Abstract
Background:
Infection by Pseudomonas aeruginosa is common in the Intensive Care Unit (ICU), leading to increased morbidity and mortality. The organism is classified into various phenotypes based on the drug resistance pattern, namely, drug-resistant (DR), multi-DR (MDR), extensively DR (XDR), and pan-DR (PDR). We aim to study the incidence of P. aeruginosa phenotypes in a tertiary level ICU.
Materials and Methods:
We conducted this prospective, observational study for 2 years (January 2014-December 2015) and collected appropriate clinical samples (blood, urine, wound discharge, etc.,) from all the patients admitted to ICU. We excluded patients with known septicemia and P. aeruginosa infection. Group 1 comprised a total 1915 patient samples and Group 2 comprised 100 active surveillance samples, collected from the medical staff and the hospital environment. The data were analyzed using appropriate statistical methods, and a P < 0.05 was considered statistically significant.
Results:
We isolated 597 pathogenic bacteria out of 1915 specimens, giving a culture positivity rate of 31.2%. Klebsiella (43%), Acinetobacter (22%), and P. aeruginosa (15%) were the top three isolated bacteria. None of the surveillance samples grew P. aeruginosa. Antibiotic resistance studies revealed that 47.7% of P. aeruginosa isolates were DR, 50% were MDR, and 2.3% were XDR phenotype. None of the strains showed PDR phenotype.
Conclusion:
Our data revealed a high prevalence of DR phenotypes of P. aeruginosa in the ICU. Judicious use of antibiotics and strict infection control measures are essential to reduce the prevalence of drug resistance.
Key words: Drug resistance, multidrug-resistant phenotype, Intensive Care Unit, Pseudomonas aeruginosa
INTRODUCTION
Pseudomonas aeruginosa is a common Gram-negative rod-shaped bacterium that leads to significant morbidity and mortality in humans. P. aeruginosa is a prototype of “multidrug-resistant pathogen” and is recognized for its ubiquitous distribution, advanced antibiotic resistance mechanisms, and nosocomial infections.[1] P. aeruginosa has the ability to survive on minimal nutrition and tolerate a variety of physical insults leading to its persistence in the community and hospital settings. Despite the wide distribution of P. aeruginosa in nature and the potential for community-acquired infections, serious infections with this bacterium are predominantly hospital-acquired infections. P. aeruginosa was identified as the fifth most frequently isolated nosocomial pathogen by the surveillance data from the United States.[2] P. aeruginosa is one of the major causes of hospital-acquired infections and is responsible for about 10–20% of nosocomial infections in patients admitted in the Intensive Care Units (ICUs).[3] P. aeruginosa can be isolated from a wide variety of sources in the hospital, including respiratory equipment, antiseptics, soaps, sinks, mops, medicines, and physiotherapy tools.[4]
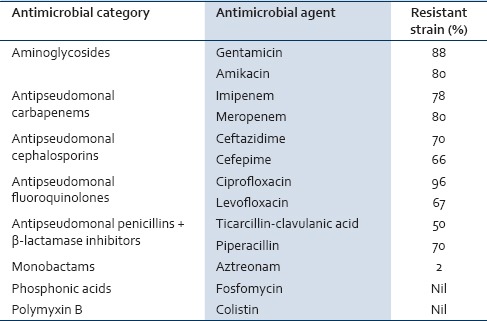
P. aeruginosa is divided into different phenotypes based on the drug resistance patterns of the organism.[5] Multidrug-resistant (MDR) phenotype is defined as P. aeruginosa, which is resistant to more than one antimicrobial agent in three or more antimicrobial categories. A similar resistance to more than one antimicrobial agent in <3 antimicrobial categories is defined as drug-resistant (DR) P. aeruginosa. The details of the antimicrobial agents and categories used to define the phenotypes are given in Table 1. Extensively DR (XDR) phenotype is defined as P. aeruginosa, which is resistant to more than one antimicrobial agent in all the antimicrobial categories, except in two or less. Pan-DR (PDR) phenotype is defined as a bacterium which is resistant to all antimicrobial agents in all antimicrobial categories.[6]
Table 1.
Antimicrobial categories and agents used to define the Pseudomonas aeruginosa phenotypes along with percentage of resistant strains
MDR, XDR, and PDR phenotypes elaborate inactivating enzymes, such as extended-spectrum beta-lactamases (ESBL) and metallo-β-lactamases (MBL), that make beta-lactams and carbapenems ineffective.[7] ESBL-producing P. aeruginosa was initially detected in Europe in the mid-1980s, and MBL-producing P. aeruginosa was first reported from Japan in 1991. They have rapidly spread over different parts of the world since then.[8] Literature is sparse regarding the incidence of different phenotypes of P. aeruginosa from our country as we could retrieve only two articles in the PubMed with the relevant (P. aeruginosa, India, phenotype, incidence) keywords.[9,10] Hence, we conducted this study to find the incidence of MDR, XDR, and PDR phenotypes of P. aeruginosa in the ICU of a tertiary care hospital.
MATERIALS AND METHODS
Hospital setting
This prospective, observational study was conducted at the Department of Microbiology of a tertiary level hospital for over a period of 2 years from January 2014 to December 2015. We included all patients who were at least 10 years of age at the time of the admission to the ICU of this hospital. As per our hospital policy, patients below 10 years of age requiring intensive are admitted to the pediatric ICU. Our study does not include data from the pediatric ICU, thereby resulting in the exclusion of patients below 10 years of age. The patient with evidence of septicemia and known diagnosis of P. aeruginosa infection at the time of admission was excluded from the study.
Sample collection and processing
Routine hematology and biochemistry tests were conducted as per the request of the clinical team. Microbiological specimens were collected from the blood, urine, and sputum samples of the patients. Other samples were collected based on the clinical suspicion from a site harboring infection, such as throat swab, wound discharge, and body fluid culture. Samples obtained after an invasive procedure and from the indwelling catheters were also included in the study. A total of 1915 patient samples were collected and are grouped as Group 1. A total of 100 environmental samples were collected as part of active surveillance of the hospital environment and staff, which were grouped as Group 2. These samples were collected from the medical staff (hand, throat, and nasal swabs) and the hospital environment (air, surgical instruments, dressing material, floors, door, walls, beds, sinks, and antiseptic solutions).
All blood culture and fluid samples with a minimum volume of 3 ml were collected in BacT/Alert® culture bottles. The BacT/Alert system is a continuously monitored blood culture system for detecting bacteremia and fungemia.[11] Bottles flagged as positive by the BacT/Alert system were subcultured and interpreted according to the standard laboratory protocols.
Samples other than the blood and body fluids of Group 1 and all Group 2 were cultured on MacConkey agar and blood agar. These plates were incubated overnight at 37°C and all the bacteria grown in these culture media were processed for identification and antibiotic susceptibility testing using automated bacterial system VITEK 2 system (BioMérieux). VITEK 2 system uses a new fluorescence-based technology used for the identification and susceptibility testing of bacteria and is compliant with the Clinical and Laboratory Standard Institute (CLSI) guidelines 2014.[12]
Microbial identification and antibiotic susceptibility testing
P. aeruginosa was identified by its colony characteristics, pigment production, grape-like odor, oxidase positivity, motility, Gram-negative character of the bacilli, ability to decarboxylate arginine and to grow at 42°C.[13] P. aeruginosa colonies were further processed and confirmed by VITEK 2 system. Antibiotic sensitivity patterns of these isolates were studied by minimal inhibitory concentration using the VITEK 2 system as per the CLSI 2014 guidelines.[12] We tested the resistance patterns to all the antibiotics as shown in Table 1. Strains which had the same types of resistance patterns (antibiotype) were considered to be from the same clone. P. aeruginosa ATCC 27853 strain was used for quality control in this study.
RESULTS
The mean age of the patients who were included in the study was 36.2 years. A total of 597 pathogenic bacteria were isolated from the 1915 clinical specimens, giving a culture positivity rate of 31.2% for the clinical specimen. The details of all the bacterial species isolated from the study samples are shown in Figure 1. There were 88 isolates of P. aeruginosa, giving a prevalence level of 14.7%. Bacteria more prevalent than P. aeruginosa were Klebsiella spp. (42.8%) and Acinetobacter spp. (22.3%). Other bacteria such as coagulase-negative staphylococci, Staphylococcus aureus, Escherichia coli, and Proteus spp. accounted for the rest of the bacterial cultures. Two of the hospital environment samples grew S. aureus. Hospital staff and Group 2 samples did not grow any pathogenic bacterium, including P. aeruginosa.
Figure 1.
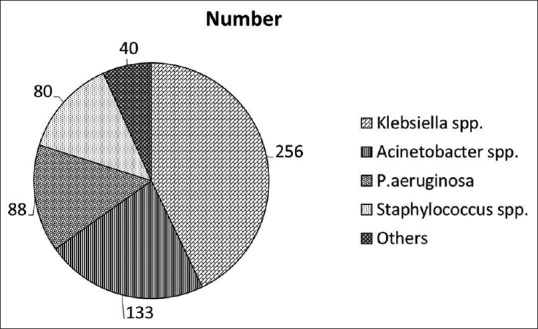
Pattern of microbes isolated in the study sample
The incidence rate of P. aeruginosa was highest (78%) in the age group of patients between 20 and 50 years. Urine (47 out of 88) and wound (38 out of 88) samples accounted for the majority of the positive isolates. The prevalence of P. aeruginosa phenotypes based on the resistance patterns is shown in Figure 2. Our data showed that 47.7% strains were DR, 50% were MDR, and 2.3% were of XDR phenotype. No strains were resistant to all antibiotics in all antimicrobial categories as all the strains were found to be sensitive to fosfomycin and colistin (100%). The percentage of resistant strains to each antibiotic is shown in Table 1.
Figure 2.
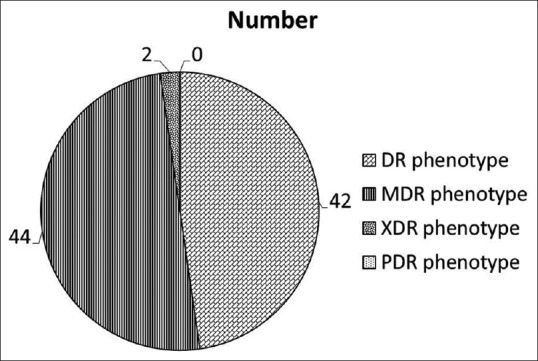
Prevalence of Pseudomonas aeruginosa drug-resistant strains
DISCUSSION
Our study showed the prevalence of drug resistance in almost 98% of the isolates of P. aeruginosa in the ICU. The only solace is the findings of the 0% presence of PDR phenotype, thereby showing the efficacy of certain antibiotics against this organism. Piperacillin resistance was found to be 70% in our study, which is comparable to a report showing it to be 73%.[14] Emergence of resistance to beta-lactams in P. aeruginosa has become a serious threat, particularly against the third- and fourth-generation cephalosporins. There are a lot of molecular mechanisms involved for resistance against these antibiotics, generation of ESBL enzyme, incorporation of bla genes in integrons, and inability of porin genes to enhance their expression level and/or alteration of antibiotic target sites.[15] Ceftazidime and cefepime are the prescribed antipseudomonal third- and fourth-generation cephalosporins, respectively. The resistance to ceftazidime and cefepime observed in our study was similar to that of a previous report from India.[14]
Our study showed that 80% of P. aeruginosa were resistant to carbapenem antibiotics such as imipenem and meropenem. Previous studies showed a similar trend from other countries and a few authors had shown 100% resistance against carbapenems.[16,17] This suggests the declining efficacy of the carbapenems as an effective antibiotic in the management of nosocomial infections. Fluoroquinolone compounds are one of the important antimicrobial agents that have been used for a variety of infections. New generation fluoroquinolones are beneficial against Gram-negative and Gram-positive bacteria, whereas older fluoroquinolones were effective against aerobic Gram-negative bacteria only.[18] Our data showed 96% resistance against ciprofloxacin and 67% resistance against levofloxacin while 100% resistance against ciprofloxacin was demonstrated earlier.[19]
Aminoglycosides are broad-spectrum antibiotics with a peculiar structure of an aminocyclitol ring. They are very effective against aerobic and facultative aerobic Gram-negative bacteria.[20] They mainly act by inhibiting protein synthesis and break cell membrane. Our data showed similar resistance pattern between amikacin and gentamicin, which was similar to that of the published literature.[21] Resistance to monobactam was seen in only 2% of the P. aeruginosa strains. Similar data were not available for comparison with other authors about the sensitivity to aztreonam. All the strains in our study were sensitive to fosfomycin and colistin. Similar data were reported from other countries where there is a high prevalence of P. aeruginosa infection.[22]
To the best of our knowledge, this is the first study from India that defines the prevalence of MDR, XDR, and PDR P. aeruginosa infection among the ICU patients. The strengths of our study include long duration of observation and the use of highly advanced automated bacteria identification system in the identification of P. aeruginosa. The limitations of our study include data derived from a single hospital may not be applicable to other geographic locations and inability to check resistance pattern to all the antipseudomonal drugs.
CONCLUSION
MDR P. aeruginosa is responsible for serious nosocomial infections among the patients admitted in the ICUs of the hospital. Epidemiological studies and strict laws regarding antibiotic policies should be constructed to limit the unnecessary use of antibiotics so that spread of multidrug resistance can be avoided.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Pollack M. Pseudomonas aeruginosa. In: Mandell GL, Dolan R, Bennett JE, editors. Principles and Practices of Infectious Diseases. New York: Churchill Livingstone; 1995. pp. 1820–2003. [Google Scholar]
- 2.National Nosocomial Infections Surveillance (NNIS) System report, data summary from October 1986-April 1998, issued June 1998. Am J Infect Control. 1998;26:522–33. doi: 10.1016/s0196-6553(98)70026-4. [DOI] [PubMed] [Google Scholar]
- 3.Nicastri E, Petrosillo N, Martini L, Larosa M, Gesu GP, Ippolito G. INF-NOS Study Group. Prevalence of nosocomial infections in 15 Italian hospitals: First point prevalance study for the INF-NOS project. Infection. 2003;31(Suppl 2):10–5. [PubMed] [Google Scholar]
- 4.Vincent JL, Bihari DJ, Suter PM, Bruining HA, White J, Nicolas-Chanoin MH, et al. The prevalence of nosocomial infection in intensive care units in Europe. Results of the European Prevalence of Infection in Intensive Care (EPIC) study. EPIC International Advisory Committee. JAMA. 1995;274:639–44. [PubMed] [Google Scholar]
- 5.Carmeli Y, Troillet N, Eliopoulos GM, Samore MH. Emergence of antibiotic-resistant Pseudomonas aeruginosa: Comparison of risks associated with different antipseudomonal agents. Antimicrob Agents Chemother. 1999;43:1379–82. doi: 10.1128/aac.43.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magiorakos AP. Multidrug Resistant (MDR), Extensively Drug Resistant (XDR) and Pandrug-1 Resistant (PDR) Bacteria in Healthcare Settings. Expert Proposal for a Standardized International Terminology. 2011 [Google Scholar]
- 7.Chaudhary U, Aggarwal R. Extended spectrum-lactamases (ESBL) - An emerging threat to clinical therapeutics. Indian J Med Microbiol. 2004;22:75–80. [PubMed] [Google Scholar]
- 8.Thuong M, Arvaniti K, Ruimy R, de la Salmonière P, Scanvic-Hameg A, Lucet JC, et al. Epidemiology of Pseudomonas aeruginosa and risk factors for carriage acquisition in an intensive care unit. J Hosp Infect. 2003;53:274–82. doi: 10.1053/jhin.2002.1370. [DOI] [PubMed] [Google Scholar]
- 9.Shalini S, Kranthi K, Gopalkrishna Bhat K. Microbiological profile of infections in ICU. J Clin Diagn Res. 2010;4:3109–12. [Google Scholar]
- 10.Mukhopadhyay C, Bhargava A, Ayyagari A. Role of mechanical ventilation and development of multidrug resistant organisms in hospital acquired pneumonia. Indian J Med Res. 2003;118:229–35. [PubMed] [Google Scholar]
- 11.Thorpe TC, Wilson ML, Turner JE, DiGuiseppi JL, Willert M, Mirrett S, et al. BacT/Alert: An automated colorimetric microbial detection system. J Clin Microbiol. 1990;28:1608–12. doi: 10.1128/jcm.28.7.1608-1612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Performance Standards for Antimicrobial Susceptibility Testing; 24th Informational Supplement. CLSI M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute; 2014. Multidrug Resistant (MDR), Extensively Drug Resistant (XDR) and Pandrug.1 Resistant (PDR) Bacteria in Healthcare Settings. [Google Scholar]
- 13.Collee JG, Duguid JP, Fraser AG, Marmion BP, Simmons A. Laboratory strategy in the diagnosis of infective syndromes. In: Collee JG, Marmion BP, Fraser AG, Simmons A, editors. Mackie and McCartney's Practical Medical Microbiology. New York: McGraw-Hill; 1996. pp. 53–94. [Google Scholar]
- 14.Javiya VA, Ghatak SB, Patel KR, Patel JA. Antibiotic susceptibility patterns of Pseudomonas aeruginosa at a tertiary care hospital in Gujarat, India. Indian J Pharmacol. 2008;40:230–4. doi: 10.4103/0253-7613.44156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfeifer Y, Cullik A, Witte W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol. 2010;300:371–9. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Rodríguez-Martínez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53:4783–8. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam VH, Chang KT, Abdelraouf K, Brioso CG, Ameka M, McCaskey LA, et al. Prevalence, resistance mechanisms, and susceptibility of multidrug-resistant bloodstream isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54:1160–4. doi: 10.1128/AAC.01446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooper DC. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin Infect Dis. 2000;31(Suppl 2):S24–8. doi: 10.1086/314056. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Kang CI, Baek JY, Cho SY, Kim SH, Ko KS, et al. Treatment failure due to induction of ciprofloxacin resistance during combination therapy with colistin and ciprofloxacin in multidrug-resistant Pseudomonas aeruginosa bacteraemia. Int J Antimicrob Agents. 2014;43:391–3. doi: 10.1016/j.ijantimicag.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Kettner M, Milosovic P, Hletková M, Kallová J. Incidence and mechanisms of aminoglycoside resistance in Pseudomonas aeruginosa serotype O11 isolates. Infection. 1995;23:380–3. doi: 10.1007/BF01713571. [DOI] [PubMed] [Google Scholar]
- 21.Vahdani M, Azimi L, Asghari B, Bazmi F, Rastegar Lari A. Phenotypic screening of extended-spectrum ß-lactamase and metallo-ß-lactamase in multidrug-resistant Pseudomonas aeruginosa from infected burns. Ann Burns Fire Disasters. 2012;25:78–81. [PMC free article] [PubMed] [Google Scholar]
- 22.Tassios PT, Gennimata V, Maniatis AN, Fock C, Legakis NJ. Emergence of multidrug resistance in ubiquitous and dominant Pseudomonas aeruginosa serogroup O: 11.The Greek Pseudomonas aeruginosa Study Group. J Clin Microbiol. 1998;36:897–901. doi: 10.1128/jcm.36.4.897-901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


