Abstract
DREB1 of the AP2/ERF superfamily plays a key role in the regulation of plant response to low temperatures. In this study, a novel DREB1/CBF transcription factor, PnDREB1, was isolated from Iceland poppy (Papaver nudicaule), a plant adaptive to low temperature environments. It is homologous to the known DREB1s of Arabidopsis and other plant species. It also shares similar 3D structure, and conserved and functionally important motifs with DREB1s of Arabidopsis. The phylogenetic analysis indicated that the AP2 domain of PnDREB1 is similar to those of Glycine max, Medicago truncatula, and M. sativa. PnDREB1 is constitutively expressed in diverse tissues and is increased in roots. qPCR analyses indicated that PnDREB1 is significantly induced by freezing treatment as well as by abscissic acid. The expression levels induced by freezing treatment were higher in the variety with higher degree of freezing tolerance. These results suggested that PnDREB1 is a novel and functional DREB1 transcription factor involved in freezing response and possibly in other abiotic stresses. Furthermore, the freezing-induction could be suppressed by exogenous gibberellins acid, indicating that PnDREB1 might play some role in the GA signaling transduction pathway. This study provides a basis for better understanding the roles of DREB1 in adaption of Iceland poppy to low temperatures.
Keywords: DREB1, expression profile, freezing stress, Iceland poppy, transcription factor
Introduction
Abiotic stress conditions, such as drought, high salinity, and cold, have adverse effects on plant growth and production. As sessile organisms, plants have developed a wide spectrum of adaptation strategies to cope with the inevitable challenges of environmental stress. Many aspects of these adaptation processes, including developmental, physiological and biochemical changes, are regulated by stress-responsive gene expression. Transcription factors (TFs) play pivotal roles in gene expression by regulating expression of downstream genes as trans-acting elements via specifically binding to cis-acting elements in the promoters of target genes. The cis- and trans-acting elements involved in the transcriptional responses of stress-responsive genes have been previously identified (Yamaguchi-Shinozaki and Shinozaki, 2006).
The APETALA 2/ethylene-responsive element binding factor (AP2/ERF) superfamily is a large group of TF, usually classified to the AP2, RAV, and ERF families (Sakuma et al., 2002; Licausi et al., 2013). The ERF family is further subdivided into ERF and DREB (dehydration-responsive element-binding protein) subfamilies based on different conserved amino acid residues within their respective AP2 domains (Nakano et al., 2006; Lata and Prasad, 2011; Lata et al., 2011). Among these, many members of DREB subfamily are involved in plant abiotic stress responses by regulating gene expression via the cis-acting dehydration-responsive element/C-repeat (DRE/CRT, A/GCCGAC) element (Yamaguchi-Shinozaki et al., 2006, Kidokoro et al., 2015) in the promoters of stress responsive genes, such as COR15A, RD29A/COR78, and COR6.6 (Stockinger et al., 1997; Liu et al., 1998; Sakuma et al., 2002; Licausi et al., 2013).
The DREB1 subgroup of DREB subfamily are major regulators of cold-stress responses. Three out of the six DREB1s of Arabidopsis, DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 are rapidly induced in response to cold stress (Stockinger et al., 1997; Liu et al., 1998; Gilmour et al., 1998; Shinwari et al., 1998). The overexpression of AtDREB1/CBF led to up-regulated expression of cold-inducible genes that function in survival at low temperatures, including those encoding late embryogenesis abundant (LEA) proteins and enzymes for sugar metabolism and fatty acid desaturation (Maruyama et al., 2004; Seki et al., 2001; Fowler and Thomashow 2002). Additionally, the expression levels of DREB1B/CBF1 and DREB1C/CBF2 are significantly correlated with freezing tolerance (Hannah et al., 2006). Heterologous expression of DREB1 was capable to improve multiple abiotic stress tolerances in agricultural crops including tobacco (Kasuga et al., 2004), wheat (Pellegrineschi et al., 2004), rice (Ito et al., 2006), chrysanthemum (Hong et al., 2006a,b,c; Hong and Kim, 2005), and Caragana korshinskii (Wang et al., 2011), etc.
Cold-inducible DREB1/CBF genes have been isolated from numerous dicotyledonous plant species, such as oilseed rape, Vaccinium myrtillus (Oakenfull et al., 2013), Caragana korshinskii (Wang et al., 2011), Capsicum annuum (Hong and Kim, 2005), grape (Xiao et al., 2006), and chrysanthemum (Tong et al., 2009), as well as monocotylous plant species, such as wheat (Triticum aestivum), rye (Secale cereale) (Jaglo et al., 2001), rice (Dubouzet et al., 2003), maize (Qin et al., 2004), etc.
Iceland poppy (Papaver nudicaule) is a dicotyledonous and boreal flowering plant, native to subpolar regions of Europe, Asia and North America, and the mountains of Central Asia. It is adapted to low temperature environments and has been widely utilized as ornamental plants because it yields large, papery, bowl-shaped, lightly fragrant flowers supported by hairy, one foot, curved stems among feathery blue-green foliage 1–6 inches long. Previous studies mainly focused on extraction and analyses of its alkaloid (Philipov et al., 2007; Istatkova et al., 2008; Tatsis et al., 2014). However, no attention has been paid on their acclimation to low temperatures. Our previous study investigated the physiological responses and tolerance of four varieties of Iceland poppy to low temperatures (from 3 to –9 °C) (unpublished). To further understand it's low temperature adaptation at molecular level and reveal novel cold responsive genes, we cloned and characterized a new DREB1 gene member, named PnDREB1, from the Iceland poppy variety Champagne Bubbles, which has prominent freezing tolerance among four varieties previously investigated (unpublished). Sequence similarity and phylogenetic relationship to the known DREB1s were comprehensively analyzed, and its spatial expression patterns and responses to freezing stress and phytohormone were also investigated.
Materials and Methods
Plant materials and treatments
A variety of Iceland poppy, Champagne Bubbles (CB), was used for gene cloning and expression analyses. Another variety, Wonderland (WL), with lower freezing performance was also used in expression analysis. Seeds were surface-sterilized with hydrogen peroxide solutions and germinated on plates containing the mixture of local soil and nutrient soil (with a ratio of 1:1). The seedlings were maintained in a greenhouse with a relative humidity of 50–70%, 12 h light at 15 °C and 12 h dark at 10 °C. After three or four leaves emerged, the plants were transferred to plastic pots with 15 cm diameter (one plant per pot).
For freezing treatment, the four-month-old plants with uniform growing status were carefully pulled out from the soil. After cleaning the roots with distilled water, the plants were cultured into Hoagland's solution for three days under normal condition and then transferred into an incubator at 0 °C with light. The leaf and root tissues were sampled at 0, 2, 4, 8, 12, 24 h post treatment; For ABA treatment, the plants were treated in 100 Hoagland's solution containing 100 μM ABA (Shan et al., 2007) under normal growth condition and the leaf and root tissues were sampled at 0, 0.5, 1, 2, 4, 8 and 12 h; For gibberellin (GA) treatment, the 80 μM GA3 solution containing 0.02% (v/v) polyoxyethylene-sorbitan monolayrate (Tween-20) were evenly sprayed onto the whole plant. Two hours later, the plants were transferred to freezing treatment and leaf tissues were sampled at 0, 0.5, 1, 2, 4, 8 and 12 h (Shan et al., 2007). Each treatment was repeated three times. Samples consisted of equal tissue quantities from 3 individual plants, which were immediately frozen in liquid nitrogen and stored at -80 °C until their use.
Nucleic acid extraction
Genomic DNA was isolated from leaves of seedlings with the cetytrimethylammonium bromide (CTAB) procedure as reported by Murray and Thompson (1980). Total RNA in various tissues was extracted according to the manual of the TRIZOL Kit (TIANGEN, Beijing). The qualities and quantities of extracted nucleotide were measured by NanoDrop 2000 (Thermo Fisher, USA).
Amplification of conserved region of DREB1
About 5 μg of total RNA was reverse transcribed with oligo18(dT) primer by using single-stranded cDNA Synthesis Kit (TaKaRa Dalian, China) following the manufacturer's introduction. To amplify the conserved region of DREB1 from Iceland poppy, a pair of degenerate primers, DREB1-F1 and DREB-F2, was designed based on the alignment of nucleotide sequences of AP2 domains of DREB1s of Arabidopsis, Glycine max, Nicotiana tabacum, Vitis vinifera, Chrysanthemum, and Prunus mume (Table 1).
Table 1. Primer sequences for expression level evaluation.
| Primer Name | Sequence (5'-3') | Target gene | Expected size (bp) | Usage |
|---|---|---|---|---|
| DREB1-F2 | CGAACAGTTCTCAACAGTTATCATC | PnDREB1 | 400 | Semi quantity RT-PCR |
| DREB1-R2 | CTCACTATATTGATAAGTTGGACTC | |||
| actin-F2 | TTGGATTCTGGTGATGGTGT | Actin1 | 300 | Semi quantity RT-PCR |
| actin-R2 | GAACCTCTGGACAACGGAACC | |||
| actin-F4 | ATGCCCTACCACATGCCATC | Actin1 | 86 | QPCR |
| actin-R4 | ACCACGCTCCGTCAAGATTT | |||
| ef1-F2 | GGAGGCTGCTGAGATGAACA | EF1 | 77 | QPCR |
| ef1-R2 | CACGCTCACGTTCAGCCTTA | |||
| DREB-F3 | GCTACACCAGAAATGGCTGC | PnDREB1 | 95 | QPCR |
| DREB-R3 | CTCCAGACGGAATCAGCGAA |
The Polymerase Chain Reaction (PCR) amplifications were performed in 25 μL reaction volume, consisting of 1 U Ex-Taq DNA polymerase (TaKaRa), 2.5 μL PCR buffer (supplied with Taq DNA polymerase), 1 μL cDNA template, 400 pmol of each primer, 1.5 mM MgCl2 and 200 μmol of each dNTP. PCR program was conducted as following: 94 °C for 5 min, 30 cycles at 94 °C for 20 s, 56 °C for 20 s, 72 °C for 20 s, followed by 72 °C for 10 min and incubation at 12 °C. Amplified fragments were separated on 1% agarose gels, and purified using agarose gel DNA purified Kit (TIANGEN, Beijing). Purified fragments were ligated onto pEASY-T1 vector (Transgene Beijing). Five positive clones were screened by PCR with M13 universal primers and sequenced on ABI 3730 sequencer (Invitrogen, Shanghai).
Amplification of 3' and 5' ends of DREB1
The Rapid Amplification of cDNA Ends (RACE) technology was employed to obtain 3' and 5' ends of the target gene. To amplify the 3' end of DREB1 from Iceland poppy, the gene-specific primers 3'RACE-GSP1 and 3'RACE-GSP2 were designed based on the sequence of conserved region of DREB1 obtained in a previous step (Table 1). Using the cDNA as template, PCR amplifications were performed using primer pair 3'RACE-GSP1 and 3UPM. The composition of the PCR mixture was the same as described above. The PCR was conducted as following program: 94 °C for 5 min, 30 cycles at 94 °C for 30 s, 54 °C for 40 s, 72 °C for 1 min, followed by 72 °C for 10 min and incubation at 12 °C. The resulting solution was 20-fold diluted and 1 μL was used as template in the second round of PCR by primer pair 3'RACE-GSP2 and 3UPM. The reaction mixture and program were the same as the first round of PCR. The final amplified products were also cloned and sequenced as previously described.
The 5' end of DREB1 was obtained by using 5' Full RACE Kit (Takara, Dalian). All reaction mixtures and programs were performed according to the protocols provided by the manufacture. The annealing temperatures for the first and second rounds of PCR amplifications were 55 °C and 53 °C, respectively. The subsequent PCR product separations, purifications, cloning, and sequencing were done as described above. The primers used are listed in Table 1.
Obtaining full sequences of DREB1 of the Iceland poppy
The two primers, DREB1-F2 and DREB1-R2, were designed based on the full length cDNA of DREB1 and were subjected to amplification of full cDNA and genomic sequences of DREB1. PCR amplifications were performed in 50 μL reaction volume, consisting of 2U Taq HiFi DNA polymerase (TRANSGENE, Beijing) with high fidelity, 5 μL of HiFi buffer (supplied with DNA polymerase), 2 μL cDNA or DNA templates, 200 pmol of dNTP mixture, and 400 pmol of each primer. The PCR program was: 94 °C for 5 min, 10 cycles at 94 °C for 30 s, 49 °C for 30 s, 72 °C for 1 min, 34 cycles at 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1min, and a final extension at 72 °C for 10 min. The resulting products were also gel-separated, purified, cloned, and sequenced.
Bioinformatics analyses
The deduced protein sequence was predicted by BioEdit (Hall, 1999). The homology modeling of DREB1 protein was performed by SWISS-MODEL with automated mode (Biasini et al., 2014). The model with the highest sequence similarity to the template and highest GMQE and QMEAN4 scores was chosen to predict the three-dimensional structure. Sequence similarity to the known DREB1s was investigated by BLASTP search against the nr protein database available on the website of National Center of Biotechnology Information and The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org).
The motifs in each protein were analyzed by Multiple Em for Motif Elicitation (MEME version 4. 10.1) (Timothy and Charles, 1994). The AP2/ERF domain in each DREB1 was identified by SMART (Letunic et al., 2012) and the corresponding sequence was retrieved. Multiple sequence alignment of amino acids of the AP2/ERF domain was conducted by using MUSCLE (Edgar, 2004) with default options. Motif logo representing the consensus sequence of AP2/ERF domains was drawn by using WebLogo (Crooks et al., 2004). MEGA5.2 software was employed to reconstruct the phylogenetic tree by maximum likelihood method, with 1000 bootstrap replications (Tamura et al., 2011). The Jones-Taylor-Thornton (JTT) model and a discrete Gamma distribution (+G) with 5 rate categories were chosen based on the test model.
Semi quantitative reverse transcription PCR (RT-PCR)
The spatial expression of PnDREB1 in petal, pedicel, leaf, petiole, and root were evaluated by semi quantitative RT-PCR. The primers used are listed in Table 1 and Actin1 was set as internal standard gene. As the sequence of Actin1 was unknown in Iceland poppy, we amplified and sequenced the Actin1 using primers (actin-F1 and actin-R1) (Table S1 (29.6KB, pdf) ) designed from known sequences of a wide range of plant species (data not shown) (Figure S1 (482.3KB, pdf) ). Then, a pair of primers (actin-F2 and actin-R2) was designed for semi quantitative RT-PCR analysis (Table 1). The 1 μL of 10 diluted cDNA reaction mixture was used as template in a 25 μL PCR volume. The PCR programs were: 94 °C for 3 min followed by 6 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and 19 (for actin1) or 24 (for PnDREB1) cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and a final 72 °C for 5 min. The amplifications for two genes were performed simultaneously in the same PCR thermal cycler with three replicates. The amplified products were separated by 1% agarose gel electrophoresis and visualized by ethidium bromide staining.
Quantitative real-time PCR (qPCR)
The cDNA templates were synthesized as mentioned previously. qPCR reactions were performed with a BioRad CFX system using the iQ SYBR Green supermix kit (Bio-Rad) according to the manufacturer's instructions. PCR procedure was: pre-incubation at 95 °C for 5 min followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 15 s, and extension at 72 °C for 15 s. The Actin1 and elongation factor 1a (EF1) (Long et al., 2010; Liang et al., 2012) were used as internal controls to quantify the relative transcript level. The sequence of EF1 was firstly obtained as mentioned above. The primers used for qPCR analyses are listed in Table 1. The amplification specificity was checked with a heat-dissociation protocol (melting curves in the 65–95 °C range) as a final step of the PCR. All primer pairs showed a single peak on the melting curve (Figure S2 (1.1MB, pdf) ). For each of the independent experiments, the target and internal control were amplified in separate wells in triplicate. The Cq values were determined automatically by BioRad CFX manager 2.1 (BioRad) and the mean Cq of triplicates was used to calculate the relative level of gene expression by using the 2–ΔΔCT method (Livak and Schmittgen, 2001). The final expression data are presented as means from three independent experiments.
Data analysis
Means and standard deviations (SD) of the expression data were calculated by using SPSS package version 16.0 (SPSS Inc.). Data were analyzed with one-way analysis of variance (ANOVA) and the mean differences were compared by the least significant difference (LSD) test.
Results
Cloning of a DREB1 gene from Iceland poppy
As no genomic resource for the Iceland poppy is available, a pair of degenerate primers, DREB1-F1 and DREB-F1, was designed based on the conserved AP2/ERF domains of DREB1s from several dicot species (data not shown). By RT-PCR, a fragment of 204 bp was obtained (Figure S3a (831.3KB, pdf) ) containing a AP2/ERF domain and showing high degree of sequence similarity to known DREB1/CBFs (Data not shown). Based on this sequence, further 3'RACE and 5'RACE were performed and a 767 bp and a 470 bp fragment were obtained, respectively (Figure S3b,c (831.3KB, pdf) ). The three fragments were assembled to a 1035 bp sequence. The sequence contains a continuous open reading frame (ORF) with an initiation codon (ATG) and a stop codon (TGA). A pair of primers was further designed to validate the obtained sequences by RT-PCR and genomic PCR (Figure S3c (831.3KB, pdf) ). The sequencing showed identical results as the assembled primer.
Sequence analyses
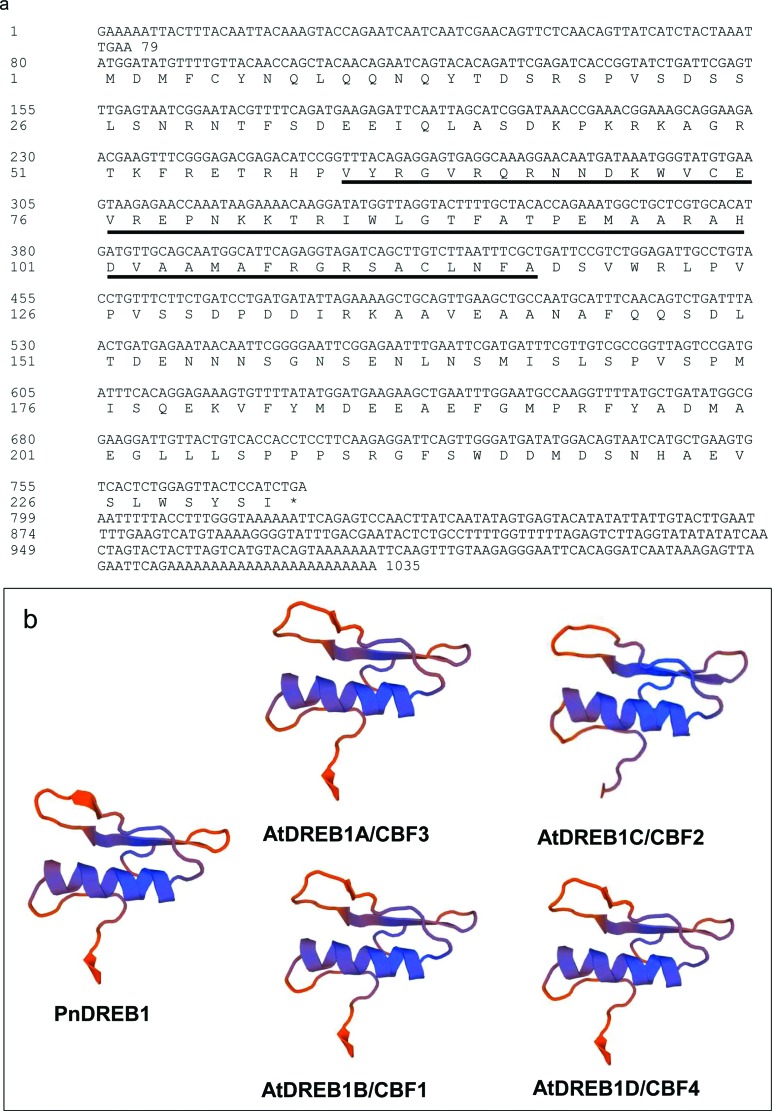
The ORF of the obtained sequence is 699 bp long and encodes a deduced protein of 232 amino acids, with 26.3 kDa molecular weight and isoelectric point of 5.33 (Figure 1a). BLAST search against Arabidopsis whole genome protein database (TAIR 10) was performed, which indicated that the obtained sequence showed the highest homology to six TFs of A-1 group of Arabidopsis DREB subfamily. Homology modeling indicated that the 3D structures of the obtained sequence and the four AtDREB1 proteins contained a conserved AP2/ERF domain with a typical three-dimensional conformation of three antiparallel β-sheets followed by a parallel α-helix (Figure 1b). These results suggested that the obtained gene belongs to DREB1 group of DREB TF subfamily, designated PnDREB1 (Table 2) (Accession No. KU500634). BLASTP search against the NCBI nr protein database indicated that PnDREB1 shares the highest sequence identity of only 58% (99% query cover and E value= 2e-80) to CBF1 of Morus alba var. multicaulis (GenBank accession number AFQ59977.1), indicating that PnDREB1 is a novel DREB1 gene.
Figure 1. Sequences of PnDREB1 (a) and comparison of 3D structures of four DREB1 proteins of Arabidopsis (b). The AP2/ERF domain is underlined.
Table 2. Homology of PnDREB1 to DREB1s in Arabidopsis.
| Gene ID | Description | Function | E value |
|---|---|---|---|
| AT5G51990.1 | DREB1D/CBF4 | Response to drought stress and ABA | 2e-50 |
| AT4G25470.1 | DREB1C/CBF2 | Response to low temperature and circadian rhythm | 4e-46 |
| AT4G25490.1 | DREB1B/CBF1 | Response to low temperature | 2e-45 |
| AT4G25480.1 | DREB1A/CBF3 | Response to low temperature | 2e-44 |
| AT1G12610.1 | DREB/DDF1 | Induce GA biosynthesis under salt stress | 4e-38 |
| AT1G63030.1 | DREB/DDF2 | Reduce ABA biosynthesis by overexpression | 5e-37 |
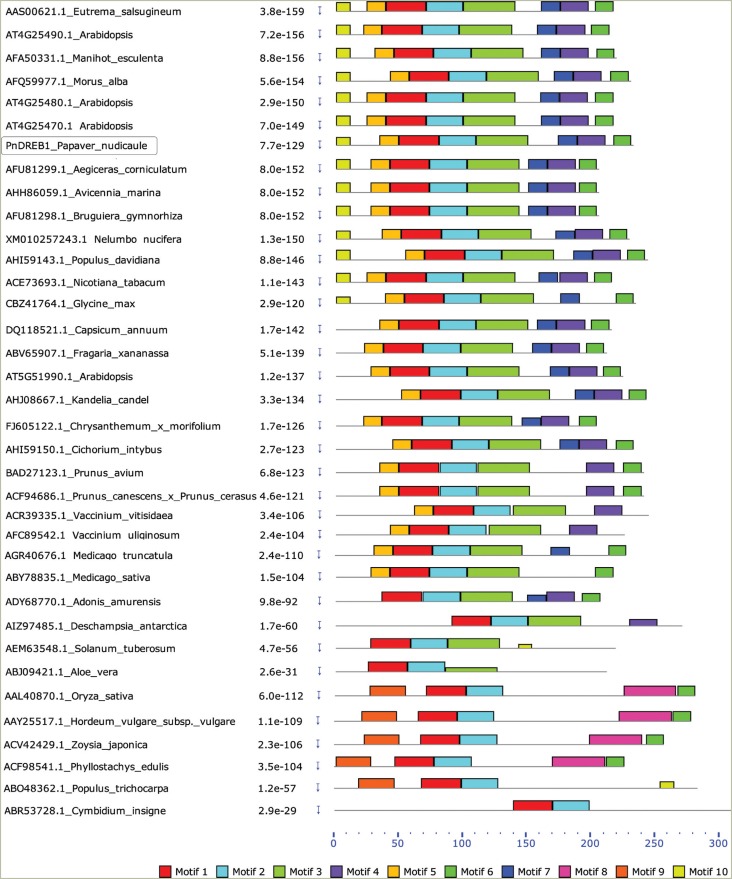
To evaluate the structural similarity, motif identification and comparison were performed between the PnDREB1 and 35 known DREB1 from 33 different dicots or monocots species (Figure 2). PnDREB1 contains eight motifs, which are similar to those of 12 DREB1s from 10 species, such as Arabidopsis, M. alba, Manihot esculenta, Avicennia marina, etc. Motif 1 and motif 2 are shared by all DREB1 proteins, covering the whole AP2/ERF domain. Motifs 3~7 are also common, present in ~79.5% to ~83.3% of the DREB1s analyzed, indicating that they might be functionally important to the DREB1s.
Figure 2. Comparison of protein motifs of 35 DREB1s from diverse dicot and monocot species. The PnDREB1 is boxed.
Comparison of AP2/ERF domain and phylogenetic analysis
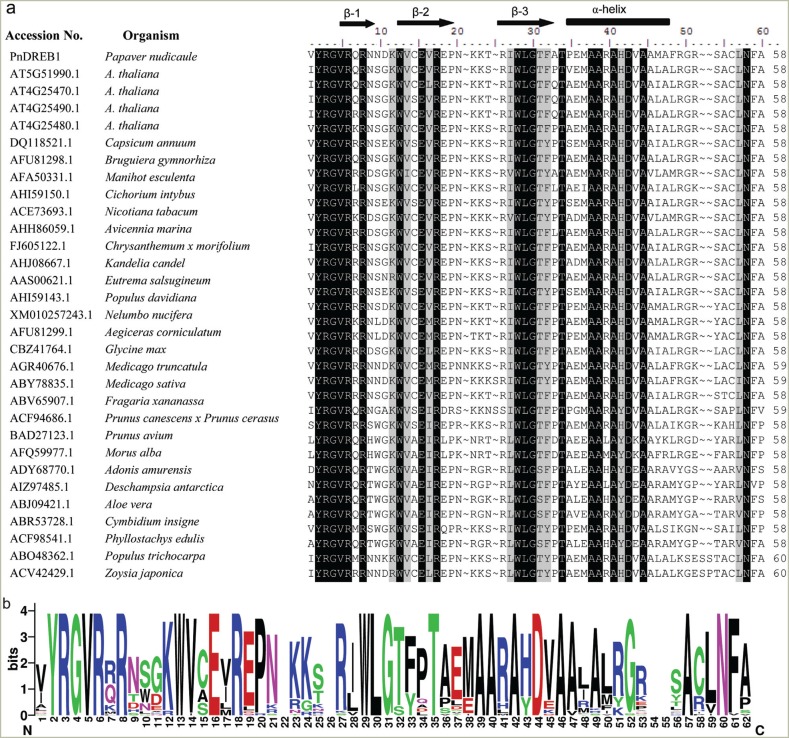
The AP2/ERF domain sequences were retrieved from the 36 DREB1s as described above. Most of the sequences are composed by 58 residues, with three exceptions, and 2 peptides containing 59 and 60 residues. The multiple sequence alignment showed that the AP2/ERF domain of PnDREB1 is highly homologous to other 35 DREB1s from divergent species (Figure 3a). A total of 19 amino acids are identical among 36 proteins, including motif YRGVR and WLG, and some other residues, such as Arg-8, Trp-13, etc. These conserved residues mainly lie in the regions comprising three β-sheets and one α-helix, which are structurally important. The residues outside these regions are rather divergent. The drawn domain logo showed the variability and conservation of each residue in AP2/ERF domain of DREB1s (Figure 3b).
Figure 3. Comparison of deduced amino acid sequences of AP2/ERF domain of 35 DREB1s from diverse dicot and monocot species. a, multiple alignment of amino acid sequences of AP2/ERF domain. Black shading indicates identical residues; gray shading indicates highly conserved residues. b, Motif logo drawn based on the multiple alignment of amino acid sequences of AP2/ERF domain. The overall height of the stack indicates the sequence conservation at that position, while the height of symbols within the stack indicates the relative frequency of each amino at that position.
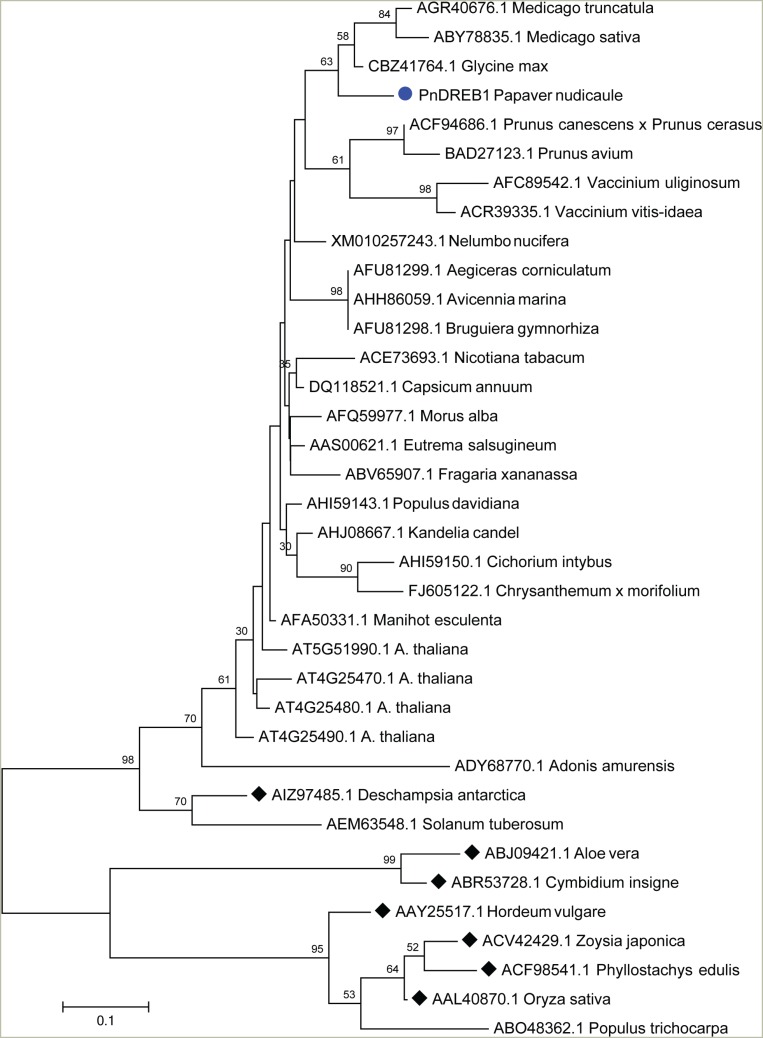
The phylogenetic tree constructed based on the alignment of amino acid sequences of AP2/ERF domain (Figure 4) showed that PnDREB1 was clustered with DREB1s of G. max, and species of Medicago, Prunus, and Vaccinium, and significantly separated from those of monocots.
Figure 4. Phylogenetic tree based on the deduced amino acid sequences of AP2/ERF domain of 35 DREB1s. The tree is constructed by maximum likelihood method with 1000 bootstrap replications. Before tree reconstruction, a model test was performed. The model with the lowest BIC scores (Bayesian Information Criterion), the Jones-Taylor-Thornton (JTT) model with parameters of Gamma distribution (+G) with 5 rate categories for Rates and Patterns were chosen. Diamonds indicate the monocot species.
Spatial expression patterns, freezing and phytohormone-induced responses
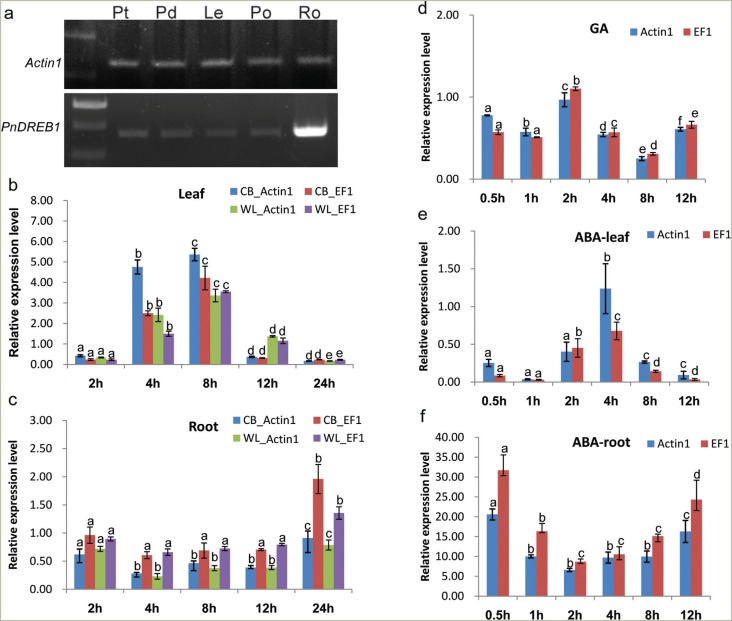
The spatial expression of PnDREB1 indicated that, under normal conditions, the expression of PnDREB1 could be detected in all analyzed tissues, including petal, pedicel, leaf, petiole, and root (Figure 5a). The root exhibited higher expression levels than other tissues.
Figure 5. Expression profiles of PnDREB1. (a), Semi quantitative RT-PCR analysis of spatial expression patterns in petal (Pt), pedicel (Pd), leaf (Le), petiole (Po), and root (Ro); b and c) show qPCR analysis results; relative expression levels of PnDREB1 at different time points (x-axis) of freezing treatment in leaf and root, respectively, in fold-change (y-axis); d) qPCR analysis results showing changes of relative expression levels of gibberellins acid- (GA3, 80 μM) treated plants under same treatment as those of b); e and f) fold changes of relative expression levels under treatment of 100 μM abscisic acid (ABA). CB, Champagne Bubbles; WL, Wonderland. Data are shown as means ± SD (n = 3). Actin1 and EF1 were used as internal controls. Different lowercase letters on rectangular columns indicate significant differences to that of previous time point (P < 0.05).
We further investigated the dynamic changes of PnDREB1 expression levels under freezing treatment (0 °C) by qPCR. In leaves, the expression level of PnDREB1 was very low at beginning of the treatment (Figure 5b). After 2h, the expression level significantly increased and reached a peak at 8 h; after 12 h, the expression decreased to similar levels as the initial stage of treatment. In roots, the expression level was slightly higher than those in leaves at the initial stage. After 2 h, the level decreased and remained low until 12 h, when it increased to a slightly higher levels than those before treatment (Figure 5c). We also evaluated the expression levels in another Iceland poppy variety (WL) with lower freezing tolerance. WL showed similar patterns in roots. However, in leaves, it increased slower and exhibited a significantly lower peak expression than CB (Figure 5b).
The responses of PnDREB1 to the phytohormones gibberellic acid (GA) and abscisic acid (ABA) were investigated by qPCR. Under freezing temperature, the GA3-treated plants exhibited lower expression levels compared to non-GA3-treated plants (Figure 5d). The ABA treatment was performed under hydroponic growth condition. In leaves, the expression level of PnDREB1 decreased to the lowest level at 1 h; one hour later, it gradually increased and reached a peak at 4 h; at 12 h, it decreased to a similar level as 1 h. At all time points, levels were lower than that of the control (untreated) (Figure 5e). In roots, the expression level intensely increased ~26-fold that of control within 0.5 h. After that, it was down-regulated to the lowest level of about ~7-fold at 2 h and then was up-regulated again ~20-fold at 12 h (Figure 5f).
Discussion
DREB1 has been characterized as an important regulator of cold response among a spectrum of plant species. In the present study, the cDNA and genomic sequences of a novel DREB1 TF, PnDREB1, with a high sequence similarity and similar predicted 3D structure to DREB1s of Arabidopsis, was isolated from the boreal ornamental plant Iceland poppy. Phylogenetic analysis indicated that the AP2 domain of PnDREB1 is close to those of G. max, and Medicago, Prunus, and Vaccinium species.
A motif is a pattern common to a set of nucleic or amino acid subsequences which share some biological property (Timothy and Charles, 1994). Thus, the motif compositions and distributions among a set of sequences reflect, to a certain extent, the structural and functional similarity. We compared the motifs of PnDREB1 to 35 known DREB1s from 33 species (Figure 2 and Figure S4 (1.5MB, pdf) ). All shared high conserved AP2/ERF domain, in which 19 residues are conserved in ~95% of DREB1s. Previous studies showed that the 14th valine (V14) and 19th glutamic acid (E19), especially the former, of the AP2/ERF domain, are conserved among the DREB protein (Liu et al., 1998). They are distinguished from alanine and aspartic acid of ERF protein and are important for its binding specificity (Sakuma et al., 2002). PnDERB1 contained the same conserved V14 and E19 at these two positions, indicating that it might possess similar binding patterns as DREB1s of Arabidopsis to DRE/CRT motif in the promoter of some downstream stress-induced genes.
Nakano et al. (2006) reported that some motifs outside the AP2/ERF domain are also conserved for DREB1 proteins. Motif CMIII-1 is common for DREB1s; CMIII-2 and CMIII-4 are conserved in C-terminal region, and CMIII-4, also known as LWSY motifs, is conserved in rice and Arabidopsis and has been shown to function as a transactivation domain (Wang et al., 2005). The CMIII-3, separated by AP2/ERF domain, is also conserved and was reported in other studies (Jaglo et al., 2001; Haake et al., 2002). Despite of different methods used for motif identification, PnDREB1 was found to contain all of these motifs: motif 3 covers CMIII-1; motif 5 and part of motif 3 is equivalent to CMIII-3; the adjacent motif 7 and motif 4 are approximate to the CMIII-2, and CMIII-4 is involved in motif 6. These results indicated that PnDREB1 might be an active stress-induced DREB1 protein.
Our further investigation of dynamic expression changes under freezing treatment showed that PnDREB1 was induced by freezing both in leaves and roots though in different patterns. The expression level in leaves was quickly upregulated and reached peak level at 8h. These results are similar to some reports in other species (Stockinger et al., 1997; Liu et al., 1998; Qin et al., 2004; Huang et al., 2007; Shan et al., 2007; Kidokoro et al., 2015). Interestingly, freezing-induced expression in leaves could be suppressed by exogenous GA3. This phenomenon was also found in cotton, indicating that it may play an important role in GA signaling (Shan et al., 2007). Our comparative analysis also indicated that expression of PnDREB1 in the CB variety with high freezing tolerance increases faster and accumulates to higher levels than those in WL variety with lower freezing tolerance. The difference in freezing inductive accumulation of PnDREB1 transcription level might partly contribute to their different performance under freezing tolerance.
In roots, PnDREB1 exhibits higher expression levels than those of other tissues under normal condition. However, in mangrove Aegiceras corniculatum, the highest expression was detected in leaves (Peng et al., 2015). This suggests that DREB1 may function diversely in plant development in different species. Under freezing stress, PnDREB1 was induced gradually and exhibited first a down- and then up-regulated pattern, which seems to be complementary to that in leaves (Figures 5b and c). Few reports individually addressed the expression changes in roots under stress. However, we speculated that this might be due to two reasons: first, our freezing treatment was performed under hydroponic condition, by which the leaves might perceive freezing stress more quickly than roots; second, there might exist a balance PnDREB1 expression between roots and leaves.
ABA is an important plant hormone that plays a regulatory role in many development processes in plants, as well as in the activation of stress-responsive genes (Agarwal and Jha, 2010). Previous studies in Arabidopsis showed that DREB1D/CBF4 is rapidly induced by drought and ABA but not by cold stress (Haake et al., 2002), whereas DREB1B/CBF1, DREB1A/ CBF3, and DREB1C/ CBF2 are strongly and transiently induced by low temperature stresses but not by ABA or dehydration (Gilmour et al., 1998; Medina et al., 1999). However, these different results come from diverse plant species. PNDREB1 of Arachis hypogaea was strongly upregulated by treatments with low temperature, and also responded to dehydration (Zhang et al., 2009); PpDBF1 of Physcomitrella patens was simultaneously induced by NaCl, cold, drought, and ABA (Liu et al., 2007). The results obtained in this study showed that besides freezing treatment, PnDREB1 is also rapidly induced by ABA, especially in roots, suggesting that PnDREB1 is possibly involved in other abiotic stress responses, such as drought and NaCl. Further research is needed to clarify this speculation.
Acknowledgments
We are grateful to Institute of Ecology and Forestry of Sichuan Agricultural University for their helpful assistance in the experiments. This work is financially supported by the Education Department of Sichuan Province, China (No. 15ZA0014).
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Adriana S. Hemerly
References
- Agarwal PK, Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol Plantarum. 2010;54:201–212. [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002;130:639–648. doi: 10.1104/pp.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK. Natural genetic variation of freezing tolerance in Arabidopsis . Plant Physiol. 2006;142:98–112. doi: 10.1104/pp.106.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Tong Z, Li QH, Ma C, Kasuga M, Yamaguchi-Shinozaki K, Gao JP. Regeneration and transformation through somatic embryogenesis, and determination of cold stress tolerance in ground cover Chrysanthemum cv. Fall color. Sci Agric Sin. 2006a;39:1443–1450. [Google Scholar]
- Hong B, Tong Z, Ma N, Kasuga M, Yamaguchi-Shinozaki K, Gao JP. Expression of Arabidopsis DREB1A gene in transgenic chrysanthemum enhances tolerance to low temperature. J Hortic Sci Biot. 2006b;81:1002–1008. [Google Scholar]
- Hong B, Tong Z, Ma N, Li JK, Kasuga M, Yamaguchi-Shinozaki K, Gao JP. Heterologous expression of the AtDREB1A gene in chrysanthemum increases drought and salt stress tolerance. Sci China C Life Sci. 2006c;49:436–445. doi: 10.1007/s11427-006-2014-1. [DOI] [PubMed] [Google Scholar]
- Hong JP, Kim WT. Isolation and functional characterization of the Ca-DREBLP1 gene encoding a dehydration-responsive element binding-factor-like protein 1 in hot pepper (Capsicum annuum L. cv Pukang) Planta. 2005;220:875–888. doi: 10.1007/s00425-004-1412-5. [DOI] [PubMed] [Google Scholar]
- Huang B, Jin L, Liu J. Molecular cloning and functional characterization of a DREB1/CBF-like gene (GhDREB1L) from cotton. Sci China Ser C-Life Sci. 2007;50:7–14. doi: 10.1007/s11427-007-0010-8. [DOI] [PubMed] [Google Scholar]
- Istatkova R, Philipov S, Yadamsurenghiin GO, Samdan J, Dangaa S. Alkaloids from Papaver nudicaule L. Nat Prod Res. 2008;22:607–611. doi: 10.1080/14786410701605315. [DOI] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001;127:910–917. [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. A combination of the Arabidopsis DREB1A gene and stress-inducible RD29A promoter improved drought and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 2004;45:346–350. doi: 10.1093/pcp/pch037. [DOI] [PubMed] [Google Scholar]
- Kidokoro S, Watanabe K, Ohori T, Moriwaki T, Maruyama K, Mizoi J, Myint PSHN, Fujita Y, Sekita S, Shinozaki K, et al. Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 2015;81:505–518. doi: 10.1111/tpj.12746. [DOI] [PubMed] [Google Scholar]
- Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot. 2011;62:4731–4748. doi: 10.1093/jxb/err210. [DOI] [PubMed] [Google Scholar]
- Lata C, Bhutty S, Bahadur RP, Majee M, Prasad M. Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet (Setaria italica L.) J Exp Bot. 2011;62:3387–3401. doi: 10.1093/jxb/err016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Deng G, Long H, Pan Z, Wang C, Cai P, Xu D, Nima Z-X, Yu M. Virus-induced silencing of genes encoding LEA protein in Tibetan hulless barley (Hordeum vulgare ssp. vulgare) and their relationship to drought tolerance. Mol Breeding. 2012;30:441–451. [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013;199:639–649. doi: 10.1111/nph.12291. [DOI] [PubMed] [Google Scholar]
- Liu N, Zhong N, Wang G, Li L, Liu X, He Y, Xia G. Cloning and functional characterization of PpDBF1 gene encoding a DRE binding transcription factor from Physcomitrella patens . Planta. 2007;226:827–838. doi: 10.1007/s00425-007-0529-8. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2 with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought and low-temperature-responsive gene expression, respectively, in Arabidopsis . Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long XY, Wang JR, Ouellet T, Rocheleau H, Wei YM, Pu ZE, Jiang QT, Lan XJ, Zheng YL. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol Biol. 2010;74:307–311. doi: 10.1007/s11103-010-9666-8. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004;38:982–993. doi: 10.1111/j.1365-313X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- Medina J, Bargues M, Tero J, Pérez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999;119:463–469. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakenfull RJ, Baxter R, Knight MR. A C-repeat binding factor transcriptional activator (CBF/DREB1) from European Bilberry (Vaccinium myrtillus) induces freezing tolerance when expressed in Arabidopsis thaliana . PLoS One. 2013;8:e54119. doi: 10.1371/journal.pone.0054119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrineschi A, Reynolds M, Pacheco M, Brito RM, Almeraya R, Yamaguchi-Shinozaki K, Hoisington D. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome. 2004;47:493–500. doi: 10.1139/g03-140. [DOI] [PubMed] [Google Scholar]
- Peng Y-L, Wang Y-S, Cheng H, Wang L-Y. Characterization and expression analysis of a gene encoding CBF/DREB1transcription factor from mangrove Aegiceras corniculatum . Ecotoxicology. 2015;24:1733–1743. doi: 10.1007/s10646-015-1485-x. [DOI] [PubMed] [Google Scholar]
- Philipov S, Istatkova R, Yadamsurenghiin GO, Samdan J, Dangaa S. A new 8,14-dihydropromorphinane alkaloid from Papaver nudicaule L. Nat Prod Res. 2007;21:852–856. doi: 10.1080/14786410701494777. [DOI] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Li J, Liu Q, Li Y-Q, Shinozaki K, Yamaguchi-Shinozaki K. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol. 2004;45:1042–1052. doi: 10.1093/pcp/pch118. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D-P, Huang J-G, Yang Y-T, Guo Y-H, Wu C-A, Yang G-D, Gao Z, Zheng C-C. Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytologist. 2007;176:70–81. doi: 10.1111/j.1469-8137.2007.02160.x. [DOI] [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun. 1998;250:161–170. doi: 10.1006/bbrc.1998.9267. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony method. Mol Bio Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsis EC, Eylert E, Maddula RK, Ostrozhenkova E, Svato A, Eisenreich W, Schneider B. Biosynthesis of Nudicaulins: A 13CO2-pulse/chase labeling study with Papaver nudicaule . Chem Biol Chem. 2014;15:1645–1650. doi: 10.1002/cbic.201402109. [DOI] [PubMed] [Google Scholar]
- Timothy LB, Charles EL. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, AAAI; Menlo Park, California. 1994. pp. 28–36. [PubMed] [Google Scholar]
- Tong Z, Hong B, Yang Y, Li Q, Ma N, Ma C, Gao J. Overexpression of two Chrysanthemum DgDREB1 group genes causing delayed flowering or dwarfism in Arabidopsis . Plant Mol Biol. 2009;71:115–129. doi: 10.1007/s11103-009-9513-y. [DOI] [PubMed] [Google Scholar]
- Wang X, Chen X, Liu Y, Gao H, Wang Z, Sun G. CkDREB gene in Caragana korshinskii is involved in the regulation of stress response to multiple abiotic stresses as an AP2/EREBP transcription factor. Mol Biol Rep. 2011;38:2801–2811. doi: 10.1007/s11033-010-0425-3. [DOI] [PubMed] [Google Scholar]
- Wang Z, Triezenberg SJ, Thomashow M, Stockinger EJ. Multiple hydrophobic motifs in Arabidopsis CBF1 COOH-terminus provide functional redundancy in trans-activation. Plant Mol Biol. 2005;58:543–559. doi: 10.1007/s11103-005-6760-4. [DOI] [PubMed] [Google Scholar]
- Xiao H, Siddiqua M, Braybrook S, Nassuth A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 2006;29:1410–1421. doi: 10.1111/j.1365-3040.2006.01524.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Zhang M, Liu W, Bi Y-P, Wang Z-Z. Isolation and identification of PNDREB1 a new DREB transcription factor from peanut (Arachis hypogaea L.) Acta Agron Sin. 2009;35:1973–1980. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.