Summary
Root branching in plants relies on the de novo formation of lateral roots (LRs). These are initiated from founder cells, triggering new formative divisions that generate lateral root primordia (LRP). The LRP size and shape depends on the balance between positive and negative signals that control cell proliferation.
The mechanisms controlling proliferation potential of LRP cells remains poorly understood. We found that Arabidopsis thaliana MYB36, which have been previously shown to regulate genes required for Casparian strip formation and the transition from proliferation to differentiation in the primary root, plays a new role in controlling LRP development at later stages.
We found that MYB36 is a novel component of LR development at later stages. MYB36 was expressed in the cells surrounding LRP where it controls a set of peroxidase genes, which maintain ROS balance. This was required to define the transition between proliferating and arrested cells inside the LRP, coinciding with the change from flat to dome-shaped primordia. Reducing the levels of hydrogen peroxide (H2O2) in myb36-5 significantly rescues the mutant phenotype.
Our results uncover a role for MYB36 outside the endodermis during LRP development through a mechanism analogous to regulating the proliferation/differentiation transition in the root meristem.
Keywords: Arabidopsis, Casparian strip, cell division, lateral root, MYB36, peroxidase, plant, transcription factor
Introduction
The post-embryonic mode of organogenesis in plants, unlike animals, involves the formation of new organs throughout adult life. This entails a continuous recruitment of cells to form organ primordia (Gutierrez, 2005; Scheres, 2007). Development of lateral roots is one example of de novo organogenesis that is particularly relevant for root architecture. Lateral root primordia (LRPs) arise from the pericycle cell layer where, at roughly regular intervals, a few cells become competent to divide in an auxin-dependent manner (De Smet et al., 2007; De Rybel et al., 2010; Moreno-Risueno et al., 2010). Thus, LRPs are initiated from founder cells, by triggering new formative divisions that increase cell number within the LRP.
Initiation and progression of LRPs require a number of divisions of the LR founder cells in the pericycle (De Smet et al., 2008; Dubrovsky et al., 2008; Petricka et al., 2012; Lucas et al., 2013; Van Norman et al., 2013; Wachsman et al., 2015; von Wangenheim et al., 2016). In the initial stages I–II, LRP founder cells divide anticlinally to increase the width of the LRP, a process restricted by ACR4 (De Smet et al., 2008). Later, cells continue dividing to produce new cell layers within the LRP. Several mechanisms contribute to the growth of LRP including a defined auxin transport (Marhavy et al., 2016). LRP emergence through the outer root cell layers involves active crosstalk with overlying layers such as the endodermis and cortex (Swarup et al., 2008; Marhavy et al., 2013; Porco et al., 2016). In addition, reactive oxygen species (ROS) have recently implicated in LR development, although the complex gene network is still largely unknown (Manzano et al., 2014; Reyt et al., 2015; Orman-Ligeza et al., 2016). The final LRP size depends on the balance between positive and negative signals that regulate cell proliferation. However, the mechanisms controlling the proliferation potential of LRP cells at these stages remain poorly understood.
MYB36 is known to regulate the genes required for Casparian strip formation (Kamiya et al., 2015; Liberman et al., 2015) and the transition from proliferation to differentiation in the primary root (Liberman et al., 2015). Here we identified MYB36 as a novel component of later stages of LR development. myb36-5 seedlings have misshaped LPRs, accumulation of LRPs at stages IV–V and fewer emerged lateral roots than wild-type seedlings. MYB36 is expressed in the cells surrounding LRP where it controls a set of peroxidase genes, which maintain the ROS balance at the LRP boundary, required for the transition from flat to dome-shaped primordia. Reducing the levels of hydrogen peroxide (H2O2) in myb36-5 significantly rescues the mutant LRP developmental phenotype. Our results uncover a role for MYB36 outside the endodermis in setting the outer boundary in LRP development through a mechanism analogous to setting the proliferation/differentiation transition in the root meristem.
Materials and Methods
Plant materials and growth conditions
The Arabidopsis thaliana (L.) Heynh. accession Col-0 was used [Author, please confirm inserted text ‘(L.) Heynh.’ is correct]. Two β-estradiol-inducible TRANSPLANTA lines (Coego et al., 2014); N2102512-283A and N2102513-283B) correspond to the MYB36 overexpression lines. A point mutation myb36-5 and a T-DNA insertion allele myb36-6 (WiscDsLox442H5) (Iyer-Pascuzzi et al., 2011) have been described (Kamiya et al., 2015; Liberman et al., 2015). The myb36-2 (GK-543B11 line) was obtained from the Nottingham Arabidopsis Stock Centre (NASC). Detection of MYB36 protein and identification of its targets was carried out using the recombineering rMYB36:GFP (GFP, Green Fluorescent Protein) and the pMYB36::MYB36:GR (GR, glucocorticoid receptor) lines (Liberman et al., 2015). CASP1 expression was analyzed using the reporter line pCASP1:mCherry (Roppolo et al., 2011). Primers used are listed in Supporting Information Table S1.
For plant growth, seeds were surface-sterilized in 20% sodium hypochlorite and 0.1% Tween-20 for 8 min and washed four times in sterile water. Seeds were stratified for 3 d at 4°C and then sowed on ½ Murashige and Skoog (MS) medium solidified with 1% (wt/vol) agar and supplemented with 1% sucrose (wt/vol). Plates were placed vertically in a controlled-environmental growth chamber (50–60% humidity, 22°C, 16 h : 8 h, light : dark photoperiod). For β-estradiol induction, 3-d-old seedlings were transferred to MS medium containing 5 μM β-estradiol (Sigma-Aldrich; E8875) for 3 d. Primary root length was measured in a minimum of 30 seedlings using the software analysis package Fiji (Schindelin et al., 2012). For the quantification of total LRPs and emerged LRs seeds were grown for 8 and 10 d, respectively. LR density was scored as the LR number cm−1 of primary root and was calculated by dividing the number of lateral roots by the primary root length for each seedling (70–100 seedlings were analyzed).
Microscopy
For GFP detection, we used confocal laser scanning microscopy on an inverted Zeiss LSM 510 confocal microscope (excitation source: argon ion laser at 488 nm and detection filters between 500 and 600 nm). To measure root cell size 7 d post sowing (dps) seedlings were fixed in 50% methanol and 10% acetic acid. After fixation, roots were incubated for 30 min in 1% periodic acid and then staining in pseudo-Schiff-propidium iodide for 1 h. Roots were cleared in chloral hydrate for 5 min and mounted in Hoyer’s solution. Confocal images were obtained with an upright Zeiss LSM 510 confocal microscope using filters settings for propidium iodide staining. Cell length was measured on a minimum of 10–15 roots per genotype using Fiji software. For visualization of LRP phenotype, roots were cleared as described and observed in an upright Axioskop2 plus (Zeiss) microscope and digital Coolsnap FX color (Roper Scientific). For detailed visualization of LRPs, roots were fixed in 4% paraformaldehyde, stained with DAPI [Author, please insert expansion for ‘DAPI’] and observed in a Zeiss LSM 710 confocal microscopy.
GUS staining
β-glucuronidase (GUS) staining of pMYB36:GUS plants was performed as previously described (Jefferson et al., 1987).
Quantitative RT-PCR Analysis
Total RNA from Col-0 and myb36 mutant, roots and the root region enriched in LRPs, (discarding the apical 0.8–1.0 cm and the differentiated root zone that already contained emerged LRs) was extracted using TRIzol reagent (Thermo-Fisher). One microgram of DNase-treated total RNA was used to generate cDNA with SuperScript III (Invitrogen) and diluted 1 : 9 for the assay. Primers were designed (Primer3 software) and tested for amplification efficiency. qRT-PCR [Author, please insert expansion for ‘qRT-PCR’] was carried out with GoTaq qPCR Master Mix (Promega) according to the manufacturer’s instructions on an ABI PRISM 7900HT Fast Real-time PCR system (Applied Biosystems). Standard curves were generated for each primer pair and values were normalized against an endogenous control gene (ACT8). Primers used are listed in Table S1.
Identification of direct targets of MYB36
To identify direct targets of MYB36 we used the inducible pMYB36::MYB36:GR line (Liberman et al., 2015) grown for 7 d and then transferred to MS medium containing 10 μM dex, with and without the protein synthesis inhibitor cycloheximide (10 μM). The relative expression of putative MYB36 targets in the LRP region was analyzed by qRT-PCR.
Results
Identification of MYB36 as a regulator of root architecture
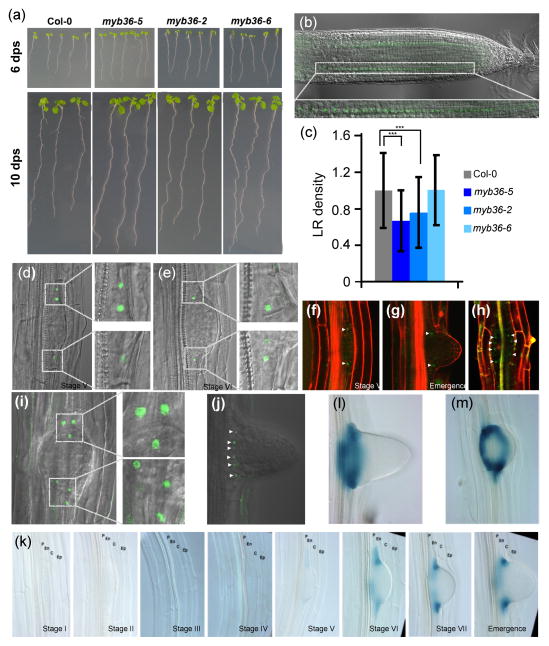
To identify novel components of the gene network controlling LR development we used of the TRANSPLANTA collection of Arabidopsis lines expressing individual transcription factors (TFs) under a β-estradiol inducible promoter (Coego et al., 2014). We screened for changes in root architecture at 3 d post induction. A line expressing MYB36 (At5g57620), which is expressed in root (Brady et al., 2007), produced a dramatic alteration of root architecture. High levels of MYB36 produced a significant shortening of the primary root (Fig. S1a,b) (Li et al., 2013; Liberman et al., 2015). To further investigate the role of MYB36 in root development we used three alleles: myb36-5, an EMS (ethyl-methane sulfonate) mutant isolated in a genetic screen for regulators of CASP expression in the endodermis (Liberman et al., 2015), myb36-2, GK-543B11 (Kamiya et al., 2015) and myb36-6, WiscDsLox442H5 (Iyer-Pascuzzi et al., 2011). All three alleles have longer primary roots (Fig. 1a). We examined MYB36 expression using a rMYB36-GFP translational fusion obtained by recombineering that complements both the Casparian strip formation phenotype (Liberman et al., 2015) and the primary root length phenotype of myb36-5. We found that MYB36 protein expressed from the rMYB36-GFP construct accumulates exclusively in the endodermis in the root tip (Fig. 1b), confirming previous observations using a different MYB36-GFP expressing line (Kamiya et al., 2015). Consistent with this expression pattern and the proposed role of MYB36 as a promoter of endodermal differentiation (Kamiya et al., 2015; Liberman et al., 2015), all myb36 alleles showed longer root meristems. We determined that this phenotype was due to an increase in meristem cell number (Figs 1, S1c,d). The balance of ROS is known to regulate root meristem size. We also found that myb36 mutants show alterations in the ROS balance associated with modifications in the root meristem boundaries (Tsukagoshi et al., 2010) (Fig. S1e; see Methods S1).
Fig. 1.
Arabidopsis thaliana MYB36 is expressed in the lateral root primordia (LRP) boundary cells at mid stages of lateral root development. (a) Images showing the length of primary root of 6 and 10 d post sowing (dps) Col-0, myb36-5, myb36-2 and myb36-6 mutants. (b) Localization of MYB36, using a rMYB36:GFP fusion protein, in the nuclei of root meristem endodermal cells. (c) Emerged lateral root density in Col-0, myb36-5, myb36-2 and myb36-6. Measurements were taken 10 dps on Murashige and Skoog (MS) agar plates. Lateral root (LR) density was calculated by dividing the number of lateral roots by the primary root length for each seedling (70–100 seedlings). The density is reported as the number of lateral roots per centimetre of primary root ± SD. Statistical significance: ***, P < 0.01. (d–g, j) Midplane and (h, i) front views of LRP at different developmental stages showing that rMYB36:GFP fusion protein localized specifically to the LRP boundary (LRPB) cells that delimit the primordia. Small white arrowheads point to the nuclei labelled with MYB36-GFP. (k) MYB36 expression determined with a transcriptional pMYB36:GUS fusion revealed that MYB36 is expressed inside the LRP, not in the endodermis, from stages V onwards, and during emergence of lateral roots. P, pericycle; En, endodermis; C, cortex; Ep, epidermis. (l, m) Midplane and front views, respectively, of LRPB cells of plants expressing pMYB36:GUS.
MYB36 is expressed in pericycle cells at the LRP boundary
Here we uncovered a novel role of MYB36 in LR development based on the observation that all myb36 mutant alleles, except myb36-6, have reduced LR density (Fig. 1c). We wanted to complement this analysis using the MYB36 overexpression lines but the lignin and perhaps suberin deposited ectopically in the endodermis, cortex and epidermis (Kamiya et al., 2015) likely precluded the emergence of LR.
MYB36 was not expressed in the endodermal cells in contact with the LRPs but within the LRP itself. MYB36 is not detected until stage V of LR development, at which time it appears in the LRP boundary (LRPB) cells (Fig. 1d–j). These outermost LRPB cells constitute an oval of cells that surrounds the emerging LRP (Fig. 1h,j; Movies S1, S2). To determine if MYB36 mRNA is produced in the LRPB cells we generated plants expressing the transcriptional reporter proMYB36:GUS. Expression from this construct fully matched rMYB36-GFP localization from stage V onwards (Fig. 1k), when the growing LRP compresses the overlying cortical cell layer. The oval of MYB36-expressing cells at the base of the LRP is also observed in the GUS marker lines (Fig. 1l,m). This indicates that the expression is cell-autonomous and that it is unlikely that MYB36 protein moves from the endodermis.
MYB36 defines the LRP boundary
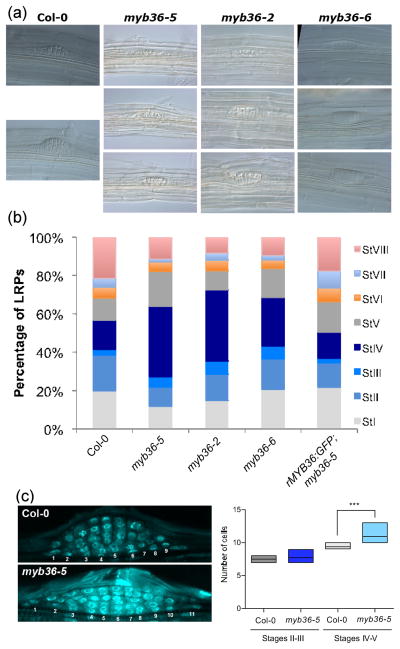
To establish the functional requirement of MYB36 for LR formation we carried out a temporal analysis of LR development in the myb36 background (Malamy & Benfey, 1997). At early stages, wild type and mutant appear similar, but at stage IV–V, the LRP of myb36 plants has a flat appearance, as if they have problems emerging through the cortex (Fig. 2a). Quantification of developmental stages indicated a statistically significant overabundance of stage IV primordia (Fig. 2b) revealing a defect in the transition from flat- to dome-shaped LRP. Importantly, the rMYB36:GFP construct was able to complement the myb36-5 mutant phenotype (Fig. 2b).
Fig. 2.
Role of Arabidopsis thaliana MYB36 in mid stage of lateral root primordia (LRP) development. (a) Representative images of myb36-5, myb36-2 and myb36-6 mutants. The lack of MYB36 produces a flat LRP phenotype, but not at early developmental stages. (b) Quantification of total number of LRP at different developmental stages of Col-0, myb36-5, myb36-2 and myb36-6 mutant roots. The complementing line expressing rMYB36:GFP in the myb36-5 mutant background was used as a control. For each genotype, 10 different roots and a total of c. 100 LRPs were scored. (c) Detail of DAPI-stained [Author, please insert expansion for ‘DAPI’] wild type (Col-0) and myb36-5 LRPs indicating the position of cells at their base (left panel) and quantification of cell number at different stages, as indicated (right panel). Differences for stages IV–V were statistically significant (***, P ≤ 0.01).
Stage IV during LRP development corresponds to the emergence of the growing LRP through the outer cell layers (Vermeer et al., 2014), concomitant with the cessation of cell divisions. To evaluate possible effects of the myb36 mutation on cell division we measured the LRP width by counting cell number in the middle plane of the LRP along the innermost cell layer. This revealed that the myb36-5 mutant, which has the strongest phenotype of the allele tested, contains more cells than the wild type (Fig. 2c). This together with the expression pattern suggests that MYB36 could control LRP width.
CASP1 has been recently reported as a direct target of MYB36 (Kamiya et al., 2015). Since MYB36 is not expressed in the endodermal cells overlying the LRP, it seemed unlikely that it plays a direct role in regulating Casparian strip remodeling at the site of LRP emergence. In support of this hypothesis, we did not detect expression of CASP1-mCherry in the LRPB cells at the time when MYB36 is expressed (Fig. S2a). Another possibility is that MYB36 may function to alter the lignin composition of LRPB cells. However, we found that lignin deposition in the LRPB cell walls was undetectable (Fig. S2b; see Methods S2).
MYB36 regulates expression of a subset of genes controlling ROS balance in LRPs
UPBEAT1 (UPB1) is a bHLH transcription factor that controls the balance of proliferation and differentiation in the RAM (root apical meristem) acting on ROS homeostasis through a subset of peroxidases, for example PER39 (AT4g11290), PER40 (AT4g16270) and PER57 (AT5g17820) (Tsukagoshi et al., 2010). UPB1 is coexpressed with several peroxidase genes also during LR development (Manzano et al., 2014). Furthermore, MYB36 is upregulated in upb1 mutants and downregulated in UPB1-overexpression plants (Tsukagoshi et al., 2010; Manzano et al., 2014). Finally, UPB1 is expressed in LRPB cells at stage III–IV (Manzano et al., 2014), suggesting the possibility of an UPB1-MYB36-PER pathway in LRP formation. Consistent with this hypothesis, a subset of UPB1-regulated PER genes showed altered expression in myb36 mutants (Kamiya et al., 2015; Liberman et al., 2015).
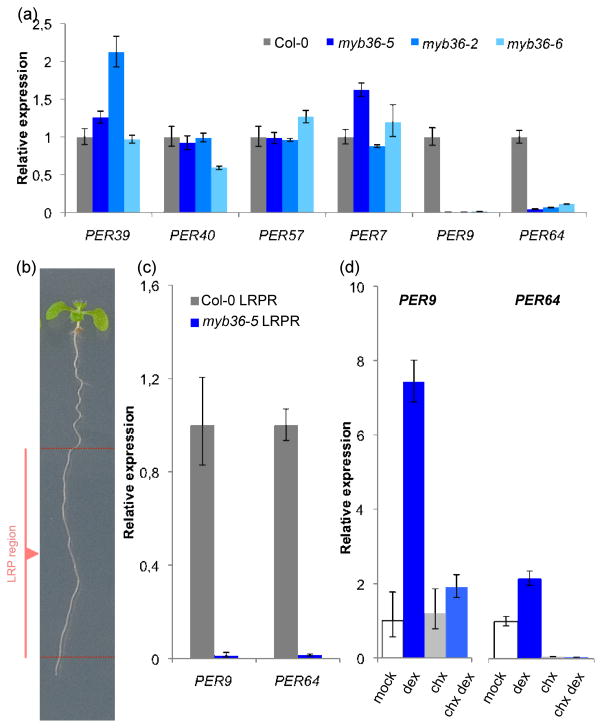
To determine if MYB36 regulates these PER genes, we analyzed mRNA levels in whole roots by RT-qPCR and found that PER39, PER40, PER57 and PER7 do not change significantly in myb36 (Fig. 3a). However, expression of PER9 (AT1G44970) and PER64 (AT5G42180), which were reported to have altered expression in myb36-5 endodermis (Kamiya et al., 2015; Liberman et al., 2015), are drastically reduced in whole roots of all three myb36 alleles studied (Fig. 3a). Since MYB36 is expressed in only a few cells within the LRP and to identify LRP-specific effects of MYB36 we determined the mRNA levels of PER9 and PER64 in the root region after removal of both the apex (0.5–1.0 cm) and the mature zone of the root that contains already emerged LRs (Fig. 3b). This strategy yielded a sample highly enriched in roots containing non-emerged LRPs that allowed us to find that PER64 and PER9 were very strongly downregulated in myb36-5 (Fig. 3c). PER64 was reported to be reduced in the esb1 mutant (Lee et al., 2013). To determine if these PER genes are direct MYB36 targets in the LRP region, we used a GR-based version of MYB36 (pMYB36::MYB36:GR) that localizes to the nuclei after treatment with dexamethasone (Dex) in combination with the protein synthesis inhibitor cycloheximide, as described previously (Hou et al., 2008). The results indicated that both PER64 and PER9 are secondary MYB36 targets, as their expression was not upregulated in the presence of cycloheximide in a MYB36-dependent manner (Fig. 3d). Other PER genes, for example PER11 and PER22, which showed an increased expression in the root meristem of myb36-5 mutant (Liberman et al., 2015), were not upregulated in the Dex-inducible MYB36 plants in the LRP region (not shown). Together, these experiments suggest that MYB36 functions at stage V–VI of LRP development by affecting the ROS balance, at least partially through regulation of PER9 and PER64.
Fig. 3.
Role of Arabidopsis thaliana MYB36 in lateral root primordia (LRP) development through various PER genes. (a) Relative expression of the indicated PER genes in roots of Col-0 and myb36 mutant plants. The only cases where differences were statistically significant and consistent in all mutants analyzed were PER9 and PER64 (P ≤ 0.01). (b) Representative image of the root highlighting the lateral root primordia region (LRPR) of the root. The LRPR of the root comprises the stages of LRP development where MYB36 is expressed (before LR emergence). (c) Abundance of PER9 and PER64 transcript levels in Col-0 and myb36-5 LRPR. Differences were statistically significant (P ≤ 0.01). (d) PER9 and PER64 mRNA levels in pMYB36::MYB36:GR in myb36-5 seedlings treated with dexamethasone (dex) in the absence or presence of cycloheximide (chx). Error bars indicate ± SD.
MYB36 restricts cell proliferation by maintaining ROS homeostasis in LRP
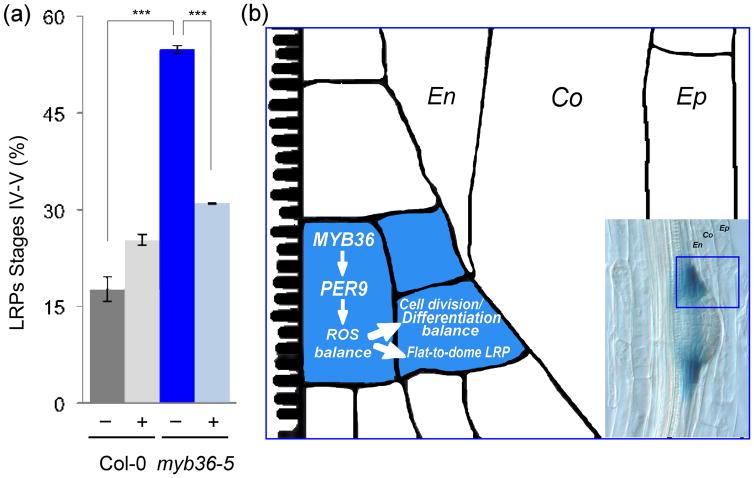
Our results support the hypothesis that defects in LRP development in myb36 may be the consequence of altered ROS balance. According to this hypothesis, increased levels of H2O2, due to a reduction of PER9 (and possibly other peroxidases) activity, could act in the LRPB cells in a manner analogous to the role of MYB36 at the proliferation/differentiation transition in the primary root meristem (Liberman et al., 2015). We tried to detect ROS at the cellular level inside the LRP of myb36 mutants but these experiments were not possible due to the inability of reagents to penetrate inside the LRPs in the mutants, likely due to the increased suberization in endodermal cells (Kamiya et al., 2015). As an alternative, we tested this hypothesis directly by treating roots with potassium iodide, a H2O2 scavenger reported to rescue the reduction of root meristem size by H2O2 excess in UPB1 overexpression lines (Tsukagoshi et al., 2010). We found that although this treatment had a measurable increase in the wild-type seedlings with stage IV–V LRP, the defect in LRP development of myb36 mutants leading to accumulation at stage IV–V, was largely rescued by KI application [Author, if appropriate, please insert expansion for ‘KI’] (Fig. 4a). This is consistent with a MYB36-PER module maintaining the correct ROS balance during LRP development. As already mentioned, the additional emergence problems of MYB36 overexpression lines precluded the study of their phenotype after H2O2 treatment.
Fig. 4.
Mechanism of action of Arabidopsis thaliana MYB36 at the lateral root boundaries. (a) Quantification of total number of lateral root primordia (LRP) at stages IV and V of Col-0 and myb36-5 roots after 3 d in the absence or presence of potassium iodide (KI). Error bars indicate ± SD. Statistical significance: ***, P < 0.01. (b) Schematic diagram describing a model for MYB36 function in LRP boundary (LRPB) cells. Inset: image of a LRP showing expression of pMYB36:GUS in the lateral root primordia (LRP) boundary cells. En, endodermis; Co, cortex; Ep, epidermis.
Discussion
In the primary root, MYB36 is expressed in the endodermis from the root apical meristem up to the differentiation zone, where it is involved in the Casparian strip biogenesis (Kamiya et al., 2015) and the transition from proliferation to differentiation (Liberman et al., 2015). Here we found that MYB36 is also expressed in a small group of cells that surround the LRPs at their base, suggesting an additional role during LRP development. Our data show that MYB36 expression in the LRP boundary cells, both at the mRNA and protein level, is cell-autonomous. This is different from the cross-signaling between LRP founder cells and the overlying endodermal cells is important for the SHY2-mediated auxin response during LRP development (Marhavy et al., 2013; Vermeer et al., 2014). LR emergence through the outer cell layers of the primary root depends on mechanical pressure of the endodermis (Lucas et al., 2013; von Wangenheim et al., 2016). In the case of myb36 mutants, the flat shape of their LRPs appears to be produced by the abnormal and ectopic deposition of lignin, also observed in the MYB36 overexpression lines (Kamiya et al., 2015; this work).
Various hormonal and developmental signals contribute to activate and maintain cell division from the early to late stages of LRP growth (Petricka et al., 2012; Atkinson et al., 2014; Vilches-Barro & Maizel, 2015). However, the final size of the organ also requires signals that inhibit cell proliferation in a localized manner to determine organ size. For example, the receptor kinase ACR4 acts at the very early stages of LRP formation to restrict anticlinal divisions of competent pericycle cells (De Smet et al., 2008). It also works in the root apical meristem to control formative cell divisions (Yue et al., 2016).
In addition, we found that the myb36-5 mutant LRPs contain more cells at the base as if MYB36 is necessary to the control the proliferative span of cells inside the LRP that determine the transition between proliferating and arrested pericycle cells. This is reminiscent of the role of MYB36 to maintain ROS balance in the root meristem where PER expression is necessary at the transition zone to limit the size of the meristematic zone (Tsukagoshi et al., 2010; Liberman et al., 2015).
We propose a model in which MYB36 is necessary to activate expression of PER9, and perhaps other peroxidases, which reduce the H2O2 levels that contribute to setting the outer boundary of the growing LRP (Fig. 4b). Thus, MYB36 is necessary before the transition from a flat to dome LRP, coinciding with the arrest of cell division to set the final width of the LRP. A highly localized expression domain in the LRP, similar to that of MYB36, has been reported for other genes such as PIN6, an intracellular auxin transporter (Benkova et al., 2003; Simon et al., 2016) and PUCHI, an auxin-regulated AP2/EREB transcription factor (Hirota et al., 2007) and several members of the RBOH family of oxidases (Orman-Ligeza et al., 2016). However, these genes are expressed earlier in LRP development and are likely not directly implicated in the flat LRP phenotype of myb36 mutants that we describe here.
Our results show that MYB36 plays a critical role in restricting cell division at later stages of LRP development by regulating a subset of PER genes, and perhaps other genes, that modulate ROS balance and cell proliferation potential. Other subsets of PER genes are implicated in regulating genes required for Casparian strip formation and in emergence of LRPs. Notably, MYB36 works in the root apical meristem by an analogous mechanism (Tsukagoshi et al., 2010; Liberman et al., 2015). These findings reinforce the importance of an adequate balance of factors acting positively and negatively to define developmental boundaries and determine the proliferation and differentiation potential during development.
Supplementary Material
Acknowledgments
We thank N. Geldner (University of Lausanne, Switzerland) for the CASP1-mCherry plants. This work was supported by grants BFU2012-34821 and BIO2011-28184-C02-01 (to C.G. and J.C.d.P., respectively) and CSD2007-0057 (TRANSPLANTA consortium) from MINECO (Spain) and grants IOS-1021619 from the NSF Arabidopsis 2010 program, R01-GM043778 from the NIH, GBMF3405 from the Gordon and Betty Moore Foundation and the Howard Hughes Medical Institute (to P.N.B.). M.F-M. was funded by the Juan de la Cierva program (MINECO). L.M.L. was funded in part by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. The institutional grant from Fundacion Ramon Areces to the CBMSO is also acknowledged. The authors declare that they have no conflicts of interest.
Footnotes
Author contributions
M.F-M. and B.D. carried out the screening to identify MYB36 and the initial characterization of primary root phenotype. M.F-M. analyzed the LRP phenotype, determined MYB36 and target expression, and carried out ROS measurements. L.M.L. and P.N.B. provided the MYB36-GFP. C.M. and J.C.d.P. quantified the developmental stages of LRPs. All authors discussed the results and suggested further experiments. C.G. coordinated the project and wrote the manuscript with the input of all authors that contributed to the final manuscript.
Additional Supporting Information may be found online in the Supporting Information tab for this article:
Fig. S1 Primary root phenotypes of MYB36oe and myb36 mutants.
Fig. S2 CASP1 and lignin production in myb36 mutants.
Table S1 Primers used in this study
Methods S1 ROS localization.
Methods S2 Lignin detection.
Movie S1 Detection of MYB36-GFP in z-stack of a LRP (lateral view).
Movie S2 Detection of MYB36-GFP in z-stack of a LRP (front view).
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Atkinson JA, Rasmussen A, Traini R, Voss U, Sturrock C, Mooney SJ, Wells DM, Bennett MJ. Branching out in roots: uncovering form, function, and regulation. Plant Physiology. 2014;166(2):538–550. doi: 10.1104/pp.114.245423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115(5):591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318(5851):801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Coego A, Brizuela E, Castillejo P, Ruiz S, Koncz C, del Pozo JC, Pineiro M, Jarillo JA, Paz-Ares J, Leon J. The TRANSPLANTA collection of Arabidopsis lines: a resource for functional analysis of transcription factors based on their conditional overexpression. Plant Journal. 2014;77(6):944–953. doi: 10.1111/tpj.12443. [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Current Biology. 2010;20(19):1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134(4):681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, et al. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science. 2008;322(5901):594–597. doi: 10.1126/science.1160158. [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benkova E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci, USA. 2008;105(25):8790–8794. doi: 10.1073/pnas.0712307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. Coupling cell proliferation and development in plants. Nature Cell Biology. 2005;7(6):535–541. doi: 10.1038/ncb0605-535. [DOI] [PubMed] [Google Scholar]
- Hirota A, Kato T, Fukaki H, Aida M, Tasaka M. The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell. 2007;19(7):2156–2168. doi: 10.1105/tpc.107.050674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Hu WW, Shen L, Lee LY, Tao Z, Han JH, Yu H. Global identification of DELLA target genes during Arabidopsis flower development. Plant Physiology. 2008;147(3):1126–1142. doi: 10.1104/pp.108.121301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN. Cell identity regulators link development and stress responses in the Arabidopsis root. Developmental Cell. 2011;21(4):770–782. doi: 10.1016/j.devcel.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T, Borghi M, Wang P, Danku JM, Kalmbach L, Hosmani PS, Naseer S, Fujiwara T, Geldner N, Salt DE. The MYB36 transcription factor orchestrates Casparian strip formation. Proc Natl Acad Sci U S A. 2015;112(33):10533–10538. doi: 10.1073/pnas.1507691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153(2):402–412. doi: 10.1016/j.cell.2013.02.045. [DOI] [PubMed] [Google Scholar]
- Li S, Liberman LM, Mukherjee N, Benfey PN, Ohler U. Integrated detection of natural antisense transcripts using strand-specific RNA sequencing data. Genome Research. 2013;23(10):1730–1739. doi: 10.1101/gr.149310.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman LM, Sparks EE, Moreno-Risueno MA, Petricka JJ, Benfey PN. MYB36 regulates the transition from proliferation to differentiation in the Arabidopsis root. Proc Natl Acad Sci U S A. 2015;112:12099–12104. doi: 10.1073/pnas.1515576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, Vobeta U, Swarup K, De Smet I, Van Damme D, Lawrence T, Peret B, Moscardi E, et al. Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci, USA. 2013;110(13):5229–5234. doi: 10.1073/pnas.1210807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124(1):33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Manzano C, Pallero-Baena M, Casimiro I, De Rybel B, Orman-Ligeza B, Van Isterdael G, Beeckman T, Draye X, Casero P, Del Pozo JC. The emerging role of reactive oxygen species signaling during lateral root development. Plant Physiology. 2014;165(3):1105–1119. doi: 10.1104/pp.114.238873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavy P, Montesinos JC, Abuzeineh A, Van Damme D, Vermeer JE, Duclercq J, Rakusova H, Novakova P, Friml J, Geldner N, et al. Targeted cell elimination reveals an auxin-guided biphasic mode of lateral root initiation. Genes and Development. 2016;30(4):471–483. doi: 10.1101/gad.276964.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavy P, Vanstraelen M, De Rybel B, Zhaojun D, Bennett MJ, Beeckman T, Benkova E. Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO Journal. 2013;32(1):149–158. doi: 10.1038/emboj.2012.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science. 2010;329(5997):1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman-Ligeza B, Parizot B, de Rycke R, Fernandez A, Himschoot E, Van Breusegem F, Bennett MJ, Perilleux C, Beeckman T, Draye X. RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development. 2016;143(18):3328–3339. doi: 10.1242/dev.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN. Control of Arabidopsis root development. Annual Review of Plant Biology. 2012;63:563–590. doi: 10.1146/annurev-arplant-042811-105501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porco S, Larrieu A, Du Y, Gaudinier A, Goh T, Swarup K, Swarup R, Kuempers B, Bishopp A, Lavenus J, et al. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development. 2016;143(18):3340–3349. doi: 10.1242/dev.136283. [DOI] [PubMed] [Google Scholar]
- Reyt G, Boudouf S, Boucherez J, Gaymard F, Briat JF. Iron- and ferritin-dependent reactive oxygen species distribution: impact on Arabidopsis root system architecture. Molecular Plant. 2015;8(3):439–453. doi: 10.1016/j.molp.2014.11.014. [DOI] [PubMed] [Google Scholar]
- Roppolo D, De Rybel B, Denervaud Tendon V, Pfister A, Alassimone J, Vermeer JE, Yamazaki M, Stierhof YD, Beeckman T, Geldner N. A novel protein family mediates Casparian strip formation in the endodermis. Nature. 2011;473(7347):380–383. doi: 10.1038/nature10070. [DOI] [PubMed] [Google Scholar]
- Scheres B. Stem-cell niches: nursery rhymes across kingdoms. Nature Reviews Cellullar and Molecular Biology. 2007;8(5):345–354. doi: 10.1038/nrm2164. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Skupa P, Viaene T, Zwiewka M, Tejos R, Klima P, Carna M, Rolcik J, De Rycke R, Moreno I, et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytologist. 2016;211(1):65–74. doi: 10.1111/nph.14019. [DOI] [PubMed] [Google Scholar]
- Swarup K, Benkova E, Swarup R, Casimiro I, Peret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nature Cell Biology. 2008;10(8):946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143(4):606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Xuan W, Beeckman T, Benfey PN. To branch or not to branch: the role of pre-patterning in lateral root formation. Development. 2013;140(21):4301–4310. doi: 10.1242/dev.090548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JE, von Wangenheim D, Barberon M, Lee Y, Stelzer EH, Maizel A, Geldner N. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science. 2014;343(6167):178–183. doi: 10.1126/science.1245871. [DOI] [PubMed] [Google Scholar]
- Vilches-Barro A, Maizel A. Talking through walls: mechanisms of lateral root emergence in Arabidopsis thaliana. Current Opinion in Plant Biology. 2015;23:31–38. doi: 10.1016/j.pbi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Wachsman G, Sparks EE, Benfey PN. Genes and networks regulating root anatomy and architecture. New Phytologist. 2015;208(1):26–38. doi: 10.1111/nph.13469. [DOI] [PubMed] [Google Scholar]
- von Wangenheim D, Fangerau J, Schmitz A, Smith RS, Leitte H, Stelzer EH, Maizel A. Rules and self-organizing properties of post-embryonic plant organ cell division patterns. Current Biology. 2016;26(4):439–449. doi: 10.1016/j.cub.2015.12.047. [DOI] [PubMed] [Google Scholar]
- Yue K, Sandal P, Williams EL, Murphy E, Stes E, Nikonorova N, Ramakrishna P, Czyzewicz N, Montero-Morales L, Kumpf R, et al. PP2A-3 interacts with ACR4 and regulates formative cell division in the Arabidopsis root. Proc Natl Acad Sci, USA. 2016;113(5):1447–1452. doi: 10.1073/pnas.1525122113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.