Abstract
Response rates to conventional chemotherapeutics remain unsatisfactory for hepatocellular carcinoma (HCC) due to the high rates of chemoresistance and recurrence. Tumor-initiating cancer stem-like cells (CSLCs) are refractory to chemotherapy, and their enrichment leads to subsequent development of chemoresistance and recurrence. To overcome the chemoresistance and stemness in HCC, we synthesized a Pt nanocluster assembly (Pt-NA) composed of assembled Pt nanoclusters incorporating a pH-sensitive polymer and HCC-targeting peptide. Pt-NA is latent in peripheral blood, readily targets disseminated HCC CSLCs, and disassembles into small Pt nanoclusters in acidic subcellular compartments, eventually inducing damage to DNA. Furthermore, treatment with Pt-NA downregulates a multitude of genes that are vital for the proliferation of HCC. Importantly, CD24+ side population (SP) CSLCs that are resistant to cisplatin are sensitive to Pt-NA, demonstrating the immense potential of Pt-NA for treating chemoresistant HCC.
Short abstract
A Pt-nanocluster assembly, by clustering extremely small-sized Pt nanoclusters with pH-sensitive polymers and a liver cancer cell targeting peptide, is demonstrated to effectively overcome the chemoresistance and stemness of hepatocellular carcinoma.
Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-associated death worldwide.1 Most HCC patients are inherently resistant to conventional chemotherapeutic drugs.2 Currently, sorafenib is the only FDA-approved target therapy drug (since 2007) for advanced HCC which increases progression-free survival by a dismal three months compared to placebo. Recently, the adjuvant sorafenib for HCC failed in a phase III, randomized, double-blind, and placebo-controlled trial.3 Therefore, the development of more effective therapeutic strategies is much needed. Moreover, treating HCC with ineffective chemotherapeutics leads to the enrichment of a rare subpopulation of tumor-initiating cancer stem-like cells (CSLCs).4 It has been reported that resistance to cisplatin is associated with the enrichment of CSLCs in ovarian,5 lung,6 and liver cancer.7 The self-renewal capacity of CSLCs plays significant roles in the progression and recurrence of tumors. Nanomedicine has emerged as a promising platform for the development of novel cancer therapy strategies8−11 to target cancer cells including CSLCs.12−14
During our massive screening of potential therapeutics against drug-resistance and stemness of HCC, we are pleasantly surprised that among many candidates, small-sized Pt nanoclusters can effectively overcome the chemoresistance and stemness in HCC. In fact, Pt nanoparticles are known to kill cancer cells15−24 by the leached Pt ions under low pH conditions such as the cell endosome.17,18 Especially when the particle size is reduced to less than 3 nm, >50% of the atoms will be located on the surface of the crystal,25,26 resulting in increased oxygen adsorption and water oxidation for surface corrosion,26 thus facilitating Pt ion release for enhanced activity. However, nonspecific targeting of Pt compound has a wide range of toxicity to normal tissues.27,28 Consequently, targeted delivery and controlled Pt ion release are essential to precise cancer therapy. Unfortunately, to our best knowledge, thus far there is no report on platinum-based nanomedicine which can achieve both effective CSLC targeting and cellular environmentally sensitive anticancer activity.
Self-assembly provides a reliable way of generating ensembles of nanoparticles with controllable properties,29−31 and stimuli-responsive nanoparticle assemblies have been thoroughly examined as not only bio- or chemosensors in vitro(32−37) but also advanced drug delivery systems in vivo.38−42 We hypothesized that the anticancer activity of platinum-based nanomedicine can be adjusted via controlled clustering of ultrasmall Pt nanoparticles using pH-sensitive surface ligands. To demonstrate the proof of concept, we herein report on the designed synthesis of a tumor pH-sensitive Pt nanocluster assembly (Pt-NA) by immobilizing ultrasmall Pt nanoclusters within pH-sensitive polymers and derivatizing with an HCC-targeting peptide. Unlike the reported platinum nanoparticle-based drugs,15−18 the resulting Pt-NA has several advanced features for tumor treatment including (i) Pt-NA is latent in peripheral blood and readily targets tumor cells including CLSC because of the surface targeting peptide; (ii) protonation of pH-sensitive polymers in an acidic intracellular environment triggers Pt-NA disassembly into extremely small Pt nanoclusters; (iii) the resulting extremely small Pt nanoclusters with large specific surface accelerate the release of toxic Pt ions inside the cells for an effective cancer treatment.
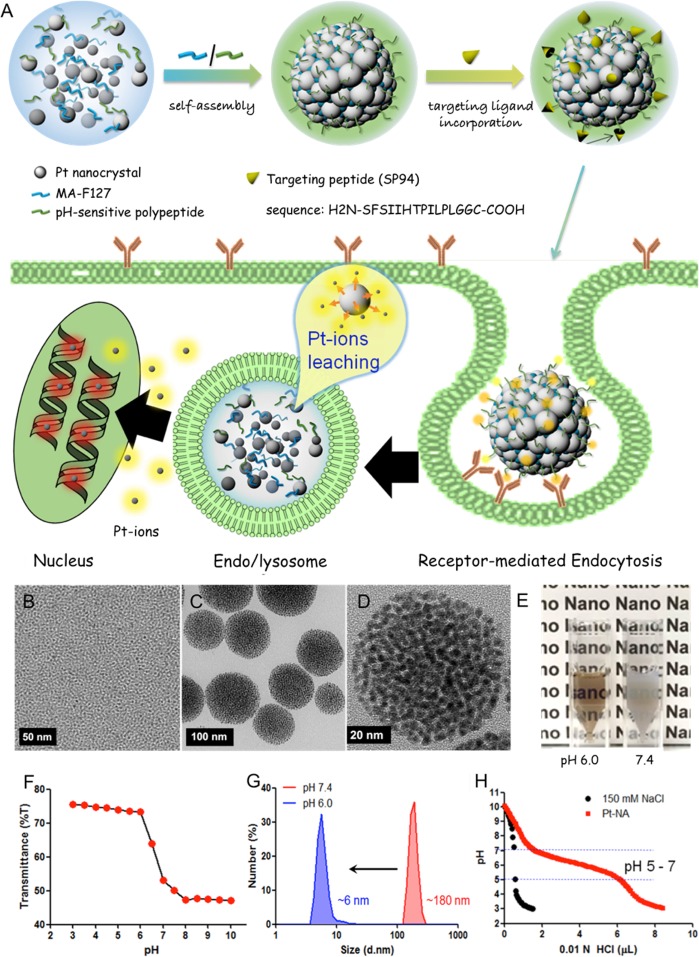
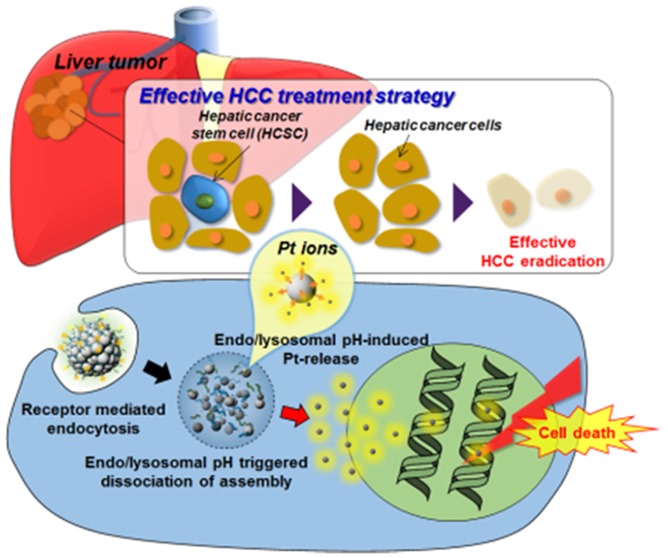
Our designed synthesis and anti-HCC strategy of Pt-NA are schematically illustrated in Figure 1A. Briefly, Pt-NA is prepared by assembling ultrasmall Pt nanoclusters in pH-sensitive polymers, followed by derivatizing with the HCC-targeting peptide. First, the assembly between Pt nanoclusters and two designed ligands (MA-F127 and octadecylamine-p(API-Asp)10) is performed via a unique hydrophobic force through the dual solvent-exchange method.41 The unique ligand design allows maleimide groups (from MA-F127) to form on the surface of the assembled structure, which can readily conjugate with the thiol group (−SH) of HCC cell-specific peptide ligand SP9443 (H2N-SFSIIHTPILPLGGC-COOH) via additional chemical reaction. We postulate that following the intravenous administration, Pt-NA accumulates in an HCC lesion via the enhanced permeability and retention (EPR) effect,44,45 and SP94 specific ligands facilitate receptor-mediated endocytosis into HCC cells.43 Furthermore, the endolysosomal acidification46−48 collapses the assembled structure to release Pt nanoclusters, by destroying the hydrophilic–lipophilic balance in Pt-NA, and subsequently increases the Pt ion release rate in HCC cells.
Figure 1.
Design and characterization of HCC-targeted pH-sensitive Pt nanocluster assembly (Pt-NA). (A) Schematic representation of Pt-NA synthesis, targeted HCC uptake and intracellular Pt ion release. (B) TEM image of the synthesized Pt nanoclusters. (C) TEM image of Pt-NA. (D) High-resolution TEM image of Pt-NA. (E) Photographs of Pt-NA in pH 6.0 and 7.4. (F) Transmittance of a suspension of Pt-NA as a function of pH. (G) DLS size measurement of Pt-NA (0.1 mg mL–1) as a function of pH. (H) pH profile of Pt-NA by acid–base titration.
Results and Discussion
Ultrasmall Pt nanoclusters of ∼2.5 nm were synthesized by thermal decomposition of Pt(acac)2 at 170 °C (Figure 1B, Figures S1 and S2). In order to obtain ultrasmall and uniform-sized Pt nanoclusters, we added 1 equiv of oleic acid in the reaction mixture containing Pt(acac)2 and oleylamine, and superhydride was injected in the reaction mixture to synthesize small-sized Pt nanoclusters (Figures S1C and S2). Interestingly, when only one ligand was added in the reaction mixture containing 1-octadecene as the solvent and Pt(acac)2, worm-like Pt nanocrystals were obtained due to insufficient capping agents (Figure S1A,D). The high concentration of oleic acid leads to particle aggregation, while colloidal uniform Pt nanocrystals are obtained when a high concentration of oleylamine as both surfactant and solvent is applied (Figure S1B,C,E). It was reported that small-sized nanoparticles generally show very high activity, because the smaller the nanoparticles are the faster the surface ion leaching is in aqueous solution.49,50 As such, Pt ions release is much faster for ∼3 nm sized Pt nanoclusters than that for ∼10 nm sized Pt nanoparticles (Figure S3). However, in order to control their activity in vivo, two polymeric ligands are designed for tumor targeted pH-sensitive Pt-NA formation: (i) maleimide-functionalized pluronic F127 (MA-F127) (Figures S4A and S5); (ii) a synthetic pH-sensitive polypeptide (octadecylamine-p(API-Asp)10) composed of aspartic acid (10-mer) modified with ionizable imidazole side chains and an octadecylamine tail (Figure S4B). The successful conjugation of SP94 with pH-sensitive Pt-NA was confirmed by Fourier transform infrared (FT-IR) spectroscopy (Figure S6).
Transmission electron microscopy (TEM, Figure 1C,D) reveals that the particle size of Pt-NA is ∼100 nm. Scanning TEM and electron energy loss spectroscopy mapping further verify the assembled structure (Figure S7). Measurement of light transmittance as a function of pH (Figure 1E,F) demonstrates a pH-dependent assembly/disassembly process and a sharp drop in transmittance (% T) at a pH higher than ∼6.0. In particular, water-dispersible Pt-NA exhibits a hydrodynamic diameter about 180 nm (Figure 1G, at pH 7.4). Besides, its stability in both pure PBS and PBS solution containing 10% FBS is studied (Figure S8); maintaining particle size in both solutions for 1 week indicates the good colloid stability of Pt-NA. A slightly wider size distribution in PBS solution containing 10% FBS seems to result from insignificant protein binding.51,52 Moreover, the assembly is highly sensitive to the acidity of the tumor and is readily dissociated upon the pH drop, as confirmed by dynamic light scattering (DLS) (Figure 1G) and TEM (Figure S9). In addition, the acid–base titration curve confirms the strong buffering capacity of aqueous Pt-NA dispersion (Figure 1H) in the physiological pH range due to the presence of imidazole rings, which is beneficial for endolysosomal escape of nanoparticles.53
This sensitivity to tumor intracellular pH is a distinct property of Pt-NA compared to the previously reported Pt drugs.15−18 We demonstrate that Pt-NA is structurally stable and latent at a physiological pH of 7.4, where Pt ion leaching is highly restrained. However, the tumor intracellular pH of <6 triggers the dissociation of Pt-NA and consequently accelerates Pt ion release (Figures S3 and S10). Moreover, these Pt nanoclusters are more robust than small-molecule drugs such as cisplatin, and they can remain inside hepatic CSLCs without being affected by ATP-binding cassette (ABC) transporters.54,55 The internalized Pt nanoclusters will constantly leak cytotoxic Pt ions only inside the HCC cells, facilitating DNA platination,56 and effectively induce DNA damage and kill the cells (Figure S11). It is known that the platinum content of only 9.0 × 106 atoms cell–1 (2.9 × 10–6 ng cell–1) can efficiently kill cancer cells.57 Consequently, according to the previous reports, the amount of Pt ions released inside HCC cells should be enough for effective cytotoxicity.57,58
To verify our rationale, the cisplatin sensitivity of a panel of >20 liver cancer cell lines was evaluated, which shows heterogeneous sensitivity to cisplatin (Figure S12). And two acquired cisplatin-resistant HCC cell populations (HuH7-Cis and PLC/PRF/5-Cis) were established by treatment of a stepwise increase in cisplatin concentrations (Figure S13). Interestingly, we found that the SP+CD24+ CSLC population is elevated in both primary resistant and acquired resistant HCC cells by side population (SP) and HCC stem cell marker CD24 analysis (Figure S14).59,60 Using a tumor sphere-forming assay, we found that the self-renewal ability of SP+CD24+ cells is much stronger than that of SP-CD24- cells (Figures S15 and S16).
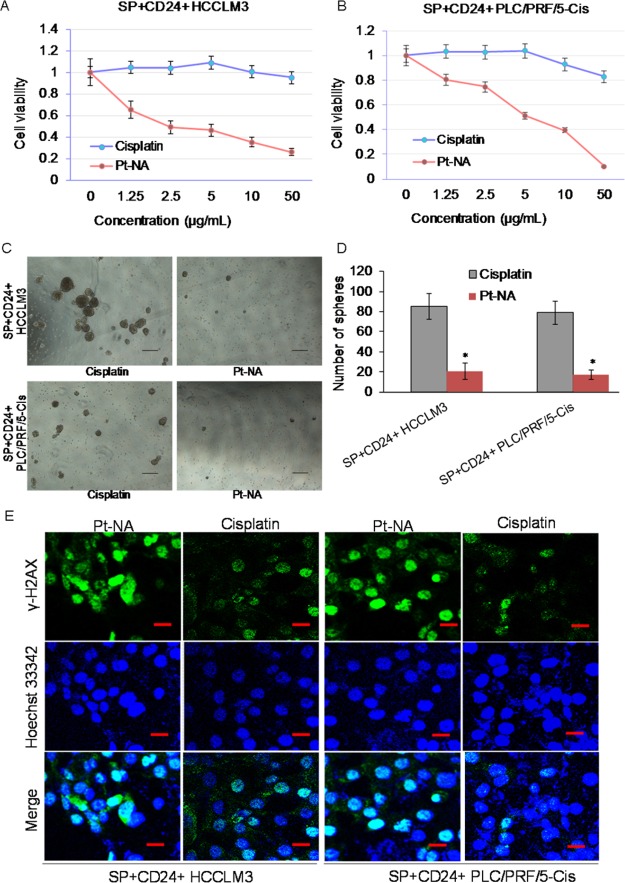
The effect of Pt-NA on overcoming the stemness of cisplatin-resistant liver cancer cells was first explored in vitro and compared to that of cisplatin (Figure 2). Pt-NA shows dose-dependent inhibition of resistant SP+CD24+ cells (Figure 2A,B) and significantly decreases the sphere-forming ability of cisplatin-resistant SP+CD24+ cells (Figure 2C,D). In contrast to cisplatin, we further found that Pt-NA can effectively induce DNA damage to resistant CSLCs (Figure 2E).
Figure 2.
Pt-NA overcomes the stemness of cisplatin-resistant HCC cells and induces DNA damage. (A, B) Dose-dependent inhibition effect of Pt-NA and cisplatin on cell growth in the cisplatin-resistant (A) HCCLM3 and (B) PLC/PRF/5-Cis cell lines. (C, D) Pt-NA significantly decreases the stemness of cisplatin-resistant SP+CD24+ HCCLM3 and PLC/PRF/5-Cis cells. (E) Pt-NA induces DNA damage in cisplatin-resistant HCC cells (scale bar, 50 μm).
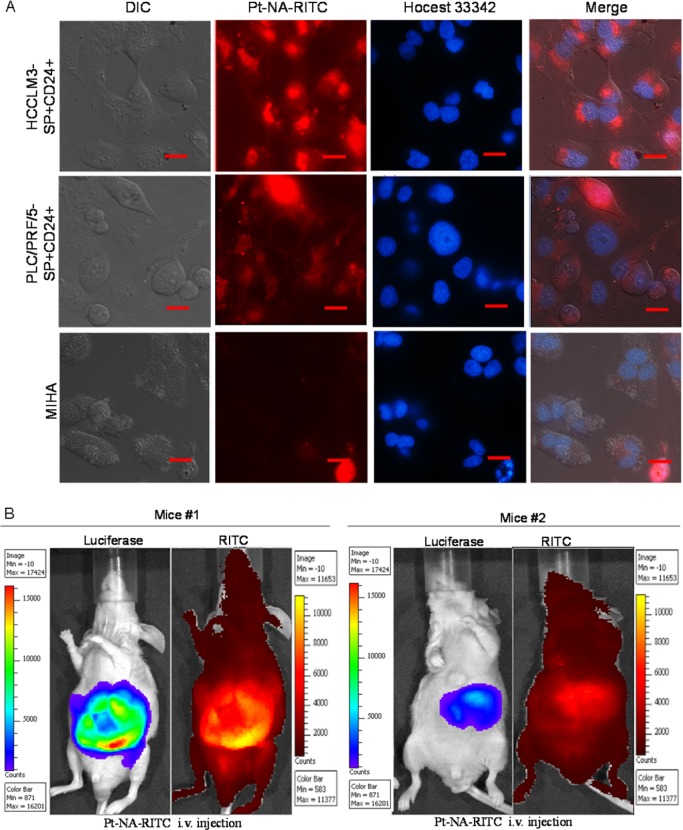
We next examined the in vitro cellular uptake efficiency of cisplatin and Pt-NA in the sorted SP–CD24– and SP+CD24+ cells from the HCCLM3 and PLC/PRF/5-Cis cell lines. The data shows that the amount of Pt taken up by sorted SP+CD24+ is much less than that by SP–CD24– cells for cisplatin, but such a difference is not observed for Pt-NA (Figure S17A–D). Importantly, Pt-NA is specifically taken up by the sorted SP+CD24+ cells rather than normal liver cells (Figure 3A). Furthermore, in contrast to pH-insensitive Pt nanoparticles, it is observed that most Pt-NA escapes from endosome (Figure S17E) due to the proton sponge effect of imidazole-containing polymeric ligands,53 facilitating the nuclei localization of released Pt ions for DNA platination. On the other hand, compared to cisplatin, Pt-NA shows significantly lower toxicity to normal liver cells (Figure S18), implying that Pt-NA is safer and shows less side effects than cisplatin. The tumor targeting ability of Pt-NA is also demonstrated in vivo (Figure 3B) using an HCC orthotopic mouse model,61 and the data are compared with those in normal mice (Figure S19). The location of tumors was further confirmed by anatomical study (Figure S20). As shown in Figure S21, Pt-NA showed a fair pharmacokinetic (PK) profile which is comparable to clinically approved cis-diammineplatinum(II) (CDDP, cisplatin),62,63 and the biodistribution (BD) result of Pt-NA is very similar to other nanomaterial-based anticancer medicines, which mainly accumulates in liver and spleen.64−66
Figure 3.
HCC targeting of Pt-NA. (A) Cellular uptake of Pt-NA in cisplatin-resistant and cancer stem-like SP+CD24+ cells. Pt-NA uptake is minimal in the normal liver cell line MIHA (scale bar, 50 μm). (B) In vivo biodistribution of Pt-NA in SP+CD24+ HCCLM3 cells of representative orthotopic HCC mice with different tumor sizes (2 h after injection, tumors are established from different number of sorted SP+CD24+ luciferase expressing HCCLM3 cells, tumor of Mouse #1 was developed from 10 000 sorted cells, while tumor of Mouse #2 was developed from 1000 sorted cells).
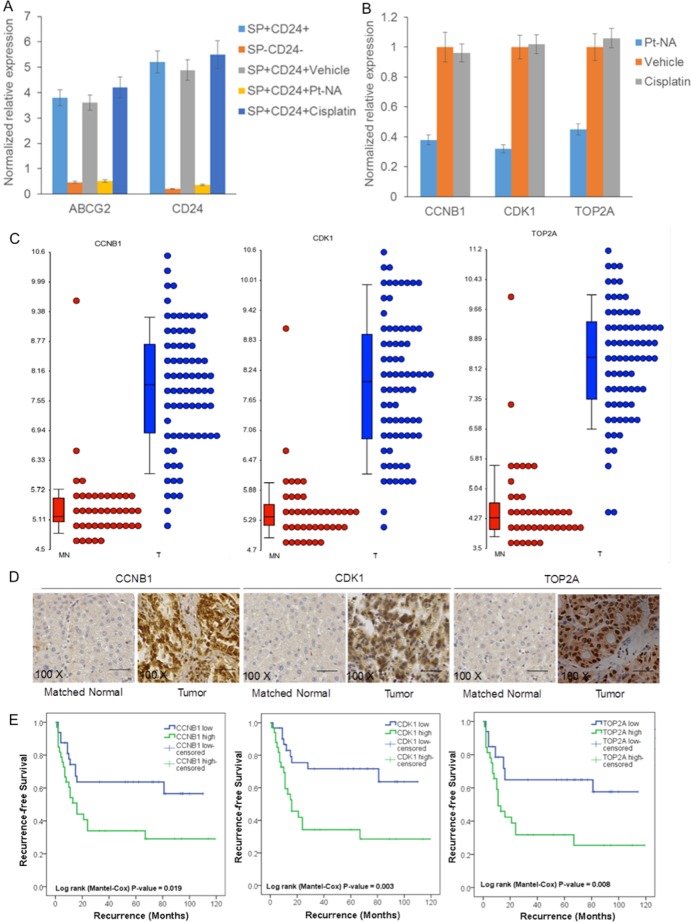
To clarify the molecular mechanism about overcoming the stemness of cisplatin-resistant liver cancer cells, we conducted a gene expression profile analysis on sorted SP+CD24+ HCCLM3 cells treated with Pt-NA. By comparing with our established global gene expression profile database of human HCC,67 many genes, which are prominently downregulated by Pt-NA, are those known to be highly expressed in liver cancer (Table S1). Ingenuity pathway analysis (IPA) demonstrates that Pt-NA mainly modulates genes related to the cell cycle and DNA damage-related pathways (Figures S22 and S23). These microarray data are further validated by RT-qPCR, showing that both ABCG2 and CD24 are highly expressed in the sorted SP+CD24+ cells and are downregulated by Pt-NA but not cisplatin (Figure 4A). Further RT-qPCR analysis also reveals that Pt-NA downregulates CCNB1, CDK1, and TOP2A expression, confirming its effects on the modulation of cell cycle and DNA damage regulation (Figure 4B). Since preclinical data regarding HCC appear to vary on a model-by-model basis,68 the expression levels of CCNB1, CDK1, and TOP2A were further examined in the clinical tissue samples from HCC patients during operation, and their expression levels are significantly overexpressed in tumor compared with matched normal tissues (Figure 4C). The immunohistochemistry staining also shows that the expression of CTNNB1, CDK1, and TOP2A is significantly upregulated in the tumor tissues compared with matched normal tissues of HCC patients (Figure 4D). The median values of CCNB1, CDK1, and TOP2A expressions are chosen as the cutoff point for determining high or low expression. Fisher’s exact test and Kaplan–Meier analysis reveal that high expression of CCNB1, CDK1, or TOP2A is correlated with poor survival in HCC patients (Figure 4E). These data suggest that the Pt-NA-mediated DNA damage can be attributed to downregulation of many genes that show high expression in liver cancer.
Figure 4.
Pt-NA induces DNA damage through downregulation of genes that are highly expressed in liver cancer. (A) The expression of ABCG2 and CD24 in SP-CD24- and SP+CD24+ HCCLM3 cells with treatment of vehicle, Pt-NA or cisplatin. The expression of ABCG2 and CD24 was are significantly higher in SP+CD24+ cells compared to SP-CD24- cells. Pt-NA (but not cisplatin) induced the downregulation of ABCG2 and CD24 expression in the SP+CD24+ cells of the cisplatin-resistant HCCLM3 cell line, as determined by RT-qPCR. (B) The expression of CCNB1, CDK1, and TOP2A in the SP+CD24+ HCCLM3 cells with treatment of vehicle, Pt-NA or cisplatin. Pt-NA (but not cisplatin) induced the downregulation of CCNB1, CDK1, and TOP2A expression in the SP+CD24+ cells of the cisplatin-resistant HCCLM3 cell line, as determined by RT-qPCR. (C) The expression of CCNB1, CDK1, and TOP2A significantly increased in tissue samples from HCC patients compared to matched adjacent nontumor tissues (T, tumorous liver tissue; MN, matched adjacent nontumor liver tissue) in our established HCC gene expression profile data set. (D) The representative images of immunohistochemistry showing that the expression of CTNNB1, CDK1, and TOP2A are significantly upregulated in the tumor tissues compared with matched normal tissues of HCC patients. (E) Fisher’s exact test and Kaplan–Meier analysis indicating that high expression of CCNB1, CDK1, or TOP2A is correlated with poor survival in HCC patients. (The median values of CCNB1, CDK1, and TOP2A expression were chosen as the cutoff points for determining high and low expression groups.)
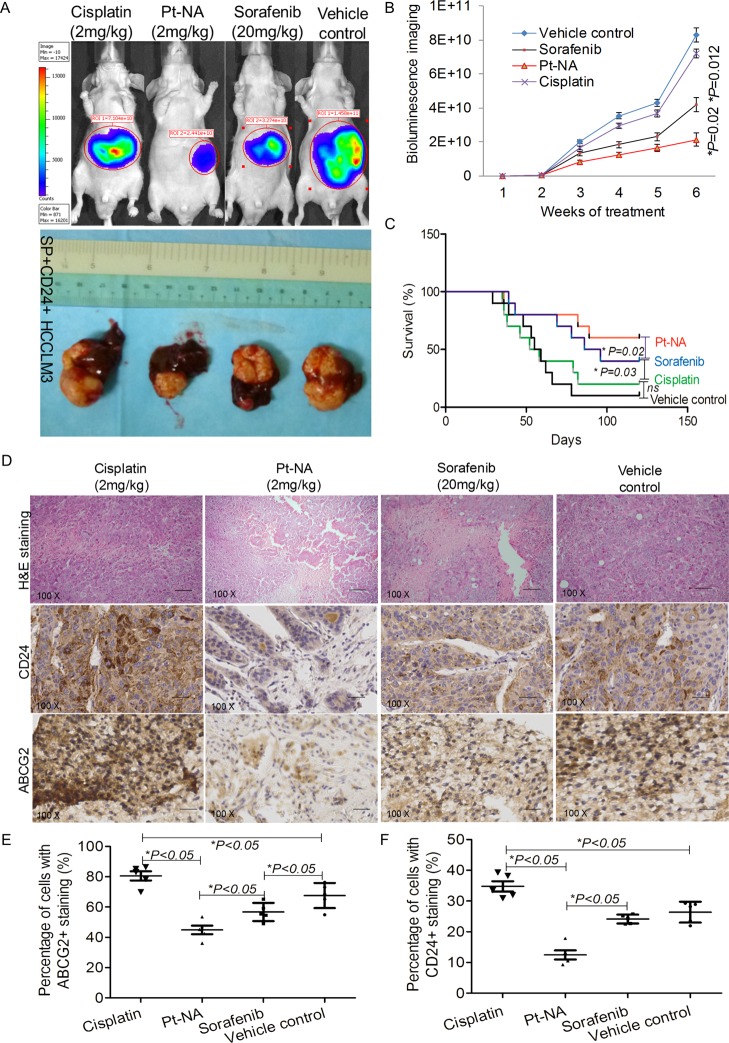
Anti-HCC study demonstrates that Pt-NA shows superior therapeutic efficacy compared to both cisplatin and sorafenib (Figure 5A,B). After 3 months, the survival rate is highest for the Pt-NA-treated mice at ∼60% (Figure 5C). Moreover, more tumors became necrosed after being treated by Pt-NA compared to other treatment groups (Figure 5D). The in vivo anti-HCC effect of Pt-NA was further compared with cisplatin-incorporating polymeric micelles (cisplatin loaded poly(l-glutamic acid)-g-methoxy poly(ethylene glycol) complex nanoparticles, nanoplatin); it is not surprising that the orthotopic tumor xenografts developed from isolated cisplatin-resistant cells were also resistant to cisplatin loaded nanoparticles of nanoplatin, and consequently Pt-NA showed significantly better tumor inhibition effects than nanoplatin at the same Pt concentration (Figure S24). Immunohistochemical staining shows that the expressions of CD24 and ABCG2 drop in the Pt-NA-treated tumor tissues, while they increase in the cisplatin-treated ones (Figure 5D–F). The real time RT-qPCR results (Figure S25) show that the expression of CCNB1, CDK1, and TOP2A of the tumors treated with Pt-NA also remarkably decreases, compared with the tumors treated with sorafenib or cisplatin. Finally, the liver/renal toxicity study together with histopathological examination prove the good biocompatibility of Pt-NA (Figure S26).
Figure 5.
Pt-NA overcomes the cisplatin resistance and stemness of HCC in vivo. (A) Representative images showing bioluminescence signals and tumor-bearing livers of the orthotopic tumor xenografts at the therapeutic end point of different treatments. (B) Quantitative analysis of bioluminescence signals of all mice in the four treatment groups measured on a weekly basis. (C) The survival analysis of all mice in the four treatment groups. (D) Immunohistochemical staining for the expression of ABCG2 and CD24 in the tumors of mice in the different treatment groups, scale bar, 100 μm. (E, F) The quantified ABCG2 (E) and CD24 (F) levels from immunohistochemical staining results according to the percentage of cells with positive nuclei.
Conclusions
In summary, we synthesized a novel Pt nanocluster assembly (Pt-NA) that effectively overcomes the cisplatin resistance and heterogeneous stemness of HCC cells. The extremely small-sized Pt nanoclusters in HCC cells lead to the high toxicity resulting from a large surface area. The assembled structures of Pt-NA are designed for HCC targeting and response to a HCC intracellular acidic stimulus, which can readily target the CSLCs of HCC cells and further disassemble into small Pt nanoclusters in acidic HCC subcellular compartments, where Pt ions are released more quickly. Pt-NA can significantly kill HCC cells via DNA damage and overcome the cisplatin resistance of CLSCs. Moreover, we demonstrate the mechanism of these effects at the molecular level, by which Pt-NA has a good potential in clinical HCC treatment through downregulating a multitude of genes that are highly expressed in liver cancer patients.
Acknowledgments
D.L. acknowledges financial support by the the National Key Research and Development Program of China (2016YFA0203600), the National Natural Science Foundation of China (51503180, 5161101036), “Thousand Talents Program” for Distinguished Young Scholars (588020*G81501/048), and Fundamental Research Funds for the Central Universities (520002*172210161). K.M.H. acknowledges financial support by the SingHealth Foundation and the National Medical Research Council, Singapore. T.H. acknowledges financial support by the Research Center Program of the Institute for Basic Science (IBS) in Korea (IBS-R006-D1).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.6b00197.
Experimental details including the synthetic scheme and characterization of two polymeric ligands for Pt nanocluster assembly, TEM images of Pt nanocrystals synthesized in different conditions, stability of Pt-NA in serum conditions, in vitro cumulative Pt ions release, DNA damage induced by Pt-NA, heterogeneity sensitivity, Pt uptake in SP−CD24− and SP+CD24+ cells, IVIS imaging, in vivo pharmacokinetic (PK) and biodistribution (BD) study, in vivo efficacy comparing to CDDP-incorporating polymeric micelles (Nanoplatin), gene expression in tumor tissues, and the liver and renal toxicity of the Pt-NA (PDF)
Author Contributions
⊥ H.X., F.L., and X.H. contributed equally to this work and all three should be considered as first authors. D.L., K.H. and T.H. designed and supervised the project. D.L., F.L., and H.X. conceived the experiments. D.L., H.X., F.L., and X.H. performed the experiments. W.P., S.W., Y.J., Y.D., S.B., S.C., T.K. and D.K. assisted with the experiments, D.L. and T.H. wrote the manuscript and analyzed the data. All authors discussed the results and commented on the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Maluccio M.; Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. Ca-Cancer J. Clin. 2012, 62, 394–399. 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- EASL–EORTC clinical practice guidelines: management of hepatocellular carcinoma. J. Hepatol. 2012, 56, 908–943. 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Bruix J.; Takayama T.; Mazzaferro V.; Chau G.-Y.; Yang J.; Kudo M.; Cai J.; Poon R. T.; Han K.-H.; Tak W. Y.; et al. Adjuvant sorafenib for hepatocellular carcinoma after resection or ablation (STORM): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2015, 16, 1344–1354. 10.1016/S1470-2045(15)00198-9. [DOI] [PubMed] [Google Scholar]
- Yu F.; Yao H.; Zhu P.; Zhang X.; Pan Q.; Gong C.; Huang Y.; Hu X.; Su F.; Lieberman J.; Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Wei X.; Dombkowski D.; Meirelles K.; Pieretti-Vanmarcke R.; Szotek P. P.; Chang H. L.; Preffer F. I.; Mueller P. R.; Teixeira J.; MacLaughlin D. T.; Donahoe P. K. Müllerian inhibiting substance preferentially inhibits stem/progenitors in human ovarian cancer cell lines compared with chemotherapeutics. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 18874–18879. 10.1073/pnas.1012667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolini G.; Roz L.; Perego P.; Tortoreto M.; Fontanella E.; Gatti L.; Pratesi G.; Fabbri A.; Andriani F.; Tinelli S.; et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 16281–16286. 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. K. W.; Castilho A.; Cheung V. C. H.; Tang K. H.; Ma S.; Ng I. O. L. CD24+ liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell 2011, 9, 50–63. 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Kim B. Y. S.; Rutka J. T.; Chan W. C. W. Nanomedicine. N. Engl. J. Med. 2010, 363, 2434–2443. 10.1056/NEJMra0912273. [DOI] [PubMed] [Google Scholar]
- Mout R.; Moyano D. F.; Rana S.; Rotello V. M. Surface functionalization of nanoparticles for nanomedicine. Chem. Soc. Rev. 2012, 41, 2539–2544. 10.1039/c2cs15294k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farokhzad O. C.; Langer R. Impact of nanotechnology on drug delivery. ACS Nano 2009, 3, 16–20. 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- Barry N. P. E.; Sadler P. J. Challenges for metals in medicine: how nanotechnology may help to shape the future. ACS Nano 2013, 7, 5654–5659. 10.1021/nn403220e. [DOI] [PubMed] [Google Scholar]
- Peer D.; Karp J. M.; Hong S.; Farokhzad O. C.; Margalit R.; Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007, 2, 751–760. 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy S.; Ke X.; Yang Y. Y. Delivery of therapeutics using nanocarriers for targeting cancer cells and cancer stem cells. Nanomedicine 2015, 10, 143–160. 10.2217/nnm.14.154. [DOI] [PubMed] [Google Scholar]
- Yoshii T.; Geng Y.; Le N. D. B.; Goel H. L.; Mercurio A. M.; Rotello V. M. Highlights from the latest articles in nanomaterial-based therapies for targeting cancer stem cells. Nanomedicine 2015, 10, 3427–3429. 10.2217/nnm.15.179. [DOI] [PubMed] [Google Scholar]
- Asharani P. V.; Xinyi N.; Hande M. P.; Valiyaveettil S. DNA damage and p53-mediated growth arrest in human cells treated with platinum nanoparticles. Nanomedicine 2010, 5, 51–64. 10.2217/nnm.09.85. [DOI] [PubMed] [Google Scholar]
- Barone M.; Sciortino M. T.; Zaccaria D.; Mazzaglia A.; Sortino S. Nitric oxide photocaging platinum nanoparticles with anticancer potential. J. Mater. Chem. 2008, 18, 5531–5536. 10.1039/b809121h. [DOI] [Google Scholar]
- Chien C.-T.; Yan J. Y.; Chiu W.-C.; Wu T. H.; Liu C. Y.; Lin S. Y. Caged Pt nanoclusters exhibiting corrodibility to exert tumor-inside activation for anticancer chemotherapeutics. Adv. Mater. 2013, 25, 5067–5073. 10.1002/adma.201302363. [DOI] [PubMed] [Google Scholar]
- Gao J.; Liang G.; Zhang B.; Kuang Y.; Zhang X.; Xu B. FePt@CoS2 yolk-shell nanocrystals as a potent agent to kill HeLa cells. J. Am. Chem. Soc. 2007, 129, 1428–1433. 10.1021/ja067785e. [DOI] [PubMed] [Google Scholar]
- Mironava T.; Simon M.; Rafailovich M. H.; Rigas B. Platinum folate nanoparticles toxicity: cancer vs. normal cells. Toxicol. In Vitro 2013, 27, 882–889. 10.1016/j.tiv.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Pelka J.; Gehrke H. M.; Turk M.; Crone M.; Brase S.; Muller T.; Blank H.; Send W.; Zibat V.; Brenner P.; Esselen M.; Schneider R.; Gerthsen D.; Marko D. Cellular uptake of platinum nanoparticles in human colon carcinoma cells and their impact on cellular redox systems and DNA integrity. Chem. Res. Toxicol. 2009, 22, 649–659. 10.1021/tx800354g. [DOI] [PubMed] [Google Scholar]
- Porcel E.; Liehn S.; Remita H.; Usami N.; Kobayashi K.; Furusawa Y.; Le Sech C.; Lacombe S. Platinum nanoparticles: a promising material for future cancer therapy?. Nanotechnology 2010, 21, 085103. 10.1088/0957-4484/21/8/085103. [DOI] [PubMed] [Google Scholar]
- Shiny P. J.; Mukherjee A.; Chandrasekaran N. DNA damage and mitochondria-mediated apoptosis of A549 lung carcinoma cells induced by biosynthesised silver and platinum nanoparticles. RSC Adv. 2016, 6, 27775–27787. 10.1039/C5RA27185A. [DOI] [Google Scholar]
- Teow Y.; Valiyaveettil S. Active targeting of cancer cells using folic acid-conjugated platinum nanoparticles. Nanoscale 2010, 2, 2607–2613. 10.1039/c0nr00204f. [DOI] [PubMed] [Google Scholar]
- Xue X.; Hall M. D.; Zhang Q.; Wang P. C.; Gottesman M. M.; Liang X. J. Nanoscale drug delivery platforms overcome platinum-based resistance in cancer cells due to abnormal membrane protein trafficking. ACS Nano 2013, 7, 10452–10464. 10.1021/nn405004f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. H.; Hackett M. J.; Park J.; Hyeon T. Synthesis, characterization, and application of ultrasmall nanoparticles. Chem. Mater. 2014, 26, 59–71. 10.1021/cm402225z. [DOI] [Google Scholar]
- Sakurai T.; Shibata M.; Horiuchi R.; Yagi I.; Kondo T. Study of platinum dissolution mechanism using a highly sensitive electrochemical quartz crystal microbalance. Chem. Lett. 2011, 40, 402–404. 10.1246/cl.2011.402. [DOI] [Google Scholar]
- Hartmann J. T.; Lipp H. P. Toxicity of platinum compounds. Expert Opin. Pharmacother. 2003, 4, 889–901. 10.1517/14656566.4.6.889. [DOI] [PubMed] [Google Scholar]
- Florea A. M.; Büsselberg D. Cisplatin as an anti-tumor drug: cellular mechanisms of activity, drug resistance and induced side effects. Cancers 2011, 3, 1351–1371. 10.3390/cancers3011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z.; Petukhova A.; Kumacheva E. Properties and emerging applications of self-assembled structures made from inorganic nanoparticles. Nat. Nanotechnol. 2010, 5, 15–25. 10.1038/nnano.2009.453. [DOI] [PubMed] [Google Scholar]
- He L.; Wang M.; Ge J.; Yin Y. Magnetic assembly route to colloidal responsive photonic nanostructures. Acc. Chem. Res. 2012, 45, 1431–1440. 10.1021/ar200276t. [DOI] [PubMed] [Google Scholar]
- Stolarczyk J. K.; Deak A.; Brougham D. F. Nanoparticle clusters: assembly and control over internal order, current capabilities, and future potential. Adv. Mater. 2016, 28, 5400–5424. 10.1002/adma.201505350. [DOI] [PubMed] [Google Scholar]
- Lee H.; Sun E.; Ham D.; Weissleder R. Chip–NMR biosensor for detection and molecular analysis of cells. Nat. Med. 2008, 14, 869–874. 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi N. L.; Mirkin C. A. Nanostructures in biodiagnostics. Chem. Rev. 2005, 105, 1547–1562. 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- Jin Y.; Gao X. Plasmonic fluorescent quantum dots. Nat. Nanotechnol. 2009, 4, 571–576. 10.1038/nnano.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.; Du X.; Zhao F.; Xu B. Magnetic nanoparticles for the manipulation of proteins and cells. Chem. Soc. Rev. 2012, 41, 2912–2942. 10.1039/c2cs15315g. [DOI] [PubMed] [Google Scholar]
- Zagorovsky K.; Chan W. C. W. A plasmonic DNAzyme strategy for point-of-care genetic detection of infectious pathogens. Angew. Chem., Int. Ed. 2013, 52, 3168–3171. 10.1002/anie.201208715. [DOI] [PubMed] [Google Scholar]
- Taton T. A.; Mirkin C. A.; Letsinger R. L. Scanometric DNA array detection with nanoparticle probes. Science 2000, 289, 1757–1760. 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- Ohta S.; Glancy D.; Chan W. C. W. DNA-controlled dynamic colloidal nanoparticle systems for mediating cellular interaction. Science 2016, 351, 841–845. 10.1126/science.aad4925. [DOI] [PubMed] [Google Scholar]
- Li H.-J.; Du J.-Z.; Du X.-J.; Xu C.-F.; Sun C.-Y.; Wang H.-X.; Cao Z.-T.; Yang X.-Z.; Zhu Y.-H.; Nie S.; Wang J. Stimuli-responsive clustered nanoparticles for improved tumor penetration and therapeutic efficacy. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 4164–4169. 10.1073/pnas.1522080113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q.; Sun X.; Ma X.; Zhou Z.; Jin E.; Zhang B.; Shen Y.; Van Kirk E. A.; Murdoch W. J.; Lott J. R.; et al. Integration of nanoassembly functions for an effective delivery cascade for cancer drugs. Adv. Mater. 2014, 26, 7615–7621. 10.1002/adma.201401554. [DOI] [PubMed] [Google Scholar]
- Ling D.; Park W.; Park S.-j.; Lu Y.; Kim K. S.; Hackett M. J.; Kim B. H.; Yim H.; Jeon Y. S.; Na K.; Hyeon T. Multifunctional tumor pH-sensitive self-assembled nanoparticles for bimodal imaging and treatment of resistant heterogeneous tumors. J. Am. Chem. Soc. 2014, 136, 5647–5655. 10.1021/ja4108287. [DOI] [PubMed] [Google Scholar]
- Lok C. N.; Zou T.; Zhang J. J.; Lin W. S.; Che C. M. Controlled-release systems for metal-based nanomedicine: encapsulated/self-assembled nanoparticles of anticancer gold(III)/platinum(II) complexes and antimicrobial silver nanoparticles. Adv. Mater. 2014, 26, 5550–5557. 10.1002/adma.201305617. [DOI] [PubMed] [Google Scholar]
- Ashley C. E.; Carnes E. C.; Phillips G. K.; Padilla D.; Durfee P. N.; Brown P. A.; Hanna T. N.; Liu J.; Phillips B.; Carter M. B.; et al. The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nat. Mater. 2011, 10, 389–397. 10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J.; Nakamura H.; Maeda H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Delivery Rev. 2011, 63, 136–151. 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Owens D. E. III; Peppas N. A. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 2006, 307, 93–102. 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Zhou K.; Huang G.; Hensley C.; Huang X.; Ma X.; Zhao T.; Sumer B. D.; DeBerardinis R. J.; Gao J. A nanoparticle-based strategy for the imaging of a broad range of tumours by nonlinear amplification of microenvironment signals. Nat. Mater. 2014, 13, 204–212. 10.1038/nmat3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling D.; Hackett M. J.; Hyeon T. Cancer imaging: lighting up tumours. Nat. Mater. 2014, 13, 122–124. 10.1038/nmat3860. [DOI] [PubMed] [Google Scholar]
- Nichols J. W.; Bae Y. H. Odyssey of a cancer nanoparticle: from injection site to site of action. Nano Today 2012, 7, 606–618. 10.1016/j.nantod.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambhy V.; Macbride M. M.; Peterson B. R.; Sen A. Silver bromide nanoparticle/polymer composites: dual action tunable antimicrobial materials. J. Am. Chem. Soc. 2006, 128, 9798–9808. 10.1021/ja061442z. [DOI] [PubMed] [Google Scholar]
- Loher S.; Schneider O. D.; Maienfisch T.; Bokorny S.; Stark W. J. Micro-organism-triggered release of silver nanoparticles from biodegradable oxide carriers allows preparation of self-sterilizing polymer surfaces. Small 2008, 4, 824–832. 10.1002/smll.200800047. [DOI] [PubMed] [Google Scholar]
- Dobrovolskaia M. A.; Patri A. K.; Zheng J.; Clogston J. D.; Ayub N.; Aggarwal P.; Neun B. W.; Hall J. B.; McNeil S. E. Interaction of colloidal gold nanoparticles with human blood: effects on particle size and analysis of plasma protein binding profiles. Nanomedicine 2009, 5, 106–117. 10.1016/j.nano.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal P.; Hall J. B.; McLeland C. B.; Dobrovolskaia M. A.; McNeil S. E. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv. Drug Delivery Rev. 2009, 61, 428–437. 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yezhelyev M. V.; Qi L.; O’Regan R. M.; Nie S.; Gao X. Proton-sponge coated quantum dots for siRNA delivery and intracellular imaging. J. Am. Chem. Soc. 2008, 130, 9006–9012. 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. E.; Shin D. M.; Chen Z. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat. Rev. Drug Discovery 2008, 7, 771–782. 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- Borst P.; Evers R.; Kool M.; Wijnholds J. A family of drug transporters: the multidrug resistance-associated proteins. J. Natl. Cancer Inst. 2000, 92, 1295–1302. 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- Wu B.; Dröge P.; Davey C. A. Site selectivity of platinum anticancer therapeutics. Nat. Chem. Biol. 2008, 4, 110–112. 10.1038/nchembio.2007.58. [DOI] [PubMed] [Google Scholar]
- Corde S.; Biston M. C.; Elleaume H.; Esteve F.; Charvet A. M.; Joubert A.; Ducros V.; Bohic S.; Simionovici A.; Brochard T.; et al. Lack of cell death enhancement after irradiation with monochromatic synchrotron X rays at the K-shell edge of platinum incorporated in living SQ20B human cells as cis-diamminedichloroplatinum (II). Radiat. Res. 2002, 158, 763–770. 10.1667/0033-7587(2002)158[0763:LOCDEA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ciccarelli R. B.; Solomon M. J.; Varshavsky A.; Lippard S. J. In vivo effects of cis- and trans-diamminedichloroplatinum(II) on SV40 chromosomes: differential repair, DNA-protein cross-linking, and inhibition of replication. Biochemistry 1985, 24, 7533–7540. 10.1021/bi00347a005. [DOI] [PubMed] [Google Scholar]
- Golebiewska A.; Brons N. H. C.; Bjerkvig R.; Niclou S. P. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell 2011, 8, 136–147. 10.1016/j.stem.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Marquardt J. U.; Raggi C.; Andersen J. B.; Seo D.; Avital I.; Geller D.; Lee Y. H.; Kitade M.; Holczbauer A.; Gillen M. C.; et al. Human hepatic cancer stem cells are characterized by common stemness traits and diverse oncogenic pathways. Hepatology 2011, 54, 1031–1042. 10.1002/hep.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling D.; Xia H.; Park W.; Hackett M. J.; Song C.; Na K.; Hui K. M.; Hyeon T. pH-sensitive nanoformulated triptolide as a targeted therapeutic strategy for hepatocellular carcinoma. ACS Nano 2014, 8, 8027–8039. 10.1021/nn502074x. [DOI] [PubMed] [Google Scholar]
- Urien S.; Brain E.; Bugat R.; Pivot X.; Lochon I.; Van M. L.; Vauzelle F.; Lokiec F. Pharmacokinetics of platinum after oral or intravenous cisplatin: a phase 1 study in 32 adult patients. Cancer Chemother. Pharmacol. 2005, 55, 55–60. 10.1007/s00280-004-0852-8. [DOI] [PubMed] [Google Scholar]
- Vermorken J. B.; van der Vijgh W. J.; Klein I.; Hart A. A.; Gall H. E.; Pinedo H. M. Pharmacokinetics of free and total platinum species after short-term infusion of cisplatin. Cancer Treat. Rep. 1984, 68, 505–513. [PubMed] [Google Scholar]
- Yu H.; Tang Z.; Zhang D.; Song W.; Zhang Y.; Yang Y.; Ahmad Z.; Chen X. Pharmacokinetics, biodistribution and in vivo efficacy of cisplatin loaded poly (L-glutamic acid)-g-methoxy poly(ethylene glycol) complex nanoparticles for tumor therapy. J. Controlled Release 2015, 205, 89–97. 10.1016/j.jconrel.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Paraskar A.; Soni S.; Basu S.; Amarasiriwardena C. J.; Lupoli N.; Srivats S.; Roy R. S.; Sengupta S. Rationally engineered polymeric cisplatin nanoparticles for improved antitumor efficacy. Nanotechnology 2011, 22, 265101–265108. 10.1088/0957-4484/22/26/265101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. N.; Poon W.; Tavares A. J.; McGilvray I. D.; Chan W. C. W. Nanoparticle–liver interactions: Cellular uptake and hepatobiliary elimination. J. Controlled Release 2016, 10.1016/j.jconrel.2016.01.020. [DOI] [PubMed] [Google Scholar]
- Xia H.; Chen J.; Shi M.; Gao H.; Sekar K.; Seshachalam V. P.; Ooi L. L. P. J.; Hui K. M. EDIL3 is a novel regulator of epithelial-mesenchymal transition controlling early recurrence of hepatocellular carcinoma. J. Hepatol. 2015, 63, 863–873. 10.1016/j.jhep.2015.05.005. [DOI] [PubMed] [Google Scholar]
- Feng G.-S. Conflicting roles of molecules in hepatocarcinogenesis: paradigm or paradox. Cancer Cell 2012, 21, 150–154. 10.1016/j.ccr.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.