Abstract
Soil salinization is a major threat to agriculture in arid and semi-arid regions, where water scarcity and inadequate drainage of irrigated lands severely reduce crop yield. Salt accumulation inhibits plant growth and reduces the ability to uptake water and nutrients, leading to osmotic or water-deficit stress. Salt is also causing injury of the young photosynthetic leaves and acceleration of their senescence, as the Na+ cation is toxic when accumulating in cell cytosol resulting in ionic imbalance and toxicity of transpiring leaves. To cope with salt stress, plants have evolved mainly two types of tolerance mechanisms based on either limiting the entry of salt by the roots, or controlling its concentration and distribution. Understanding the overall control of Na+ accumulation and functional studies of genes involved in transport processes, will provide a new opportunity to improve the salinity tolerance of plants relevant to food security in arid regions. A better understanding of these tolerance mechanisms can be used to breed crops with improved yield performance under salinity stress. Moreover, associations of cultures with nitrogen-fixing bacteria and arbuscular mycorrhizal fungi could serve as an alternative and sustainable strategy to increase crop yields in salt-affected fields.
Keywords: salinity, tolerance mechanisms, transport of sodium, detoxification pathways, beneficial soil microorganisms, engineering of plant salinity tolerance
Introduction
It is expected that world population will continue to grow and exceed nine billion by 2050 (Department of Economic and Social affairs of the United Nations, 20151. Therefore, the global food production must increase substantially to ensure food security for the growing population. However, food production is seriously threatened by various environmental factors and soil salinity is one of the major stresses adversely affecting plant growth and crop productivity, especially in arid and semi-arid regions. Worryingly, these regions continue to expand and they represent today 40% of the world’s land surface where two billion people are living, mostly in developing countries (UNEP, 1992; Flowers and Yeo, 1995). As a result, more irrigation with brackish water is unavoidable and salinization becomes a serious agricultural concern worldwide. Therefore, engineering crops with enhanced salt stress tolerance traits is one of the most important challenges for modern agriculture.
This review focuses on the state of the art regarding the effect of soil salinity on plant growth and gives an overview of mechanisms controlling salt stress tolerance from sensing and signaling to gene expression and adaptive plant responses. Such knowledge is primordial for setting molecular approaches to enhance plant salinity tolerance. We will also discuss the potential of beneficial soil microorganisms in providing sustainable alternatives for improving crop production in saline soils.
Salinization in Arid and Semi-Arid Region and the Problem of Land Degradation
According to standard definition, saline soils are those which have an electrical conductivity (EC) of the saturation soil-paste extract of more than 4 dS/m at 25°C, which corresponds to approximately 40 mM NaCl and generates an osmotic pressure of approximately 0.2 MPa (Munns and Tester, 2008; USDA-ARS, 2008). When grown on soils with an EC value above 4, crops significantly reduce their yield. Salts may include chlorides, sulfates, carbonates and bicarbonates of sodium, potassium, magnesium, and calcium, the diverse ionic composition of salt-affected soils results in a wide range of physiochemical properties. In the case of saline-sodic soils growth is hindered by a combination of high alkalinity, high Na+, and high salt concentration (Eynard et al., 2005). In this regard, it is important to distinguish between soil salinization and soil sodicity.
Soil salinization is referred as the accumulation of soluble salts in the soils (Bockheim and Gennadiyev, 2000). This is particularly favored by arid and semi-arid climates with evapotranspiration volumes being greater than precipitation volumes along the year.
Soil sodicity is a term given to the amount of Na+ detained in the soil. High sodicity (more than 5% of Na+ of the overall cation content) causes clay to swell excessively when wet, hence limiting severely air and water movements and resulting in poor drainage.
Salts may arise naturally in subsoil (primary salinization) or maybe be introduced (secondary salinization) by soil amendments, inorganic fertilizers, and most importantly irrigation with brackish water (Carillo et al., 2011). As a result, the total area of salt-affected lands in the world is estimated at more than 800 million hectares (ha), which account for more than 6% of the world’s total land area. Of the current 230 million ha of irrigated land, 45 million ha (19.5%) have been already damaged by salt (FAO, 2016).
As NaCl is the most soluble and widespread salt, plants have evolved mechanisms to tolerate/exclude it while allowing acquisition of other nutrients available at low concentrations, such as phosphate, potassium, and nitrate.
Impact of Soil Salinization on Plant Growth and Survival
High soil salinity impacts the growth of numerous plant species especially glycophytes (salt-sensitive compared to salt-tolerant halophytes species), wherein fall major crops. Salt stress tolerance level varies from one species to another and, for cereal crops, bread wheat is a moderately salt-tolerant crop (Maas and Hoffman, 1977). In field conditions, the wheat crop yield will be reduced in the presence of 100 mM NaCl (10 dS/m), whereas rice cannot survive up to maturity under such conditions. Barley (Hordeum vulgare), the most tolerant cereal, can tolerate up to 250 mM NaCl (equivalent to 50% seawater), beyond which the survival rates drop drastically. Other cereals, such as durum wheat (Triticum turgidum ssp.), maize (Zea mays), and sorghum (Sorghum bicolor) are less tolerant to salinity (Maas and Hoffman, 1977).
The reduction in plant growth following salt exposure is due to two phases, osmotic stress and ionic toxicity (Munns and Tester, 2008). Upon a salt stress, the first phase is a rapid response to an increase in the osmotic pressure of the soil solution, whereas the second one is a slower response and takes place after the accumulation of Na+ in photosynthetic tissues. Although they can be clearly identified in most plants and under various salt stress conditions, these two phases are not obvious under high salinity or in the case of Na-hypersensitive plant species such as rice (Munns and Tester, 2008).
It is noteworthy that the overall leaf/shoot development is more sensitive to salinity than root growth and it is assumed that less expanded leaves would decrease the water use by the plant, hence allowing it to conserve soil moisture and prevent a further rise in the salt concentration in the soil. Knowledge about the mechanism by which leaf growth and shoot development are down-regulated under salt stress is relatively scarce, but within days following a salt stress, there is evidence for the involvement, in this inhibitory mechanism, of long distance signals consisting of mainly hormones, as it was previously reported in barley (Munns et al., 2000).
Moreover, salt may affect plant growth indirectly by decreasing the rate of photosynthesis and stomatal conductance (Brugnoli and Lauteri, 1991). Decrease in stomatal aperture is considered as the most dramatic response that occurs soon after plant exposure to salinity owing to the osmotic effect of salt outside the roots (Munns and Tester, 2008).
Stomata are the main structures responsible for gas exchange control, and salt stress affects not only stomatal opening but also their size and density, resulting in a decrease in stomatal conductance. Consequently, rates of transpiration (i.e., water loss) and photosynthesis (CO2 uptake) are also reduced. Indeed, in cotton plants submitted to salt stress treatments, a substantial reduction in photosynthesis has been associated with a decrease in total chlorophyll content and distortion in chlorophyll ultrastructures (Zhang L. et al., 2014). It has been also evoked that sink to source feedback inhibition would moderate the rate of photosynthesis to match the reduced demand arising from growth inhibition (Paul and Foyer, 2001).
On the other hand, the reduced rate of photosynthesis increases the formation of reactive oxygen species (ROS), and enzymatic antioxidant activities such as superoxide dismutase (SOD), catalase (CAT), and various peroxidases (Apel and Hirt, 2004; Foyer and Noctor, 2005; Logan, 2005). These functionally inter-related enzymes act in a coordinated manner to ensure a balance between the rate of formation and removal of ROS. However, previous studies in Arabidopsis have suggested that the mode of coordination between different components of the ROS removal network is complex, since ROS are also used by plants as signaling molecules controlling different processes such as growth, development, and stress responses (Mittler et al., 2004).
Salt Tolerance Mechanisms in Plants
Sensing and Signaling to Regulate Plant Salt Stress Response
To tolerate salt in soil solution, plants deploy a variety of traits to control cell function and development that relies on signal perception, signal integration, and processing. The development of high-throughput sequencing technologies during the last few years has generated huge quantity of data and led to the discovery of several signaling molecules. Among them, are those involved in the activation of signaling pathways that can enhance plant ability to tolerate salt stress. ROS, despite their potential toxicity, have the advantage of being versatile signaling molecules with regard to their properties and mobility within cells. The mitogen-activated protein kinase (MAPK) can trigger plant response to biotic and abiotic stresses by activating the antioxidant enzymes. Many different MAPKs cascades are activated upon ROS accumulation. These include the ROS-responsive MAPKKK, MEKK1, MPK4, and MPK6 (Xing et al., 2008; Jammes et al., 2009). ROS signaling is tightly linked to cellular homeostasis and highly integrated with hormonal signaling networks, allowing plants to regulate development processes, as well as adaptive responses to environmental constraints (Miller et al., 2010). Increased generation and accumulation of ROS such as superoxide (O2-), hydrogen peroxide (H2O2), and nitric oxide (NO) cause oxidative damages in the apoplastic compartment and lipid peroxidation of cellular membranes, and have an extensive impact on ion homeostasis by interfering with ion fluxes (Baier et al., 2005). Excess of ROS levels are particularly scavenged by antioxidant metabolites such as ascorbate, glutathione, tocopherols, and by ROS detoxifying enzymes such as SOD, ascorbate peroxidase (APX), and CAT. Moreover, increased ROS levels can cause salicylic acid accumulation contributing to plant defense, cell death, and induced stomatal closure (Khokon et al., 2011). Recent advance in considering the important role of ROS in plant salt responses was the discovery of a coupled function of plastid heme oxygenases and ROS production in salt acclimation (Xie et al., 2011). These findings strongly suggest involvement of the chloroplast to nucleus signaling pathway in plant salt adaptation. In depth study on cross-species expression of a SUMO conjugating enzyme has provided considerable insight into the links between ROS, ABA (abscisic acid) dependent signaling and the sumoylation pathways in plant salt and drought tolerance (Karan and Subudhi, 2012).
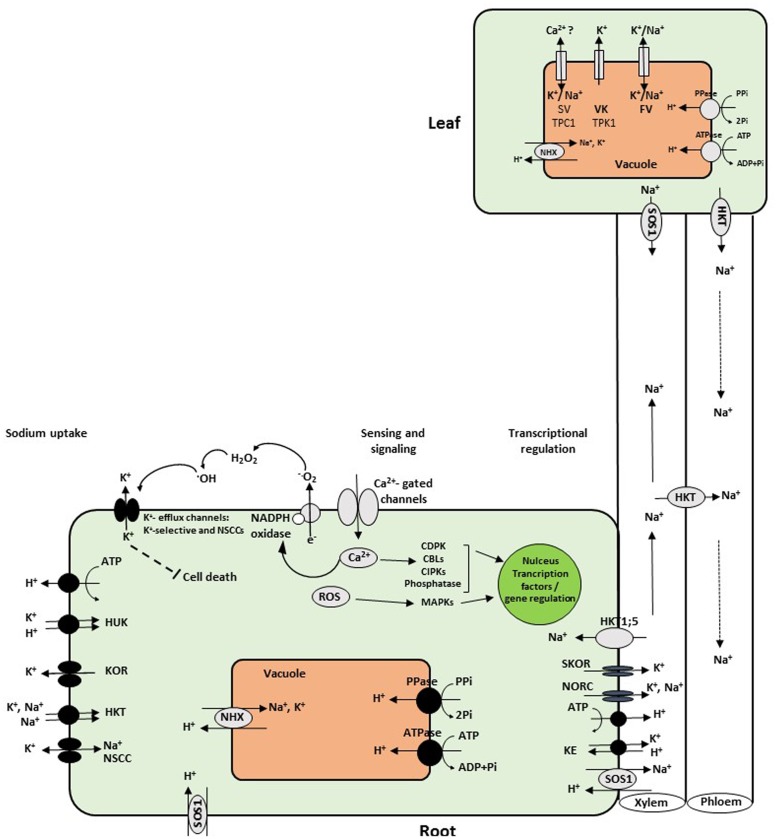
To cope with salt stress, plants have developed the ability to sense both the hyperosmotic and the ionic Na+ components of the stress. However, the molecular identities of the plant Na+ and hyperosmotic sensors remain unknown until now. Nevertheless, plant hyperosmotic sensors are likely to be coupled with Ca2+ channels due to the rapid rise in cytosolic Ca2+ levels within seconds of exposure to NaCl or mannitol (Knight et al., 1997; Figure 1). The Ca2+ signal occurs in roots and in several cell types (Martí et al., 2013). This finding indicates that hyperosmotic stress may be sensed by a mechanically gated Ca2+ channel (Kurusu et al., 2013). In a recent study, Choi et al. (2014) suggested that Ca2+-dependent signaling plays a role in the process of systemic signaling in response to salt stress. Thus, local salt stress of the root tip leads to the spreading of a Ca2+ wave that propagates preferentially through cortical and endodermal cells to distal shoot tissues at speeds of up to 400 μm/s. Salt stress-induced long-distance Ca2+ wave is dependent on the activity of the ion channel protein Two Pore Channel 1 (TPC1), which appears to contribute to whole-plant stress tolerance. In Arabidopsis, the vacuolar cation channel TPC1 is involved in propagation of calcium waves and mediates the passage of K+ and Na+. TPC1 plays an important role for cation homeostasis and vacuolar storage function (Larisch et al., 2016). These results suggest that plants do possess a sensory network that uses ion fluxes moving through defined cell types to rapidly transmit information between distant sites within the organism. Downstream of Ca2+ signaling, calcium-dependent protein kinases (CDPKs), and calcineurin B-like proteins (CBLs) with CBL-interacting protein kinases (CIPKs) may become active and transduce the hyperosmotic signal to downstream protein activity and gene transcription (Weinl and Kudla, 2008; Boudsocq and Sheen, 2013).
FIGURE 1.
Schematic overview of sodium uptake into roots and transport mechanisms into leaves. Na+ enters root cells and cross the plasma membrane via NSCC, CNGC, members of the HKT gene subfamily 1, and apoplastic pathways. To cope with salt stress, Na+ is sensed by the hyperosmotic and ionic sensors leading to activated Ca2+, ROS, and hormone signaling pathways. CDPKs, CBLs, CIPKs, MAPK become active and transduce signal downstream gene transcription in the nucleus. This signaling pathways result in activation of detoxification mechanisms, including the plasma membrane Na+/H+ antiporter (SOS1), HKT, and the tonoplast Na+, K+/H+ exchanger (NHX). SOS1 extrudes Na+ from the cortex cells at the root–soil interface, while at the xylem parenchyma cells; it loads Na+ into xylem sap. The HKT1 protein mediates the reverse flux and unloads Na+ from the xylem vessels to prevent overaccumulation in photosynthetic tissues. Other candidates for loading Na+ to the xylem are the outward-rectifying K+ channels KORC and NORC. At the tonoplast membrane, NSCCs include the slow vacuolar (SV) and fast vacuolar (FV) conductances, whereas the vacuolar K+ (VK) channel is selective for K+, while TPC1 is perfectly leaky for Na+ (Maathuis and Amtmann, 1999; Shabala and Cuin, 2007; Kronzucker and Britto, 2011). To maintain low concentration of Na+ in leaves, it is either retranslocated with HKT gene through the phloem to lower leaves and down to the roots, or detoxified by sequestration into the vacuole with NHX proteins. NSCCs, nonselective cation channels; HUK, HKT, high potassium affinity transporter; ROS, reactive oxygen species; CDPKs, calcium-dependent protein kinases; CBLs, calcineurin B-like proteins; CIPKs, CBL-interacting protein kinases; MAPK, mitogen-activated protein kinase; KOR, outward-rectifying K+channels; NORC, nonselective outward-rectifying channels.
On another hand, potassium as an essential macroelement, is needed at large amounts to be taken up from the soil and transported throughout the plant and enable efficient growth and development (Ahmad and Maathuis, 2014). Under salinity the increase in cytoplasmic Na+ and reduction of K+ result in changes of membrane potential, osmotic pressure, turgor pressure, calcium signaling, ROS signaling, etc. Recent results on ion fluxes in glycophyte Arabidopsis thaliana and the halophytic relative Thellungiella halophila showed lower Na+ fluxes and higher K+/Na+ selectivity of ion currents in the roots and root protoplasts of the halophyte under salt treatment (Volkov and Amtmann, 2006; Amtmann, 2009). Maintenance of K+ homeostasis is essential for enzyme activities, ionic and pH homeostasis, and cytosolic K+ is considered to be an attribute of plant adaptive responses to a broad range of environmental constraints (Shabala and Pottosin, 2014). In addition, a strong correlation between the root’s K+ retention ability and plant salinity stress tolerance was reported for several species including wheat (Cuin et al., 2012), barley (Wu et al., 2015), and Brassica species (Chakraborty et al., 2016). In fact, this strong correlation between K+ retention and net Na+ uptake observed in Brassica species argue toward involvement of GORK (outward-rectifying potassium selective) channels as a major pathway for the salt stress-induced K+ efflux from Brassica roots. GORK is central for Na+-induced K+ loss from root epidermis. GORK channels are activated by membrane depolarization (Véry et al., 2014), and their gating is strongly dependent upon the extracellular K+ concentration (Anschütz et al., 2014).
On another hand, it is well established that electrolyte leakage which is considered as a hallmark of plant cell response to abiotic (including salinity) and biotic stresses, is based mainly on K+ efflux (Palta et al., 1977). This stress induced K+ leakage is often accompanied by ROS generation and leads to cell death (Demidchik et al., 2014). In addition, there are K+ outwardly rectifying channels that are induced by ROS and especially hydroxyl radicals (Demidchik et al., 2003, 2010). Therefore, under stress K+ leakage, ROS and plant cell-death (PCD) seem to be intimately connected. Such hypothesis was elegantly strengthen by Demidchik et al. (2010) who showed through pharmacological and genetic approaches that blocking K+-channel and the lack of functional GORK both inhibit the stress-induced activation proteases and endonucleases that lead to PCD.
Transport of Sodium and Detoxification Pathways
Na+-influx pathways into roots occur via different channels and transporters. Na+ may cross the plasma membrane through nutrient channels and calcium-permeable nonselective cation channels (NSCCs), including the cyclic nucleotide-gated channel (CNGC) and the glutamate-like receptor (GLR), which represent a likely entry point of Na+ into the cell (Tyerman and Skerrett, 1999; Guo et al., 2008; Deinlein et al., 2014). The role of NSCC is not restricted to mediate Na+ influx, which triggers K+ efflux via KORG, but they can also contribute to K+ efflux directly. Under salt stress and for efficient storage of Na+ to the vacuole, NSCC may play important role by preventing Na+ leak to the cytosol and without perturbing K+ release (Pottosin and Dobrovinskaya, 2014). This leads to the activation of K+-efflux channels (GORK) and the loss of K+ from the cell, stimulating cell-death enzymes (Demidchik et al., 2010). These functions might be regulated by ROS-activated ion channels in plant roots. Salt-induced K+ efflux in relation to Na+ influx, which depolarizes the membrane, increasing the driving force for K+ efflux and causing the activation of outward-rectifying K+ channels (Shabala et al., 2006; Cuin et al., 2008).
The several HKT-type transporters were shown to be involved in Na+/K+ symport in different plant species including Arabidopsis, rice and wheat. Based on sequence and transport analyses, HKT transporters can be classified into two distinct subgroups class I and II, with the first being more Na+-selective transport and the second as Na+–K+ co-transporter (Munns and Tester, 2008; Deinlein et al., 2014). The rice Na+ transporter OsHKT2;1 (previously named OsHKT1) has been shown to mediate Na+ influx into roots under K+ starvation (Horie et al., 2007). Other members of the HKT gene subfamily 1, are thought to mediate Na+ influx in root cells and to regulate the Na+ distribution between roots and shoots (Sunarpi et al., 2005; Horie et al., 2009).
The Na+/H+ antiporter SOS1 extrudes Na+ from the cortex cells at the root–soil interface, thereby reducing the net uptake of Na+. In contrast, at the xylem parenchyma cells, SOS1 loads Na+ into xylem sap, whereas HKT1-like proteins mediate the reverse flux and unload Na+ from xylem vessels to prevent Na+ overaccumulation in photosynthetic tissues (Figure 1). Na+ enters the xylem by efflux out of stellar cells and is subsequently transported to aerial plant tissues. HKT1-like proteins may facilitate the translocation of Na+ to the upper shoot or back to the roots by unloading ions from the xylem and transported to the phloem via symplastic diffusion (Berthomieu et al., 2003; Sunarpi et al., 2005). Studies on xylem parenchyma-localized class I HKT transporters have led to the identification of an essential mechanism for plants to protect photosynthetic organs from Na+ overaccumulation and balancing to high K+/Na+ ratio in plants (Sunarpi et al., 2005). In Arabidopsis, the phloem recirculation model proposed by Berthomieu et al. (2003) suggests that Na+ is loaded into shoot phloem cells by AtHKT1;1 and then transferred to roots via the downstream phloem flow. In fact, it is generally accepted that AtHKT1;1 mediates the retrieval of Na+ from the xylem sap, thereby restricting the amount of Na+ reaching the young photosynthetic tissues (Berthomieu et al., 2003; Sunarpi et al., 2005).
Potential candidates for the control of xylem loading of Na+ are the outward-rectifying K+ channels KORC and NORC (Wegner and de Boer, 1997).
To balance the toxic effect of Na+ accumulation and to control ion homeostasis during salinity stress, plants require the maintenance of stable K+ acquisition and distribution (Schroeder et al., 1994). The tonoplast-localized Na+/H+ exchangers (NHX1 and NHX2) and the plasma membrane-localized Na+/H+ antiporter (SOS1), are thought to play important roles in osmoregulation and to maintain low cytoplasmic Na+ concentration in plant cells (Shi et al., 2002; Apse et al., 2003; Brini et al., 2007a; Olias et al., 2009). Most NHX proteins are essential for Na+ detoxification through sequestration into the vacuole, whereas SOS signaling pathways were responsible to export Na+ outside the cell. The concept that vacuolar NHX proteins were capable of exchanging Na+ and H+ across the tonoplast as claimed by Apse et al. (1999) and Blumwald (2000), was challenged by the biochemistry of NHX proteins. Indeed, NHX proteins were shown not to discriminate between Na+ and K+ nor having preference for K+ transport (Jiang et al., 2010). Moreover, it has been shown that NHX1 and NHX2 proteins of Arabidopsis play a comparatively greater role in K+ homeostasis, rather than in Na+ sequestration (Leidi et al., 2010; Bassil et al., 2011; Barragan et al., 2012). In addition, NHX1 overexpression in tomato conferred tolerance to NaCl, which was related to the preferential accumulation of K+ in vacuoles and improved K+ retention after stress imposition, but did not enhance the ability to compartmentalize toxic Na+ ions into the vacuole (Leidi et al., 2010).
Adaptive Mechanisms of Salt Tolerance at the Cell and Organ Level
Research carried out on salinity tolerance mechanisms was mainly performed on model plants (i.e., A. thaliana) and only few cases were reported on crop plants. In glycophytes, salinity tolerance is mainly achieved through more than one strategy operating either simultaneously or separately, depending on the duration and intensity of the stress. According to Chakraborty et al. (2016), the overall superior salinity tolerance observed in Brassica napus was achieved by the high osmotolerance matched by the moderate tissue tolerance and superior K+ retention ability in the leaf mesophyll. Moreover, Chakraborty et al. (2016) provide strong evidence that higher salt tolerance in B. napus is conferred by at least three complementary physiological mechanisms: (i) higher Na+ extrusion ability from roots correlated with increased expression and activity of plasma membrane SOS1-like Na+/H+ exchangers; (ii) better root K+ retention ability resulting from stress-inducible activation of H+-ATPase and the ability to maintain a more negative membrane potential under saline conditions; and (iii) reduced sensitivity of B. napus root K+-permeable channels to ROS. Shabala et al. (2015) has argued that changes in SOS1 activity strongly correlated with changes in net K+ and H+ fluxes, making the involvement of the H+-ATPase/GORK tandem system as a potential sensor a plausible hypothesis. Therefore, it was suggested that Na+ exclusion from uptake plays an important but not a crucial role as a determinant of genetic variability in salinity stress tolerance in Brassica. At the root level, exclusion of ∼95% of salt entering the roots back to the soil solution constitutes the major adaptive trait that plants will undertake to avoid the toxic effect of high salinity in the shoots. Species exhibiting significant genotypic variation in Na+ accumulation in leaves have shown a correlation between salt tolerance and Na+ exclusion. This is the case in sensitive species like rice and durum wheat (Munns, 2005), but also in more salt-tolerant species like barley (Chen et al., 2005). Indeed, a strong correlation between salt exclusion and salt tolerance does exist in many species (Munns and James, 2003). Further removal of sodium from xylem to translocate back into roots is another way to prevent Na+ overaccumulation in photosynthetic tissues. QTL analyses for salinity resistance have suggested that similar xylem Na+-unloading mechanisms are essential for salt tolerance in rice and wheat (James et al., 2006). In both cases, major salt tolerance QTL map to regions that include HKT1;5 orthologs, encoding a more Na+-selective class I HKT transporter (Byrt et al., 2007). When reaching the stem, sodium will either be stored or controlled in its long distance transport. Partitioning of sodium into leaf sheath/petiole during its export through xylem to the leaves, could conceivably help to maintain low salt concentration in the transpiration stream (James et al., 2006; Byrt et al., 2007). However, its retranslocation through the phloem in the lower leaves and down to the roots is considered as relatively limited. To avoid raising cytosolic Na+ concentration and balance the toxic effect of Na+ accumulation, Garcia de la Garma et al. (2015) reported extensive vesicle trafficking of Na+ between the plasma membrane and Na+-rich vacuolar compartment in salt-acclimated tobacco BY2 cells. However, and as mentioned above, the enhancement of salt tolerance observed on transgenic tomato plants overexpressing AtNHX1 was related to larger K+ vacuolar pools and improved K+ retention rather than a compartmentalization of toxic Na+ ions into the vacuole (Leidi et al., 2010). The lack of correlation between greater salt tolerance and the enhancement of Na+ accumulation in different species overexpressing NHX proteins has been specified by Rodríguez-Rosales et al. (2009) and Jiang et al. (2010). The maintenance of K+ acquisition with the exclusion of Na+ from photosynthetic leaves has been indeed found to be highly correlated with plant salt tolerance (Hauser and Horie, 2010).
Strategies to Improve Salt Tolerance in Crops
Interaction with Beneficial Soil Microorganisms to Improve Salinity Tolerance
Interactions with beneficial soil microorganisms including symbiotic nitrogen-fixing bacteria (Frankia and rhizobia) and mycorrhizal fungi can have a large impact on plant tolerance to salt stress. The impact of salt stress on these symbioses has been reviewed elsewhere and will be not covered in this review (Swaraj and Bishnoi, 1999; Zahran, 1999; Serraj and Adu-Gyamfi, 2004; Evelin et al., 2009; Porcel et al., 2012; Ngom et al., 2016b).
Symbiotic associations with arbuscular mycorrhizal fungi (AMF) are found in roughly 80% of terrestrial plant species (Smith and Smith, 2011). These microsymbionts play a critical role in plant nutrition and profit from plant carbon in return (Smith and Read, 2010). In addition, they enhance plant performance and resistance to abiotic stresses (Sadhana, 2014). AMF can alleviate salt stress in host plants by enhancing water absorption capacity, nutrient uptake and accumulation of osmoregulators to increase osmotic potential of cells. Studies have reported that mycorrhizal colonization can reduce the uptake of Cl- ions while preventing Na+ translocation to shoot tissues under salinity (Evelin et al., 2009). AMF have been known to occur naturally in saline environments (Evelin et al., 2009). For example, AMF (Glomus intraradices, Glomus versiform, and Glomus etunicatum predominantly) were observed in the severely saline soils of the Tabriz plains of Iran, where soil salinity levels range from 7.3 to 92.0 dS/m (Aliasgharzadeh et al., 2001). The effects of mycorrhizal symbiosis on plant salinity tolerance have been studied in many species including Medicago sativa (Azcon and El-Atrash, 1997), Sesbania aegyptiaca, and Sesbania grandiflora (Giri and Mukerji, 2004), Z. mays (Feng et al., 2002; Krishnamoorthy et al., 2016), Capsicum annum (Kaya et al., 2009), Olea europaea (Porras-Soriano et al., 2009), Citrus tangerine (Wu et al., 2010), Gossypium arboreum (Tian et al., 2004), and Lycopersicon esculentum (Al-Karaki, 2000, 2006; Al-Karaki and Hammad, 2001; Latef and Chaoxing, 2011). In all these species, AMF improved plant salinity tolerance, leading to enhanced plant growth and yield (Azcon and El-Atrash, 1997; Giri and Mukerji, 2004; Diouf et al., 2005; Kaya et al., 2009; Porras-Soriano et al., 2009; Wu et al., 2010), nutrient acquisition (Feng et al., 2002; Giri and Mukerji, 2004; Diouf et al., 2005; Kaya et al., 2009; Porras-Soriano et al., 2009; Wu et al., 2010; Krishnamoorthy et al., 2016), chlorophyll content (Feng et al., 2002; Giri and Mukerji, 2004; Kaya et al., 2009), proline concentration (Diouf et al., 2005; Kaya et al., 2009), and promoting higher accumulation of soluble sugars in roots (Feng et al., 2002; see Table 1). In tomato, mycorrhizal colonization significantly improved fruit fresh weight and fruit yield under salt stress (Latef and Chaoxing, 2011). The fruit yield (kg/plant) increased by 33.3 and 106% at 50 and 100 mM salinity levels, respectively. A similar positive effect was reported in the squash Cucurbita pepo leading to better fruit yield production in saline conditions (Colla et al., 2008). These benefits of mycorrhizal fungi under saline conditions depend on the symbiotic associations and could therefore be improved by selection of efficient fungal strains.
Table 1.
Example of beneficial soil microorganisms enhancing plant salinity tolerance.
| Beneficial microorganisms inoculum | Plant species | Reference |
|---|---|---|
| Mycorrhizal fungi | ||
| Glomus claroideum | Olea europaea | Porras-Soriano et al., 2009 |
| Glomus clarum | Capsicum annum | Kaya et al., 2009 |
| Glomus intraradices | Acacia auriculiformis | Diouf et al., 2005 |
| A. mangium | Diouf et al., 2005 | |
| Cucurbita pepo | Colla et al., 2008 | |
| O. europaea | Porras-Soriano et al., 2009 | |
| Glomus macrocarpum | Sesbania aegyptiaca | Giri and Mukerji, 2004 |
| S. grandiflora | Giri and Mukerji, 2004 | |
| Glomus mosseae | Citrus tangerine | Wu et al., 2010 |
| Medicago sativa | Azcon and El-Atrash, 1997 | |
| Zea mays | Feng et al., 2002 | |
| O. europaea | Sheng et al., 2008 | |
| Gossypium arboreum | Porras-Soriano et al., 2009 | |
| Lycopersicon esculentum | Tian et al., 2004 | |
| Al-Karaki, 2000, 2006 | ||
| Al-Karaki and Hammad, 2001 | ||
| Latef and Chaoxing, 2011 | ||
| Paraglomus occultum | C. tangerine | Wu et al., 2010 |
| Rhizophagus intraradices | Zea mays | Krishnamoorthy et al., 2016 |
| Rhizobia | ||
| Rhizobium spp. strain AC-2 | A. nilotica | Bala et al., 1990 |
| Rhizobium spp. strain AC-1 | A. nilotica | |
| Rhizobium spp. strain L-10 | L. leucocephala | |
| Rhizobium spp. strain P-4 | Prosopis juliflora | |
| Rhizobium PMA63/1 | A. ampliceps | Zou et al., 1995 |
| Rhizobium tropici CIAT899 | Phaseolus vulgaris | Dardanelli et al., 2008 |
| Rhizobium etli ISP42 | ||
| Rhizobium strain USDA 208 | Glycine max | Elsheikh and Wood, 1995 |
| Bradyrhizobium strain RCR 3407 | ||
| Frankia | ||
| Crushed nodule suspension | Casuarina equisetifolia | Ng, 1987 |
| Crushed nodule suspension | Alnus glutinosa | Oliveira et al., 2005 |
| CcI3 strain | C. equisetifolia | Ngom et al., 2016a |
| C. glauca | ||
| CeD strain | C. equisetifolia | Ngom et al., 2016a |
| C. glauca | ||
| Dual inoculation | ||
| Mixed spores from Glomus, Gigaspora, and Acaulospora genera + Sinorhizobium terangae | A. saligna | Soliman et al., 2014 |
| Glomus mosseae + Mesorhizobium mediterraneum | Lathyrus sativus | Jin et al., 2010 |
| Glomus intraradices + Bradyrhizobium strains Aust 11c and Aust 13c | A. auriculiformis | Diouf et al., 2005 |
| A. mangium | ||
| Glomus clarum + Azospirillum brasilense | Vigna sinensis | Rabie et al., 2005 |
| Vicia faba | Rabie and Almadini, 2005 | |
| Rhizophagus intraradices + Massilia sp. RK4 | Zea mays | Krishnamoorthy et al., 2016 |
| Crushed nodule suspension + Glomus intraradices | A. glutinosa | Oliveira et al., 2005 |
Rhizobia are a group of Gram-negative soil heterogeneous bacteria that are also able to form nitrogen-fixing nodules on the roots, or occasionally the shoots, of legumes (Young, 1996) and Parasponia species (Akkermans et al., 1978). Bacteria are accommodated intracellularly in nodule cells and fix dinitrogen for plant growth, while being supplied with carbon sources by the plant (Pawlowski and Demchenko, 2012). Many studies have demonstrated that inoculation with suitable Rhizobium sp. increase plant dry weight in legumes including Acacia nilotica, Leucaena leucocephala, Prosopis juliflora (Bala et al., 1990), Acacia ampliceps (Zou et al., 1995), Phaseolus vulgaris (Dardanelli et al., 2008), and soybean (Elsheikh and Wood, 1995) under salt stress. These beneficial effects on plant growth result from an effective N2-fixing symbiosis, as acetylene reduction activities were detected even at high salinity levels, depending on the Rhizobium–legume associations (Bala et al., 1990; Elsheikh and Wood, 1995; Zou et al., 1995). Indeed, under saline conditions, the salt-tolerant strains of Rhizobium sp. formed more effective N2-fixing symbiosis with A. nilotica, L. leucocephala, P. juliflora (Bala et al., 1990), A. ampliceps (Zou et al., 1995), and soybean (Elsheikh and Wood, 1995) than did the salt-sensitive strains. These results indicate that biological N2-fixation under saline conditions may be improved by inoculation with a salt-tolerant Rhizobium strain. However, the tolerance of the legume host is the most important factor determining the success of compatible Rhizobium strains in forming effective symbioses under conditions of high soil salinity (Craig et al., 1991). Thus, a screening of both symbiotic partners is necessary for obtaining an efficient N2-fixing symbiosis under saline soils (Zahran, 1999). Salinity tolerance of legumes could be better improved by associated Rhizobium with mycorrhizal fungi (Diouf et al., 2005; Jin et al., 2010; Soliman et al., 2014) and/or plant growth promoting rhizobacteria (PGPR; Rabie and Almadini, 2005; Rabie et al., 2005; Bano and Fatima, 2009). Compared to control treatments, dual inoculation of Rhizobium bacteria and mycorrhizal fungi under salt stress, enhanced plant nutrition, growth parameters and proline concentration in Acacia saligna (Soliman et al., 2014), Lathyrus sativus (Jin et al., 2010), Acacia auriculiformis, and Acacia mangium (Diouf et al., 2005). Co-inoculation with Rhizobium and PGPR including Azospirillum brasilense and Pseudomonas species showed the same beneficial effects in Z. mays (Bano and Fatima, 2009), Vigna sinensis (Rabie et al., 2005), and Vicia faba (Rabie and Almadini, 2005), which were more pronounced in plant inoculated with AMF, in addition to Rhizobium and PGPR (Rabie and Almadini, 2005; Rabie et al., 2005). In Cowpea plants, dual inoculation with AMF and nitrogen-fixing bacteria such as A. brasilense increased plant nitrogen content by 230% against 151 and 94% in plants inoculated separately with nitrogen-fixing bacteria and AMF, respectively, at 7.2 dS/m salinity (Rabie et al., 2005).
Frankia is a genus of Gram-positive filamentous actinobacteria that can induce the formation of nitrogen-fixing nodulation the roots of 260 species, belonging to eight dicotyledonous families (Betulaceae, Casuarinaceae, Myricaceae, Rosaceae, Eleagnaceae, Rhamnaceae, Datiscaceae, and Coriariaceae), collectively called actinorhizal plants (Benson and Silvester, 1993). Like rhizobia–legume symbioses, bacteria are hosted in root nodules and fix atmospheric nitrogen (Perrine-Walker et al., 2011). Some actinorhizal plants such as Casuarinaceae trees are largely used in land reclamation pros including salinized land (reviewed in Diagne et al., 2013). Several studies have therefore reported strategies to improve the tolerance of these plants to salt stress. Similarly to what occurs in legume–rhizobia symbioses, it has been reported that inoculation with the microsymbiont Frankia improves the host plant salinity tolerance (Reddell et al., 1986; Ng, 1987; Oliveira et al., 2005; Ngom et al., 2016a). Frankia strains CcI3 and CeD significantly improved Casuarina glauca and Casuarina equisetifolia plant growth, shoot, root, and total dry weight, proline and chlorophyll contents according to the symbiotic association (Ngom et al., 2016a). According to Ng (1987), inoculated C. equisetifolia plants exhibited greater growth (shoot, root, and total dry weight) compared to uninoculated plants under saline conditions. This increase in dry weight was associated with increase in the total nitrogen content of the nodulated plants even at 500 mM NaCl. In saline anthropogenic sediment (conductivity of 5,980 μS/cm), Alnus glutinosa plants inoculated with Frankia spp. alone significantly increased the growth parameters (total leaf area, shoot height, root collar diameter, and total dry weight), the leaf N content by 197% and the chlorophyll a + b content by 478%, as compared to uninoculated controls (Oliveira et al., 2005). The increased levels of N indicates the effectiveness of the nitrogen fixation process under salinity, and depends on the plant–Frankia isolate associations (Reddell et al., 1986). Furthermore, these beneficial effects were significantly greater when A. glutinosa plants were inoculated with both Frankia spp. and a mycorrhizal fungi, G. intraradices. Dual inoculation increased the leaf N, P, and K contents and chlorophyll a + b by 277, 240, 129, and 531%, respectively, suggesting the ability of both microsymbionts in improving actinorhizal plants nutrition under salt-stressed conditions (Oliveira et al., 2005). Frankia isolates exhibited diversity in their response to salt stress. Salinity affected their in vitro growth and N2 fixation depending on the isolate. Among strains studied, Casuarina isolates seem to be more tolerant to salinity than others (Ngom et al., 2016b). Under saline conditions, the effects of inoculation of actinorhizal plants by salt-tolerant vs salt-sensitive Frankia strains remain poorly studied. Nevertheless, Reddell et al. (1986) showed that under salt stress, Frankia ys, collected from Casuarina obesa at 1.3 mg Cl-/g soil, was able to increase the dry weight of shoot and nodule and the nitrogen content in C. obesa plants better than Frankia cb, collected from Casuarina cunninghamiana at 0.1 mg Cl-/g soil. Among three Frankia isolates used separately as inoculums of C. glauca, Thr which was more in vitro sensitive to salt stress, was the most effective strain in planta. A recent study showed that inoculation of C. glauca plants with the salt-sensitive CcI3 strain improved plants growth under saline conditions while in C. equisetifolia plants the salt-tolerant strain CeD was more effective (Ngom et al., 2016a). These results suggest there is no correlation between in vitro salt tolerance of Frankia strains and their effectiveness in association with plants under salt stressed conditions (Girgis et al., 1992; Ngom et al., 2016a). Thus, as what was observed in Rhizobium–legumes symbiosis, effectiveness of established actinorhizal symbioses to saline conditions is primary dependent on the salt tolerance of the host plant (Girgis et al., 1992).
Taken together, these studies indicate that AMF and nitrogen-fixing bacteria could be used to increase salt tolerance both in crops and in plants used for saline soils rehabilitation. Co-inoculation with different beneficial microbes has the potential to further increase tolerance. These beneficial microbes improve plant growth and nutrition and promote higher accumulation of osmolytes such as proline and sugars in saline environments. This leads to higher crop yield but also to products with better nutritional properties (Latef and Chaoxing, 2011). To fully exploit the beneficial effect of these symbioses, they need to be taken into account in breeding programs for salt-resistant crop varieties. Breeder will need to identify QTLs that control the response of the plant to these beneficial microbes in normal and salt-stress conditions. These QTLs can then be included in breeding programs along more classical salt-resistance QTLs. Furthermore, the development of industrial scale microbial inoculants for salinized soils will be required.
Salt Tolerance in Crops through Marker-Assisted Selection and Genetic Engineering
Conventional plant breeding approaches through which beneficial traits can be introgressed into elite varieties have been adopted since a long time to generate stress tolerant varieties (for review, see Ashraf, 2010; Varshney et al., 2011). Hence, traditional breeding allowed the development of new salt-tolerant rice and wheat varieties (Munns et al., 2006; Singh et al., 2010). However, as salinity tolerance is a multigenic trait, such approaches have only limited success, which can explain the absence of commercially available salt-tolerant crops. Moreover, as a procedure, plant breeding is time consuming and labor intensive, relies on well characterized germplasms and can result in introducing undesirable traits along with the selected one. Therefore, biotechnological approaches including molecular breeding and genetic engineering seem to be more attractive alternatives.
Plant Salinity Tolerance through Genetic Engineering
In the last 30 years, tremendous progress has been made toward the isolation and molecular characterization of genes involved in plant salt stress responses. These candidate genes can be classified into two main groups: effectors and regulatory genes. The first group includes mainly genes encoding ion transporters, channels, enzymes involved in osmolyte biosynthesis, antioxidant systems and protective proteins such as heat shock proteins and late embryogenesis abundant (LEA) proteins. The second group is composed of genes involved in transcriptional and post-transcriptional regulation as well as in signaling pathways. Within this group we find essentially transcription factors, protein kinases, phosphatases, and proteases. Numerous reports described how the overexpression of these genes can improve plant tolerance to various abiotic stresses including salinity (for review, see Ashraf, 2009; Türkan and Demiral, 2009; Cominelli et al., 2013; Roy et al., 2014; Fita et al., 2015; see Table 2).
Table 2.
Example of genes leading to improvement of salt stress tolerance of crop plants through genetic engineering.
| Transgene | Function | Donor | Transgenic plant | Description | Reference |
|---|---|---|---|---|---|
| Na+/H+ antiporter (AtNHX1) | Vacuolar sequestration of Na+ and K+? | Arabidopsis | Tomato | Enhanced salt tolerance with higher accumulation in leaves but not in fruits | Zhang and Blumwald, 2001 |
| Brassica napus | Maintenance of seed yield and seed oil quality under high salinity | Zhang et al., 2001 | |||
| Wheat | Improved grain yield in saline soils | Xue et al., 2004 | |||
| Cotton | Increased fiber yield under salt stress | He et al., 2005 | |||
| H+-pyrophosphatase (AVP1) AtNHX + AVP1 | Vacuolar membrane-bound proton pump | Arabidopsis | Cotton | Increased fiber yield under salt stress in field conditions | Pasapula et al., 2011 |
| Barley | Higher biomass production and grain yield in saline field | Schilling et al., 2013 | |||
| Cotton | Further enhancement of salt tolerance compared to single-gene overexpressing plants | Shen et al., 2014 | |||
| Tomato | Increased salt stress tolerance compared to single gene overexpression | Gouiaa and Khoudi, 2015 | |||
| Ncl | homologous to NHX gene family | Soybean | Soybean | Improved salt tolerance | Do et al., 2016 |
| Mannitol-1-phosphate dehydrogenase mt1D | Mannitol biosynthesis | E. coli | Tobacco | Increased salt tolerance | Tarczynski et al., 1992 |
| Choline synthase codA | Betaine biosynthesis | E. coli | Rice | Enhanced tolerance to salinity and cold | Sakamoto et al., 1998 |
| delta 1-pyrroline-5-carboxylate synthase (P5CS) | Proline biosynthesis | Arabidopsis | Tobacco | Increased tolerance to drought and salt stress | Kishor et al., 1995 |
| ascorbate peroxidase (AtAPX) | ROS-scavenging | Arabidopsis | Tobacco | Enhanced tolerance to salt and osmotic stress | Badawi et al., 2004 |
| Late embryogenesis abundant protein (HVA7) | Osmoprotection | Barley | Rice | Enhanced tolerance to salt and osmotic stress | Xu et al., 1996 |
| Transcription factor DREB1B/CBF1 | Transcription regulation | Arabidopsis | Rice | Enhanced tolerance to salinity and drought | Oh et al., 2005 |
| Transcription factor DREB1A/CBF3 | Transcription regulation | Arabidopsis | Rice | Increase in salinity tolerance | Hu et al., 2006 |
| SNAC1 | Transcription regulation | Rice | Rice | Increase in salinity tolerance | Hu et al., 2008. |
| Transcription factor SlAREB1 | Transcription regulation | Tomato | Tomato | Enhanced tolerance to salinity and water stress | Orellana et al., 2010 |
| Transcription factor ZFP179 | Transcription regulation | Rice | Rice | Increase in salinity tolerance | Sun et al., 2010 |
| calcium-dependent protein kinase OsCDPK21 | Calcium signaling | Rice | Rice | Increase in salinity tolerance | Asano et al., 2011 |
| MAP kinase GhMPK2 | MAPK signaling | Cotton | Tobacco | Enhanced tolerance to salinity and drought | Zhang et al., 2011 |
Ion transporters
Ion transporters are obvious candidates among effectors since they participate in salt detoxification and ion homeostasis. Overexpression of genes involved in Na+ transport was therefore largely investigated. The Arabidopsis vacuolar Na+/H+ antiporter (AtNHX1) was among the first candidate genes which when overexpressed lead to enhanced salinity tolerance (Apse et al., 1999). These transgenic plants were reported to exhibit higher ability for vacuolar sequestration of Na+ to avoid its toxic accumulation into the cytoplasm. Later on, overexpression of AtNHX1 and related NHX proteins from various sources have been shown to increase salt tolerance in other plant species including tomato (Zhang and Blumwald, 2001), B. napus, wheat and cotton (Pardo et al., 2006; Munns and Tester, 2008). Interestingly, the tomato AtNHX1 overexpressing plants are more salt stress tolerant, with an accumulation of NaCl in leaves but not in fruits. However, and as was indicated above, its role in salt stress tolerance may also result from its capacity to facilitate K+ uptake at the tonoplast (Leidi et al., 2010). Nevertheless, engineering of vacuolar cation/H+ antiporter coupled with its H+-translocating pyrophosphatase (H+-PPase) often leads to increased salt stress tolerance. Therefore, overexpression of the wheat TNHX1 and TVP1 improves salt stress tolerance in Arabidopsis (Brini et al., 2007a), tobacco (Gouiaa et al., 2012), and tomato (Gouiaa and Khoudi, 2015). Alone the Arabidopsis H+-PPase (AtAVP1) was also demonstrated to increase stress tolerance in a crop plant. Indeed, Schilling et al. (2013) reported that barley transgenic plants overexpressing AtAVP1 are more tolerant to salinity under greenhouse conditions but they also showed increased shoot biomass production and grain yield in a saline field.
On another hand, the HKT family and the SOS pathway play also a relevant role in controlling Na+ transport within the plant and have been considered in transgenic approaches to increase salt stress tolerance in crops. However, knowing their role in the removal of Na+ from the xylem sap into the surrounding xylem parenchyma cells, improving salinity tolerance using HKTs can be successful only when its expression is targeted to the stele or driven by salt-inducible promoter (Møller et al., 2009; Roy et al., 2014). Recently, the overexpression of Ncl gene (homologous to the Na+/H+ antiporter gene family) into a Japanese soybean salt-sensitive cultivar Kariyutaka, resulted in improved salt tolerance in transgenic soybean. A close association was observed between the high expression of the salt tolerance gene Ncl in the root, the lower accumulation of Na+, K+, and Cl- in the shoot under salt stress and salt tolerance in the transgenic lines (Do et al., 2016).
Osmolyte accumulation
Under salt stress, along with Na+ exclusion from the cytoplasm, plant cell accumulates a wide range of compatible solutes or osmolytes to balance the osmotic pressure of ions in vacuoles. Sucrose, proline, and glycine betaine are among the most studied osmolytes accumulating upon salt stress in some plant species including halophytes (Flowers et al., 1977). Therefore, plant engineering for higher accumulation of these compounds was considered as a possible way in improving crop tolerance to salinity. The bacterial mt1D gene encoding the mannitol-1-phosphate dehydrogenase, enzyme involved in mannitol biosynthesis was, early on, successful in enhancing salt tolerance after its ectopic expression in tobacco (Tarczynski et al., 1992, 1993). Similarly, transgenic rice overexpressing choline oxidase showed increased levels of glycine betaine and enhanced tolerance to salinity and cold (Sakamoto et al., 1998). Also, plants such as tobacco transformed with delta 1-pyrroline-5-carboxylate synthase (P5CS) gene exhibited higher proline production, correlated with increased tolerance to drought and salt stress (Kishor et al., 1995). Similarly, transgenic rice plants expressing the mothbean P5CS gene under constitutive or stress inducible promoter showed significant tolerance to high levels of NaCl (Su and Wu, 2004).
Antioxidant systems and protective proteins
Despite their importance as signaling molecules regulating cellular responses to various stresses (for review, see Apel and Hirt, 2004), ROS can also damage plant tissues during salinity stress by perturbing enzyme, cell wall and membrane function. Plants detoxify then ROS generated by salt stress by up-regulating antioxidative enzymes such as SOD, CAT, APX, and glutathione peroxidase. Therefore, overexpressing ROS-scavenging enzymes were shown to promote tolerance of plants to various stresses including salinity (Rodriguez-Rosales et al., 1997; Roxas et al., 1997; McKersie et al., 1999; Badawi et al., 2004; Miller et al., 2008). For example, the overexpression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit (Badawi et al., 2004).
Other proteins like osmotin and LEA proteins contribute in alleviating salt stress by protecting macromolecules from damages caused by ion toxicity and/or water deficit. HVA7, a LEA from barley, when transferred to rice, confers water and salt stress tolerance (Xu et al., 1996). It is worth to note that, among the different groups, the group 2 of LEA proteins known as dehydrins are particularly interesting and were shown to enhance plant tolerance to various stresses (Hanin et al., 2011). Indeed, Brini et al. (2007b) showed that the expression of the wheat dehydrin DHN-5 in A. thaliana led to an increase in salt and osmotic stress tolerance, but also evidence is provided for the involvement of DHN-5 in other abiotic and biotic stress responses (Brini et al., 2011; Drira et al., 2015).
Transcription factors and signaling proteins
Plant response to salinity is complex and involves multiple genes involved in distinct or overlapping regulatory pathways. Therefore, the engineering of a single downstream effector gene as indicated above, albeit efficient in some circumstances might have limited success when one considers multiple stress combination as occurring in the field. In contrast, regulatory proteins such as transcription factors and signaling proteins will gain increasing interest as they are expected to modulate the expression of numerous downstream genes involved in stress responses. It is well documented that transcription factors belonging to the families of DREB, NAC, MYB, MYC, Cys2/His2 zinc finger, bZIP, AP2/ERF, and WRKY are relevant in salt stress tolerance (Golldack et al., 2011, 2014).
In this regard, several transcription factors such as DREBs, MYCs, AP2/ERFs, and NACs were tested in model plant species and few crops. In some cases, the overexpression of these transcription factors was successful to enhance salinity tolerance in crops (for review, see Lata and Prasad, 2011; Lata et al., 2011; Turan et al., 2012; Nuruzzaman et al., 2013).
The expression of DREB1B/CBF1 or DREB1A/CBF3 under the control of the cauliflower mosaic virus 35S promoter in Arabidopsis plants increases significantly tolerance to freezing, drought, and high salinity stresses (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999). Similarly, transgenic rice plants constitutively expressing DREB1A/CBF3 were reported to be more tolerant to drought and salinity (Oh et al., 2005). Also, the overexpression of SNAC1 or SNAC2 (stress responsive NAC) resulted in an enhanced salinity tolerance (Hu et al., 2006, 2008).
SlAREB1 is a bZIP transcription factor from tomato (Solanum lycopersicum), member of the ABA-responsive element binding protein (AREB)/ABA-responsive element binding factor (ABF) subfamily. Its overexpression in tomato was reported to improve tolerance to water and salt stresses (Orellana et al., 2010). Likewise, Sun et al. (2010) have shown that the overexpression of ZFP179, a salt responsive gene encoding a Cys2/His2 zinc finger protein enhanced salt tolerance in rice.
Using strong and constitutive promoters to drive the expression of transcription factors is still, however, considered as a controversial strategy. The constitutive expression of transcription factors caused in some cases growth defects under standard conditions as was reported for the 35S:TaDREB1 rice transgenic plants overexpressing the bread wheat DREB1 gene that showed a dwarf phenotype (Shen et al., 2003). Therefore, one should reconsider the use of constitutive promoters and rather employ stress inducible/tissue-specific promoters to avoid secondary deleterious effects (Munns and Tester, 2008).
Moreover, effects of the overexpression of genes encoding signaling proteins such as kinases and phosphatases on salt tolerance were reported. As conserved signaling proteins at the crossroads of several signaling cascades, MAPKs play pivotal role in plant responses to various stresses. Many transgenic plants that have been engineered with MAPK cascade were reported to be tolerant to salt stress. The ectopic expression of cotton GhMPK2 improves salinity and drought tolerance in tobacco (Zhang et al., 2011). In addition, CDPKs, which are involved in salt stress response, were found efficient in transgenesis approaches, as was reported for the overexpression of OsCDPK21 that resulted in increased salinity tolerance of transgenic rice (Asano et al., 2011). However, other MAPKs can have opposite effects and this was described for the rice OsMAPK33, the overexpression of which caused higher sensitivity to drought and salinity compared to wild-type plants (Lee et al., 2011). Regulators of MAPKs, the MAPK phosphatases (MKPs) can also be involved in the control of stress responses. Interestingly, while the Arabidopsis AtMKP1 acts as a negative regulator, the wheat counterpart acts as a positive regulator of salt stress responses (Ulm et al., 2002; Zaidi et al., 2016).
Noteworthy, it is still until now problematic to generate and commercialize crops more tolerant to salinity or to any other stresses. This can be due to several factors. First of all, the laboratory growth conditions are far different from field conditions. Field tests are often forbidden or restricted in several countries but mainly one needs to consider the reproducibility of the field tests and be able to measure and monitor any environmental changes that may affect crop yield (humidity, soil composition, light intensity,…). Second, often the stress tolerance of transgenic crop grown in a greenhouse or a growth chamber is assessed by measuring survival and or recovery rates, while for farmers; the main trait for stress tolerance is crop yield. Moreover, in the first case the evaluation of stress tolerance level is limited to particular traits (plant height, leaf or root size) or development stages during the vegetative phase and only occasionally covers the reproductive stage. The need to conduct field trials under monitored conditions over several years in distinct environmental conditions is a necessary condition to ensure the development of sustainable genetically improved salt-tolerant crops.
Marker-Assisted Selection and Salt Stress Tolerance
One of the major limitations for the use of conventional breeding to improve salt tolerance in crops is its slowness which is closely linked to the complexity of this polygenic trait. Traditionally, the selection of crops raised from backcrosses of genetically diverse germplasms is limited to their phenotype analyses in the field. As an alternative to streamline this process, breeders use QTL analyses coupled with marker-assisted selection (MAS), referred as an approach linking a quantitative trait with a genetic marker that is polymorphic between parental lines (Ashraf and Foolad, 2013). Thus still, accumulating knowledge on plant salt stress response is a must to be able to develop confident and efficient markers. The fact that maize, rice, barley, sorghum, and soybean genomes are sequenced and the advances made in next generation sequencing (NGS) helps nowadays to develop high-resolution genetic maps to produce salt-tolerant crops. A quick search on ene database for QTL related to salt revealed 17 QTLs in Oryza sativa covering six chromosomes and delimited by several markers2. “Saltol” was reported as a promising QTL for salinity tolerance in rice that was identified after searching for more than 100 SSR markers in 140 recombinant inbred lines between Pokkali and IR29 (Thomson et al., 2010). “Saltol” is located on chromosome 1 and is important for the maintenance of the shoot ratio of Na+/K+ (Thomson et al., 2010). Other QTL analyses performed in wheat for Na+ tolerance allowed the identification of Nax1 locus, which maps to the region of the TaHKT1;4 gene that contributes to Na+ removal from xylem in the leaf sheath avoiding its over-accumulation in leaf blades (Huang et al., 2006). In soybean, genetic variation for salt tolerance has been described and was observed in wild and cultivated soybean species, suggesting that genetic improvement of salt tolerance is feasible (Lee et al., 2008; Qi et al., 2014). A major QTL for salt tolerance was constantly detected on soybean chromosome 3 (linkage group N) in different populations (Lee et al., 2004; Hamwieh et al., 2011). This QTL is likely to be the Ncl locus based on pedigree tracing (Lee et al., 2004). Recently, the introgression of the tolerance allele Ncl into soybean cultivar Jackson, using DNA MAS, produced an improved salt-tolerant line (Do et al., 2016).
Despite these successful examples, MAS based breeding pros are still limited because undesirable traits may be transferred with the QTL when wild relatives are used as donors and the results raised from QTL analyses must be confirmed in different conditions and genetic backgrounds. In fact, the cultivated soybean germplasm may be more efficiently used in breeding cultivars with improved salt tolerance compared with that of wild soybean, which generally possesses several undesirable agronomic traits.
Nevertheless, NGS technologies will significantly contribute into discovering molecular markers to obtain high density genetic maps, a prerequisite for a precise location and quicker cloning of new QTLs. Moreover, the advent of genomic selection should speed up the production of varieties combining several salt-resistance QTLs.
Conclusion
Over the last two decades, research made on Arabidopsis and a few crops shed light on several aspects of the molecular mechanisms controlling the salt stress tolerance. However, many challenges still lie ahead before successfully improving crop yield under saline conditions. Hopefully, available tools including molecular breeding and advanced biotechnology methods combined to the exploitation of the potential of soil microorganisms can speed up the release of salt-tolerant crop varieties. A combination of approaches will accelerate the identification and characterization of specific loci involved in tolerance to salinity that can be introgressed into elite sensitive varieties through molecular marker-assisted breeding. To achieve this goal, we need to have diversity in germplasm resources, high-throughput phenotyping platforms, genome sequencing of crops and their relatives. Furthermore, molecular genetic resources including mutation detection, gene discovery and expression profile, genome wide association studies, and powerful omics databases are also needed. Many traits related to salt stress tolerance have been identified and shown effective for engineering stress tolerance in model plants. The most impressive results were obtained when manipulating signaling factors, as they control a broad range of downstream events, which results in superior tolerance. Effective expression systems, including cell type-specific and stress-inducible promoters will be required to adapt the plant response to stress, lower the energy cost, according to the environmental constraints as using constitutive promoters can have severe drawbacks on plant growth or yield. Finally, the targeted genome editing using CRISPR-Cas9 technology has emerged as an alternative to classical plant breeding and transgenic (GMO) methods. CRISPR-Cas9 technology enables precision design of alleles that aid stress tolerance (Zhang H. et al., 2014), but in depth study of genome editing to engineer mechanisms of salt stress tolerance needs to be pursued in the coming years.
Author Contributions
KM handled the entire process of preparing this paper and prepared the topic on salt tolerance mechanisms in plants. MH and CE prepared the topics related to salinization in arid and semi arid region and the problem of land degradation, the impact of soil salinization on plant growth and survival and salt tolerance in crops through marker-assisted selection and genetic engineering. Finally, LL and MN prepared the topic related to the interaction with beneficial soil microorganisms to improve salinity tolerance. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. We are grateful to the College of Food and Agriculture, United Arab Emirates University (UAEU) for funding support of this publication to KM. MN and LL acknowledge support from the IRD and the Joint Genome Institute (Community Sequencing Programme n° CSP-580 to LL).
References
- Ahmad I., Maathuis F. (2014). Cellular and tissue distribution of potassium: physiological relevance, mechanisms and regulation. J. Plant Physiol. 171 708–714. 10.1016/j.jplph.2013.10.016 [DOI] [PubMed] [Google Scholar]
- Akkermans A. D. L., Abdulkadir S., Trinick M. J. (1978). N2-fixing root nodules in Ulmaceae: Parasponia or (and) Trema spp.? Plant Soil 49 711–715. 10.1007/BF02183301 [DOI] [Google Scholar]
- Al-Karaki G. N. (2000). Growth, water use efficiency, and sodium and potassium acquisition by tomato cultivars grown under salt stress. J. Plant Nutr. 23 1–8. 10.1080/01904160009381992 [DOI] [Google Scholar]
- Al-Karaki G. N. (2006). Nursery inoculation of tomato with arbuscular mycorrhizal fungi and subsequent performance under irrigation with saline water. Sci. Hortic. 109 1–7. 10.1016/j.scienta.2006.02.019 [DOI] [Google Scholar]
- Al-Karaki G. N., Hammad R. (2001). Mycorrhizal influence on fruit yield and mineral content of tomato grown under salt stress. J. Plant nutr. 24 1311–1323. 10.1081/PLN-100106983 [DOI] [Google Scholar]
- Aliasgharzadeh N., Rastin S. N., Towfighi H., Alizadeh A. (2001). Occurrence of arbuscular mycorrhizal fungi in saline soils of the Tabriz Plain of Iran in relation to some physical and chemical properties of soil. Mycorrhiza 11 119–122. 10.1007/s005720100113 [DOI] [PubMed] [Google Scholar]
- Amtmann A. (2009). Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant 2 3–12. 10.1093/mp/ssn094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anschütz U., Becker D., Shabala S. (2014). Going beyond nutrition: regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. J. Plant Physiol. 171 670–687. 10.1016/j.jplph.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Apse M. P., Aharon G. S., Snedden W. A., Blumwald E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285 1256–1258. 10.1126/science.285.5431.1256 [DOI] [PubMed] [Google Scholar]
- Apse M. P., Sottosanto J. B., Blumwald E. (2003). Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36 229–239. 10.1046/j.1365-313X.2003.01871.x [DOI] [PubMed] [Google Scholar]
- Asano T., Hakata M., Nakamura H., Aoki N., Komatsu S., Ichikawa H., et al. (2011). Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 75 179–191. 10.1007/s11103-010-9717-1 [DOI] [PubMed] [Google Scholar]
- Ashraf M. (2009). Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol. Adv. 27 84–93. 10.1016/j.biotechadv.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Ashraf M. (2010). Inducing drought tolerance in plants: recent advances. Biotechnol. Adv. 28 169–183. 10.1016/j.biotechadv.2009.11.005 [DOI] [PubMed] [Google Scholar]
- Ashraf M., Foolad M. R. (2013). Crop breeding for salt tolerance in the era of molecular markers and marker-assisted selection. Plant Breed. 132 10–20. 10.1111/pbr.12000 [DOI] [Google Scholar]
- Azcon R., El-Atrash F. (1997). Influence of arbuscular mycorrhizae and phosphorus fertilization on growth, nodulation and N2 fixation (15N) in Medicago sativa at four salinity levels. Biol. Fertil. Soils 24 81–86. 10.1007/BF01420225 [DOI] [Google Scholar]
- Badawi G. H., Kawano N., Yamauchi Y. (2004). Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiol. Plant 121 231–238. 10.1111/j.0031-9317.2004.00308.x [DOI] [PubMed] [Google Scholar]
- Baier M., Kandlbinder A., Golldack D., Dietz K. J. (2005). Oxidative stress and ozone: perception, signalling and response. Plant Cell Environ. 28 1012–1020. 10.1111/pce.12288 [DOI] [Google Scholar]
- Bala N., Sharma P. K., Lakshminarayana K. (1990). Nodulation and nitrogen fixation by salinity-tolerant rhizobia in symbiosis with tree legumes. Agric. Ecosyst. Environ. 33 33–46. 10.1016/0167-8809(90)90142-Z [DOI] [Google Scholar]
- Bano A., Fatima M. (2009). Salt tolerance in Zea mays (L).following inoculation with Rhizobium and Pseudomonas. Biol. Fertil. Soils 45 405–413. 10.1007/s00374-008-0344-9 [DOI] [Google Scholar]
- Barragan V., Leidi E. O., Andres Z., Rubio L., Deluca A., Fernandez J. A., et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24 1127–1142. 10.1105/tpc.111.095273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E., Tajima H., Liang Y. C., Ohto M. A., Ushijima K., Nakano R., et al. (2011). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23 3482–3492. 10.1105/tpc.111.089581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. R., Silvester W. B. (1993). Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol. Rev. 57 293–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthomieu P., Conejero G., Nublat A., Brackenbury W. J., Lambert C., Savio C., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 22 2004–2014. 10.1093/emboj/cdg207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E. (2000). Sodium transport and salt tolerance in plants. Curr. Opin. Cell. Biol. 12 431–434. 10.1016/S0955-0674(00)00112-5 [DOI] [PubMed] [Google Scholar]
- Bockheim J. G., Gennadiyev A. N. (2000). The role of soil-forming processes in the definition of taxa in soil taxonomy and the world soil reference base. Geoderma 95 53–72. 10.1016/S0016-7061(99)00083-X [DOI] [Google Scholar]
- Boudsocq M., Sheen J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18 30–40. 10.1016/j.tplants.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brini F., Hanin M., Mezghani I., Berkowitz G. A., Masmoudi K. (2007a). Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt-and drought stress tolerance in Arabidopsis thaliana plants. J. Exp. Bot. 58 301–308. 10.1093/jxb/erl251 [DOI] [PubMed] [Google Scholar]
- Brini F., Hanin M., Lumbreras V., Amara I., Khoudi H., Hassairi A., et al. (2007b). Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Rep. 26 2017–2026. 10.1007/s00299-007-0412-x [DOI] [PubMed] [Google Scholar]
- Brini F., Yamamoto A., Jlaiel L., Takeda S., Hobo T., Dinh H. Q., et al. (2011). Pleiotropic effects of the wheat dehydrin DHN-5 on stress responses in Arabidopsis. Plant Cell Physiol. 52 676–688. 10.1093/pcp/pcr030 [DOI] [PubMed] [Google Scholar]
- Brugnoli E., Lauteri M. (1991). Effects of salinity on stomatal conductance, photo-synthetic capacity, and carbon isotope discrimination of salt-tolerant (Gossypium hirsutum L.) and salt-sensitive (Phaseolus vulgaris L.) C(3) non-halophytes. Plant Physiol. 95 628–635. 10.1104/pp.95.2.628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrt C. S., Platten J. D., Spielmeyer W., James R. A., Lagudah E. S., Dennis E. S., et al. (2007). HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 143 1918–1928. 10.1104/pp.106.093476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carillo P., Annunziata M. G., Pontecorvo G., Fuggi A., Woodrow P. (2011). “Salinity stress and salt tolerance,” in Abiotic Stress in Plants- Mechanisms and Adaptations, eds Shanker A., venkateswaralu B. (Rijeka: In Tech; ). [Google Scholar]
- Chakraborty K., Bose J., Shabala L., Shabala S. (2016). Difference in root K+ retention ability and reduced sensitivity of K+permeablechannels to reactive oxygenspecies confer differential salt tolerance in three Brassica species. J. Exp. Bot. 67 4611–4625. 10.1093/jxb/erw236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Newman I., Zhou M., Mendham N., Zhang G., Shabala S. (2005). Screening plants for salt tolerance by measuring K+ flux: a case study for barley. Plant Cell Environ. 28 1230–1246. 10.1111/j.1365-3040.2005.01364.x [DOI] [Google Scholar]
- Choi W. G., Toyota M., Kim S. H., Hilleary R., Gilroy S. (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 111 6497–6502. 10.1073/pnas.1319955111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colla G., Rouphael Y., Cardarelli M., Tullio M., Rivera C. M., Rea E. (2008). Alleviation of salt stress by arbuscular mycorrhizal in zucchini plants grown at low and high phosphorus concentration. Biol. Fertil. Soils 44 501–509. 10.1007/s00374-007-0232-8 [DOI] [Google Scholar]
- Cominelli E., Conti L., Tonelli C., Galbiati M. (2013). Challenges and perspectives to improve crop drought and salinity tolerance. Nat. Biotechnol. 30 355–361. 10.1016/j.nbt.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Craig G. F., Atkins C. A., Bell D. T. (1991). Effect of salinity on growth of four strains of Rhizobium and their infectivity and effectiveness on two species of Acacia. Plant Soil 133 253–262. 10.1007/BF00009197 [DOI] [Google Scholar]
- Cuin T. A., Betts S. A., Chalamandrier R., Shabala S. (2008). A root’s ability to retain K+ correlates with salt tolerance in wheat. J. Exp. Bot. 59 2697–2706. 10.1093/jxb/ern128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuin T. A., Zhou M., Parsons D., Shabala S. (2012). Genetic behaviour of physiological traits conferring K+/Na+ homeostasis in wheat. Plant Biol. 14 438–446. 10.1111/j.1438-8677.2011.00526.x [DOI] [PubMed] [Google Scholar]
- Dardanelli M. S., de Cordoba F. J. F., Espuny M. R., Carvajal M. A. R., Díaz M. E. S., Serrano A. M. G., et al. (2008). Effect of Azospirillum brasilense coinoculated with Rhizobium on Phaseolus vulgaris flavonoids and Nod factor production under salt stress. Soil Biol. Biochem. 40 2713–2721. 10.1016/j.soilbio.2008.06.016 [DOI] [Google Scholar]
- Deinlein U., Stephan A. B., Horie T., Luo W., Xu G., Schroeder J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19 371–379. 10.1016/j.tplants [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V., Cuin T. A., Svistunenko D. (2010). Arabidopsis root K+ effluxconductance activated by hydroxyl radicals: single-channel properties, genetic basis and involvement in stress-induced cell death. J.Cell Sci. 123 1468–1479. 10.1242/jcs.064352 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Shabala S. N., Coutts K. B., Tester M. A., Davies J. M. (2003). Free oxygen radicals regulate plasma membrane Ca2+- and K+-permeablechannels in plant root cells. J. Cell Sci. 116 81–88. 10.1242/jcs.00201 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Straltsova D., Medvedev S. S., Pozhvanov G. A., Sokolik A., Yurin V. (2014). Stress-induced electrolyte leakage:the role of K+permeable channels andinvolvement in promed cell death and metabolic adjustment. J. Exp. Bot. 65 1259–1270. 10.1093/jxb/eru004 [DOI] [PubMed] [Google Scholar]
- Diagne N., Arumugam K., Ngom M., Nambiar-Veetil M., Franche C., Narayanan K. K., et al. (2013). Use of Frankia and actinorhizal plants for degraded lands reclamation. Biomed. Res. Int. 2013 948258 10.1155/2013/948258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diouf D., Duponnois R., Ba A. T., Neyra M., Lesueur D. (2005). Symbiosis of Acacia auriculiformis and Acacia mangium with mycorrhizal fungi and Bradyrhizobium spp. improves salt tolerance in greenhouse conditions. Funct. Plant Biol. 32 1143–1152. 10.1071/FP04069 [DOI] [PubMed] [Google Scholar]
- Do T. D., Chen H., Vu H. T. T., Hamwieh A., Yamada T., Sato T., et al. (2016). Ncl synchronously regulates Na+, K+, and Cl- in soybean greatly increases the grain yield in saline field conditions. Sci. Rep. 6: 19147 10.1038/srep19147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drira M., Saibi W., Amara I., Masmoudi K., Hanin M., Brini F. (2015). Wheat dehydrin K-segments ensure bacterial stress tolerance, antiaggregation and antimicrobial effects. Appl. Biochem. Biotechnol. 175 3310–3321. 10.1007/s12010-015-1502-9 [DOI] [PubMed] [Google Scholar]
- Elsheikh E. A. E., Wood M. (1995). Nodulation and N2 fixation by soybean inoculated with salt-tolerant rhizobia or salt-sensitive bradyrhizobia in saline soil. Soil Biol. Biochem. 27 657–661. 10.1016/0038-0717(95)98645-5 [DOI] [Google Scholar]
- Evelin H., Kapoor R., Giri B. (2009). Arbuscular mycorrhizal fungi in alleviation of salt stress: a review. Ann. Bot. 104 1263–1280. 10.1093/aob/mcp251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eynard A., Lal R., Wiebe K. (2005). Crop response in salt-affected soils. J. Sust. Agric. 27 5–50. 10.1300/J064v27n01_03 [DOI] [Google Scholar]
- FAO (2016). FAO Soils Portal. Available at: http://www.fao.org/soils-portal/soil-management/management-of-some-problem-soils/salt-affected-soils/more-information-on-salt-affected-soils/en/ [Google Scholar]
- Feng G., Zhang F., Li X., Tian C., Tang C., Rengel Z. (2002). Improved tolerance of maize plants to salt stress by arbuscular mycorrhiza is related to higher accumulation of soluble sugars in roots. Mycorrhiza 12 185–190. 10.1007/s00572-002-0170-0 [DOI] [PubMed] [Google Scholar]
- Fita A., Rodríguez-Burruezo A., Boscaiu M., Prohens J., Vicente O. (2015). Breeding and domesticating crops adapted to drought and salinity: a new paradigm for increasing food production. Front. Plant Sci. 6:978 10.3389/fpls.2015.00978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers T. J., Troke P. F., Yeo A. R. (1977). The mechanism of salt tolerance in halophytes. Ann. Rev. Plant Physiol. 28 89–121. 10.1146/annurev.pp.28.060177.000513 [DOI] [Google Scholar]
- Flowers T. J., Yeo A. R. (1995). Breeding for salinity resistance in crop plants: where next? Aust. J. Plant Physiol. 22 875–884. 10.3109/07388551.2015.1004521 [DOI] [Google Scholar]
- Foyer C. H., Noctor G. (2005). Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 28 1056–1071. 10.1111/j.1365-3040.2005.01327.x [DOI] [Google Scholar]
- Garcia de la Garma J., Fernandez-Garcia N., Bardisi E., Pallol B., Salvador Asensio-Rubio J., Bru R., et al. (2015). New insights into plant salt acclimation: the roles of vesicle trafficking and reactive oxygen species signaling in mitochondria and the endomembrane system. New Phytol. 205 216–239. 10.1111/nph.12997 [DOI] [PubMed] [Google Scholar]
- Girgis M. G. Z., Ishac Y. Z., Diem H. G., Dommergues Y. R. (1992). Selection of salt tolerant Casuarina glauca and Frankia. Acta Oecol. 13 443–451. [Google Scholar]
- Giri B., Mukerji K. G. (2004). Mycorrhizal inoculant alleviates salt stress in Sesbania aegyptiaca and Sesbania grandiflora under field conditions: evidence for reduced sodium and improved magnesium uptake. Mycorrhiza 14 307–312. 10.1007/s00572-003-0274-1 [DOI] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H., Probst N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151 10.3389/fpls.2014.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Lüking I., Yang O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30 1383–1391. 10.1007/s00299-011-1068-0 [DOI] [PubMed] [Google Scholar]
- Gouiaa S., Khoudi H. (2015). Co-expression of vacuolar Na+/H+ antiporter and H+-pyrophosphatase with an IRES-mediated dicistronic vector improves salinity tolerance and enhances potassium biofortification of tomato. Phytochemistry 117 537–546. 10.1016/j.phytochem.2015.05.016 [DOI] [PubMed] [Google Scholar]
- Gouiaa S., Khoudi H., Leidi E. O., Pardo J. M., Masmoudi K. (2012). Expression of wheat Na(+)/H(+) antiporter TNHXS1 and H(+)- pyrophosphatase TVP1 genes in tobacco from a bicistronic transcriptional unit improves salt tolerance. Plant Mol. Biol. 79 137–155. 10.1007/s11103-012-9901-6 [DOI] [PubMed] [Google Scholar]
- Guo K. M., Babourina O., Christopher D. A., Borsics T., Rengel Z. (2008). The cyclic nucleotide-gated channel, AtCNGC10, influences salt tolerance in Arabidopsis. Physiol. Plant. 134 499–507. 10.1111/j.1399-3054.2008.01157.x [DOI] [PubMed] [Google Scholar]
- Hamwieh A., Tuyen D. D., Xu D. H., Cong H., Benitez E. R., Takahashi R. E. (2011). Identification and validation of a major QTL for salt tolerance in soybean. Euphytica 79 451–459. 10.1007/s10681-011-0347-8 [DOI] [Google Scholar]
- Hanin M., Brini F., Ebel C., Toda Y., Takeda S., Masmoudi K. (2011). Plant dehydrins and stress tolerance: versatile proteins for complex mechanisms. Plant Signal. Behav. 6 1503–1509. 10.4161/psb.6.10.17088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F., Horie T. (2010). A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 33 552–565. 10.1111/j.1365-3040.2009.02056.x [DOI] [PubMed] [Google Scholar]
- He C. X., Yan J. Q., Shen G. X., Fu L. H., Holaday A. S., Auld D., et al. (2005). Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performanceunder salt conditions and increases fiber yield in the field. Plant Cell Physiol. 46 1848–1854. 10.1093/pcp/pci201 [DOI] [PubMed] [Google Scholar]
- Horie T., Costa A., Kim T. H., Han M. J., Horie R., Leung H.-Y., et al. (2007). Rice OsHKT2;1 transporter mediates large Na+ influx component into K+- starved roots for growth. EMBO J. 26 3003–3014. 10.1038/sj.emboj.7601732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Hauser F., Schroeder J. I. (2009). HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14 668 10.1016/j.tplants.2009.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Dai M., Yao J., Xiao B., Li X., Zhang Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103 12987–12992. 10.1073/pnas.0604882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., You J., Fang Y., Zhu X., Qi Z., Xiong L. (2008). Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 67 169–181. 10.1007/s11103-008-9309-5 [DOI] [PubMed] [Google Scholar]
- Huang S., Spielmeyer W., Lagudah E. S., James R. A., Platten J. D., Dennis E. S., et al. (2006). A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 142 1718–1727. 10.1104/pp.106.088864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O., Thomashow M. F. (1998). Arabidopsis CBF1 overexpression induces cor genes and enhances freezing tolerance. Science 280 104–106. 10.1126/science.280.5360.104 [DOI] [PubMed] [Google Scholar]
- James R. A., Davenport R. J., Munns R. (2006). Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiol. 142 1537–1547. 10.1104/pp.106.086538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes F., Song C., Shin D., Munemasa S., Takeda K., Gu D., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. U.S.A. 106 20520–20525. 10.1073/pnas.0907205106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Leidi E. O., Pardo J. M. (2010). How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal. Behav. 7 792–795. 10.4161/psb.5.7.11767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Sun X., Wang X., Shen Y., Hou F., Chang S., et al. (2010). Synergistic interactions of arbuscular mycorrhizal fungi and rhizobia promoted the growth of Lathyrus sativus under sulphate salt stress. Symbiosis 50 157–164. 10.1007/s13199-010-0058-2 [DOI] [Google Scholar]
- Karan R., Subudhi P. K. (2012). A stress inducible SUMO conjugating enzyme gene (SaSce9) from a grass halophyte Spartina alterniflora enhances salinity and drought stress tolerance in Arabidopsis. BMC Plant Biol. 12:187 10.1186/1471-2229-12-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17 287–291. 10.1038/7036 [DOI] [PubMed] [Google Scholar]
- Kaya C., Ashraf M., Sonmez O., Aydemir S., Tuna A. L., Cullu M. A. (2009). The influence of arbuscular mycorrhizal colonisation on key growth parameters and fruit yield of pepper plants grown at high salinity. Sci. Hortic. 121 1–6. 10.1016/j.scienta.2009.01.001 [DOI] [Google Scholar]
- Khokon M. A. R., Okuma E., Hossain M. A., Munemasa S., Uraji M., Nakamura Y., et al. (2011). Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 34 434–443. 10.1111/j.1365-3040.2010.02253.x [DOI] [PubMed] [Google Scholar]
- Kishor P. B. K., Hong Z., Miao G.-H., Hu C.-A. A., Verma D. P. S. (1995). Overexpression of Δ1-pyrroline-5-carboxilase synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 108 1387–1394. 10.1104/pp.108.104/pp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Trewavas A. J., Knight M. R. (1997). Calcium signaling in Arabidpsis thaliana responding to drought and salinity. Plant J. 12 1067–1078. 10.1046/j.1365-313X.1997.12051067.x [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy R., Kim K., Subramanian P., Senthilkumar M., Anandham R., Sa T. (2016). Arbuscular mycorrhizal fungi and associated bacteria isolated from salt-affected soil enhances the tolerance of maize to salinity in coastal reclamation soil. Agric. Ecosyst. Environ. 231 233–239. 10.1016/j.agee.2016.05.037 [DOI] [Google Scholar]
- Kronzucker H. J., Britto D. T. (2011). Sodium transport in plants: a critical review. New Phytol. 189 54–81. 10.1111/j.1469-8137.2010.03540.x [DOI] [PubMed] [Google Scholar]
- Kurusu T., Kuchitsu K., Nakano M., Nakayama Y., Lida H. (2013). Plant mechanosensing and Ca2+ transport. Trends Plant Sci. 18 227–233. 10.1016/j.tplants.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Larisch N., Kirsch S. A., Schambony A., Studtrucker T., Böckmann R. A., Dietrich P. (2016). The function of the two-pore channel TPC1 depends on dimerization of its carboxy-terminal helix. Cell Mol. Life Sci. 73 2565–2581. 10.1007/s00018-016-2131-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata C., Prasad M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62 4731–4748. 10.1093/jxb/err210 [DOI] [PubMed] [Google Scholar]
- Lata C., Yadav A., Prasad M. (2011). “Role of plant transcription factors in abiotic stress tolerance,” in Abiotic Stress Response in Plants – Physiological, Biochemical and Genetic Perspectives, eds Shanker A., Venkateshwarulu B. (Rijeka: InTech; ). [Google Scholar]
- Latef A. A. H. A., Chaoxing H. (2011). Effect of arbuscular mycorrhizal fungi on growth, mineral nutrition, antioxidant enzymes activity and fruit yield of tomato grown under salinity stress. Sci. Hortic. 127 228–233. 10.1016/j.scienta.2010.09.020 [DOI] [Google Scholar]
- Lee G. J., Carter T. E., Jr., Villagarcia M. R., Li Z., Zhou X., Gibbs M. O., et al. (2004). A major QTL conditionning salt tolerance in S-100 soybean and descendent cultivars. Theor. Appl. Genet. 109 1610–1619. 10.1007/s00122-004-1783-9 [DOI] [PubMed] [Google Scholar]
- Lee J. D., Smothers S. L., Dunn D., Villagarcia M., Shumway C. R., Carter T. E., et al. (2008). Evaluation of a simple method to screen soybean genotypes for salt tolerance. Crop Sci. 48 2194–2200. 10.2135/cropsci2008.02.0090 [DOI] [Google Scholar]
- Lee S. K., Kim B. G., Kwon T. R., Jeong M. J., Park S. R., Lee J. W., et al. (2011). Overexpression of the mitogen-activated protein kinase gene OsMAPK33 enhances sensitivity to salt stress in rice (Oryza sativa L.). J. Biosci. 36 139–151. 10.1007/s12038-011-9002-8 [DOI] [PubMed] [Google Scholar]
- Leidi E. O., Barragan V., Rubio L., El-Hamdaoui A., Ruiz M. T., Cubero B., et al. (2010). The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 61 495–506. 10.1111/j.1365-313X.2009.04073.x [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Goda H., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 391–406. 10.1105/tpc.10.8.1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan B. A. (2005). “Reactive oxygen species and photosynthesis,” in Antioxidants and Reactive Oxygen Species in Plants, ed. Smirnoff N. (Oxford: Blackwell; ), 250–267. [Google Scholar]
- Maas E. V., Hoffman G. J. (1977). Crop salt tolerance – current assessment. J. Irrig. Drainage Div. Am. Soc. Civil Eng. 103 115–134. [Google Scholar]
- Maathuis F. J. M., Amtmann A. (1999). K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann. Bot. 84 123–133. 10.1006/anbo.1999.0912 [DOI] [Google Scholar]
- Martí M. C., Stancombe M. A., Webb A. A. (2013). Cell- and stimulus type-specific intracellular free Ca2+ signals in Arabidopsis. Plant Physiol. 163 625–634. 10.1104/pp.113.222901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKersie B. D., Bowley S. R., Jones K. S. (1999). Winter survival of transgenic alfalfa overexpressing superoxide dismutase. Plant Physiol. 119 839–848. 10.1104/pp.119.104/pp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Shulaev V., Mittler R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133 481–489. 10.1111/j.1399-3054.2008.01090.x [DOI] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33 453–467. 10.1111/j.1365-3040.2009.02041.x [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 490–498. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Møller I. S., Gilliham M., Jha D., Mayo G. M., Roy S. J., Coates J. C., et al. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21 2163–2178. 10.1105/tpc.108.064568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R. (2005). Genes and salt tolerance: bringing them together. New Phytol. 167 645–663. 10.1111/j.1469-8137.2005.01487.x [DOI] [PubMed] [Google Scholar]
- Munns R., Guo J., Passioura J. B., Cramer G. R. (2000). Leaf water status controls day-time but not daily rates of leaf expansion in salt-treated barley. Aust. J. Plant Physiol. 27 949–957. [Google Scholar]
- Munns R., James R. A. (2003). Screening methods for salt tolerance: a case study with tetraploid wheat. Plant Soil 253 201–218. 10.1023/A:1024553303144 [DOI] [Google Scholar]
- Munns R., James R. A., Läuchli A. (2006). Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 57 1025–1043. 10.1093/jxb/erj100 [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Ann. Rev. Plant Biol. 59 651–681. 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- Ng B. H. (1987). The effects of salinity on growth, nodulation and nitrogen fixation of Casuarina equisetifolia. Plant Soil 103 123–125. 10.1007/BF02370676 [DOI] [Google Scholar]
- Ngom M., Gray K., Diagne N., Oshone R., Fardoux J., Gherbi H., et al. (2016a). Symbiotic performance of diverse Frankia strains on salt-stressed Casuarina glauca and Casuarina equisetifolia plants. Front. Plant Sci. 7:1331 10.3389/fpls.2016.01331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngom M., Oshone R., Diagne N., Cissoko M., Svistoonoff S., Tisa L. S., et al. (2016b). Tolerance to environmental stress by the nitrogen-fixing actinobacterium Frankia and its role in actinorhizal plants adaptation. Symbiosis 70 17–29. 10.1007/s13199-016-0396-9 [DOI] [Google Scholar]
- Nuruzzaman M., Sharoni A. M., Kikuchi S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248 10.3389/fmicb.2013.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. J., Song S. I., Kim Y. S., Jang H. J., Kim S. Y., Kim M., et al. (2005). Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 138 341–351. 10.1104/pp.104.059147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olias R., Eljakaoui Z., Li J., De Morales P. A., Marin-Manzano M. C., Pardo J. M., et al. (2009). The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 32 904–916. 10.1111/j.1365-3040.2009.01971.x [DOI] [PubMed] [Google Scholar]
- Oliveira R. S., Castro P. M. L., Dodd J. C., Vosátka M. (2005). Synergistic effect of Glomusintraradices and Frankia spp. on the growth and stress recovery of Alnus glutinosa in an alkaline anthropogenic sediment. Chemosphere 60 1462–1470. 10.1016/j.chemosphere.2005.01.038 [DOI] [PubMed] [Google Scholar]
- Orellana S., Yañez M., Espinoza A., Verdugo I., González E., Ruiz-Lara S., et al. (2010). The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 33 2191–2208. 10.1111/j.1365-3040.2010.02220.x [DOI] [PubMed] [Google Scholar]
- Palta J., Levitt J., Stadelmann E. J. (1977). Freezing injury in onion bulbcells. I. Evaluation of the conductivity method and analysis of ion and sugarefflux. Plant Physiol. 60 393–397. 10.1104/pp.60.3.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo J. M., Cubero B., Leidi E. O., Quintero F. J. (2006). Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 57 1181–1199. 10.1093/jxb/erj114 [DOI] [PubMed] [Google Scholar]
- Pasapula V., Shen G., Kuppu S., Paez-Valencia J., Mendoza M., Hou P., et al. (2011). Expression of an Arabidopsis vacuolar H+ – pyrophosphatase gene (AVP1) in cotton improves drought- and salt tolerance and increases fibre yield in the field conditions. Plant Biotechnol. J 9 88–99. 10.1111/j.1467-7652.2010.00535.x [DOI] [PubMed] [Google Scholar]
- Paul M. J., Foyer C. H. (2001). Sink regulation of photosynthesis. J. Exp. Bot. 52 1383–1400. 10.1093/jexbot/52.360.1383 [DOI] [PubMed] [Google Scholar]
- Pawlowski K., Demchenko K. N. (2012). The diversity of actinorhizal symbiosis. Protoplasma 249 967–979. 10.1007/s00709-012-0388-4 [DOI] [PubMed] [Google Scholar]
- Perrine-Walker F., Gherbi H., Imanishi L., Hocher V., Ghodhbane-Gtari F., Lavenus J., et al. (2011). Symbiotic signaling in actinorhizal symbioses. Curr. Protein Pept. Sci. 12 156–164. 10.2174/1389211213488422037 [DOI] [PubMed] [Google Scholar]
- Porcel R., Aroca R., Ruiz-Lozano J. M. (2012). Salinity stress alleviation using arbuscular mycorrhizal fungi. A review. Agron. Sust. Dev. 32 181–200. 10.1007/s13593-011-0029-x [DOI] [Google Scholar]
- Porras-Soriano A., Soriano-Martín M. L., Porras-Piedra A., Azcón R. (2009). Arbuscular mycorrhizal fungi increased growth, nutrient uptake and tolerance to salinity in olive trees under nursery conditions. J. Plant Physiol. 166 1350–1359. 10.1016/j.jplph.2009.02.010 [DOI] [PubMed] [Google Scholar]
- Pottosin I., Dobrovinskaya O. (2014). Non-selective cation channels in plasma and vacuolar membranes and their contribution to K+ transport. J. Plant Physiol. 171 732–742. 10.1016/j.jplph.2013.11.013 [DOI] [PubMed] [Google Scholar]
- Qi X., Li M. W., Xie M., Liu X., Ni M., Shao G., et al. (2014). Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 5:4340 10.1038/ncomms5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie G. H., Aboul-Nasr M. B., Al-Humiany A. (2005). Increased salinity tolerance of cowpea plants by dual inoculation of an arbuscular mycorrhizal fungus Glomus clarum and a nitrogen-fixer Azospirillum brasilense. Mycobiology 33 51–60. 10.4489/MYCO.2005.33.1.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabie G. H., Almadini A. M. (2005). Role of bioinoculants in development of salt-tolerance of Vicia faba plants under salinity stress. Afr. J. Biotechnol. 4: 210–222. [Google Scholar]
- Reddell P., Foster R. C., Bowen G. D. (1986). The effects of sodium chloride on growth and nitrogen fixation in Casuarina obesa Miq. New Phytol. 102 397–408. 10.1111/j.1469-8137.1986.tb00817.x [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rosales M. P., Galvez F. J., Huertas R., Aranda M. N., Baghour M., Cagnac O., et al. (1997). Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 15 988–991. 10.1038/nbt1097-988 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rosales M. P., Gálvez F. J., Huertas R., Aranda M. N., Baghour M., Cagnac O., et al. (2009). Plant NHX cation/proton antiporters. Plant Signal. Behav. 4 265–276. 10.4161/psb.4.4.7919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxas V. P., Smith R. K., Jr., Allen E. R., Allen R. D. (1997). Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 15 988–991. 10.1038/nbt1097-988 [DOI] [PubMed] [Google Scholar]
- Roy S. J., Negrão S., Tester M. (2014). Salt resistant crop plants. Curr. Opin. Biotechnol. 26 115–124. 10.1016/j.copbio.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Sadhana B. (2014). Arbuscular Mycorrhizal Fungi (AMF) as a biofertilizer. A review. Int. J. Curr. Microbiol. Appl. Sci. 3 384–400. [Google Scholar]
- Sakamoto A., Murata A., Murata N. (1998). Metabolic engineering of rice leading to biosynthesis of glycine betaine and tolerance to salt and cold. Plant Mol. Biol. 38 1011–1019. 10.1023/A:1006095015717 [DOI] [PubMed] [Google Scholar]
- Schilling R. K., Marschner P., Shavrukov Y., Berger B., Tester M., Roy S. J., et al. (2013). Expression of the Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) improves the shoot biomass of transgenic barley and increases grain yield in a saline field. Plant Biotechnol. J. 12 378–386. 10.1111/pbi.12145 [DOI] [PubMed] [Google Scholar]
- Schroeder J. I., Ward J. M., Gassmann W. (1994). Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: biophysical implications for K+ uptake. Annu. Rev. Biophys. Biomol. Struct. 23 441–471. 10.1146/annurev.bb.23.060194.002301 [DOI] [PubMed] [Google Scholar]
- Serraj R., Adu-Gyamfi J. (2004). Role of symbiotic nitrogen fixation in the improvement of legume productivity under stressed environments. West Afr. J. Appl. Ecol. 6 95–109. [Google Scholar]
- Shabala S., Cuin T. A. (2007). Potassium transport and plant salt tolerance. Physiol. Plant. 133 651–669. 10.1111/j.1399-3054.2007.01008.x [DOI] [PubMed] [Google Scholar]
- Shabala S., Demidchik V., Shabala L., Cuin T. A., Smith S. J., Miller A. J., et al. (2006). Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+- permeable channels. Plant Physiol. 141 1653–1665. 10.1104/pp.106.082388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S., Pottosin I. (2014). Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiol. Plant. 151 257–279. 10.1111/ppl.12165 [DOI] [PubMed] [Google Scholar]
- Shabala S., Wu H., Bose J. (2015). Salt stress sensing and early signalling events in plant roots: current knowledge and hypothesis. Plant Sci. 241 109–119. 10.1016/j.plantsci.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Shen G., Wei J., Qiu X., Hu R., Kuppu S., Auld D., et al. (2014). Co-overexpression of AVP1 and AtNHX1 in cotton further improves drought and salt tolerance in transgenic cotton plants. Plant Mol. Biol. Rep. 33 167–177. 10.1007/s11105-014-0739-8 [DOI] [Google Scholar]
- Shen Y. G., Zhang W. K., He S. J., Zhang J. S., Liu Q., Chen S. Y. (2003). An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor. Appl. Genet. 106 923–930. 10.1007/s00122-002-1131-x [DOI] [PubMed] [Google Scholar]
- Sheng M., Tang M., Chen H., Yang B. W., Zhang F. F., Huang Y. H. (2008). Influence of arbuscular mycorrhizae on photosynthesis and water status of maize plants under salt stress. Mycorrhiza 18 287–296. 10.1007/s00572-008-0180-7 [DOI] [PubMed] [Google Scholar]
- Shi H., Quintero F. J., Pardo J. M., Zhu J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14 465–477. 10.1105/tpc.010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. K., Redoña E., Refuezo L. (2010). “Varietal improvement for abiotic stress tolerance in crop plants: special reference to salinity in rice,” in Abiotic Stress Adaptation in Plants: Physiological, Molecular and Genomic Foundation, eds Pareek A., Sopory S. K., Govindjee B. H. (New York, NY: Springer; ), 387–415. 10.1007/978-90-481-3112-9_18 [DOI] [Google Scholar]
- Smith S. E., Read D. J. (2010). Mycorrhizal Symbiosis. New York, NY: Academic press. [Google Scholar]
- Smith S. E., Smith F. A. (2011). Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 62 227–250. 10.1146/annurev-arplant-042110-103846 [DOI] [PubMed] [Google Scholar]
- Soliman A. S., Shanan N. T., Massoud O. N., Swelim D. M. (2014). Improving salinity tolerance of Acacia saligna (Labill.) plant by arbuscular mycorrhizal fungi and Rhizobium inoculation. Afr. J. Biotechnol. 11 1259–1266. [Google Scholar]
- Su J., Wu R. (2004). Stress-inducible synthesis of proline in transgenic rice confers faster growth under stress conditions than that with constitutive synthesis. Plant Sci. 166 941–948. 10.1016/j.plantsci.2003.12.004 [DOI] [Google Scholar]
- Sun S. J., Guo S. Q., Yang X., Bao Y. M., Tang H. J., Sun H., et al. (2010). Functional analysis of a novel Cys2/His2 type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 61 2807–2818. 10.1093/jxb/erq120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi Horie T., Motoda J., Kubo M., Yang H., Yoda K., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 44 928–938. 10.1111/j.1365-313X.2005.02595.x [DOI] [PubMed] [Google Scholar]
- Swaraj K., Bishnoi N. R. (1999). Effect of salt stress on nodulation and nitrogen fixation in legumes. Indian J. Exp. Biol. 37 843–848. [PubMed] [Google Scholar]
- Tarczynski M. C., Jensen R. G., Bohnert H. J. (1992). Expression of a bacterial mtlD gene in transgenic tobacco leads to production and accumulation of mannitol. Proc. Natl. Acad. Sci. U.S.A. 89 2600–2604. 10.1073/pnas.89.7.2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarczynski M. C., Jensen R. G., Bohnert H. J. (1993). Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science 259 508–510. 10.1126/science.259.5094.508 [DOI] [PubMed] [Google Scholar]
- Tian C. Y., Feng G., Li X. L., Zhang F. S. (2004). Different effects of arbuscular mycorrhizal fungal isolates from saline or non-saline soil on salinity tolerance of plants. Appl. Soil Ecol. 26 143–148. 10.1016/j.apsoil.2003.10.010 [DOI] [Google Scholar]
- Thomson M. J., deOcampo M., Egdane J., Rahman M. A., Sajise A. G., Adorada D. L., et al. (2010). Characterizing the saltol quantitative trait locus for salinity tolerance in rice. Rice 3 148–160. 10.1007/s12284-010-9053-8 [DOI] [Google Scholar]
- Turan S., Cornish K., Kumar S. (2012). Salinity tolerance in plants: breeding and genetic engineering. Austr. J. Crop Sci. 6 1337–1348. [Google Scholar]
- Türkan I., Demiral T. (2009). Recent developments in understanding salinity tolerance. Environ. Exp. Bot. 67 2–9. 10.1016/j.envexpbot.2009.05.008 [DOI] [Google Scholar]
- Tyerman S. D., Skerrett I. M. (1999). Root ion channels and salinity. Sci. Hort. 78 175–235. 10.1016/S0304-4238(98)00194-0 [DOI] [Google Scholar]
- Ulm R., Ichimura K., Mizoguchi T., Peck S. C., Zhu T., Wang X., et al. (2002). Distinct regulation of salinity and genotoxic stress responses by ArabidopsisMAP kinase phosphatase 1. Eur. Mol. Biol. Organ. J. 21 6483–6493. 10.1093/emboj/cdf646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNEP (1992). ‘World Atlas of Desertification. London: Edward Arnold. [Google Scholar]
- USDA-ARS (2008). “Research databases,” in Bibliography on Salt Tolerance, ed. Brown G. E., Jr. (Riverside, CA: United States Department of Agriculture Agricultural Research Service; ). [Google Scholar]
- Varshney R. K., Bansal K. C., Aggarwal P. K., Datta S. K., Craufurd P. Q. (2011). Agricultural biotechnology for crop improvement in a variable climate: hope or hype. Trends Plant Sci. 16 363–371. 10.1016/j.tplants.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Véry A. A., Nieves-Cordones M., Daly M., Khan I., Fizames C., Sentenac H. (2014). Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J. Plant Physiol. 171 748–769. 10.1016/j.jplph.2014.01.011 [DOI] [PubMed] [Google Scholar]
- Volkov V., Amtmann A. (2006). Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion-channel features supporting K+/Na+ homeostasis under salinity stress. Plant J. 48 342–353. 10.1111/j.1365-313X.2006.02876.x [DOI] [PubMed] [Google Scholar]
- Wegner L. H., de Boer A. H. (1997). Properties of two outwards rectifying channels in root xylem parenchyma cells suggest a role in K+ homeostasis and long distance signaling. Plant Physiol. 115 1707–1719. 10.1104/pp.115.4.1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinl S., Kudla J. (2008). The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 179 675–686. 10.1111/j.1469-8137.2009.02938.x [DOI] [PubMed] [Google Scholar]
- Wu H., Zhu M., Shabala L., Zhou M., Shabala S. (2015). K+ retention in leaf mesophyll, an overlooked component of salinity tolerance mechanism: a case study for barley. J. Integr. Plant Biol. 57 171–185. 10.1111/jipb.12238 [DOI] [PubMed] [Google Scholar]
- Wu Q. S., Zou Y. N., He X. H. (2010). Contributions of arbuscular mycorrhizal fungi to growth, photosynthesis, root morphology and ionic balance of citrus seedlings under salt stress. Acta Physiol. Plant. 32 297–304. 10.1007/s11738-009-0407-z [DOI] [Google Scholar]
- Xie Y. J., Xu S., Han B., Wu M. Z., Yuan X. X., Han Y., et al. (2011). Evidence of Arabidopsis salt acclimation induced by up-regulation of HY1 and the regulatory role of RbohD-derived reactive oxygen species synthesis. Plant J. 66 280–292. 10.1111/j.1365-313X.2011.04488.x [DOI] [PubMed] [Google Scholar]
- Xing Y., Jia W., Zhang J. (2008). AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54 440–451. 10.1111/j.1365-313X.2008.03433.x [DOI] [PubMed] [Google Scholar]
- Xu D., Duan X., Wang B., Hong B., Ho T. H. D., Wu R. (1996). Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110 249–257. 10.1104/pp.110.1.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z. Y., Zhi D. Y., Xue G. P., Zhang H., Zhao Y. X., Xia G. M. (2004). Enhanced salt tolerance of transgenic wheat (Triticum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci. 167 849–859. 10.1016/j.plantsci.2004.05.034 [DOI] [Google Scholar]
- Young J. P. W. (1996). Phylogeny and taxonomy of rhizobia. Plant Soil 186 45–52. 10.1007/BF00035054 [DOI] [Google Scholar]
- Zahran H. H. (1999). Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. Rev. 63 968–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi I., Ebel C., Belgaroui N., Ghorbel M., Amara I., Hanin M. (2016). The wheat MAP kinase phosphatase 1 alleviates salt stress and increases antioxidant activities in Arabidopsis. J. Plant Physiol. 193 12–21. 10.1016/j.jplph.2016.01.011 [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Wei P., Zhang B., Gou F., Feng Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12 797–807. 10.1111/pbi.12200 [DOI] [PubMed] [Google Scholar]
- Zhang H. X., Blumwald E. (2001). Transgenic salt tolerant tomato plants accumulate salt in the foliage but not in the fruits. Nat. Biotechnol. 19 765–768. 10.1038/90824 [DOI] [PubMed] [Google Scholar]
- Zhang H. X., Hodson J. N., Williams J. P., Blumwald E. (2001). Engineering salt-tolerant Brassica plants: characterization of yieldand seed oil quality in transgenic plants with increased vacuolarsodium accumulation. Proc. Natl. Acad Sci. U.S.A. 98 12832–12836. 10.1073/pnas.231476498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ma H., Chen T., Pen J., Yu S., Zhao X. (2014). Morphological and physiological responses of cotton (Gossypium hirsutum L.) plants to salinity. PLoS ONE 9:e112807 10.1371/journal.pone.0112807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Xi D., Li S., Gao Z., Zhao S., Shi J., et al. (2011). A cotton group C MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant Mol. Biol. 77 17–31. 10.1007/s11103-011-9788-7 [DOI] [PubMed] [Google Scholar]
- Zou N., Dart P. J., Marcar N. E. (1995). Interaction of salinity and rhizobial strain on growth and N2-fixation by Acacia ampliceps. Soil Biol. Biochem. 27 409–413. 10.1016/0038-0717(95)98611-Q [DOI] [Google Scholar]