse cellular activities, such as signal transduction, kinase, motor activity, transport, metabolic process and DNA binding.
Table 4. Validation of RNA-Seq data by RT-PCR.
| RNA-Seq-Grey goat | RNA-Seq-Black goat | q-PCR-Grey goat | q-PCR-Black goat | q-PCR-Grey goat SD | q-PCR-Black goat SD | |
|---|---|---|---|---|---|---|
| chi-miR-183 | 0.310417431 | 1 | 0.0416 | 1.0000 | 0.004 | 0.1503 |
| chi-miR-493-3p | 2.049366037 | 1 | 1.593 | 1.0000 | 0.1396 | 0.04933 |
| chi-miR-145-3p | 2.464514141 | 1 | 1.307 | 1.0000 | 0.1798 | 0.1026 |
| chi-miR-187 | 3.241859467 | 1 | 1.7 | 1.0000 | 0.294 | 0.05 |
| bta-miR-2478 | 2.072571248 | 1 | 1.43 | 0.9967 | 0.0781 | 0.05783 |
| chi-miR-655 | 2.399380397 | 1 | 5.997 | 1.0000 | 0.363 | 0.115 |
Figure 4. qPCR validation of 6 miRNAs.
The gene U6 was used as the reference gene.
Discussion
With the use of RNA-Seq technology, here, we performed a genome-wide analysis on the miRNAs in Jining Grey and Laiwu Black goats and characterized 30 differentially expressed miRNAs. Because these two breeds of goats demonstrate different fecundity, the differentially regulated miRNAs may contribute to this process. Previous studies indicate that several miRNAs are important for the follicular-luteal transition in the ruminant ovary and for fetal gonadal development27,28. Homologs of the identified differentially expressed miRNAs function in various cellular activities. For instance, miR-9-3p is an important regulatory factor in the osteoblastic differentiation of mouse iPS cells and also targets β1 integrin to sensitize claudin-low breast cancer cells to MEK inhibition29,30. miR-449a regulates cell proliferation and causes Rb-dependent cell cycle arrest and senescence31,32. Mature miR-183 promotes 3T3-L1 adipogenesis through the inhibition of the canonical Wnt/β-catenin signaling pathway by targeting LRP633. miR-542-3p also has many important functions, including the induction of growth arrest through the survival pathway, the suppression of osteoblast cell proliferation and differentiation, the targeting of BMP-7 signaling and the inhibition of bone formation34,35. Given the broad roles of the miRNA homologs, the differentially expressed miRNA molecules might also potentially be involved in the regulation of goat growth and development.
miRNAs function mainly by pairing with mRNAs, and by an in silico analysis, we predicted the potential target genes for the differentially expressed miRNAs identified in our RNA-Seq analysis. Using the predicted target genes, the Gene Ontology (GO) enrichment analysis revealed several significantly enriched biological processes that were associated with the steroid biosynthetic process, the BMP signaling pathway and the transforming growth factor beta receptor signaling pathway (Fig. 1). The glutathione metabolic process is involved in a variety of protective processes, and we found that, in this process, 15 differentially expressed miRNAs potentially target 16 genes (Fig. 2A), which include the homologs of miR-9-3p, miR-183, miR-449a and miR-542-3p, which were mentioned in the prior section. In addtion, more favorable cellular processes in the ovaries, such as the steroid biosynthetic process, the BMP signaling pathway, the transforming growth factor beta receptor signaling pathway, contribute to the high prolificacy observed in the Jining Grey goat. Moreover, among the differentially expressed miRNAs that were identified to be involved in the glutathione metabolic process, 15 of the predicted target genes are also involved in the bone development (Fig. 2B). A recent study reported36 that certain genes in ovaries were involved in cartilage development and bone healing, which is consistent with our findings. The KEGG pathway analysis was consistent with the GO analysis, indicating that the miRNA targeted genes were significantly enriched for glutathione metabolism and the steroid biosynthesis pathways.
Moreover, the relationship between the miRNAs and the genes BMPR1B, SMAD1, BMP7, ACVR1, CHRD, BMPR2, BMP88, ACVR18, SMAD4, THBS1, TGFB1, MAPK3 and SMAD3, which are related with reproduction37, is complicated. The BMPR1B gene, which is regulated by chi-miR-493-3P alone, was a very important finding. BMPR1B (FecB), was the first major prolificacy gene identified in sheep. The FecB locus is autosomal with a codominant expression and has an additive effect for ovulation rate when it is associated with a mutation (Q249R) in the BMPR1B gene38,39. BMPR1B is a member of the bone morphogenetic protein (BMP) receptor family of transmembrane serine/threonine kinases. BMPs are involved in endochondral bone formation and embryogenesis. BMPR1B is a member of the family of receptors for transforming growth factor (TGF) ligands, such as TGF, activins, BMPs and growth and differentiation factors (GDFs), which play a role in folliculogenesis40.
In conclusion, we identified 30 differentially expressed miRNAs in the ovaries of the two goats and most of these had defined homologs that were key miRNAs related to reproduction. This genome-wide miRNA analysis study will serve as a resource for understanding goat proliferacy. The differential miRNAs identified here are predicted to contribute to different prolificacies of the two goat breeds through a number of biological processes and pathways. In particular, Chi-miR-187, chr12_10768_star, and chi-miR-874-3P may play an important role in the reproductive regulation processes. This study provides a unique resource to address important issues related to goat fecundity, which will aid in elucidating the specific mechanisms responsible for differential gene expression in the ovaries between two distinctive breeds of goats. Moreover, these data also highlight the important roles of miRNAs during genetic regulation in different goat breeds.
Materials and Methods
Ethics statement
All of the experiments were performed in accordance with the relevant guidelines and regulations and were approved by the Institutional Animal Care and Use Committee of Institute of Animal Sciences, Chinese Academy of Agricultural Sciences.
Goat sample preparation
All of the samples in this study were from female adult goats between the ages of 1.5 to 2 years old. The age-matched healthy female Jining Grey goats and female Laiwu Black goats (5 per breed) were obtained from the Qingdao Aote Farm (Shandong, China). The goats were treated with intravaginal sponges (Intervet, Mexico) to achieve estrous synchronization as previously described41. Pregnant mare serum gonadotropin (PMSG) (Ningbo Sansheng Pharmaceutical Co., LTD., Zhejiang, China) was injected after the removal of the sponge. Estrus was checked by observing the goats’ reactions to an adult male goat every 12 h after the sponge was removed. The female goats were determined to be in estrus when they showed standing estrous behavior. The goats were sacrificed 4–5 hours after estrus was determined, and the whole ovaries were excised. The samples were collected to obtain ovulation points on the surface of the ovaries. All of the samples were immediately snap-frozen in liquid nitrogen and stored at −70 °C for RNA extraction41.
Construction of small RNA libraries and sequencing
Total RNA was extracted from ovaries with better ovulation points on the surfaces from 5 Jining Grey and 5 Laiwu Black goats using Trizol (Life Technologies/Invitrogen, California, USA) according to the manufacturer’s instructions42. The extracted RNA from each breed was pooled. The quality and quantity of the RNA samples were assessed on a Bioanalyzer 2100 system using an RNA 6000 Nano kit (Agilent Technologies, Palo Alto, CA). After the total RNA was extracted, two small RNA libraries were prepared as described in the Illumina® TruSeq™ Small RNA Sample Preparation protocol. First, the 3′ adaptor and 5′ adaptor were ligated to the RNA by T4 RNA ligase, and then, reverse transcription was carried out with SuperScriptII reverse transcriptase (Invitrogen) to generate the cDNA. The resulting cDNA was then amplified to generate the small RNA library. The DNA size and the purity of the cDNA library were checked using a high sensitivity DNA 1000 kit on a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA), and the quantification of the cDNA libraries was performed with Qubit™ dsDNA HS kit on a Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). The cDNA libraries were then subjected to single-end sequencing on the Illumina Genome AnalyzerIIx (GAIIx) using the proprietary Solexa sequencing-by-synthesis method at the Shanghai Biotechnology Corporation (Shanghai, China) according to the manufacturer’s recommended cycling parameters. The image analysis and the base calling were performed with the Illumina built-in SCS2.8/RTA1.8 software.
Workflow of the bioinformatics analysis
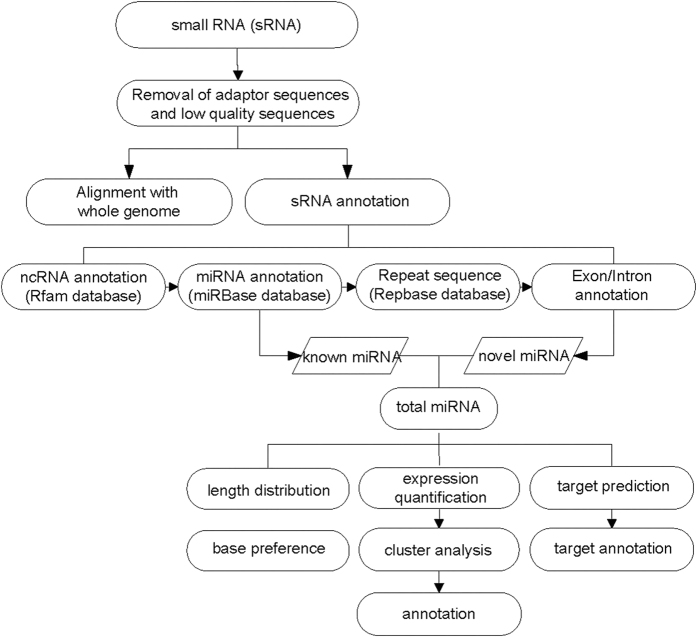
The workflow of bioinformatics analysis of the RNA-Seq results is shown in Fig. 5. Briefly, the adaptor sequences and the low quality sequences were removed from the original reads by fastx (fastx_toolkit-0.0.13.2) (http://hannonlab.cshl.edu/fastx_toolkit/), and the quality read data were aligned with the whole reference genome and were compared with multiple databases for the annotation of the ncRNA, miRNA, repeat sequences, and exon/intron sequences. The differential expression of the miRNAs was identified using IDEG6 software43. For the unannotated small RNA sequences, a miRNA prediction process was carried out, and then, the novel miRNAs and the targets of the differentially expressed miRNAs were annotated.
Figure 5. Workflow of the bioinformatics analysis of the small RNA sequencing results.
Quality control was performed followed by further analysis.
Programs used for the bioinformatics analysis
To predict potential novel miRNAs, Mireap software23 was used to analyze the unannotated reads. The inverted repeats (step loops or hairpin structure) described by Jones-Rhoades and Bartel44 were searched, and the secondary structure of the inverted repeat was predicted by RNA fold45. To predict the targets of the miRNAs, the programs TargetFinder25 and miRanda26 were used. The differentially expressed miRNAs were identified using IDEG6 software43. A rigorous significance test for the digital gene expression profiling was used46. The differentially expressed gene contigs were analyzed by gene ontology (GO)47. Three structured controlled vocabularies (ontologies), including cellular component, molecular function, and biological process, were used to create a consistent description of the gene products as previously described48,49. Like the GO enrichment, the association of the genes with different pathways was computed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg)50,51. Moreover, based on the String database (http://string-db.org/), the interaction networks of the miRNAs and their target genes, related with reproduction, were predicted to draw relationships between the miRNAs and their target genes.
Validation of RNA-Seq data
To validate the RNA-Seq results, the expression of several of the miRNAs was confirmed by quantitative real-time RT-PCR. Briefly, the cDNA for the miRNA was produced using a miScript II Reverse Transcription Kit (Qiagen, US) in a GeneAmp® PCR System 9700 (Applied Biosystems, USA). Next, the cDNA was used as the template for the qPCR, which was performed using a SYBR Green I Master (Roche, Swiss). The primer sequences were designed in the laboratory and were synthesized by Generay Biotech (Generay, PRC) and are based on the mRNA sequences obtained from the NCBI database. The gene U6 was used as the internal control, and the expression levels were calculated using the 2 delta delta Ct method (2−ΔΔCt).
Additional Information
How to cite this article: Miao, X. et al. Genome-wide analysis of miRNAs in the ovaries of Jining Grey and Laiwu Black goats to explore the regulation of fecundity. Sci. Rep. 6, 37983; doi: 10.1038/srep37983 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by a grant from The Major Science and Technology Project of New Variety Breeding of Genetically Modified Organisms (nos. 2009ZX08008-004 and 2008ZX08008-003), the National High Technology Research Development Programme of China (863 Programme No. 2008AA10Z140), the National Natural Science Foundation of China (No. 30571339), the Agricultural Science and Technology Innovation Programme (ASTIPIAS05), the Innovation Research Foundation of CAAS (No. 2004-CAAS-1) and the Basic Research Fund for Central Public Research Institutes of CAAS (2013ywf-yb-5, 2013ywf-zd-2).
Footnotes
Author Contributions X.Y.M. conceived, designed and performed the experiments and wrote the paper; Q.M.L., H.J.Z. and X.Y.Q. performed the experiments. All of the authors read and approved the final manuscript.
References
- Miao X., Luo Q. & Qin X. Genome-wide transcriptome analysis in the ovaries of two goats identifies differentially expressed genes related to fecundity. Gene 582, 69–76 (2016). [DOI] [PubMed] [Google Scholar]
- Tu Y. R. Small Tailed Han sheep. The sheep and goat breeds in China. [ Tu Y. R. ed.] [88–90, 98–101] (Shanghai Science and Technology Press, Shanghai, 1989). [Google Scholar]
- Liu S. S., Wang X. B., Liu Q. B., Zhang Y., Gao J. R. et al. Comprehensive evaluation of germplasm resources of Laiwu black goat. The Chinese Livest. Poultry Breeding 10, 64–65 (2015). [Google Scholar]
- Ahlawat S. et al. Current status of molecular genetics research of goat fecundity. Samll Ruminant Res. 125, 34–42 (2015). [Google Scholar]
- Davis G. H. Major genes affecting ovulation rate in sheep. Genet. Sel. Evol. 37, S11–23 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulsant P. et al. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc. Natl. Acad. Sci. USA 98, 5104–5109 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. et al. Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol. Reprod. 64, 1225–1235 (2001). [DOI] [PubMed] [Google Scholar]
- Dong Y. et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 31, 135–141 (2013). [DOI] [PubMed] [Google Scholar]
- Mak H. C. Goat genome sequence by optical mapping. Nat. Biotechnol. 31, 123 (2013). [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L. & Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993). [DOI] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M., Martello G. & Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol. Cell. Biol. 11, 252–263 (2010). [DOI] [PubMed] [Google Scholar]
- Bushati N. & Cohen S. M. microRNA functions. Annu. Rev. Cell Dev. Biol. 23, 175–205 (2007). [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M. & Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao X., Luo Q. & Qin X. Genome-wide transcriptome analysis of mRNAs and microRNAs in Dorset and Small Tail Han sheep to explore the regulation of fecundity. Mol. Cell. Endocrinol. 402, 32–42 (2015). [DOI] [PubMed] [Google Scholar]
- Miao X., Luo Q., Zhao H. & Qin X. Ovarian transcriptomic study reveals the differential regulation of miRNAs and lncRNAs related to fecundity in different sheep. Sci. Rep. 6, 35299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao X., Luo Q. & Qin X. Genome-wide analysis reveals the differential regulations of mRNAs and miRNAs in Dorset and Small Tail Han sheep muscles. Gene 562, 188–196 (2015). [DOI] [PubMed] [Google Scholar]
- Miao X., Luo Q., Qin X. & Guo Y. Genome-wide analysis of microRNAs identifies the lipid metabolism pathway to be a defining factor in adipose tissue from different sheep. Sci. Rep. 5, 18470 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao X., Luo Q., Qin X., Guo Y. & Zhao H. Genome-wide mRNA-Seq profiling reveals predominant down-regulation of lipid metabolic processes in adipose tissues of Small Tail Han than Dorset sheep. Biochem. Biophys. Res. Commun. 467, 413–420 (2015). [DOI] [PubMed] [Google Scholar]
- Peng Z. et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 30, 253–260 (2012). [DOI] [PubMed] [Google Scholar]
- Hu S. et al. Sensitization of sodium channels by cystathionine beta-synthetase activation in colon sensory neurons in adult rats with neonatal maternal deprivation. Exp. Neurol. 248, 275–285 (2013). [DOI] [PubMed] [Google Scholar]
- Shi W., Jiang J. X., Miao X. G. & Kong X. Y. The complete mitochondrial genome sequence of Heteromycteris japonicus (Pleuronectiformes: Soleidae). Mitochondr. DNA 25, 257–258 (2014). [DOI] [PubMed] [Google Scholar]
- Jin X., Sun J., Miao X., Liu G. & Zhong D. Inhibitory effect of geraniol in combination with gemcitabine on proliferation of BXPC-3 human pancreatic cancer cells. J. Int. Med. Res. 41, 993–1001 (2013). [DOI] [PubMed] [Google Scholar]
- John B. et al. Human MicroRNA targets. PLoS Biol. 2, e363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride D. et al. Identification of miRNAs associated with the follicular-luteal transition in the ruminant ovary. Reproduction 144, 221–233 (2012). [DOI] [PubMed] [Google Scholar]
- Torley K. J. et al. Expression of miRNAs in ovine fetal gonads: potential role in gonadal differentiation. Reprod. Biol. Endocrinol. 9, 2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H. et al. Involvement of microRNAs in regulation of osteoblastic differentiation in mouse induced pluripotent stem cells. PloS One 7, e43800 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawistowski J. S. et al. MicroRNA 9-3p targets beta1 integrin to sensitize claudin-low breast cancer cells to MEK inhibition. Mol. Cell. Biol. 33, 2260–2274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan E. J., Place R. F., Basak S., Pookot D. & Li L. C. miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget 1, 349–358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Fang Y., Cao Y., Qin R. & Chen Q. miR-449a Regulates proliferation and chemosensitivity to cisplatin by targeting cyclin D1 and BCL2 in SGC7901 cells. Digest Dis. Sci. 59, 336–345 (2014). [DOI] [PubMed] [Google Scholar]
- Chen C., Xiang H., Peng Y. L., Peng J. & Jiang S. W. Mature miR-183, negatively regulated by transcription factor GATA3, promotes 3T3-L1 adipogenesis through inhibition of the canonical Wnt/beta-catenin signaling pathway by targeting LRP6. Cell. Signal. 26, 1155–1165 (2014). [DOI] [PubMed] [Google Scholar]
- Yoon S. et al. Induction of growth arrest by miR-542-3p that targets survivin. FEBS Lett. 584, 4048–4052 (2010). [DOI] [PubMed] [Google Scholar]
- Kureel J. et al. miR-542-3p suppresses osteoblast cell proliferation and differentiation, targets BMP-7 signaling and inhibits bone formation. Cell Death Dis. 5, e1050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager M. et al. Composite transcriptome assembly of RNA-Seq data in a sheep model for delayed bone healing. BMC Genomics 12, 158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Yefen, Li Erlin, Han Yedong, Chen Ling & Xie Zhuang. Differential expression of mRNAs encoding BMP/Smad pathway molecules in antral follicles of high- and low-fecundity Hu sheep. Anim. Reprod. Sci. 120, 47–55 (2010). [DOI] [PubMed] [Google Scholar]
- Davis G. H., Galloway S. M., Ross I. K. et al. DNA test in prolific sheep from eight countries provide new evidence on origin of the Booroola (FecB) mutation. Biol. Reprod. 66, 1869–1874 (2002). [DOI] [PubMed] [Google Scholar]
- Souza C. J., MacDougal C., Campbell B. K., McNeilly A. S. & Baird D. T. The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1B (BMPR1B) gene. J. Endocrinol. 2, R1–R6 (2001). [DOI] [PubMed] [Google Scholar]
- Derynck R. & Feng X. H. TGF-beta receptor signaling. Biochim. Biophys. Acta 1333, F105–F150 (1997). [DOI] [PubMed] [Google Scholar]
- Miao X. & Luo Q. Genome-wide transcriptome analysis between small-tail Han sheep and the Surabaya fur sheep using high-throughput RNA sequencing. Reproduction 145, 587–596 (2013). [DOI] [PubMed] [Google Scholar]
- Rio D. C., Ares M. Jr, Hannon G. J. & Nilsen T. W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. (6), pdb.prot 5439 (2010). [DOI] [PubMed] [Google Scholar]
- Romualdi C., Bortoluzzi S., D’Alessi F. & Danieli G. A. IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol. Genomics 12, 159–162 (2003). [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M. W. & Bartel D. P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14 (2004). [DOI] [PubMed] [Google Scholar]
- Hofacker I. L. et al. Fast folding and comparison of RNA Secondary structures. Monatshefte für Chemie 125, 167–188 (1994). [Google Scholar]
- Audic S. & Claverie J. M. The significance of digital gene expression profiles. Genome Res. 7, 986–995 (1997). [DOI] [PubMed] [Google Scholar]
- Ashburner M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. A. et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–261 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhao L. J., Tan Y. X., Ren H. & Qi Z. T. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis 33, 1113–1120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H., Goto S., Fujibuchi W. & Kanehisa M. Computation with the KEGG pathway database. Bio Systems 47, 119–128 (1998). [DOI] [PubMed] [Google Scholar]
- Aoki-Kinoshita K. F. & Kanehisa M. Gene annotation and pathway mapping in KEGG. Methods Mol. Biol. 396, 71–91 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.