Abstract
Background:
Candida infection is a major cause of morbidity and mortality in immunocompromised patients; an accurate and early identification is a prerequisite need to be taken as an effective measure for the management of patients. The purpose of this study was to compare the conventional identification of Candida species with identification by Vitek-2 system and the antifungal susceptibility testing (AST) by broth microdilution method with Vitek-2 AST system.
Materials and Methods:
A total of 172 Candida isolates were subjected for identification by the conventional methods, Vitek-2 system, restriction fragment length polymorphism, and random amplified polymorphic DNA analysis. AST was carried out as per the Clinical and Laboratory Standards Institute M27-A3 document and by Vitek-2 system.
Results:
Candida albicans (82.51%) was the most common Candida species followed by Candida tropicalis (6.29%), Candida krusei (4.89%), Candida parapsilosis (3.49%), and Candida glabrata (2.79%). With Vitek-2 system, of the 172 isolates, 155 Candida isolates were correctly identified, 13 were misidentified, and four were with low discrimination. Whereas with conventional methods, 171 Candida isolates were correctly identified and only a single isolate of C. albicans was misidentified as C. tropicalis. The average measurement of agreement between the Vitek-2 system and conventional methods was >94%. Most of the isolates were susceptible to fluconazole (88.95%) and amphotericin B (97.67%). The measurement of agreement between the methods of AST was >94% for fluconazole and >99% for amphotericin B, which was statistically significant (P < 0.01).
Conclusion:
The study confirmed the importance and reliability of conventional and molecular methods, and the acceptable agreements suggest Vitek-2 system an alternative method for speciation and sensitivity testing of Candida species infections.
Key words: Antifungal agents, antifungal resistance, Candida species, fungi, mycology, opportunistic infections
INTRODUCTION
Candida infections often associated with high morbidity and mortality have increased remarkably during the couple of decades.[1] The incidence of Candida infections is on the rise with the increase in number of immunocompromised patients due to excessive use of immunosuppressive drugs as well as the use of medical and surgical interventions.[2] Although Candida albicans is the most prevalent species,[3] an epidemiological shift in Candida pathogens has been recently noted by the increasing number of infections caused by nonalbicans Candida species (NAC).[1,3,4,5] The increased species diversity and incidence of infections have resulted in the need for an accurate and rapid identification of Candida isolates and have become important for proper patient management as various species respond differently to antifungals and for the prevention of emergence of drug resistance.[1,2,6]
Conventional methods of identification are time-consuming.[6,7] Commercially available biochemical and molecular methods, which allow identification within several hours, have been developed and evaluated.[8] The Biomerieux Vitek-2 system includes the Vitek-2 cards that allow species identification by comparison of the biochemical profile with an extensive database. The system also incorporates the antifungal susceptibility testing (AST) cards, which is designed for AST.[9] Recently, molecular genotyping methods have become more popular for epidemiological analysis.[10]
AST has been increasingly required in clinical practice. It is already well established that the outcome of invasive fungal infections could be improved by early initiation of appropriate antifungal agent based on the susceptibility profile of infecting Candida species.[11,12] Recently, Biomerieux Vitek-2 expanded its role in this area with a yeast susceptibility test that determines Candida growth spectrophotometrically using Vitek-2 microbiology systems, performing fully automated testing of susceptibility to flucytosine, amphotericin B, fluconazole, and voriconazole.[12]
The earliest possible identification and drug susceptibility profiling of Candida infections in immunocompromised patients allow for prompt optimization of antimicrobial therapy and diminished need for additional diagnostic studies, helping timely in saving the life of many patients. Hence, the objectives of this study were to compare the Vitek-2 yeast identification system with conventional and molecular methods of identification and Vitek-2 AST system with the broth microdilution method.
MATERIALS AND METHODS
The study was conducted at the Mycology Laboratory, Department of Microbiology, Maulana Azad Medical College and Associated Lok Nayak Hospitals, New Delhi, India, which is a 1500-bedded tertiary care hospital where patients came from all over the India. All the isolates of Candida spp. were recovered from various clinically available specimens, namely, oropharyngeal swab, blood culture, sputum, urine, cerebrospinal fluid, and stool.
The following strains were used as controls for the evaluation of various methods: C. albicans ATCC90028, Candida parapsilosis ATCC22019, Candida krusei ATCC6258, Candida glabrata ATCC90030, and Candida tropicalis ATCC750.
Identification and speciation of clinical isolates were done by conventional methods, Vitek-2 system, and the molecular methods. For the purpose of comparison, molecular methods were taken as gold standard method.
Identification by conventional methods
Identification and speciation of Candida isolates were done on the basis of germ tube production, morphology on corn meal agar with Tween 80 (Hi Media, India), HiCrome Candida agar morphology (Hi Media, India), carbohydrate fermentation, and assimilation tests using yeast nitrogen base agar (Difco, Becton Dickinson, India) as per the standard recommended procedures[13,14,15] and using the above control strains.
Identification by Vitek-2 system
Pure subcultures suspended in aqueous 0.45% (wt/vol) NaCl to achieve a turbidity equivalent to a McFarland 2.0 standard were measured on the DensiChek turbidity meter (Biomerieux, India), an instrument designed to measure the optical density of an organism suspension. The reading range of the DensiChek turbidity meter is 0.0-4.0 McFarland. The Vitek-2 instrument was automatically filled, sealed, and incubated by individual test cards with prepared culture suspension. Cards were held at 35.5°C for 18 h, with optical density readings taken automatically at every 15 min. Based on these readings, an identification profile was established and interpreted according to a specific algorithm.[16]
Identification by molecular methods
Molecular identification was performed by the Southern blot hybridization and random amplified polymorphic DNA (RAPD) analysis. Before genotyping, chromosomal DNA was isolated from each isolate using Xu et al. method.[17] After evaluating the quality of DNA on agarose gel, the DNA concentrations of each sample were measured and were subsequently subjected for further tests as described below:
DNA fingerprinting of the isolates by Southern blot hybridization
For DNA fingerprinting, around 2 μg of chromosomal DNA from each isolate was digested with restriction enzyme EcoR1. Digested DNA was separated on agarose gel (0.8%) in 1XTBE buffer (89 mM Tris borate, 1 mM ethylenediaminetetraacetic acid [EDTA]) by applying a voltage gradient of 2 V/cm for 20 h, stained with 0.5 μg/ml, visualized under ultraviolet (UV) light, and photographed. In the next step, separated DNA fragments were denatured in situ using alkali and neutralized with acid. The denatured DNA fragments were transferred to nylon membrane by capillary action. Transferred DNA fragments were then cross-linked to the membrane prehybridized in 300 mM phosphate buffer containing 7% sodium dodecyl sulfate (SDS) and 1 mM EDTA at 65°C for 2-4 h. In the next step, immobilized fragment of DNA was hybridized with32 P labeled C. albicans-specific probe CARE-2 at the same temperature for 16 h.[18] The nylon membrane containing hybridized DNA fragment was washed several times with 2 × SSC containing 0.1% SDS, dried and exposed to X-ray film at −80°C for 16-24 h, and developed.
Random amplified polymorphic DNA
For RAPD analysis, the DNA was purified by the method described by Makimura et al.[19] with slight modifications. Twenty random oligonucleotides (Sigma) were used as a primer for the PCR reaction. Different polymerase chain reaction (PCR) parameters were standardized to optimize the conditions for achieving better results. Finally, PCR was carried out with 50 ng DNA; 200 μM (each) dATP, dCTP, dTTP, and dGTP; 50 pmol oligonucleotides; 0.25 U Taq polymerase; and PCR buffer. The final volume of the reaction mixture was 30 μl. The cycling conditions were 94°C for 2 min, then thirty cycles of denaturation at 94°C for 1 min, thirty cycles of annealing at 42°C for 1 min, and extension at 72°C for 2 min. Final extension was given for 5 min at 72°C. Amplified products (30 μl) were resolved by agarose gel electrophoresis (1%) at 100 V for 1.5 h. The gel was stained with ethidium bromide, visualized under UV light, and photographed.
Antifungal susceptibility testing
Clinical and laboratory standards institute broth microdilution method
Susceptibility of Candida isolates to antifungal fluconazole and amphotericin B was done by the broth microdilution method as per the Clinical and Laboratory Standards Institute (CLSI) M27-A3 document using Roswell Park Memorial Institute medium and morpholinepropanesulfonic acid buffer.[20] The concentration range was tested between 0.125 and 128 μg/ml for fluconazole and 0.016-16 μg/ml for amphotericin B.[12] Minimum inhibitory concentration (MIC) was recorded as the lowest concentration of the drug that produced a visible decrease in turbidity compared to drug-free growth control according to the CLSI standards. The MIC breakpoints recommended by CLSI guidelines were followed. For fluconazole, MIC breakpoints were as follows: Susceptible, MIC ≤8 μg/ml; susceptible dose-dependent, MIC 16-32 μg/ml; and resistant, MIC ≥64 μg/ml. For amphotericin B, isolates with MICs of ≥1 μg/ml were categorized as resistant.[20]
Vitek-2 antifungal susceptibility testing method
Inoculum suspensions for Vitek-2 cards were obtained from the overnight cultures, with the turbidity being adjusted to a 1.8-2.2 McFarland standard according to the manufacturer's recommendations.[12] A standardized inoculum suspension was placed into a Vitek-2 cassette along with a sterile polystyrene test tube and an antifungal susceptibility test card for each organism. The loaded cassettes were placed into the Vitek-2 instrument, and the respective inoculum suspensions were diluted appropriately by the instrument, after which the cards were filled, incubated, and read automatically. The incubation time varied from 9.1 to 27.1 h based on the rate of growth in the drug-free control well.[21] In accordance with the M27-A3 document, the results from the 48 h reading were used. Complete data (from the CLSI and Vitek-2 methods) for each fungal isolate were recorded.
Statistical analyses
The reproducibility of AST was assessed by nonparametric correlation coefficient, and AST was considered reproducible if the correlation coefficient was P < 0.05. All statistical analyses were done with the Statistical Package for the Social Sciences (version 17.0; SPSS S.L., Madrid, Spain). All tests of statistical significance were two tailed.
RESULTS
With Vitek-2 ID system, 155 of 172 Candida isolates including C. albicans (n = 126), C. tropicalis (n = 12), C. krusei (n = 10), C. glabrata (n = 4), and C. parapsilosis (n = 3) were correctly identified. Eleven C. albicans were misidentified (6.39%) as C. famata (n = 6), C. tropicalis (n = 1), C. glabrata (n = 1), C. krusei (n = 1), G. capitatum (n = 1), and K. ohmeri (n = 1). One each isolate of C. tropicalis and C. parapsilosis was misidentified as K. ohmeri and C. famata, respectively. Four isolates (2.32%) such as C. albicans (n = 2), C. parapsilosis (n = 1), and C. tropicalis (n = 1) were identified with low discrimination [Table 1].
Table 1.
Comparison of Vitek-2 ID system with conventional and molecular methods of identification (n = 172)

On the other hand by conventional methods, 171 of 172 Candida isolates including C. albicans 138 (80.23%), C. tropicalis 14 (8.14%), C. krusei 10 (5.8%), C. parapsilosis 5 (2.9%), and C. glabrata 4 (2.3%) were correctly identified. Only one C. albicans was misidentified as C. tropicalis.
All the isolates tested by Vitek-2 ID system and conventional methods were subjected to DNA fingerprinting analysis by using a widely used C. albicans specific probe, the CARE-2 probe. Two low discriminated and 11 misidentified isolates by Vitek-2 ID system and one misidentified isolate by conventional methods were identified as C. albicans by restriction fragment length polymorphism (RFLP) analysis with CARE-2 probe hybridization [Figure 1]. However, NAC isolates did not show any fingerprinting pattern when probed with CARE-2.
Figure 1.

Restriction fragment length polymorphism patterns of Candida isolates, Lanes: M (1 kb ladder), Lane 01–06, 08, 09, and 11–15 are Candida albicans isolates and Lane 07, 10, and 16 are nonalbicans isolates (no hybridization or nontypical hybridization with CARE-2 probe)
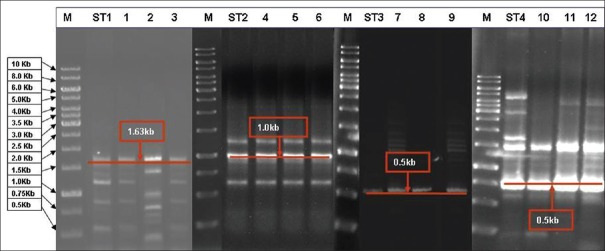
The isolates of NAC were subjected to RAPD analysis which produced Candida species-specific RAPD patterns distinct for individual ATCC standard strains. All the isolates tested to be NAC by conventional methods (except one C. albicans isolate which was misidentified as C. tropicalis) exhibited similar results by showing similar typical RAPD patterns to their respective ATCC strains [Figure 2]. Misidentified (n = 2) and low discriminated isolates (n = 2) with Vitek-2 ID system were identified as C. parapsilosis (n = 2) and C. tropicalis (n = 2) by RAPD patterns.
Figure 2.
Random amplified polymorphic DNA analysis of Candida isolates. Lane M 1 kb ladder; ST1, Candida parapsilosis ATCC; ST2, Candida krusei ATCC; ST3, Candida glabrata ATCC; and ST4, Candida tropicalis ATCC; Lane 1–3 are Candida parapsilosis; Lane 4–6 are Candida krusei; Lane 7–9 are Candida glabrata; and 10–12 are Candida tropicalis
Measurement of percentage agreement between the Vitek-2 ID identification system and conventional methods of identification was >94% for all Candida isolates. Measurement of percentage agreement between the Vitek-2 ID system and conventional methods by Kappa was 70% for C. albicans, 97.8% for C. tropicalis, 97% for C. krusei, 96.7% for C. glabrata, and 98% for C. parapsilosis.
In AST for fluconazole, Vitek-2 AST system showed that 92.4% isolates of the Candida species were susceptible. All the isolates of C. albicans were susceptible while among NAC, 66.6% isolates were susceptible and remaining 33.4% were resistant. While by the CLSI broth microdilution method, 88.95% of Candida species isolates were susceptible with 97.1% C. albicans and 54.5% NAC were susceptible for fluconazole. All the isolates of C. parapsilosis were found to be susceptible while all the C. krusei (100%) were resistant to fluconazole by both the methods [Table 2]. The measurement of percentage agreement between the Vitek-2 AST system and CLSI broth microdilution method by Kappa was 94% for fluconazole.
Table 2.
Antifungal susceptibility testing pattern of the Candida spp. isolates by the Vitek-2 system and Clinical Laboratory Standards Institute broth microdilution method (n = 172)
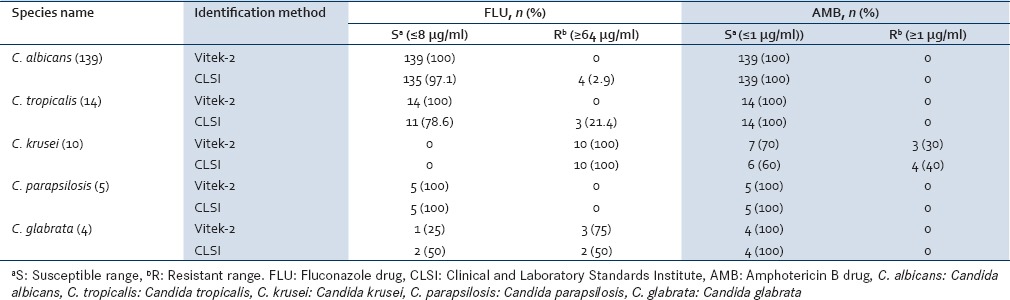
For amphotericin B drug, Vitek-2 AST system showed that 98.3% isolates of Candida species were susceptible while 1.7% were resistant. All the isolates of C. albicans (100%) and 90.9% of NAC were susceptible [Table 2]. While when tested by CLSI broth micro-dilution method, 97.7% isolates of Candida species were susceptible to amphotericin B. All the isolates of C. albicans (100%), C. tropicalis (100%), C. glabrata (100%), C. parapsilosis (100%), and 60% isolates of C. krusei were susceptible with remaining 40% isolates of C. krusei being resistant. However, using the Vitek-2 AST system, 70% isolates of C. krusei were found to be susceptible. Except C. krusei, all the other Candida species isolates were susceptible to amphotericin B by both methods. The MIC of the two quality control strains was within the range of expected values and showed reproducibility by both methods. For amphotericin B drug susceptibility testing, the measurement of percentage agreement between the Vitek-2 AST system and CLSI broth microdilution method by Kappa was 99%.
The correlation coefficient index (CCI) between Vitek-2 ID system and conventional methods of identification was 0.938, and it was statistically significant (P < 0.05). The CCIs values between the Vitek-2 AST system and the CLSI broth microdilution method for the antifungal agents (fluconazole and amphotericin B) were also highly significant [Table 3]. Correlation coefficient indices were expressed to a maximum value of 1. All the correlation coefficient indices were statistically significant (P < 0.05).
Table 3.
Nonparametric correlation coefficient indices between antifungal (fluconazole drug and amphotericin B drug) susceptibility testing by Vitek-2 system and Clinical Laboratory Standards Institute broth microdilution method

DISCUSSION
Candida species is an important cause of systemic mycosis in hospitalized patients, and morbidity and mortality worldwide, especially in critically ill patients.[22] Among Candida species, C. albicans, C. glabrata, C. tropicalis, C. parapsilosis, and C. krusei were the most common species encountered in routine clinical laboratory samples.[23] In our study, we have found C. albicans (80.8%) as the predominant species followed by C. tropicalis (8.13%), C. krusei (5.8%), C. parapsilosis (2.9%), and C. glabrata (2.3%), which is consistent with a previous study of Jha et al.,[24] in which the majority of Candida species were C. albicans (70%) followed by C. tropicalis (13.33%), C. krusei (10%), C. parapsilosis (3.33%), and C. stellatoidea (3.33%).[24] A study from South India by Kumari et al. in 2014 reported with overall predominance of NAC spp. and the predominant species identified was C. albicans.[25]
In this study, we compared the fully automated Vitek-2 ID system with conventional methods for identification of Candida species. Of 172 Candida isolates, Vitek-2 identified 155 (90.12%) Candida isolates correctly, 13 (7.56%) were misidentified, and 4 (2.32%) were identified with low discrimination. Massonet et al.[8] in their prospective study reported that Vitek-2 identified 41 (67.21%) Candida isolates correctly, 10 (16.39%) were not identified, 3 (4.91%) were misidentified, and 7 (11.47%) isolates were identified with low discrimination[8] whereas other studies reported that Vitek-2 system to correctly identify 98.5%[16] and 100%[26] of clinical isolates.
In our study with Vitek-2 ID system, most problems were encountered with the identification of C. albicans: 11 isolates were misidentified and two isolates were identified with low discrimination. Graf et al.[23] compared the results of the ID 32C system with Vitek-2 system, out of 241 Candida isolates, 222 (92.1%) were unequivocally identified to the species level by the Vitek-2 system, including 11 strains (4.6%) with low discrimination resolved by simple additional tests, 10 (4.1%) of which could not be definitely identified to the species level by additional tests. Four strains (1.7%) were misidentified and five strains (2.1%) could not be identified.[23]
In this study, the strains were simultaneously tested by the molecular techniques. Molecular techniques are excellent tools for identification and methods are highly reproducible, more discriminatory, high throughput, easy-to-use, digitally portable, and amenable to standardization.[27] These techniques have been used in a number of studies with Candida species.[27] An advantage of the method described here is the stable and easy-to-read RFLP patterns.[28] RAPD assay has become one of the most favorable choices for DNA fingerprinting of medically important Candida species.[29]
Baires-Varguez et al.[30] found that RAPD sensitivity for total isolates was 91% (84 of 92 isolates being correctly identified), reinforcing the previously described RAPD procedures for Candida species identification.[30] RAPD fingerprints generated from a single primer correctly identified the species of most (>98%) of the isolates identified with CHROMagar Candida plates as NAC.[31] However, there were certain limitations in this study; there was no isolate of cryptic species which is very difficult to distinguish with RAPD fingerprints.
In this study, we also evaluated the Vitek-2 AST system with the CLSI broth microdilution method for Candida species. A majority of Candida isolates were susceptible to both antifungal drugs tested by AST-YS06 Vitek-2 cards and the CLSI M27-A3 method. All the isolates of C. krusei (100%) were resistant to fluconazole drug by both the methods also seen by other workers,[32] emphasizing its intrinsic resistance toward azoles[33] and poor susceptibility to other antifungals, including amphotericin B.[34] In current clinical management practices, fluconazole is not recommended as a treatment option for C. Krusei infection[35] or susceptibility testing.[36]
The measurement of percentage agreement between the Vitek-2 AST system and CLSI broth microdilution method by Kappa was 94% for fluconazole, quite in concordance with a study by Bourgeois et al.,[36] where the agreements were 94.6%. Earlier in a study, the overall essential agreement between the Vitek-2 AST system and the broth microdilution MICs has been found to range from 97.9% with 24 h broth microdilution result compared to 93.7% with the 48 h bone mineral density result used as reference.[21] However, we found that for amphotericin B drug susceptibility testing, the measurement of percentage agreement between the Vitek-2 AST system and CLSI broth microdilution method by Kappa was 99%. While the overall essential agreement between the Vitek-2 AST system and the broth microdilution, MICs were found to range from 96.7% (voriconazole) to 99.1% (amphotericin B and flucytosine) with the 24 h broth microdilution result used as the reference in another study.[21]
The CCIs values between all the methods were statistically significant (P < 0.05). The CCIs values for the Vitek-2 AST system and the CLSI broth microdilution method were also statistically significant. However, the CCIs values for the Vitek-2 AST system were lower than those observed for the CLSI broth microdilution method; this may be because the ranges of antifungal agents in the Vitek-2 AST system do not match exactly with the ranges of the CLSI broth microdilution method.
CONCLUSION
The present study revealed that Vitek-2 system reduces the period required for identification and improves the rate of identification of Candida species isolates. However, conventional identification methods are still considered reference standard methods, in spite of time-consuming along with ambiguities results, and hence, suited better for research purposes. Therefore, Vitek-2 system appeared to be an alternative method for identification and AST for the Candida species to prescribe appropriate antifungal agents for the better management of opportunistic infection among immunosuppressed patients.
Financial support and sponsorship
Indian Council of Medical Research (ICMR), Government of India, New Delhi.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors would like to specially thank the Indian Council of Medical Research (ICMR), Government of India, New Delhi, for providing financial assistance to perform this work comfortably. The CARE-2 probe was a kind gift from B. A. Lasker.
REFERENCES
- 1.Costa AR, Silva F, Henriques M, Azeredo J, Oliveira R, Faustino A. Candida clinical species identification: Molecular and biochemical methods. Ann Microbiol. 2010;60:105–12. [Google Scholar]
- 2.Sood P, Mishra B, Dogra V, Mandal A. Comparison of Vitek Yeast Biochemical Card with conventional methods for speciation of Candida. Indian J Pathol Microbiol. 2000;43:143–5. [PubMed] [Google Scholar]
- 3.Fridkin SK, Jarvis WR. Epidemiology of nosocomial fungal infections. Clin Microbiol Rev. 1996;9:499–511. doi: 10.1128/cmr.9.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oberoi JK, Wattal C, Goel N, Raveendran R, Datta S, Prasad K. Non-albicans Candida species in blood stream infections in a tertiary care hospital at New Delhi, India. Indian J Med Res. 2012;136:997–1003. [PMC free article] [PubMed] [Google Scholar]
- 5.Deorukhkar SC, Saini S, Mathew S. Non-albicans Candida infection: An emerging threat. Interdiscip Perspect Infect Dis. 2014:7. doi: 10.1155/2014/615958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wadlin JK, Hanko G, Stewart R, Pape J, Nachamkin I. Comparison of three commercial systems for identification of yeasts commonly isolated in the clinical microbiology laboratory. J Clin Microbiol. 1999;37:1967–70. doi: 10.1128/jcm.37.6.1967-1970.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crist AE, Jr, Johnson LM, Burke PJ. Evaluation of the microbial identification system for identification of clinically isolated yeasts. J Clin Microbiol. 1996;34:2408–10. doi: 10.1128/jcm.34.10.2408-2410.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massonet C, Van Eldere J, Vaneechoutte M, De Baere T, Verhaegen J, Lagrou K. Comparison of VITEK 2 with ITS2-fragment length polymorphism analysis for identification of yeast species. J Clin Microbiol. 2004;42:2209–11. doi: 10.1128/JCM.42.5.2209-2211.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuenca-Estrella M, Gomez-Lopez A, Alastruey-Izquierdo A, Bernal-Martinez L, Cuesta I, Buitrago MJ, et al. Comparison of the Vitek 2 antifungal susceptibility system with the Clinical and Laboratory Standards Institute (CLSI) and European committee on antimicrobial susceptibility testing (EUCAST) broth microdilution reference methods and with the sensititre yeast one and Etest techniques for in vitro detection of antifungal resistance in yeast isolates. J Clin Microbiol. 2010;48:1782–6. doi: 10.1128/JCM.02316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marol S, Yücesoy M. Molecular epidemiology of Candida species isolated from clinical specimens of intensive care unit patients. Mycoses. 2008;51:40–9. doi: 10.1111/j.1439-0507.2007.01435.x. [DOI] [PubMed] [Google Scholar]
- 11.Morrell M, Fraser VJ, Kollef MH. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: A potential risk factor for hospital mortality. Antimicrob Agents Chemother. 2005;49:3640–5. doi: 10.1128/AAC.49.9.3640-3645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borghi E, Iatta R, Sciota R, Biassoni C, Cuna T, Montagna MT, et al. Comparative evaluation of the Vitek 2 yeast susceptibility test and CLSI broth microdilution reference method for testing antifungal susceptibility of invasive fungal isolates in Italy: The GISIA3 study. J Clin Microbiol. 2010;48:952–10. doi: 10.1128/JCM.00952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore GS, Jaciow DM. Mycology for the Clinical Laboratory. Reston, VA: Prentice-Hall; 1979. [Google Scholar]
- 14.Koneman EW, Allen SD, Janda WM, Schreckenberger PC. Color Atlas and Textbook of Diagnostic Microbiology. 5th ed. Philadelphia, PA: Lippincott Williams and Wilkins; 1997. Mycology; pp. 983–1057. [Google Scholar]
- 15.Forbes BA, Sahm DF, Weissfeld AS. Bailey and Scott's Diagnostic Microbiology. 11th ed. St. Louis: Mosby; 2002. Laboratory methods in basic mycology; pp. 711–98. [Google Scholar]
- 16.Hata DJ, Hall L, Fothergill AW, Larone DH, Wengenack NL. Multicenter evaluation of the new VITEK 2 advanced colorimetric yeast identification card. J Clin Microbiol. 2007;45:1087–92. doi: 10.1128/JCM.01754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Ramos AR, Vilgalys R, Mitchell TG. Clonal and spontaneous origins of fluconazole resistance in Candida albicans. J Clin Microbiol. 2000;38:1214–20. doi: 10.1128/jcm.38.3.1214-1220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasker BA, Page LS, Lott TJ, Kobayashi GS. Isolation, characterization, and sequencing of Candida albicans repetitive element 2. Gene. 1992;116:51–7. doi: 10.1016/0378-1119(92)90628-3. [DOI] [PubMed] [Google Scholar]
- 19.Makimura K, Murayama SY, Yamaguchi H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J Med Microbiol. 1994;40:358–64. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 20.Reference Method for Broth Microdilution Antifungal Susceptibility Testing of Yeasts; Approved Standard. CLSI Document M27-A3. 3rd ed. Vol. 28. Wayne, Pennsylvania, USA: Clinical and Laboratory Standards Institute (CLSI); 2008. Clinical and Laboratory Standards Institute. [Google Scholar]
- 21.Pfaller MA, Diekema DJ, Procop GW, Rinaldi MG. Multicenter comparison of the Vitek 2 antifungal susceptibility test with the CLSI broth microdilution reference method for testing amphotericin B, flucytosine, and voriconazole against Candida species. J Clin Microbiol. 2007;45:3522–8. doi: 10.1128/JCM.00403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sardi JC, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJ. Candida species: Current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol. 2013;62(Pt 1):10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 23.Graf B, Adam T, Zill E, Göbel UB. Evaluation of the VITEK 2 system for rapid identification of yeasts and yeast-like organisms. J Clin Microbiol. 2000;38:1782–5. doi: 10.1128/jcm.38.5.1782-1785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jha BJ, Dey S, Tamang MD, Joshy ME, Shivananda PG, Brahmadatan KN. Characterization of Candida species isolated from cases of lower respiratory tract infection. Kathmandu Univ Med J (KUMJ) 2006;4:290–4. [PubMed] [Google Scholar]
- 25.Kumari KS, Raghunath P, Harshavardhan B, Chaudhury A. Distribution of Candida albicans and the non-albicans Candida species in different clinical specimens from South India. Int J Microbiol Res. 2014;5:1–5. [Google Scholar]
- 26.Melhem MS, Bertoletti A, Lucca HR, Silva RB, Meneghin FA, Szeszs MW. Use of the VITEK 2 system to identify and test the antifungal susceptibility of clinically relevant yeast species. Braz J Microbiol. 2014;44:1257–66. doi: 10.1590/S1517-83822014005000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soll DR. The ins and outs of DNA fingerprinting the infectious fungi. Clin Microbiol Rev. 2000;13:332–70. doi: 10.1128/cmr.13.2.332-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge YP, Wang L, Lu GX, Shen YN, Liu WD. A simple and reliable PCR-restriction fragment length polymorphism assay to identify Candida albicans and its closely related Candida dubliniensis. Braz J Microbiol. 2012;43:873–9. doi: 10.1590/S1517-83822012000300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metzgar D, Belkum AV, Field D, Haubrich R, Wills C. Random amplification of polymorphic DNA and microsatellite genotyping of pre- and posttreatment isolates of Candida spp. from human immunodeficiency virus-infected patients on different fluconazole regimens. J Clin Microbiol. 1998;36:2308–13. doi: 10.1128/jcm.36.8.2308-2313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baires-Varguez L, Cruz-García A, Villa-Tanaka L, Sánchez-García S, Gaitán-Cepeda LA, Sánchez-Vargas LO, et al. Comparison of a randomly amplified polymorphic DNA (RAPD) analysis and ATB ID 32C system for identification of clinical isolates of different Candida species. Rev Iberoam Micol. 2007;24:148–51. doi: 10.1016/s1130-1406(07)70031-1. [DOI] [PubMed] [Google Scholar]
- 31.Steffan P, Vazquez JA, Boikov D, Xu C, Sobel JD, Akins RA. Identification of Candida species by randomly amplified polymorphic DNA fingerprinting of colony lysates. J Clin Microbiol. 1997;35:2031–9. doi: 10.1128/jcm.35.8.2031-2039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pahwa N, Kumar R, Nirkhiwale S, Bandi A. Species distribution and drug susceptibility of Candida in clinical isolates from a tertiary care centre at Indore. Indian J Med Microbiol. 2014;32:44–8. doi: 10.4103/0255-0857.124300. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Xiao M, Watts MR, Wang H, Fan X, Kong F, et al. Development of fluconazole resistance in a series of Candida parapsilosis isolates from a persistent candidemia patient with prolonged antifungal therapy. BMC Infect Dis. 2015;15:340. doi: 10.1186/s12879-015-1086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernst JF, Schmidt A. Dimorphism in Human Pathogenic and Apathogenic Yeasts. 1st ed. Basel, Switzerland: Karger, S, Inc.; 2000. [Google Scholar]
- 35.Pappas PG, Kauffman CA, Andes D, Benjamin DK, Jr, Calandra TF, Edwards JE, Jr, et al. Clinical practice guidelines for the management of candidiasis: 2009 update by the infectious diseases society of America. Clin Infect Dis. 2009;48:503–35. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourgeois N, Dehandschoewercker L, Bertout S, Bousquet PJ, Rispail P, Lachaud L. Antifungal susceptibility of 205 Candida spp. isolated primarily during invasive Candidiasis and comparison of the Vitek 2 system with the CLSI broth microdilution and Etest methods. J Clin Microbiol. 2010;48:154–61. doi: 10.1128/JCM.01096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]