Abstract
This study used qRT-PCR to examine variation in the expression of 13 myogenes during muscle development in four prenatal periods (21, 40, 70 and 90 days post-insemination) in commercial (the three-way Duroc, Landrace and Large-White cross) and local Piau pig breeds that differ in muscle mass. There was no variation in the expression of the CHD8, EID2B, HIF1AN, IKBKB, RSPO3, SOX7 and SUFU genes at the various prenatal ages or between breeds. The MAP2K1 and RBM24 genes showed similar expression between commercial and Piau pigs but greater expression (p < 0.05) in at least one prenatal period. Pair-wise comparisons of prenatal periods in each breed showed that only the CSRP3, LEF1, MRAS and MYOG genes had higher expression (p < 0.05) in at least one prenatal period in commercial and Piau pigs. Overall, these results identified the LEF1 gene as a primary candidate to account for differences in muscle mass between the pig breeds since activation of this gene may lead to greater myoblast fusion in the commercial breed compared to Piau pigs. Such fusion could explain the different muscularity between breeds in the postnatal periods.
Keywords: commercial line, gene expression, myogenesis, pig, Piau breed
Introduction
Myogenesis is a prenatal process that involves the formation of muscle fibers through changes in gene expression and cell phenotype, and is influenced by genetic and environmental factors (Te Pas et al., 2005). The size and number of muscle fibers determine muscle mass since skeletal muscle growth depends on the number of fibers formed during myogenesis and on postnatal muscle hypertrophy, which is limited by physiological and genetic factors (Rehfeldt et al., 2000).
During prenatal development, two waves of myoblast proliferation and fusion give rise to primary and secondary muscle fibers (Wigmore and Evans, 2002). Primary muscle fibers are formed de novo in the early stages of myoblast fusion (first wave of differentiation) and secondary fibers use the primary fibers as a template in a second wave of differentiation (Rehfeldt et al., 2000; Te Pas et al., 2005); the latter fibers account for the majority of fibers in skeletal muscle (Beermann et al., 1978). The number and size of primary myotubes are intrinsic factors that affect the number of secondary fibers. The number of secondary myotubes is sensitive to external factors such as nutrition, while the number of primary myotubes is genetically programmed and is unaffected by exogenous influences (Maltin et al., 2001). In pigs, the waves of muscle fiber formation involve relatively long periods of time, i.e., ~30-60 days and 54-90 days of gestation for the first and second waves, respectively (Wigmore and Stickland, 1983).
The changes in gene expression associated with muscle development and growth have been examined in different breeds of pigs at various prenatal ages (Te Pas et al., 2005; Cagnazzo et al., 2006; Murani et al., 2007; Sollero et al., 2011; Zhao et al., 2011). The analysis of changes in myogenic gene expression during the prenatal period, when the two waves of myoblast fusion occur, can be very important for understanding the biochemical differences that contribute to distinct interbreed variations in the degree of muscularity and meat quality.
In this study, we analysed the expression of myogenes in a commercial pig line (the three-way Duroc, Landrace and Large-White cross) and Piau pigs, which differ in muscularity from the commercial breed, in order to assess possible differences in gene expression during myogenesis.
Material and Methods
Biological material
Embryos and fetuses were obtained by cesarean section from three unrelated pregnant gilts for each genetic group of pigs (local Piau and commercial breeds) at 21, 40, 70 and 90 days post-insemination (dpi) maintained at the Pig Breeding Farm of the Departamento de Zootecnia at the Universidade Federal de Viçosa (UFV). Embryos and fetuses collected from the commercial breed were obtained from gilts of the three-way Duroc, Landrace and Large-White cross. Samples from three unrelated fetuses and embryos were used as biological replicates in gene expression analysis for each breed at 21, 40, 70 and 90 dpi. The procedures for obtaining the embryos and fetuses were approved by the Ethics Committee for Animal Use at UFV (protocol no. CEUA-UFV 85/2013), in accordance with current Brazilian federal legislation.
Longissimus dorsi (LD) muscle was collected at all periods, except from 21 dpi embryos, for which the whole individual was collected and used in RNA extraction. Samples were immediately placed in Falconer tubes containing 10 ml of RNA holder solution (BioAgency Laboratórios, Brazil) and sent to the Animal Biotechnology Laboratory at the Departamento de Zootecnia (UFV) where they were stored overnight at 4 °C and then transferred to a freezer at −80 °C until RNA extraction.
RNA extraction
Total RNA was extracted using TRIzol® reagent (Life Technologies, USA). The RNA was quantified in a NanoVue Plus spectrophotometer (GE Healthcare, Germany) and RNA A260/A280 ratios of 1.8-2.0 were used as an indicator of purity. The quality and intactness of extracted RNA were verified by electrophoresis in a 1% agarose gel. Total RNA was stored at −70 °C until cDNA synthesis.
cDNA synthesis
Prior to the reverse transcription step, RNA was treated with DNase I amplification grade (Invitrogen, USA) to remove contaminant DNA, according to the manufacturer's instructions. The first strand of complementary DNA (cDNA) was synthesized using a GoScriptTM reverse transcription system (Promega, USA), according to the manufacturer's recommendations. cDNA concentrations were estimated spectrophotometrically in NanoVueTM plus (GE Healthcare) and single stranded cDNA was stored at −20 °C until used in qPCR assays.
Gene selection and primer design for qRT-PCR
The genetic data for differentially expressed genes (DEG) from an RNAseq experiment were used to compare gene expression between breeds in embryos and fetuses of a commercial pig breed (a two-way Landrace and Large-White cross) and Piau pigs (unpublished data). HUGO Gene Nomenclature Committee (HGNC) symbols for genes were obtained with the BIOMART/ENSEMBL program using ENSEMBL transcript identifications for DEG in conjunction with the pig (Sus scrofa) database. When HGNC symbols were not available for pigs, they were obtained by orthology using the Homo sapiens database. The HGNC symbols for genes were subsequently subjected to gene ontology analysis.
Information on gene ontology for the genes was obtained using the ToppCluster program (Kaimal et al., 2010). The terms related to muscle development (myogenesis) were identified and their genes was used in metabolic pathway analysis. Metabolic pathway maps from KEGG (Kyoto Encyclopedia of Genes and Genomes) were obtained using DAVID software (Dennis et al., 2003; Huang et al., 2009). Cytoscape software (Shannon et al., 2003) was used to view and edit the biological processes, molecular functions and metabolic pathways identified with ToppCluster (Kaimal et al., 2010) and DAVID (Dennis Jr. et al., 2003; Huang et al., 2009) software. In addition, gene functions were evaluated using a Gene Cards database (Safran et al., 2010) that provided concise information on all known and predicted human genes, in addition to information on gene ontologies that was not obtained by Toppcluster (Kaimal et al., 2010).
Based on these findings, 13 genes were selected for analysis of their expression profiles using RT-qPCR. This work focused on genes that were differentially expressed between breeds because they represented primary candidates for information on the source of variation in muscularity and meat quality. The selected genes were related to myogenesis and were chosen based on gene ontology, the identification of metabolic pathways and their function.
qPCR primers were designed using PrimerQuest® software (Owczarzy et al., 2008) and nucleotide sequences obtained from the S. scrofa transcriptome database at GenBank (Benson et al., 2013). The only nucleotide sequence not available for pigs was that of RSPO3, for which a homologous sequence from humans (Homo sapiens) was used. Table 1 summarizes relevant information for the genes that were studied, including the accession numbers of the transcript sequences used in primer design, the nucleotide sequences of the primers and amplicon size.
Table 1. GenBank accession numbers, primer sequences and amplicon sizes of the genes analyzed in this study.
| Gene | Accession number | Primer sequences (5’ → 3’) | Amplicon size1 |
|---|---|---|---|
| CHD8 | XM_003482263.1 | F: AGTGAGGACGAGAAGGAAGA | 104 |
| R: GGGAATCCATCTTGGGACATAG | |||
| CSRP3 | NM_001172368.1 | F: CAGCAACCCTTCCAAGTTCA | 91 |
| R: CATCACCTTCTCAGCAGCATAG | |||
| EID2B | XM_003127131.1 | F: CGCCACTATCTGGAACACTAC | 122 |
| R: CGCTGATATTCGGCATCAAAC | |||
| HIF1AN | XM_003359328.1 | F: GTACTGGTGGCATCACATAGAG | 118 |
| R: CTGATGGGCTTTGAGAGGATATT | |||
| IKBKB | NM_001099935.1 | F: GATGGCGACAGTCAGGAAAT | 107 |
| R: TTGCAAACCACCGTCTTACT | |||
| LEF1 | NM_001129967.1 | F: CTATTGTAACGCCTCAGGTCAA | 99 |
| R: TTGGCTCTTGCTCCTTTCTC | |||
| MAP2K1 | NM_001143716.1 | F: GGAGCTGGAGCTGATGTTT | 110 |
| R: GTCGGCTGTCCATTCCATAA | |||
| MRAS | XM_003358570.2 | F: GGTCGATTTGATGCATTTGAGG | 96 |
| R: TCCTTGGCACTGGTTTCTATG | |||
| MYOG | NM_001012406.1 | F: CAGGCTCAAGAAGGTGAATGA | 118 |
| R: GCACTCGATGTACTGGATGG | |||
| RBM24 | XM_001925447.3 | F: TACCTGCCCACTATGTCTATCC | 118 |
| R: GCAGCTCCCGTGTAATCAAT | |||
| RSPO3 | NM_032784.4 | F: GAAACACGGGTCCGAGAAATA | 110 |
| R: CCCTTCTGACACTTCTTCCTTT | |||
| SOX7 | XM_003359052.1 | F: TCTCCACTCCAACCTCCA | 120 |
| R: TCATTGCGATCCATGTCCTC | |||
| SUFU | XM_001928912.4 | F: GGAGCCCTCATTCCTCTTTG | 83 |
| R: GCCATGTCACCTGTGATACTT | |||
| ACTB2 | XM_003124280.3 | F: AAGATCAAGATCATCGCGCCTCCA | 108 |
| R: ACTCCTGCTTGCTGATCCACATCT | |||
| GAPDH2 | NM_001206359.1 | F: ACAGTCTTCTGGGTGGCAGTGAT | 176 |
| R: CATGTTTGTGATGGGCGTGAACAA | |||
| HPRT12 | NM_001032376.2 | F: GCTGACCTGCTGGATTACAT | 101 |
| R: CTGGTCATTACAGTAGCTCTTCAG |
Amplicon size in nucleotide number,
Reference gene. CHD8 – chromodomain helicase DNA binding protein 8, CSRP3 – cysteine and glycine-rich protein 3, EID2B – EP300 interacting inhibitor of differentiation 2B, HIF1AN – hypoxia inducible factor 1, α subunit inhibitor, IKBKB - inhibitor of κ light polypeptide gene enhancer in B-cells, kinase β, LEF1 – lymphoid enhancer-binding factor 1, MAP2K1 – mitogen-activated protein kinase kinase 1, MRAS – muscle RAS oncogene homolog, MYOG – myogenin (myogenic factor 4), RBM24 – RNA binding motif protein 24, RSPO3 – R-spondin 3, SOX7 – SRY (sex determining region Y)-box 7, SUFU – suppressor of fused homolog, ACTB – β-actin, GAPDH – glyceraldehyde-3-phosphate dehydrogenase, HPRT1 – hypoxanthine phosphoribosyltransferase 1.
Testing of the designed primers
Primer amplification was assessed with the polymerase chain reaction (PCR) using 75 ng of pooled cDNA derived from embryo and fetal tissues of commercial and Piau pigs. Primers were tested at 200 nM and an annealing temperature of 60 °C in a Veriti® 96-well thermal cycler (Applied Biosystems, USA). PCR was done with a GoTaq® Green master mix kit (Promega) according to the manufacturer's protocol. The amplification products were screened for reaction specificity and the presence of primer dimers by electrophoresis on 8% polyacrylamide gels at 100 V for 2 h.
Real time qPCR
Real time qPCR reactions were run in an ABI Prism 7300 Sequence Detection Systems thermocycler (Applied Biosystems) using a Gotaq® qPCR master mix kit (Promega) according to the manufacturer's protocol. cDNA (25, 75 or 225 ng) or nucleic acid-free water (negative control) was added to each well of the plate along with upstream and downstream primers at 100, 200 or 400 nM.
The amplification conditions were: 95 °C for 2 min, 40 denaturation cycles at 95 °C for 15 s, and annealing and extension at 60 °C or 61 °C for 60 s. The efficiency of amplification was assessed after 40 amplification cycles by including an additional step in which the temperature was gradually raised from 60 °C to 94 °C to obtain the primer dissociation curve.
Target and reference gene amplifications were done in different wells of the same plate. The assays were run with three biological and two technical replicates for each treatment in a single 96-well plate. The coefficient of variation, used as an indicator of precision and reproducibility, was less than 5%, which was adequate for the Ct (threshold cycle) values of the technical replicates within each sample.
Amplification efficiency
To calculate the amplification efficiency of target and reference genes, all reactions were done in 96-well plates using pooled cDNAs containing biological replicates of treatments and two technical replicates for each treatment. From the real time qPCR raw data, the Ct values and log10 amount of cDNA (25, 75 and 225 ng) were plotted in graphs for primers at 100, 200 and 400 nM. The slope of the resulting relationship was determined by linear regression and was used to calculate the amplification efficiency with the following equation, modified from Rasmussen (2001), in which efficiencies equal to 1 represent 100%:
Based on the results obtained in the preceding step, the highest amplification efficiency of target and reference genes was chosen, along with the appropriate primer concentration and quantity of cDNA for each gene to be used in the final qPCR reactions. Amplification efficiencies of 0.80 to 1.0 were considered appropriate (Table S1 (448.2KB, pdf) ). The suitability of three reference genes (ACTB, GAPDH and HPRT1) for qRT-PCR was investigated using NormFinder software (Andersen et al., 2004), and GAPDH was selected for data normalization since the gene used for this procedure should show no change in expression along the treatments. Ct values for a specific gene were normalized to the Ct value of highest expression (Ct minimum value) for that gene. The normalized Ct values were then used in the following equation (McCulloch et al., 2012):
where: Q = normalized Ct value for a gene in the current sample, E = calculated amplification efficiency (ranging from 1 to 2, in which 100% = 2), min Ct = minimum Ct value for a gene among all samples, and Ct sample = Ct value for current sample and gene.
Statistical analysis
The experimental design was a completely randomized 2 (breeds) x 4 (prenatal ages) factorial design with six repetitions (three biological and two technical replicates) per treatment. ANOVA was done using the following statistical model:
Yijkl is the expression level of gene k, in animal l, breed i and prenatal age j, in which i = 1 or 2 (commercial or Piau breed, respectively) and j = 1, 2, 3 or 4 (21, 40, 70 or 90 dpi, respectively),
μ is the general constant,
A(ij)l is the random effect of animal l in breed i and age j,
A(ij)l ~ N(0, σ2 A),
Dijk is the sample-specific random effect (common to both genes), Dijk ~ N(0, σ2 D),
(RIG)ijk is the interaction effect between breed i and age j in gene k, and
εijkl is the random error, i.e., εijkl ~ N(0, σ2 e).
This model was fitted to the data using the %QPCR_MIXED macro in SAS (Statistical Analysis System Institute Inc., USA), which is based on linear mixed models (Steibel et al., 2009). The significance of contrast estimate values was assessed using Student's t-test. Contrast estimate values correspond to ΔΔCt and were used to assess relative expression (fold-change) by using the formula 2–ΔΔCt (Livak and Schmittgen, 2001). In all cases, the level of significance was set at p < 0.05.
Results
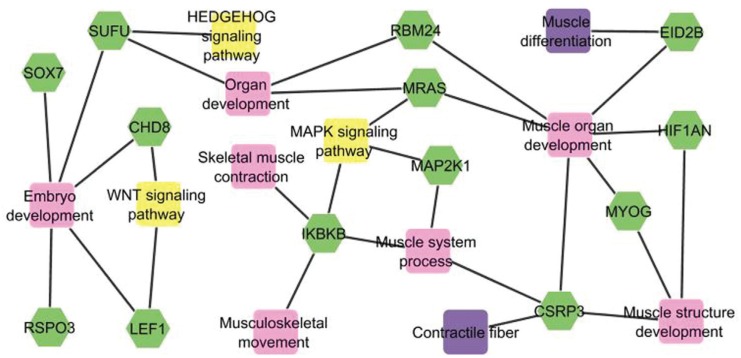
To understand the role of the 13 selected genes, information on gene ontology was obtained using Toppcluster software and metabolic pathways were investigated using DAVID software. These 13 genes are part of relevant functional metabolic networks for muscle development (Figure 1). These networks are for skeletal muscle contraction, muscle structure development, embryo development, organ development, muscle organ development, muscle differentiation, contractile fiber, musculoskeletal movement, muscle system process, and HEDGEHOG, MAPK and WNT signaling pathways. In addition, Table 2 describes the function of these 13 genes as defined in gene ontology terms.
Figure 1. Functional gene networks and their interactions, showing the relationship between 13 genes (green). Twelve important subnets related to muscle development were included in biological process (pink), molecular function (violet) and metabolic pathway (yellow).
Table 2. Metabolic pathways and gene ontologies for genes represented in the gene network.
| Gene ontologies | ||||
|---|---|---|---|---|
| Gene | Metabolic pathway | Cellular component | Molecular function | Biological process |
| CHD8 | WNT signaling pathway | Nuclear lumen | Histone and DNA binding | Embryo development |
| CSRP3 | – | Cytoskeleton | Contractile fiber | Muscle organ development/Muscle structure development/Muscle system process |
| EID2B | – | Nucleus | Muscle differentiation | Muscle organ development |
| HIF1AN | – | Nucleus/cytosol | Protein binding | Muscle organ development/Muscle structure development |
| IKBKB | MAPK signaling pathway | Nucleus/cytosol | Protein binding | Muscle system process/Musculoskeletal movement/Skeletal muscle contraction |
| LEF1 | WNT signaling pathway | Nucleus | Chromatin and DNA binding | Embryo development |
| MAP2K1 | MAPK signaling pathway | Cytoskeleton/cytosol/nucleus | Protein kinase activity | Muscle system process |
| MRAS | MAPK signaling pathway | Intracellular | GTPase activity/nucleotide binding | Muscle organ development/Organ development |
| MYOG | – | Nucleus | Chromatin and DNA binding | Muscle organ development/Muscle structure development |
| RBM24 | – | Nucleus/cytoplasm | Nucleotide binding | Muscle organ development/Organ development |
| RSPO3 | – | Extracellular region | Receptor binding | Embryo development |
| SOX7 | – | Nucleus/cytoplasm | Nucleic acid binding | Embryo development |
| SUFU | Hedgehog signaling pathway | Nucleus/cytoplasm | Transcription corepressor activity | Embryo development/Organ development |
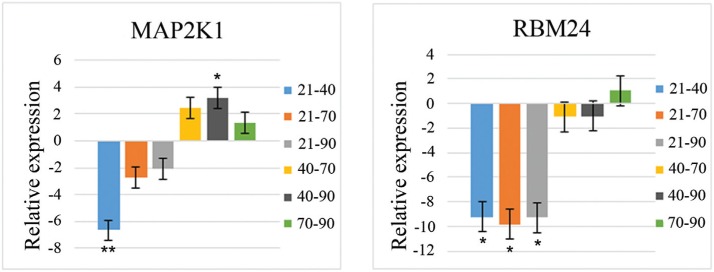
qRT-PCR assays were done at four prenatal ages (21, 40, 70 and 90 dpi) in commercial and Piau pigs and the data were analyzed using ANOVA (Table 3). Seven genes (CHD8, EID2B, HIF1AN, IKBKB, RSPO3, SOX7 and SUFU) showed no significant alterations, indicating that their expression was constant over time and between breeds. Two genes (MAP2K1 and RBM24) showed significant changes (p < 0.05) in relation to the prenatal period, but there was no significant Breed x Period interaction for these genes. Table 4 shows the Student's t-test results for pair-wise comparisons among the prenatal periods for these genes. The relative gene expression (fold-change) for pair-wise comparisons of prenatal periods is shown in Figure 2. Of the genes analyzed, MAP2K1 showed greater expression at 40 dpi (period of primary fiber formation) and 70 dpi (period of secondary fiber formation), whereas RBM24 showed greater expression at 40 dpi (period of primary fiber formation) and at 70 and 90 dpi (periods of secondary fiber formation).
Table 3. P-values for ANOVA in relation to Breed, Period and interaction Breed x Period for the genes studied.
| Genes | Factors | ||
|---|---|---|---|
| Breed | Period | Breed x Period | |
| CHD8 | 0.9764 | 0.3615 | 0.6094 |
| CSRP3 | 0.8615 | < 0.0001 | < 0.0001 |
| EID2B | 0.9072 | 0.1615 | 0.4284 |
| HIF1AN | 0.5757 | 0.2535 | 0.5533 |
| IKBKB | 0.7473 | 0.6656 | 0.6948 |
| LEF1 | 0.9772 | < 0.0001 | 0.0004 |
| MAP2K1 | 0.4445 | 0.0125 | 0.0712 |
| MRAS | 0.6557 | 0.0019 | 0.0205 |
| MYOG | 0.7314 | < 0.0001 | < 0.0001 |
| RBM24 | 0.9866 | 0.0284 | 0.1756 |
| RSPO3 | 0.7198 | 0.0885 | 0.2230 |
| SOX7 | 0.3451 | 0.4870 | 0.6915 |
| SUFU | 0.6504 | 0.2974 | 0.4175 |
Values in bold were statistically significant (p < 0.05) by F-Test;
Table 4. P-values for two-period comparisons for the genes MAP2K1 and RBM24. The ANOVA results (F-test) for these genes were significant for the factor Period.
| Genes | Comparisons | |||||
|---|---|---|---|---|---|---|
| 21d x 40d a | 21d x 70d | 21d x 90d | 40d x 70d | 40d x 90d | 70d x 90d | |
| MAP2K1 | 0.0027 | 0.0772 | 0.1882 | 0.1177 | 0.0456 | 0.6143 |
| RBM24 | 0.0192 | 0.0165 | 0.0188 | 0.9412 | 0.9917 | 0.9495 |
21d, 40d, 70d and 90d indicate the prenatal ages. Values in bold were statistically significant (p < 0.05).
Figure 2. Relative expression levels for two genes (MAP2K1 and RMB24) in pair-wise comparisons of prenatal ages (21, 40, 70 and 90 days post-insemination). These genes differed significantly in relation to the factor ‘Period’ (p < 0.05, F-test in ANOVA), but showed no significant difference for the interaction ‘Breed x Period’. *p < 0.05 and **p < 0.01 indicates significant pair-wise comparisons by Student's t-test. A positive fold-change means that the first period in the comparison shows greater expression than the second period. Negative fold-change means that the second period in comparison presents greater expression than the first period.
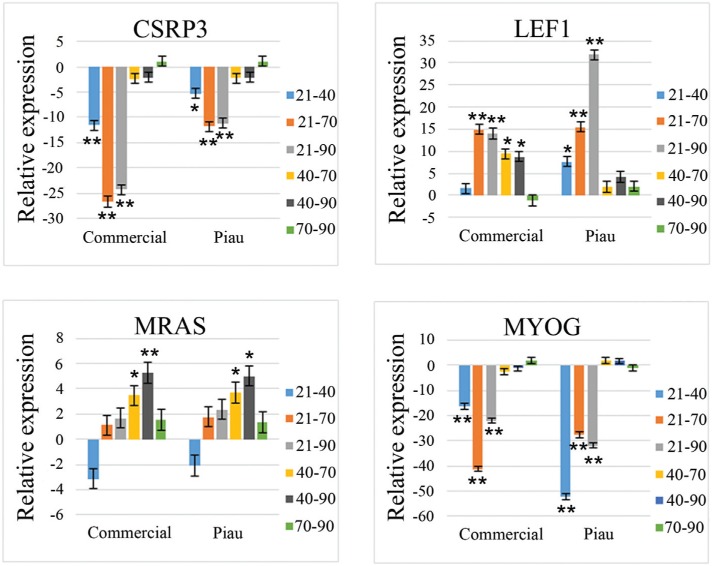
Four genes (CSRP3, MRAS, LEF1 and MYOG) showed a significant (p < 0.05) Breed x Period interaction and Period factor based on ANOVA. Table 5 shows the Student's t-test results for comparisons that assessed a Breed x Period interaction for these genes. The relative gene expression (fold-change) for pair-wise comparisons of prenatal periods is shown in Figure 3 and revealed Breed x Period interactions (p < 0.05).
Table 5. P-values for two-period comparisons in commercial and Piau pigs. The ANOVA results (F-test) for these genes showed a significant Breed x Period interaction.
| Genes | Breed | Comparisons | |||||
|---|---|---|---|---|---|---|---|
| 21d x 40d a | 21d x 70d | 21d x 90d | 40d x 70d | 40d x 90d | 70d x 90d | ||
| CSRP3 | Commercial | 0.0032 | 0.0003 | 0.0003 | 0.2524 | 0.3070 | 0.8964 |
| Piau | 0.0309 | 0.0029 | 0.0035 | 0.2719 | 0.3062 | 0.9365 | |
| LEF1 | Commercial | 0.5880 | 0.0049 | 0.0058 | 0.0155 | 0.0183 | 0.9356 |
| Piau | 0.0257 | 0.0045 | 0.0007 | 0.4091 | 0.1052 | 0.3973 | |
| MRAS | Commercial | 0.0575 | 0.8639 | 0.3663 | 0.0412 | 0.0089 | 0.4609 |
| Piau | 0.1992 | 0.3261 | 0.1414 | 0.0318 | 0.0107 | 0.6007 | |
| MYOG | Commercial | 0.0026 | 0.0002 | 0.0012 | 0.2565 | 0.7099 | 0.4366 |
| Piau | 0.0001 | 0.0006 | 0.0004 | 0.4341 | 0.5396 | 0.8629 | |
21d, 40d, 70d and 90d indicate the prenatal ages. Values in bold were statistically significant (p < 0.05) by Student's t-test.
Figure 3. Relative expression for four genes (CSRP3, LEF1, MRAS and MYOG) in pair-wise comparisons of prenatal ages (21, 40, 70 and 90 days post-insemination) in commercial and Piau pigs. These genes showed a significant interaction for Breed x Period (p < 0.05, F-test in ANOVA). *p < 0.05 and **p < 0.01 indicates significant pair-wise comparisons by Student's t-test. A positive fold-change means that the first period in the comparison shows greater expression than the second period. Negative fold-change means that the second period in the comparison presents greater expression than the first period.
Figure 3 and Table 5 show that commercial and Piau pigs had similar expression patterns for CSRP3, MRAS and MYOG. CSRP3 showed greater expression during the two waves of myoblast fusion that gave rise to primary fibers at 40 dpi and secondary fibers at 70 and 90 dpi. MRAS showed greater expression during somite formation and proliferation at 21 dpi and during primary fiber formation at 40 dpi. MYOG had a greater expression at 40 dpi (period of primary fiber formation) and at 70 and 90 dpi (period of secondary fiber formation). LEF1 showed a divergent expression pattern for commercial and Piau pigs: expression was greater at 21 dpi (period of somite formation and proliferation) and 40 dpi (period of primary fiber formation) in commercial pigs, whereas in Piau pigs this gene showed greater expression only at 21 dpi.
Discussion
Thirteen genes related to important gene networks for muscle development and structure were analyzed by qRT-PCR to determine their expression profiles at 21 dpi (embryos) and at 40, 70 and 90 dpi (fetuses) in commercial (three-way Duroc, Landrace and Large-White cross) and local Piau pigs. In 21 dpi embryos we examined the region that would give rise to muscle, and which should contain only undifferentiated mesenchymal stem cells (also known as somite cells), since a histochemical study of pig embryos at 20 dpi found only somites in this region (Swatland and Cassens, 1973). In the other periods analyzed, primary myotubes were formed at 40 dpi and secondary myotubes developed at 70 and 90 dpi after myoblast differentiation. The analysis of myogene expression in these periods shed light on possible differences in myogenesis and subsequent muscularity in commercial and Piau pigs. Local Piau pigs are expected to have a higher level of fat than commercial pigs (Serao et al., 2011), which have a greater muscle mass.
Muscle mass is influenced by the number and size of its muscle fibers (Rehfeldt et al., 2000). We therefore focused on the expression of genes involved in the formation of myogenic cells in four periods since differential gene expression between genetically distinct lineages and periods could explain differences in muscle mass between breeds. Muscle development is a complex biological process regulated by various genes that interact with each other and a series of signal transduction pathways (Zhao et al., 2011). Myogenic regulatory factors are controlled by regulatory pathways that activate or repress their activity, although additional factors are also probably involved in various transcription circuits that control myogenesis (Kong et al., 1997).
MAP2K1 showed similar expression in both breeds, with greater expression during primary and secondary fiber formation (40 and 70 dpi, respectively). This gene is involved in the MAPK signaling pathway that is important for muscle development since it can activate transcription factors involved in differentiation (Keren et al., 2006), such as MEF2A, MEF2C and MYOD (Wu et al., 2000) that can accelerate myoblast differentiation (Ulloa et al., 2007). MAP2K1 can inhibit and activate myogenesis, depending on the developmental stage (Jo et al., 2009), and can negatively control the switch from myoblast proliferation to differentiation by suppressing MYOD activity in the early stages of myogenesis (Perry et al., 2001). In contrast, the presence of MAP2K1 protein in the nucleus of proliferating myoblasts can have a stimulatory role on muscle differentiation (Jo et al., 2009). Thus, the differential expression at 40 dpi and 70 dpi suggests that MAP2K1 may have a stimulatory role in myoblast differentiation in myotubes of both breeds.
RBM24 showed a similar expression pattern in commercial and Piau pigs, with greater expression during primary and secondary fiber formation (at 40 dpi and 70-90 dpi, respectively). RBM24 is involved in biological processes related to muscle organ development. RBM24 protein interacts with MYOG mRNA to regulate the stability and expression of the latter through a post-transcriptional regulatory pathway (Jin et al., 2010), but can also promote myogenic differentiation by modulating the cell cycle (Miyamoto et al., 2009). The RBM24 expression profile confirmed its role in controlling the stability and expression of MYOG mRNA and may therefore be involved in promoting primary and secondary fiber formation in both breeds. MYOG expression was also evaluated and showed a similar level of expression to RBM24 throughout prenatal periods, as described below.
MYOG showed greater expression during the periods of primary (40 dpi) and secondary (70 dpi and 90 dpi) fiber formation in commercial and Piau pigs. This gene is related to the biological processes of muscle structure development and muscle organ development. MYOG is an important myogenic regulatory factor that is necessary for the formation of multinucleated myotubes (Keren et al., 2006). These results confirmed the importance of this transcription factor for primary and secondary fiber formation during myogenic differentiation and showed that MYOG mRNA can be stabilized by RBM24 proteins since the synthesis of this mRNA is also high during primary and secondary fiber formation.
CSRP3 showed greater expression during primary fiber (40 dpi) and secondary fiber (70 and 90 dpi) formation in commercial and Piau pigs. This gene is included in the molecular function of contractile fiber and in the biological processes of muscle system process, muscle organ development and muscle structure development. CSRP3 promotes myoblast differentiation and it is first expressed and accumulated in the nucleus when there is myotube formation and growth (Arber et al., 1994). As shown here, CSRP3 expression was enhanced during the two waves of myoblast differentiation in both breeds, in agreement with its role in primary and secondary fiber formation.
LEF1 showed greater expression at 21 dpi and 40 dpi in commercial and at 21 dpi in Piau pigs, with lower expression thereafter. This gene belongs to the WNT signaling pathway and is related to the biological process of embryo development. The WNT pathway is important for muscle development because it can control the expression of myogenic regulatory factors such as MYF5 and MYOD, thereby influencing myogenic differentiation and survival (Cossu and Borello, 1999; von Maltzahn et al., 2012). LEF1 can induce cellular cycle progression, cellular differentiation and apoptosis through transcriptional activation of E2F1 (Zhou et al., 2008). Based on the expression profile observed here, LEF1 is more important in the early stages of muscle development, mainly at 21 dpi when somites are formed and proliferate. The additional peak of expression seen at 40 dpi in commercial pigs indicates that LEF1 is possibly involved in the greater proliferation and fusion of myoblasts in this breed, which could account for the greater number of primary fibers in commercial pigs. Indeed, as mentioned above, LEF1 can induce cell cycle progression and cellular differentiation (Zhou et al., 2008).
MRAS showed greater expression during the period of somite formation and proliferation (21 dpi) and primary fiber formation (40 dpi), with lower expression thereafter. This gene is related to the biological processes of muscle organ development and organ development and is involved in the MAPK signaling pathway. MRAS is a negative regulator of myoblast differentiation during myogenesis (Yokoyama et al., 2007). Thus, we suggest that this gene has a greater role at 21 and 40 dpi, possibly by controlling somite formation and proliferation, as well as myoblast differentiation in primary fibers. MRAS can negatively regulate the expression and function of muscle-specific transcription factors such as MYOD and MEF2 family (Lassar et al., 1989; Winter and Arnold, 2000; Tortorella et al., 2001) that are essential for controlling myoblast development and fusion to give rise to primary myotubes in both breeds. Myoblast differentiation into myotubes thus involves a balance between genes that activate and inhibit the process.
CHD8, EID2B, HIF1AN, IKBKB, RSPO3, SOX7 and SUFU showed no changes in expression during the various periods or between breeds. Since there were no changes in gene expression, studies at the protein level are necessary in order to assess the roles of these genes in myogenesis in commercial and Piau pigs; differences in the amount of activated protein could be an important factor in distinguishing between the two breeds.
CHD8, RSPO3, SOX7 and SUFU are related to embryo development. CHD8 belongs to the WNT signaling pathway and can negatively regulate the transcriptional activity of various genes induced by activation of the WNT/β-catenin signaling pathway (Nishiyama et al., 2012), in addition to preventing apoptosis (Nishiyama et al., 2009). RSPO3 is a positive regulator of myogenesis in skeletal muscle by activating the WNT/β-catenin signaling pathway and can induce MYF5 expression (Han et al., 2011). SOX7 is a negative regulator of the WNT/β-catenin signaling pathway (Chan et al., 2012) and can also be a tumor suppressor (Takash et al., 2001). SUFU is related to the biological process of organ development and is included in the Hedgehog signaling pathway, of which it is a negative regulator by suppressing the activity and function of GLI transcription factors (Ding et al., 1999). This suppression can increase the expression of specific muscle genes such as MYOD (Voronova et al., 2013). The pathway is important for myogenesis because it is involved in cell proliferation and differentiation, tissue remodeling (Heretsch et al., 2010), and specifies cellular growth and differentiation patterns (Rossi et al., 2007).
EID2B and HIF1AN are involved in the biological process muscle organ development. EID2B is also related to muscle differentiation and can prevent myoblast differentiation into myotubes (Sasajima et al., 2005). HIF1AN is also involved in the biological process of muscle structure development. This gene negatively regulates apoptosis (Yan et al., 2011) and is important for myoblast differentiation, in which it acts as a crucial transcription factor that regulates myogenesis (Li et al., 2007). Unaltered HIF1AN mRNA expression has already been observed during myogenesis (Wagatsuma et al., 2011).
IKBKB is included in the MAPK signaling pathway and is related to the biological processes of skeletal muscle contraction, muscle system process and musculoskeletal movement. This gene can negatively regulate myoblast differentiation during myogenesis (Bakkar et al., 2008).
Overall, the results of this study indicate that the LEF1 gene is a primary candidate to explain the difference in muscularity between commercial and Piau pigs. Although this gene is expressed in both breeds, in commercial pigs this expression probably leads to greater fusion of myoblasts than in Piau pigs. In commercial pigs, LEF1 showed an additional peak of greater expression at 40 dpi that corresponds to a critical period of myoblast proliferation and fusion during the first wave of myofiber formation. Thus, the greater number of fibers formed in commercial pigs compared to Piau pigs accounts for the greater muscularity seen in the former breed during postnatal development. The potential growth of skeletal muscle depends on the number of muscle fibers formed during the prenatal period and their postnatal hypertrophy (Rehfeldt et al., 2000).
The findings reported here contribute to our understanding of the molecular mechanisms involved in muscle tissue formation in a commercial pig breed (the three-way Duroc, Landrace and Large-White cross) and Piau pigs. The new information on gene expression analyzed by qRT-PCR for the first time in pigs should be useful in understanding myogenesis and the possible mechanisms involved in the differences in muscularity between genetically distinct breeds.
Acknowledgments
The authors thank the trainees and employees of the Swine Breeding Farm at Universidade Federal de Viçosa for help in sample collection. EPR was supported by a scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior(CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). This work was supported by grants from CNPq (472858/2010-6 and 478181/2012-4).
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Alexandre Rodrigues Caetano
References
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- Bakkar N, Wang J, Ladner KJ, Wang H, Dahlman JM, Carathers M, Acharyya S, Rudnicki MA, Hollenbach AD, Guttridge DC. IKK/NF-κΒ regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J Cell Biol. 2008;180:787–802. doi: 10.1083/jcb.200707179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beermann DH, Cassens RG, Hausman GJ. A second look at fiber type differentiation in porcine skeletal muscle. J Anim Sci. 1978;46:125–132. doi: 10.2527/jas1978.461125x. [DOI] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnazzo M, te Pas MF, Priem J, de Wit AA, Pool MH, Davoli R, Russo V. Comparison of prenatal muscle tissue expression profiles of two pig breeds differing in muscle characteristics. J Anim Sci. 2006;84:1–10. doi: 10.2527/2006.8411. [DOI] [PubMed] [Google Scholar]
- Chan DW, Mak CS, Leung TH, Chan KK, Ngan HY. Down-regulation of Sox7 is associated with aberrant activation of Wnt/b-catenin signaling in endometrial cancer. Oncotarget. 2012;3:1546–1556. doi: 10.18632/oncotarget.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossu G, Borello U. Wnt signaling and the activation of myogenesis in mammals. EMBO J. 1999;18:6867–6872. doi: 10.1093/emboj/18.24.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3–P3. [PubMed] [Google Scholar]
- Ding Q, Fukami S, Meng X, Nishizaki Y, Zhang X, Sasaki H, Dlugosz A, Nakafuku M, Hui C. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9:1119–1122. doi: 10.1016/s0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- Han XH, Jin YR, Seto M, Yoon JK. A WNT/β-catenin signaling activator, R-spondin, plays positive regulatory roles during skeletal myogenesis. J Biol Chem. 2011;286:10649–10659. doi: 10.1074/jbc.M110.169391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heretsch P, Tzagkaroulaki L, Giannis A. Modulators of the hedgehog signaling pathway. Bioorg Med Chem. 2010;18:6613–6624. doi: 10.1016/j.bmc.2010.07.038. [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jin D, Hidaka K, Shirai M, Morisaki T. RNA-binding motif protein 24 regulates myogenin expression and promotes myogenic differentiation. Genes Cells. 2010;15:1158–1167. doi: 10.1111/j.1365-2443.2010.01446.x. [DOI] [PubMed] [Google Scholar]
- Jo C, Jang BG, Jo SA. MEK1 plays contrary stage-specific roles in skeletal myogenic differentiation. Cell Signal. 2009;21:1910–1917. doi: 10.1016/j.cellsig.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. ToppCluster: A multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 2010;38:W96–W102. doi: 10.1093/nar/gkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren A, Tamir Y, Bengal E. The p38 MAPK signaling pathway: A major regulator of skeletal muscle development. Mol Cell Endocrinol. 2006;252:224–230. doi: 10.1016/j.mce.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Kong Y, Flick MJ, Kudla AJ, Konieczny SF. Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol Cell Biol. 1997;17:4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar AB, Thayer MJ, Overell RW, Weintraub H. Transformation by activated ras or fos prevents myogenesis by inhibiting expression of MyoD1. Cell. 1989;58:659–667. doi: 10.1016/0092-8674(89)90101-3. [DOI] [PubMed] [Google Scholar]
- Li X, Zhu L, Chen X, Fan M. Effects of hypoxia on proliferation and differentiation of myoblasts. Med Hypotheses. 2007;69:629–636. doi: 10.1016/j.mehy.2006.12.050. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maltin CA, Delday MI, Sinclair KD, Steven J, Sneddon AA. Impact of manipulations of myogenesis in utero on the performance of adult skeletal muscle. Reproduction. 2001;122:359–374. doi: 10.1530/rep.0.1220359. [DOI] [PubMed] [Google Scholar]
- McCulloch RS, Ashwell MS, O’Nan AT, Mente PL. Identification of stable normalization genes for quantitative real-time PCR in porcine articular cartilage. J Anim Sci Biotechnol. 2012;3:36–36. doi: 10.1186/2049-1891-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Hidaka K, Jin D, Morisaki T. RNA-binding proteins Rbm38 and Rbm24 regulate myogenic differentiation via p21-dependent and -independent regulatory pathways. Genes Cells. 2009;14:1241–1252. doi: 10.1111/j.1365-2443.2009.01347.x. [DOI] [PubMed] [Google Scholar]
- Murani E, Muraniova M, Ponsuksili S, Schellander K, Wimmers K. Identification of genes differentially expressed during prenatal development of skeletal muscle in two pig breeds differing in muscularity. BMC Dev Biol. 2007;7:e109. doi: 10.1186/1471-213X-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Oshikawa K, Tsukada Y, Nakagawa T, Iemura S, Natsume T, Fan Y, Kikuchi A, Skoultchi AI, Nakayama KI. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol. 2009;11:172–182. doi: 10.1038/ncb1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Skoultchi AI, Nakayama KI. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-β-catenin signaling pathway. Mol Cell Biol. 2012;32:501–512. doi: 10.1128/MCB.06409-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J, McEntaggart NO, et al. IDT SciTools: A suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:W163–W169. doi: 10.1093/nar/gkn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RL, Parker MH, Rudnicki MA. Activated MEK1 binds the nuclear MyoD transcriptional complex to repress transactivation. Mol Cell. 2001;8:291–301. doi: 10.1016/s1097-2765(01)00302-1. [DOI] [PubMed] [Google Scholar]
- Rasmussen R. Quantification on the lightcycler. In: Meuer S, Wittwer C, Nakagawara K, editors. Rapid Cycle Real-Time PCR: Methods and Applications. Springer; Heidelberg: 2001. pp. 21–34. [Google Scholar]
- Rehfeldt C, Fiedler I, Dietl G, Ender K. Myogenesis and postnatal skeletal muscle cell growth as influenced by selection. Livest Prod Sci. 2000;66:177–188. [Google Scholar]
- Rossi D, Murayama T, Manini I, Franci D, Ogawa Y, Sorrentino V. Expression and functional activity of ryanodine receptors (RyRs) during skeletal muscle development. Cell Calcium. 2007;41:573–580. doi: 10.1016/j.ceca.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al. GeneCards Version 3: The human gene integrator. Database. 2010 doi: 10.1093/database/baq020. 2010. baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasajima Y, Tanaka H, Miyake S, Yuasa Y. A novel EID family member, EID-3, inhibits differentiation and forms a homodimer or heterodimer with EID-2. Biochem Biophys Res Commun. 2005;333:969–975. doi: 10.1016/j.bbrc.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Serão NV, Veroneze R, Ribeiro AM, Verardo LL, Braccini J, Neto, Gasparino E, Campos CF, Lopes PS, Guimarães SE. Candidate gene expression and intramuscular fat content in pigs. J Anim Breed Genet. 2011;128:28–34. doi: 10.1111/j.1439-0388.2010.00887.x. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollero BP, Guimarães SE, Rilington VD, Tempelman RJ, Raney NE, Steibel JP, Guimarães JD, Lopes PS, Lopes MS, Ernst CW. Transcriptional profiling during foetal skeletal muscle development of Piau and Yorkshire-Landrace cross-bred pigs. Anim Genet. 2011;42:600–612. doi: 10.1111/j.1365-2052.2011.02186.x. [DOI] [PubMed] [Google Scholar]
- Steibel JP, Poletto R, Coussens PM, Rosa GJ. A powerful and flexible linear mixed model framework for the analysis of relative quantification RT-PCR data. Genomics. 2009;94:146–152. doi: 10.1016/j.ygeno.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Swatland HJ, Cassens RG. Prenatal development, histochemistry and innervation of porcine muscle. J Anim Sci. 1973;36:343–354. doi: 10.2527/jas1973.362343x. [DOI] [PubMed] [Google Scholar]
- Takash W, Cañizares J, Bonneaud N, Poulat F, Mattéi M-G, Jay P, Berta P. SOX7 transcription factor: Sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res. 2001;29:4274–4283. doi: 10.1093/nar/29.21.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Pas MF, De Wit AA, Priem J, Cagnazzo M, Davoli R, Russo V, Pool MH. Transcriptome expression profiles in prenatal pigs in relation to myogenesis. J Muscle Res Cell Motil. 2005;26:157–165. doi: 10.1007/s10974-005-7004-6. [DOI] [PubMed] [Google Scholar]
- Tortorella LL, Milasincic DJ, Pilch PF. Critical proliferation-independent window for basic fibroblast growth factor repression of myogenesis via the p42/p44 MAPK signaling pathway. J Biol Chem. 2001;276:13709–13717. doi: 10.1074/jbc.M100091200. [DOI] [PubMed] [Google Scholar]
- Ulloa F, Itasaki N, Briscoe J. Inhibitory Gli3 activity negatively regulates Wnt/ β-catenin signaling. Curr Biol. 2007;17:545–550. doi: 10.1016/j.cub.2007.01.062. [DOI] [PubMed] [Google Scholar]
- von Maltzahn J, Chang NC, Bentzinger CF, Rudnicki MA. Wnt signaling in myogenesis. Trends Cell Biol. 2012;22:602–609. doi: 10.1016/j.tcb.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronova A, Coyne E, Al Madhoun A, Fair JV, Bosiljcic N, St-Louis C, Li G, Thurig S, Wallace VA, Wiper-Bergeron N, et al. Hedgehog signaling regulates MyoD expression and activity. J Biol Chem. 2013;288:4389–4404. doi: 10.1074/jbc.M112.400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagatsuma A, Kotake N, Yamada S. Spatial and temporal expression of hypoxia-inducible factor-1α during myogenesis in vivo and in vitro . Mol Cell Biochem. 2011;347:145–155. doi: 10.1007/s11010-010-0622-3. [DOI] [PubMed] [Google Scholar]
- Wigmore PM, Evans DJ. Molecular and cellular mechanisms involved in the generation of fiber diversity during myogenesis. Int Rev Cytol. 2002;216:175–232. doi: 10.1016/s0074-7696(02)16006-2. [DOI] [PubMed] [Google Scholar]
- Wigmore PM, Stickland NC. Muscle development in large and small pig fetuses. J Anat. 1983;137:235–245. [PMC free article] [PubMed] [Google Scholar]
- Winter B, Arnold HH. Activated raf kinase inhibits muscle cell differentiation through a MEF2-dependent mechanism. J Cell Sci. 2000;113:4211–4220. doi: 10.1242/jcs.113.23.4211. [DOI] [PubMed] [Google Scholar]
- Wu Z, Woodring PJ, Bhakta KS, Tamura K, Wen F, Feramisco JR, Karin M, Wang JYJ, Puri PL. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol Cell Biol. 2000;20:3951–3964. doi: 10.1128/mcb.20.11.3951-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B, Kong M, Chen YH. Prevention of apoptosis by the interaction between FIH1 and Bax. Mol Cell Biochem. 2011;348:1–9. doi: 10.1007/s11010-010-0631-2. [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Takano K, Yoshida A, Katada F, Sun P, Takenawa T, Andoh T, Endo T. DA-Raf1, a competent intrinsic dominant-negative antagonist of the Ras-ERK pathway, is required for myogenic differentiation. J Cell Biol. 2007;177:781–793. doi: 10.1083/jcb.200703195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Mo D, Li A, Gong W, Xiao S, Zhang Y, Qin L, Niu Y, Guo Y, Liu X, et al. Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness. PLoS One. 2011;6:e19774. doi: 10.1371/journal.pone.0019774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Zhang L, Gong K, Lu G, Sheng B, Wang A, Zhao N, Zhang X, Gong Y. LEF-1 activates the transcription of E2F1. Biochem Biophys Res Commun. 2008;365:149–153. doi: 10.1016/j.bbrc.2007.10.138. [DOI] [PubMed] [Google Scholar]
Internet resources
- DAVID Feb 02, 2015. http://david.abcc.ncifcrf.gov/tools.jsp.
- BIOMART/ ENSEMBL Feb 02, 2015. http://www.ensembl.org/biomart/martview/
- GenBank Feb 02, 2015. https://www.ncbi.nlm.nih.gov/genbank/
- Gene Cards Database Feb 02, 2015. http://www.genecards.org/
- PrimerQuest Feb 02, 2015. http://www.idtdna.com/Primerquest/Home/Index.
- Normfinder Feb 02, 2015. http://moma.dk/normfinder-software.
- %QPCR_MIXED Feb 02, 2015. https://www.msu.edu/~steibelj/JP_files/QPCR.html.
- Toppcluster Feb 02, 2015. https://toppcluster.cchmc.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- BIOMART/ ENSEMBL Feb 02, 2015. http://www.ensembl.org/biomart/martview/
- GenBank Feb 02, 2015. https://www.ncbi.nlm.nih.gov/genbank/
- Gene Cards Database Feb 02, 2015. http://www.genecards.org/