Abstract
Periodontal infections contribute to HIV-associated co-morbidities in the oral cavity and provide a model to interrogate the dysregulation of macrophage function, inflammatory disease progression, and HIV replication during co-infections. We investigated the effect of Porphyromonas gingivalis on the establishment of HIV infection in monocyte-derived macrophages. HIV replication in macrophages was significantly repressed in the presence of P. gingivalis. This diminished viral replication was due partly to a decrease in the expression of integrated HIV provirus. HIV repression depended upon signaling through TLR4 as knock-down of TLR4 with siRNA rescued HIV expression. Importantly, HIV expression was reactivated upon removal of P. gingivalis. Our observations suggest that exposure of macrophages to Gram-negative bacteria influence the establishment and maintenance of HIV persistence in macrophages through a TLR4-dependent mechanism.
1. Introduction
Periodontal disease is one of the most common chronic infections in the US, estimated to affect more than 100 million people (Eke et al., 2012; Slade and Beck, 1999). Periodontal disease is aggravated in HIV-infected patients and leads to oral complications such as necrotizing ulcerative gingivitis and ulcerative periodontitis. Though anti-retroviral therapy (ART) decreases the prevalence of periodontal disease, it does not ameliorate oral complications in many patients. Thus, oral disease is a major co-morbidity that affects the quality of life of HIV-infected individuals (Patton et al., 2000; Schmidt-Westhausen et al., 2000; Tappuni and Fleming, 2001). HIV target cells are present in the oral cavity (Cutler and Jotwani, 2006) and how the mouth microenvironment and the associated dysregulated inflammatory axis influence HIV replication has not been thoroughly investigated.
We are interested in determining how oral pathobionts influence the course of HIV infection and replication. Although, the microbiology of HIV-associated periodontal disease is complex, the Gram-negative bacterium, Porphyromonas gingivalis is an important etiological agent of disease (Hajishengallis et al., 2012; Holt et al., 1988; Socransky et al., 1998; Ximenez-Fyvie et al., 2000). P. gingivalis is present in sub-gingival plaques of HIV-infected individuals (Cross and Smith, 1995; Pereira et al., 2014; Tenenbaum et al., 1997) and it has been shown to activate HIV transcription in some cell models through TLR2 and TLR9 and through the expression of short-chain fatty acids (Das et al., 2015; Gonzalez et al., 2010; Imai et al., 2009). These observations suggest that HIV and P. gingivalis co-infections exacerbate oral disease. Therefore, P. gingivalis provides a model pathogen to explore the interactions between periodontal disease-associated microbes and HIV infection.
Macrophages represent a diverse population of immune cells residing within all anatomical compartments and are important effectors of the innate immune response against microbes, modulate inflammatory responses and bridge the innate and adaptive immune system through antigen presentation to CD4+ T cells (Varol et al., 2015). In the case of periodontal disease, macrophages represent up to 30% of the infiltrating immune cells in lesions (Okada and Murakami, 1998). Furthermore, macrophages within many anatomical compartments are susceptible to direct infection by HIV-1 and this infection correlates with macrophage dysfunction including altered cytokine production, phagocytosis, pathogen killing and chemotaxis (Pugliese et al., 2005). HIV replication is dependent on the regulation of transcription factors, cytokines and anti-viral restriction factors associated with macrophage activation and the cytokine microenvironment (Cassetta et al., 2013; Cassol et al., 2009; Herbein and Varin, 2010; Koyanagi et al., 1988; Schlaepfer et al., 2014; Schuitemaker et al., 1994; Schuitemaker et al., 1992). However, it remains unclear to what extent these signals affect long term HIV pathogenesis or possibly HIV persistence. Despite the success of highly active anti-retroviral therapy (HAART) in controlling HIV replication and preventing the onset of AIDS in infected patients, the infection cannot be fully eliminated due to the persistence of HIV-infected cells resistant to therapy (Siliciano and Greene, 2011).
The population of persistently infected cells is primarily comprised of latently-infected resting CD4+ T cells (Brenchley et al., 2004; Chomont et al., 2009; Chun et al., 1997; Douek et al., 2002; Maldarelli et al., 2014; Ostrowski et al., 1999; Wagner et al., 2014), but it has been hypothesized that macrophages and other myeloid cells contribute to long-term viral persistence (Coleman and Wu, 2009). Two potential mechanisms could mediate persistent HIV infection in macrophages. Macrophages are known to resist the cytopathic effects of HIV replication and survive infection to produce low levels of viral particles (Gartner et al., 1986; Koenig et al., 1986; Nicholson et al., 1986; Salahuddin et al., 1986; Sonza et al., 2002). This form of persistence would be especially important if infected macrophages reside within anatomical sanctuaries where therapies are not present at effective concentrations. The other possibility is that HIV provirus following infection is transcriptionally silenced and provirus is reactivated upon cell stimulation (Brown et al., 2006; Coleman and Wu, 2009).
To gain insights into how existing oral bacteria influence HIV infection of macrophages, we tested whether P. gingivalis altered the course of infection in primary monocyte-derived macrophages (MDMs). We found that exposure to P. gingivalis at the time of HIV infection led to repression of HIV expression despite efficient viral entry, integration and transcription. Importantly, this repression was reversible upon removing bacteria and dependent on the recognition of bacterial lipopolysaccharide (LPS) through TLR4 signaling. These results suggest that exposure to P. gingivalis at the time of HIV infection induces persistence in macrophages and suggest that pre-existing infections play a role in the establishment of HIV persistence within macrophages.
2. Results
2.1. P. gingivalis influences macrophage phenotypes
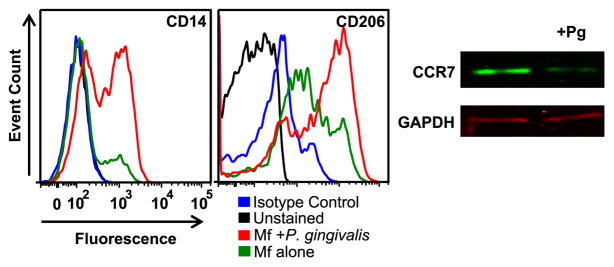
In order to appreciate how P. gingivalis influences HIV replication in macrophages, it was necessary to establish whether P. gingivalis skews macrophage phenotypes. Monocyte-derived-macrophages (MDM) were enriched from peripheral blood and matured on plastic for one week prior to co-culturing with heat-killed P. gingivalis strain 381. Although this species of bacteria is found among the normal flora of the oral cavity, P. gingivalis can be pathogenic and it is a major contributor to periodontal disease. An advantage of using heat-killed bacteria over live bacteria is that it reduces the complexity of the culture system by eliminating variables associated with bacterial growth and virulence factors; any effect on HIV replication can be directly attributed to the recognition of bacteria structure by macrophages. We examined whether bacteria altered MDM phenotype by measuring the expression of CD14, CD206 and CCR7. MDMs exposed to bacteria up-regulated CD14 and CD206 and down-regulated CCR7 (Fig. 1). Expression of these surface molecules has been associated with anti-inflammatory functions (Martinez et al., 2006).
Fig. 1.
MDMs polarize in response to P. gingivalis. (A) MDMs were exposed to P. gingivalis at an MOI of 100 for 3 days and monitored surface molecule expression by flow cytometry. CCR7 expression was assessed by Western blotting. Data are representative of 3 individual experiments.
2.2. Exposure of MDMs to P. gingivalis reduces HIV replication
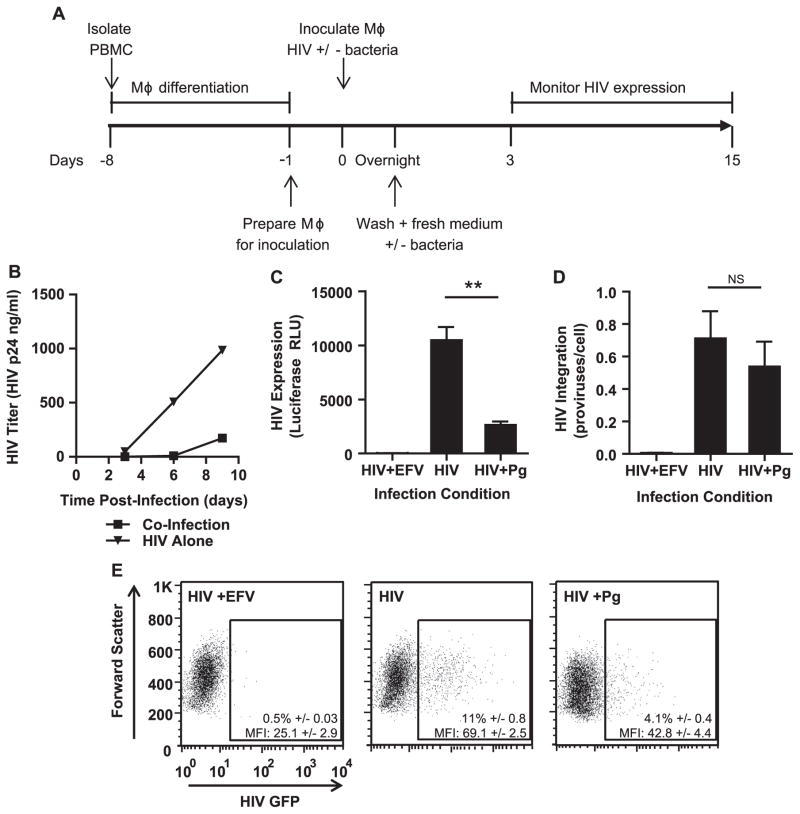
To test whether HIV replication in MDMs was affected by the presence of P. gingivalis, we inoculated MDMs with HIV in the presence or absence of bacteria and monitored HIV replication by p24 ELISA over time (Fig. 2A and B). HIV replication was inhibited by up to 10-fold when P. gingivalis was present in the culture at the time of infection with HIV (Fig. 2B). To investigate whether this effect in viral replication was due in part to defects in post-integration steps in the viral life cycle, we used single round envelope-deleted HIV pseudotyped with VSV-G encoding firefly luciferase or GFP. Any defects in steps post-viral integration would indicate that exposure to P. gingivalis at the time of HIV infection enables the establishment of persistent infection in macrophages. We found that HIV was expressed less efficiently in MDMs co-exposed to heat-killed bacteria despite similar levels of integrated proviral DNA as measured by a nested Alu-quantitative PCR (Fig. 2C–E). Together, these results suggest that steps post-integration are repressed when macrophages are exposed to heat killed P. gingivalis at the time of HIV infection.
Fig. 2.
The presence of P. gingivalis at the time of infection represses HIV replication in MDMs. (A) General experimental outline to test the effect of heat-killed P. gingivalis on HIV replication in MDMs. (B) MDMs were infected with HIV-WT/BaL with or without exposure to P. gingivalis at an MOI of 10. HIV spread was monitored by HIV-p24 ELISA. Data is representative of 3 independent experiments. To test HIV expression in the context of single round infection, MDMs were infected with HIV-NL4-3ΔEnv-Luc(VSV-G) with or without P. gingivalis (MOI 30) and monitored HIV infection by (C) luciferase expression (D) and Alu-PCR for HIV integration at 3 days post-infection. Cultures treated with 1 μM efavirenz (EFV) served as a negative control. Error bars represent the standard deviation of triplicate measurements. Both graphs were generated from the same experiment and are representative of 3 individual experiments. Asterisks and NS represent p<0.01 and p>0.05 respectively as assessed by Student’s T test. (E) MDMs were infected with HIV-NL4-3ΔEnv-GFP(VSV-G) and monitored GFP expression 3 days post-infection by flow cytometry. Percentages are displayed±the standard deviation of triplicate infections of cells from the same donor. The geometric mean intensity of GFP (MFI) is displayed±the standard deviation of triplicate inoculations of cells from the same donor. The experiment is representative of 3 individual experiments.
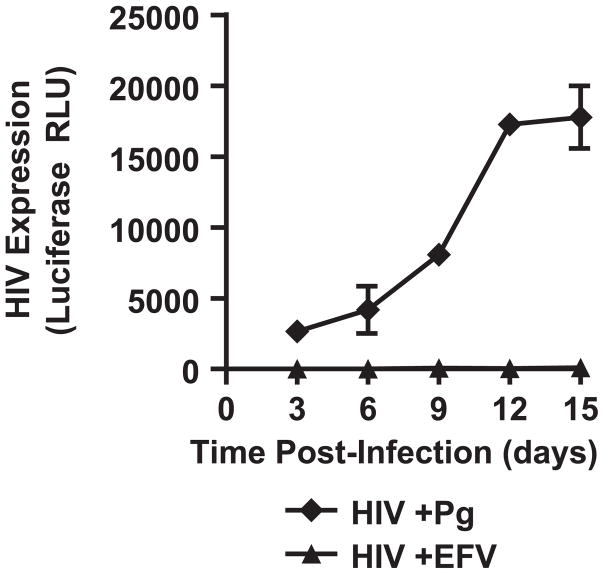
To determine whether P. gingivalis-mediated repression was reversible, bacteria were washed out 3 days post-infection and HIV expression was monitored over 12 days (Fig. 3). While there was a lag of 3–6 days in HIV expression in MDMs after the removal of bacteria, by day 12 a greater than a log increase in HIV expression was observed (Fig. 3). These results suggest that the repression induced by P. gingivalis is reversible and that MDMs can be persistently infected.
Fig. 3.
HIV expression rebounds after removal of bacteria. MDMs were infected with HIV-NL4-3ΔEnv-Luc(VSV-G) with P. gingivalis for 3 days followed by removal of bacteria. HIV expression was monitored by luciferase expression every 3 days. Error bars represent the standard deviation of infections done in triplicate.
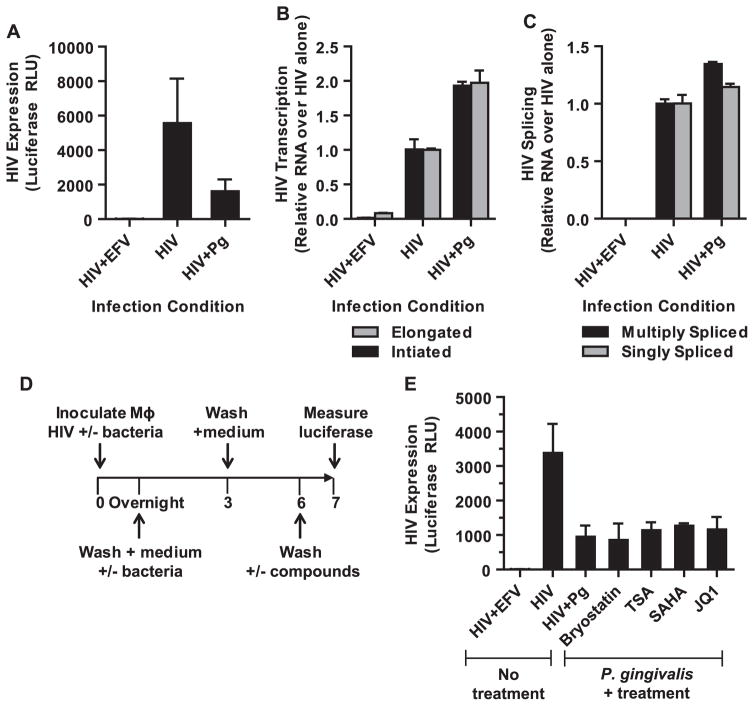
To gain further insight into the mechanisms of repression, we examined the level of proviral gene transcription. We found that P. gingivalis exposure did not decrease the proportion of initiated or elongated HIV transcripts relative to MDMs infected with HIV alone despite a 3–5-fold reduction in HIV expression as determined by luciferase expression from a parallel inoculation (Fig. 4A and B). Instead, a ~2-fold increase in the levels of both initiated and elongated transcripts was detected. In addition, no significant changes in transcript splicing were observed (Fig. 4C). This suggests that HIV expression and production is repressed at a step post-proviral transcription. In agreement with this observation, we found that HIV expression could not be re-activated by common latency reversing agents that target transcriptional cofactors and chromatin remodeling complexes (Fig. 4D and E). Together, these results suggest that P. gingivalis induces a state in macrophages that represses HIV expression through a mechanism that differs from latency described in T cells.
Fig. 4.
P. gingivalis mediated repression of HIV is post-transcriptional. MDMs were infected with HIV-NL4-3ΔEnv-Luc(VSV-G) for 4 h in the presence or absence P. gingivalis at an MOI of 30. (A) The cells were collected 3 days post-infection to measure HIV expression by luciferase. Error bars represent the standard deviation of 3 infections. In a parallel infections, the cells were harvested to measure the levels of (B) initiated and elongated HIV transcripts and (C) multiply- and singly-spliced HIV transcripts by RT-PCR. Error bars represent the standard deviation of measurements done in triplicate. The data shown is representative of 2 independent experiments. (D) Latency reversing compounds were used to reactivate repressed HIV in MDMs exposed to P. gingivalis. After removing the bacteria and culturing MDMs in the absence of bacteria for 3 days, the cells were exposed to a panel of stimulating agents for 24 h and followed by analysis of luciferase expression (E): bryostatin (10 nM), trichostatin A (100 nM), SAHA (10 μM), and JQ1 (10 μM). Error bars represent the standard deviation of measurements done in triplicate. The data is representative of more than 3 experiments.
2.3. TLR4 mediates repression of HIV replication in MDMs exposed to P. gingivalis
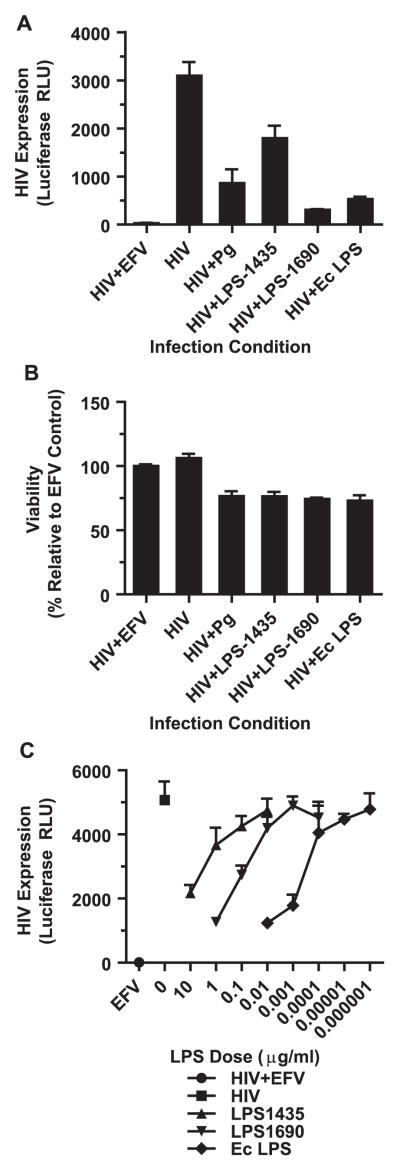
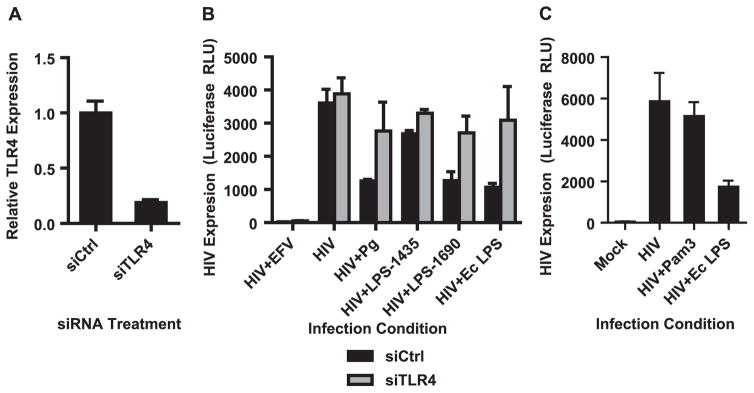
Since components of the bacterial surface are likely factors to interact with macrophages during HIV infection in our culture system, we investigated whether bacterial LPS is sufficient to induce the observed repression of HIV expression. P. gingivalis has two dominant forms of LPS based on the acylation status of Lipid A. LPS1435 is tetra-acylated while LPS1690 is penta-acylated (Darveau et al., 2004). LPS1435 and LPS1690 differentially repressed HIV expression, with LPS 1435 having a modest effect on HIV replication but LPS1690 inhibiting HIV replication by greater than 90% (Fig. 5A). This reduction in HIV expression could not be attributed to cell death in culture and it was dependent on the LPS dose (Fig. 5B and C). LPS derived from E. coli also significantly repressed HIV expression consistent with previous reports (Ahmed et al., 2010; Equils et al., 2006; Kornbluth et al., 1989; Simard et al., 2008). These results suggest that a common LPS-mediated pathway is responsible for the repression of HIV in macrophages. Both forms of the P. gingivalis LPS are recognized by TLR2 and TLR4, but LPS1435 has a stronger bias toward TLR2 whereas LPS1690 strongly activates TLR4 mediated NF-κB signaling (Darveau et al., 2004; Herath et al., 2013). Since both E. coli LPS and LPS1690 repressed HIV expression most robustly, while LPS1435 had a weaker repressive capacity, this implicates TLR4 in the recognition of P. gingivalis and its ability to repress HIV. To test the role of TLR4 in limiting HIV replication, siRNA was used to knock-down TLR4 in MDMs. We achieved approximately 80% decrease in TLR4 mRNA expression (Fig. 6A). This knockdown of TLR4 was sufficient to prevent P. gingivalis-mediated repression of HIV expression (Fig. 6B). This is consistent with the inability of Pam3CSK4, a TLR2 agonist, to repress HIV expression (Fig. 6C).
Fig. 5.
LPS from P. gingivalis is sufficient to repress HIV expression. (A) MDMs were infected with HIV-NL4-3ΔEnv-Luc(VSV-G) with or without P. gingivalis (MOI 30), purified P. gingivalis LPS1435 (1 μg/ml), purified P. gingivalis LPS1690 (1 μg/ml) or E. coli LPS (10 ng/ml) following the outline in Fig. 2A and monitored luciferase expression at 3 days post-infection. Cultures treated with 1 μM efavirenz (EFV) served as a negative control. Error bars represent the standard deviation of triplicate measurements. Data is representative of more than 3 individual experiments. (B) Cell viability was monitored using CellTiter-Glo and represents the viability relative to the EFV-treated cells. Graph was generated from a parallel infections as the experiment in (A). Error bars represent the standard deviation from triplicate measurements. The data is representative of 2 individual experiments. (C) MDMs were infected with HIV-NL4-3ΔEnv-Luc(VSV-G) with or without serial dilutions of P. gingivalis and E. coli LPS. Error bars represent the standard deviation of measurements done in triplicate. The data is representative of 2 independent experiments.
Fig. 6.
Knock-down of TLR4 rescues HIV expression. MDMs were transfected with siRNA against TLR4 or a non-targeting control. (A) Confirmed TLR4 down-regulation by RT-PCR 48 h post-transfection. Error bars represent the standard deviation of triplicate measurements. (B) Cells were infected with HIV-NL4-3ΔEnv-Luc(VSV-G) 72 h post-transfection under the conditions shown following the experiment outline in Fig. 1A and measured HIV expression by luciferase 3 days post-infection. Error bars represent the standard deviation of triplicate infections. (C) Engaging TLR2 with the agonist Pam3CSK4 (1 μg/ml) does not repress HIV expression to a similar extent as E. coli LPS (1 μg/ml). Error bars represent the standard deviation of infections done in triplicate and the data is representative of more than 3 independent experiments.
3. Discussion
Macrophages are sentinel cells involved in the first line of defense during the innate immune response to the microbial microenvironment. How these interactions with commensal and pathobiont organisms influence HIV infection and replication in macrophages has not been thoroughly explored. We report that the presence of the periodontal pathobiont P. gingivalis at the time of HIV infection of macrophages significantly represses viral replication. This repression is reversible, correlates with phenotypic changes of the macrophages and dependent on TLR4 signaling.
Our observation that HIV expression is suppressed by P. gingivalis LPS is consistent with published work using E. coli LPS and other bacteria (Ahmed et al., 2010; Bernstein et al., 1991; Kornbluth et al., 1989; Liu et al., 2006; Simard et al., 2008). E. coli LPS has been suggested to interfere with HIV replication at multiple steps of the HIV replication cycle in macrophages including interference of efficient viral entry (Franchin et al., 2000; Herbein et al., 1995; Herbein and Varin, 2010; Mikulak et al., 2009; Verani et al., 1997; Verani et al., 2002) and a restriction after entry but before proviral integration (Devadas et al., 2010; Donninelli et al., 2016; Pushkarsky et al., 2001; Schlaepfer et al., 2014; Wang et al., 2011). Our data suggests that a step post-proviral integration and transcription are targeted in MDMs when infected with HIV in the presence of P. gingivalis. Although the precise mechanism for this P. gingivalis mediated restriction has not been identified, one candidate is the anti-viral activity of interferons secreted in response to bacterial LPS (Ahmed et al., 2010; Kornbluth et al., 1989; Meylan et al., 1993; Mosoian et al., 2010; Simard et al., 2008). However, in our preliminary experiments P. gingivalis did not significantly induce type 1 interferons and HIV expression in the presence of P. gingivalis was not rescued by blocking INF-R1 with antibodies (data not shown). Additional work is required to validate these initial observations and determine the repressive mechanisms initiated by P. gingivalis. The similarities between the suppression of HIV expression by E. coli-derived LPS and P. gingivalis-derived LPS1690, reflect a common TLR4-dependent mechanism for the repression of HIV. This signaling through TLR4 is specific since engaging TLR2 with the agonist Pam3CSK4 was not capable of repressing HIV replication. This is also consistent with the mild repressive state induced by P. gingivalis LPS1435 which tends to signal more strongly through TLR2 rather than through TLR4 (Darveau et al., 2004; Herath et al., 2013). These results suggest that HIV repression is not a general property of Gram-negative bacteria.
TLR4 signaling occurs through two cascades: MyD88 and TRIF. Signaling through MyD88 promotes pro-inflammatory cytokine production (e.g. TNF-α and IL-β) whereas TRIF is upstream of type 1 IFN induction (Kawai and Akira, 2007; Mosoian et al., 2010). The outcome of HIV expression will depend on how these two signaling pathways are activated and integrated in different cell types, and may explain contradictory observations reported on the response of HIV expression to TLR4 ligands (Bergamaschi and Pancino, 2010; Equils et al., 2001, 2004, 2006; Kadoki et al., 2010; Lodie et al., 1998; Pomerantz et al., 1990). Macrophages in mucosal tissues encounter a combination of stimuli ranging from host-derived cytokines to various pathogen-derived TLR ligands. These stimuli lead to various outcomes in macrophage phenotype and behavior. For example, we found that macrophages exposed to P. gingivalis adopt a phenotype consistent with anti-inflammatory macrophages (Fig. 1). Therefore, TLR4 in specific contexts may protect against ectopic inflammation. Other receptors that maintain tissue macrophage anti-inflammatory functions have been shown to repress HIV transcription, such as the receptor tyrosine kinase RON (Cary et al., 2013; Klatt et al., 2008; Lee et al., 2004). We propose that the stimuli encountered by tissue macrophages will skew the behavior of these cells, which will directly influence the ability of the macrophages to support HIV infection, replication or persistence. This in turn will affect HIV pathogenesis and the resolution of opportunistic infections such as periodontitis in HIV-infected individuals.
One important observation is that P. gingivalis as well as E. coli LPS suppressed HIV replication in macrophages at a step after HIV entry and integration. This repression of HIV replication in macrophages was different than latency in T cells since it could not be reverse with a number of latency reversing compounds (Fig. 4E). This result is consistent with our observation that the P. gingivalis mediated block is post-transcriptional and this overrides efficient HIV transcription in P. gingivalis-treated macrophages. This has important implications for the development of “shock and kill” approaches to cure, since persistent HIV infection in macrophages induced by TLR signaling would not be susceptible to latency reversing compounds.
In conclusion, our studies suggest that exposure of MDM to bacterial products promotes HIV infection but inhibits replication through a TLR4-mediated mechanism. Since this repression is reversible and macrophages are long lived cells that are resistant to the cytopathic effects of HIV replication, this may provide a mechanism for macrophages to contribute to the viral load of different tissues upon anti-retroviral treatment interruption. These studies also provide insights into the complex interplay between macrophage function, pre-existing microbiota and HIV and their role in HIV infection and inflammation.
4. Materials and methods
4.1. Purification and differentiation of monocyte derived macrophages
Peripheral blood mononuclear cells (PBMC) were isolated from de-identified human leukapheresis-processed blood (New York Biologics, Inc) by centrifugation through Histopaque-1077 (Sigma Aldrich, Inc.) density gradient. Briefly, room temperature blood was diluted with an equal volume of room temperature PBS (Invitrogen). 35 ml of diluted blood was then layered over 15 ml of room temperature Histopaque-1077 and spun at 1500 rpm for 30 min at room temperature in a bench top centrifuge. PBMC layer was removed and washed 3 times with PBS. Excess red blood cells were lysed by resuspending the cell pellet in room temperature lysis buffer (155 mM NH4Cl, 12 mM NaHCO3, 0.1 mM EDTA in water) for 1–2 min. To purify and differentiate MDMs, PBMC were resuspended in RPMI (Invitrogen) without serum at a density of 5 × 106 cells/ml and plated 10 ml per 10 cm tissue culture dish. Cells were incubated at 37 °C for 1–2 h to allow binding of monocytes to the plates. The plates were then swirled to resuspend unbound cells and these cells were discarded. The bound cells were cultured in RPMI supplemented with 10% fetal bovine serum (FBS) (Corning, Inc.), 10% human AB serum (Gemini Bio-Products), 100U/ml of penicillin/streptomycin (Invitrogen) and 2 mM L-glutamine (Invitrogen). MDMs were allowed to differentiate for a week at 37 °C and spent medium containing unbound cells was removed and replaced with fresh medium every 2–3 days. To prepare differentiated MDMs for inoculations, the cells were washed twice with PBS to remove contaminating lymphocytes followed by treatment with Versene (Invitrogen) at 37 °C for 10–20 min. To lift the MDMs from the plates, we pipeted Versene directly onto the cells and gently scraped the cells that were still attached.
4.2. Viruses
Viruses were generated by transfecting HEK293T cells (cultured in DMEM supplemented with 10% FBS, 100U/ml of penicillin/streptomycin and 2 mM L-glutamine) by calcium-phosphate precipitation using plasmids encoding the following HIV-1 clones: pNL4-3. Luc. R-E- (NIH AIDS Reagents Program), NL4-3ΔEnv-GFP or pWT/BaL (NIH AIDS Reagents Program). Viruses defective in the envelope glycoprotein were pseudotyped with VSV-G. Viruses were typically concentrated by ultracentrifugation over a cushion of 10% sucrose solution in PBS at 19,000 rpm for 1.5 h at 4 °C.
4.3. Cultivation of P. gingivalis
P. gingivalis strain 381 was used in these studies and was cultured as previously described (Shaik-Dasthagirisaheb et al., 2014). In brief, bacteria were grown on anaerobic blood agar plates for 4–6 days, and placed into brain-heart infusion (BHI) broth for 24 h growth. Bacteria were harvested by centrifugation, washed, and suspended in PBS to OD660 of 1.0 (1 × 109 colony forming units (CFU)/ml). Bacterial culture purity was confirmed by Gram staining. Heat-killed P. gingivalis preparations were generated by incubating an aliquot at 60 °C for 30 min; plate counts were used to confirm killing.
4.4. HIV infection of MDMs
MDMs were plated at 30,000 cells per 100 μl of medium per well in 96-well plates the day prior to infection with HIV. Cells were typically infected overnight by adding 50 μl of virus at roughly an MOI of 1 based on TZMbl infectivity (NIH AIDS Reagents Program) with or without heat-killed P. gingivalis, purified LPS from P. gingivalis (LPS1435 or LPS1690 from Astarte Biologics) or purified E. coli LPS (Sigma). MDMs infected in the presence of 1 μM of the reverse transcriptase inhibitor efavirenz (NIH AIDS Reagents Program) served as negative controls. After overnight incubation, the wells were washed with 200 μl of PBS and added fresh medium with or without bacterial products. The cultures were incubated for another 3 days and harvested for HIV infection analysis. When longer incubation periods were necessary, half of the medium was replaced with fresh medium every 2–3days.
4.5. Luciferase expression and viability
For measuring HIV expression based on luciferase, the cells were lysed with 20 μl of 1x Luciferase Cell Culture Lysis Reagent (Promega) at room temperature for 20 min. The lysate was transferred to clear bottom, black-walled 96-well plates, added 20 μl of luciferase substrate (Promega) and read with a BioTek Synergy HT plate reader at 1 s per well. Cell viability was assessed in infection done in parallel in clear bottom black-walled 96-well plates. 3 days post-infection, the supernatant was removed from the wells, the wells were washed with PBS once and added 20 μl of fresh medium with 20 μl of CellTiter-Glo (Promega). The cells were incubated at 37 °C for 10 min followed by reading luminescence with a BioTek Synergy HT plate reader at 1 s per well.
4.6. Measuring HIV titers with ELISA
Culture supernatants were diluted in PBS supplemented with 20%FBS and 1% Triton-X and subsequently bound to a 96-well plate coated with anti-HIV immunoglobulin (AIDS Reagent Program catalog number 3957). Antibodies from the culture supernatant of hybridoma cells specific for HIV p24 were used as primary antibodies (AIDS Reagent Program catalog number 183-H12-5C). This was followed by a HRP coupled anti-mouse secondary antibody (Santa Cruz). Detection of color change of TMB SureBlue (KPL) solution was measured in a TECAN microplate reader at 450 nm, followed by quantification against a commercially available p24 standard (eEnzyme).
4.7. Flow cytometry
MDMs were cultured in a 24-well plate in the presence or absence of P. gingivalis at an MOI of 100 for 3 days. The cells were then treated directly in the culture plate with 100 μl of FACS buffer (PBS supplemented with 2 mM EDTA and 0.5% bovine serum albumin) supplemented with human serum at 10% for 10 min at room temperature to block Fc-receptors. The cells were then stained with anti-CD206 (clone 19.2, BD Biosciences), anti-CD14 (clone M5E2, BD Biosciences) or isotype controls (BD Biosciences), and incubated for 15 min at room temperature. The cells were then washed twice with FACS buffer, resuspended in FACS buffer supplemented with 1% paraformaldehyde and transferred to flow cytometry tubes for analysis with a FACSCalibur instrument (BD Biosciences).
4.8. Western blotting
Cells were lysed with RIPA buffer (Thermo Scientific). Proteins were separated on a 12% SDS-PAGE gel and transferred to a nitrocellulose membrane. CCR7 was detected with a monoclonal antibody (Abcam) and a secondary antibody coupled to a fluorophore using a Li-cor Odyssey system. The amount of protein for each lane was normalized to GAPDH (cell signaling).
4.9. Measuring HIV integration
Macrophages infected with HIV in vitro were treated with DNAse-I (50 μg/ml) in the presence of 10 mM of MgCl2 for 30 min to remove residual HIV plasmid DNA from the medium. The cells were then removed from the plates and the genomic DNA was purified using the DNeasy Blood and Tissue kit (Qiagen). Genomic DNA was eluted with 10 mM Tris-HCl (pH8.0) buffer. Prior to measuring HIV integration, the number of cells represented in the DNA samples was estimated by quantitative PCR amplification of human albumin as similarly done previously (Agosto et al., 2007). DNA from uninfected PBMC was used as a standard with the estimation that 10 μl of 24 μg/ml of DNA sample equals 34,800 cells. The reaction conditions were 10 μl of DNA sample diluted in 10 mM Tris-HCl buffer (pH 8.0) with 10 μl of master mixture at 2x concentration: 2x PCR buffer (40 mM KCl, 40 mM Tris-HCl (pH 8), 10 mM (NH4)2SO4), 11 mM MgCl2, 2.4 mM of dNTP mix (0.6 mM of each dATP, dCTP, dGTP and dTTP), 0.52 μM of forward primer 5′-GCTGTCATCTCTTGTGGGCTGT-3′ (Invitrogen), 0.52 μM of reverse primer 5′-AAACTCATGGGAGCTGCTGGTT-3′ (Invitrogen), 0.4 μM of probe 5′-FAM-CCTGTCATGCCCACACAAATCTCTCC-TAMRA-3′ (Integrated DNA Technologies) and 1.25U of Maxima Hot Start Taq Polymerase (Fisher Scientific). The reaction was run in a Quant-Studio 3 (Applied Biosystems) with the following program: hot-start at 95 °C for 2 min, followed by 50 cycles of denaturation for 15 s at 95 °C, annealing for 30 s at 60 °C, plate read and extension for 1 min at 72 °C.
HIV integration was measured by nested Alu-PCR as similarly described (Agosto et al., 2007). The DNA samples were diluted in 10 mM Tris-HCl (pH 8.0) and adjusted to a total DNA concentration of 2 μg/ml using uninfected PBMC DNA. This corresponds to roughly 3,000 cells per 10 μl of DNA sample. Samples were pre-amplified alongside an integration standard made from a polyclonal cell line carrying a single integrated copy per cell starting at a total DNA concentration of 3 μg/ml (Liszewski et al., 2009). The integration standard was serially diluted 5-fold in 10 mM Tris-HCl (pH 8.0) containing 2 μg/ml of uninfected PBMC DNA in order to maintain the total DNA concentration constant. The samples are pre-amplified with primers specific for cellular Alu and viral gag: genomic Alu forward 5′ GCC TCC CAA ACT GCT GGG ATT ACA G-3′ and HIV gag reverse 5′-GCT CTC GCA CCC ATC TCT CTC C-3′. The reaction conditions were 10 μl of DNA sample with 10 μl of master mixture at 2x concentration: 2x PCR buffer (40 mM KCl, 40 mM Tris-HCl (pH 8), 10 mM (NH4)2SO4), 11 mM MgCl2, 1.68 mM of dNTP mix (0.42 mM of each dATP, dCTP, dGTP and dTTP), 0.24 μM of Alu forward primer, 1.2 μM of gag reverse primer and 1.25U of Maxima Hot Start Taq Polymerase (Fisher Scientific). A parallel reaction without forward primer for Alu was done for each sample as a control for unintegrated HIV DNA. The reaction was conducted in a Biometra T-Professional thermocycler with the following program: hot start at 95 °C for 4 min and 20 cycles of 93 °C for 15 s, 50 °C for 15 s and 70 °C for 2.5 min 10 μl of the reaction products from samples and standard were transferred to another plate containing 10 μl of the following 2x master mixture specific for the RU5 region of the HIV LTR: 2x PCR buffer (40 mM KCl, 40 mM Tris-HCl (pH 8), 10 mM (NH4)2SO4), 11 mM MgCl2, 2.4 mM of dNTP mix (0.6 mM of each dATP, dCTP, dGTP and dTTP), 0.52 μM of forward 5′-GCC TCA ATA AAG CTT GCC TTG A-3′, 0.52 μM of reverse 5′-TCC ACA CTG ACT AAA AGG GTC TGA-3′ primers, 0.4 μM of probe 5′-FAM-CCAGAGTCACACAACAGACG-TAMRA-3′ (Integrated DNA Technologies) and 1.25U of Maxima Hot Start Taq Polymerase (Fisher Scientific). The transferred samples and standard were amplified alongside a pNL4-3 copy standard and integration standard that had not been pre-amplified. The PCR was conducted in a QuantStudio 3 (Applied Biosystems) with the same program as the albumin assay described above. The data was analyzed by plotting the total number of molecules calculated for each dilution of the not-pre-amplified integration standard versus the cycle threshold determined for each dilution of the pre-amplified integration standard in a natural log scale. The linear curve generated was used to calculate the total number of integration events per sample based on reaction cycle threshold. To calculate the number of integration events per cell, used the cell number pre-determined based on the albumin assay described above.
4.10. siRNA knock-down
ON-TARGETplus SMART-pool siRNA (GE Dharmacon) was used to knock-down human TLR4 in MDMs. The siRNA pool targets the following regions of TLR-4 transcripts: 5′-UGGUGGAAGUU-GAACGAAU-3′, 5′-GUUUAGAAGUCCAUCGUUU-3′, 5′-CAUUGAA-GAAUUCCGAUUA-3′, and 5′-GGAAAAUGGUGUAGCCGUU-3′. ON-TARGETplus Non-targeting Pool siRNA (GE Dharmacon) was used as the negative control. The control siRNA targets the following sequences: 5′-uggUUUACAUGUCGACUAA-3′, 5′-UGGUUUACAU-GUUGUGUGA-3′, 5′-UGGUUUACAUGUUUUCUGA-3′, and 5′-UG-GUUUACAUGUUUUCCUA-3′. For transfection of siRNA, MDMs were plated in 10 cm dishes at 3–7 × 106 cells per dish the night before transfection. The following morning, the medium was replaced with 10 ml of medium without antibiotics. In the afternoon, the cells were transfected using Lipofectamine 2000 as follows. 600 pmol of siRNA duplexes were diluted in 900 μl of OptiMEM (Invitrogen) in a tube. In a separate tube, added 60 μl of Lipofectamine 2000 (Invitrogen) to 900 μl of OptiMEM. Incubated both tubes at room temperature for 5 min. The diluted siRNA was added to the contents of tube 2 and mixed by pipeting gently 3 times. Incubated the transfection mixture at room temperature for 10 min. Added to cultured cells and swirled the plate to mix. After overnight incubation at 37 °C, the supernatant was removed and added fresh medium containing antibiotics. The cells were harvested 48 h later and plated for HIV infection the following day. A subset of the cells was lysed with TRIzol (Invitrogen) for RNA analysis of TLR4 expression.
4.11. RNA extractions
MDM pellets were resuspended in 1 ml of TRIzol, added 200 μl of chloroform and mixed by vortexing. The samples were then incubated at room temperature for 5 min and spun at 12,000g for 15 min at 4 °C. The top aqueous layer was transferred to a separate tube and mixed with 500 μl of isopropanol (AmericanBio, Inc.) and mixed gently by inverting the tube to precipitate RNA. The samples were incubated at room temperature for 5 min followed by centrifugation at 12,000g for 10 min at 4 °C. The supernatant was removed and the pellet was washed with 70% ethanol solution (AmericanBio, Inc.). The supernatant was removed and let the RNA pellet dry at room temperature. Once dried, the pellet was re-suspended in typically 30 μl of nuclease-free water (Ambion). The RNA concentration in the samples was determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific).
4.12. Generation of cDNA
500 ng of RNA was typically used to generate cDNA libraries from each sample. The RNA was mixed with 8.3 μg/ml of random primers (Promega) and 0.83 mM of dNTP mix (Thermo Fisher Scientific) in a final volume of 12 μl. Parallel aliquots for each sample were prepared which did not receive reverse transcriptase and were used as negative controls for genomic DNA contamination. The samples were then incubated at 65 °C for 5 min followed by a 10 min incubation at room temperature prior to adding 8 μl of the reverse transcriptase reaction mixture. The reverse transcriptase mixture comprised the following: 2.5x First Strand Buffer (Invitrogen), 0.025 M DTT (Invitrogen), 0.5U/μl of RNaseOUT (Invitrogen) and 12.5U/μl of SuperScript-II enzyme (Invitrogen) all in nuclease-free water. The same mixture without SuperScript-II enzyme was used for the genomic DNA control samples. The reaction was conducted in a Biometra T-Professional thermocycler with the following program: 42 °C for 50 min followed by 70 °C for 15 min. The cDNA was stored at −20 °C until needed.
4.13. Quantitative PCR analysis of cDNA
2 μl of cDNA samples were mixed with 10 μl of GoTaq master mixture (Promega) for PCR amplification and analysis. The master mixture was prepared as follows: 7.5 μl of GoTaq master mixture, 1.25 μl of each forward and reverse primers pre-diluted at 5 μM. The primer sets used were the following: actin-forward – 5′-TGGGACGACATGGAGAA-3′, actin-reverse – 5′-GGGTGTTGAAGGT CTCAAA-3′, HIV initiated transcription-forward – 5′-GGGTCTCT CTGGTTAGA-3′, HIV initiated transcription-reverse – 5′-AGAGCTCCCAGGCTCA-3′, HIV elongated transcription-forward – 5′ GGAGCCAGTAGATCCTAGAC-3′, HIV elongated transcription-reverse – 5′-CTTGGCAATGAAAGCAACAC-3′, TLR-4-forward – 5′-CCTCAGTCTGCTTGTAGTATC-3′, TLR-4-reverse – 5′-CATCCTGGCTT-GAGTAGATAAC-3′. The PCR program was the following: hot start for 15 min at 94 °C, followed by 45 cycles of 15 s at 94 °C, 30 s at 60 °C, 30 s at 72 °C and plate read. Relative HIV transcription and TLR-4 expression was calculated using the ΔΔCt method (Livak and Schmittgen, 2001).
Acknowledgments
This work was in part supported by the amfAR Mathilde Krim Fellowship in Basic Biomedical Research awarded to L.A. (award 109263-59-RKRL), NIH grants awarded to A.H. and F.G. (awards DE023950, AI097117, AI118682), Providence-Boston CFAR (P30-AI042853) and BU-CTSI/Department of Medicine pilot grant in support of Y.S.-D. We also thank Kingsley Ozongwu and Xavier de Luna for their contribution to the development of this project as well as the Boston University School of Medicine Flow Cytometry Core.
Contributor Information
Luis M. Agosto, Email: agosto@bu.edu.
Andrew J. Henderson, Email: andrew.henderson@bmc.org.
References
- Agosto LM, Yu JJ, Dai J, Kaletsky R, Monie D, O’Doherty U. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology. 2007;368:60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N, Hayashi T, Hasegawa A, Furukawa H, Okamura N, Chida T, Masuda T, Kannagi M. Suppression of human immunodeficiency virus type 1 replication in macrophages by commensal bacteria preferentially stimulating Toll-like receptor 4. J Gen Virol. 2010;91:2804–2813. doi: 10.1099/vir.0.022442-0. [DOI] [PubMed] [Google Scholar]
- Bergamaschi A, Pancino G. Host hindrance to HIV-1 replication in monocytes and macrophages. Retrovirology. 2010;7:31. doi: 10.1186/1742-4690-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein MS, Tong-Starksen SE, Locksley RM. Activation of human monocyte–derived macrophages with lipopolysaccharide decreases human immunodeficiency virus replication in vitro at the level of gene expression. J Clin Invest. 1991;88:540–545. doi: 10.1172/JCI115337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol. 2004;78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Zhang H, Lopez P, Pardo CA, Gartner S. In vitro modeling of the HIV-macrophage reservoir. J Leukoc Biol. 2006;80:1127–1135. doi: 10.1189/jlb.0206126. [DOI] [PubMed] [Google Scholar]
- Cary DC, Clements JE, Henderson AJ. RON receptor tyrosine kinase, a negative regulator of inflammation, is decreased during simian immunodeficiency virus-associated central nervous system disease. J Immunol. 2013;191:4280–4287. doi: 10.4049/jimmunol.1300797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassetta L, Kajaste-Rudnitski A, Coradin T, Saba E, Della Chiara G, Barbagallo M, Graziano F, Alfano M, Cassol E, Vicenzi E, Poli G. M1 polarization of human monocyte-derived macrophages restricts pre and postintegration steps of HIV-1 replication. AIDS. 2013;27:1847–1856. doi: 10.1097/QAD.0b013e328361d059. [DOI] [PubMed] [Google Scholar]
- Cassol E, Cassetta L, Rizzi C, Alfano M, Poli G. M1 and M2a polarization of human monocyte-derived macrophages inhibits HIV-1 replication by distinct mechanisms. J Immunol. 2009;182:6237–6246. doi: 10.4049/jimmunol.0803447. [DOI] [PubMed] [Google Scholar]
- Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Coleman CM, Wu L. HIV interactions with monocytes and dendritic cells: viral latency and reservoirs. Retrovirology. 2009;6:51. doi: 10.1186/1742-4690-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DL, Smith GL. Comparison of periodontal disease in HIV seropositive subjects and controls (II). Microbiology, immunology and predictors of disease progression. J Clin Periodontol. 1995;22:569–577. doi: 10.1111/j.1600-051x.1995.tb00806.x. [DOI] [PubMed] [Google Scholar]
- Cutler CW, Jotwani R. Oral mucosal expression of HIV-1 receptors, co-receptors, and alpha-defensins: tableau of resistance or susceptibility to HIV infection? Adv Dent Res. 2006;19:49–51. doi: 10.1177/154407370601900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, Howald WN, Way SS, Hajjar AM. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect Immun. 2004;72:5041–5051. doi: 10.1128/IAI.72.9.5041-5051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Dobrowolski C, Shahir AM, Feng Z, Yu X, Sha J, Bissada NF, Weinberg A, Karn J, Ye F. Short chain fatty acids potently induce latent HIV-1 in T-cells by activating P-TEFb and multiple histone modifications. Virology. 2015;474:65–81. doi: 10.1016/j.virol.2014.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devadas K, Hewlett IK, Dhawan S. Lipopolysaccharide suppresses HIV-1 replication in human monocytes by protein kinase C-dependent heme oxygenase-1 induction. J Leukoc Biol. 2010;87:915–924. doi: 10.1189/jlb.0307172. [DOI] [PubMed] [Google Scholar]
- Donninelli G, Gessani S, Del Corno M. Interplay between HIV-1 and Toll-like receptors in human myeloid cells: friend or foe in HIV-1 pathogenesis? J Leukoc Biol. 2016;99:97–105. doi: 10.1189/jlb.4VMR0415-160R. [DOI] [PubMed] [Google Scholar]
- Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91:914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- Equils O, Faure E, Thomas L, Bulut Y, Trushin S, Arditi M. Bacterial lipopolysaccharide activates HIV long terminal repeat through Toll-like receptor 4. J Immunol. 2001;166:2342–2347. doi: 10.4049/jimmunol.166.4.2342. [DOI] [PubMed] [Google Scholar]
- Equils O, Madak Z, Liu C, Michelsen KS, Bulut Y, Lu D. Rac1 and Toll-IL-1 receptor domain-containing adapter protein mediate Toll-like receptor 4 induction of HIV-long terminal repeat. J Immunol. 2004;172:7642–7646. doi: 10.4049/jimmunol.172.12.7642. [DOI] [PubMed] [Google Scholar]
- Equils O, Salehi KK, Cornateanu R, Lu D, Singh S, Whittaker K, Baldwin GC. Repeated lipopolysaccharide (LPS) exposure inhibits HIV replication in primary human macrophages. Microbes Infect. 2006;8:2469–2476. doi: 10.1016/j.micinf.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Franchin G, Zybarth G, Dai WW, Dubrovsky L, Reiling N, Schmidtmayerova H, Bukrinsky M, Sherry B. Lipopolysaccharide inhibits HIV-1 infection of monocyte- derived macrophages through direct and sustained down-regulation of CC chemokine receptor 5. J Immunol. 2000;164:2592–2601. doi: 10.4049/jimmunol.164.5.2592. [DOI] [PubMed] [Google Scholar]
- Gartner S, Markovits P, Markotitz DM, Kaplan MH, Gallo RC, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- Gonzalez OA, Li M, Ebersole JL, Huang CB. HIV-1 reactivation induced by the periodontal pathogens Fusobacterium nucleatum and Porphyromonas gingivalis involves Toll-like receptor 2 [corrected] and 9 activation in monocytes/macrophages. Clin Vaccin Immunol. 2010;17:1417–1427. doi: 10.1128/CVI.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herath TD, Darveau RP, Seneviratne CJ, Wang CY, Wang Y, Jin L. Tetra-and penta-acylated lipid A structures of Porphyromonas gingivalis LPS differentially activate TLR4-mediated NF-kappaB signal transduction cascade and immuno-inflammatory response in human gingival fibroblasts. PLoS One. 2013;8:e58496. doi: 10.1371/journal.pone.0058496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbein G, Doyle AG, Montaner LJ, Gordon S. Lipopolysaccharide (LPS) down-regulates CD4 expression in primary human macrophages through induction of endogenous tumour necrosis factor (TNF) and IL-1 beta. Clin Exp Immunol. 1995;102:430–437. doi: 10.1111/j.1365-2249.1995.tb03801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbein G, Varin A. The macrophage in HIV-1 infection: from activation to deactivation? Retrovirology. 2010;7:33. doi: 10.1186/1742-4690-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole J, Felton J, Brunsvold M, Kornman KS. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science. 1988;239:55–57. doi: 10.1126/science.3336774. [DOI] [PubMed] [Google Scholar]
- Imai K, Ochiai K, Okamoto T. Reactivation of latent HIV-1 infection by the periodontopathic bacterium Porphyromonas gingivalis involves histone modification. J Immunol. 2009;182:3688–3695. doi: 10.4049/jimmunol.0802906. [DOI] [PubMed] [Google Scholar]
- Kadoki M, Choi BI, Iwakura Y. The mechanism of LPS-induced HIV type I activation in transgenic mouse macrophages. Int Immunol. 2010;22:469–478. doi: 10.1093/intimm/dxq032. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Klatt A, Zhang Z, Kalantari P, Hankey PA, Gilmour DS, Henderson AJ. The receptor tyrosine kinase RON represses HIV-1 transcription by targeting RNA polymerase II processivity. J Immunol. 2008;180:1670–1677. doi: 10.4049/jimmunol.180.3.1670. [DOI] [PubMed] [Google Scholar]
- Koenig S, Gendelman HE, Orenstein JM, Dal Canto MC, Pezeshkpour GH, Yungbluth SS, Janotta F, Aksamit A, Martin MA, Fauci AS. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- Kornbluth RS, Oh PS, Munis JR, Cleveland PH, Richman DD. Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J Exp Med. 1989;169:1137–1151. doi: 10.1084/jem.169.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi Y, O’Brien WA, Zhao JQ, Golde DW, Gasson JC, Chen IS. Cytokines alter production of HIV-1 from primary mononuclear phagocytes. Science. 1988;241:1673–1675. doi: 10.1126/science.241.4873.1673. [DOI] [PubMed] [Google Scholar]
- Lee ES, Kalantari P, Tsutsui Section S, Klatt A, Holden J, Correll PH, Power Section C, Henderson AJ. RON receptor tyrosine kinase, a negative regulator of inflammation, inhibits HIV-1 transcription in monocytes/macrophages and is decreased in brain tissue from patients with AIDS. J Immunol. 2004;173:6864–6872. doi: 10.4049/jimmunol.173.11.6864. [DOI] [PubMed] [Google Scholar]
- Liszewski MK, Yu JJ, O’Doherty U. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods. 2009;47:254–260. doi: 10.1016/j.ymeth.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Mosoian A, Li-Yun Chang T, Zerhouni-Layachi B, Snyder A, Jarvis GA, Klotman ME. Gonococcal lipooligosaccharide suppresses HIV infection in human primary macrophages through induction of innate immunity. J Infect Dis. 2006;194:751–759. doi: 10.1086/506360. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lodie TA, Reiner M, Coniglio S, Viglianti G, Fenton MJ. Both PU. 1 and nuclear factor-kappa B mediate lipopolysaccharide- induced HIV-1 long terminal repeat transcription in macrophages. J Immunol. 1998;161:268–276. [PubMed] [Google Scholar]
- Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- Meylan PR, Guatelli JC, Munis JR, Richman DD, Kornbluth RS. Mechanisms for the inhibition of HIV replication by interferons-alpha, -beta, and -gamma in primary human macrophages. Virology. 1993;193:138–148. doi: 10.1006/viro.1993.1110. [DOI] [PubMed] [Google Scholar]
- Mikulak J, Gianolini M, Versmisse P, Pancino G, Lusso P, Verani A. Biological and physical characterization of the X4 HIV-1 suppressive factor secreted by LPS-stimulated human macrophages. Virology. 2009;390:37–44. doi: 10.1016/j.virol.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Mosoian A, Teixeira A, Burns CS, Sander LE, Gusella GL, He C, Blander JM, Klotman P, Klotman ME. Prothymosin-alpha inhibits HIV-1 via Toll-like receptor 4-mediated type I interferon induction. Proc Natl Acad Sci USA. 2010;107:10178–10183. doi: 10.1073/pnas.0914870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Cross GD, Callaway CS, McDougal JS. In vitro infection of human monocytes with human T lymphotropic virus type III/lymphadenopathy-associated virus (HTLV-III/LAV) J Immunol. 1986;137:323–329. [PubMed] [Google Scholar]
- Okada H, Murakami S. Cytokine expression in periodontal health and disease. Crit Rev Oral Biol Med. 1998;9:248–266. doi: 10.1177/10454411980090030101. [DOI] [PubMed] [Google Scholar]
- Ostrowski MA, Chun TW, Justement SJ, Motola I, Spinelli MA, Adelsberger J, Ehler LA, Mizell SB, Hallahan CW, Fauci AS. Both memory and CD45RA(+)/CD62L(+) naive CD4(+) T cells are infected in human immunodeficiency type 1-infected individuals. J Virol. 1999;73:6430–6435. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton LL, McKaig R, Strauss R, Rogers D, Eron JJ., Jr Changing prevalence of oral manifestations of human immunodeficiency virus in the era of protease inhibitor therapy. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2000;89:299–304. doi: 10.1016/s1079-2104(00)70092-8. [DOI] [PubMed] [Google Scholar]
- Pereira VT, Pavan P, Souza RC, Souto R, Vettore MV, Torres SR, Colombo AP, de Uzeda M, Sansone C, Goncalves LS. The association between detectable plasmatic human immunodeficiency virus (HIV) viral load and different subgingival microorganisms in Brazilian adults with HIV: a multilevel analysis. J Periodontol. 2014;85:697–705. doi: 10.1902/jop.2013.130273. [DOI] [PubMed] [Google Scholar]
- Pomerantz RJ, Feinberg MB, Trono D, Baltimore D. Lipopolysaccharide is a potent monocyte/macrophage-specific stimulator of human immunodeficiency virus type 1 expression. J Exp Med. 1990;172:253–261. doi: 10.1084/jem.172.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugliese A, Vidotto V, Beltramo T, Torre D. Phagocytic activity in human immunodeficiency virus type 1 infection. Clin Vaccin Immunol. 2005;12:889–895. doi: 10.1128/CDLI.12.8.889-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarsky T, Dubrovsky L, Bukrinsky M. Lipopolysaccharide stimulates HIV-1 entry and degradation in human macrophages. J Endotoxin Res. 2001;7:271–276. [PubMed] [Google Scholar]
- Salahuddin SZ, Rose RM, Groopman JE, Markham PD, Gallo RC. Human T lymphotropic virus type III infection of human alveolar macrophages. Blood. 1986;68:281–284. [PubMed] [Google Scholar]
- Schlaepfer E, Rochat MA, Duo L, Speck RF. Triggering TLR2, -3, -4, -5, and -8 reinforces the restrictive nature of M1- and M2-polarized macrophages to HIV. J Virol. 2014;88:9769–9781. doi: 10.1128/JVI.01053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Westhausen AM, Priepke F, Bergmann FJ, Reichart PA. Decline in the rate of oral opportunistic infections following introduction of highly active antiretroviral therapy. J Oral Pathol Med. 2000;29:336–341. doi: 10.1034/j.1600-0714.2000.290708.x. [DOI] [PubMed] [Google Scholar]
- Schuitemaker H, Kootstra NA, Fouchier RAM, Hooibrink B, Miedema F. Productive HIV-1 infection of macrophages restricted to the cell fraction with proliferative capacity. EMBO J. 1994;13:5929–5936. doi: 10.1002/j.1460-2075.1994.tb06938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuitemaker H, Kootstra NA, Koppelman MH, Bruisten SM, Huisman HG, Tersmette M, Miedema F. Proliferation-dependent HIV-1 infection of monocytes occurs during differentiation into macrophages. J Clin Investig. 1992;89:1154–1160. doi: 10.1172/JCI115697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik-Dasthagirisaheb YB, Huang N, Gibson FC., 3rd Inflammatory response to Porphyromonas gingivalis partially requires interferon regulatory factor (IRF) 3. Innate Immun. 2014;20:312–319. doi: 10.1177/1753425913492180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano RF, Greene WC. HIV latency. Cold Spring Harb Perspect Med. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard S, Maurais E, Gilbert C, Tremblay MJ. LPS reduces HIV-1 replication in primary human macrophages partly through an endogenous production of type I interferons. Clin Immunol. 2008;127:198–205. doi: 10.1016/j.clim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Slade GD, Beck JD. Plausibility of periodontal disease estimates from NHANES III. J Public Health Dent. 1999;59:67–72. doi: 10.1111/j.1752-7325.1999.tb03237.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Sonza S, Mutimer HP, O’Brien K, Ellery P, Howard JL, Axelrod JH, Deacon NJ, Crowe SM, Purcell DF. Selectively reduced tat mRNA heralds the decline in productive human immunodeficiency virus type 1 infection in monocyte-derived macrophages. J Virol. 2002;76:12611–12621. doi: 10.1128/JVI.76.24.12611-12621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tappuni AR, Fleming GJ. The effect of antiretroviral therapy on the prevalence of oral manifestations in HIV-infected patients: a UK study. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;92:623–628. doi: 10.1067/moe.2001.118902. [DOI] [PubMed] [Google Scholar]
- Tenenbaum H, Elkaim R, Cuisinier F, Dahan M, Zamanian P, Lang JM. Prevalence of six periodontal pathogens detected by DNA probe method in HIV vs non-HIV periodontitis. Oral Dis. 1997;3(Suppl 1):S153–S155. doi: 10.1111/j.1601-0825.1997.tb00350.x. [DOI] [PubMed] [Google Scholar]
- Varol C, Mildner A, Jung S. Macrophages: development and tissue specialization. Annu Rev Immunol. 2015;33:643–675. doi: 10.1146/annurev-immunol-032414-112220. [DOI] [PubMed] [Google Scholar]
- Verani A, Scarlatti G, Comar M, Tresoldi E, Polo S, Giacca M, Lusso P, Siccardi AG, Vercelli D. C-C chemokines released by lipopolysaccharide (LPS)-stimulated human macrophages suppress HIV-1 infection in both macrophages and T cells. J Exp Med. 1997;185:805–816. doi: 10.1084/jem.185.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verani A, Sironi F, Siccardi AG, Lusso P, Vercelli D. Inhibition of CXCR4-tropic HIV-1 infection by lipopolysaccharide: evidence of different mechanisms in macrophages and T lymphocytes. J Immunol. 2002;168:6388–6395. doi: 10.4049/jimmunol.168.12.6388. [DOI] [PubMed] [Google Scholar]
- Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chao W, Saini M, Potash MJ. A common path to innate immunity to HIV-1 induced by Toll-like receptor ligands in primary human macrophages. PLoS One. 2011;6:e24193. doi: 10.1371/journal.pone.0024193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J Clin Periodontol. 2000;27:722–732. doi: 10.1034/j.1600-051x.2000.027010722.x. [DOI] [PubMed] [Google Scholar]