Abstract
The term filaminopathy was introduced after a truncating mutation in the dimerization domain of filamin C (FLNc) was shown to be responsible for a devastating muscle disease. Subsequently, the same mutation was found in patients from diverse ethnical origins, indicating that this specific alteration is a mutational hot spot. Patients initially present with proximal muscle weakness, while distal and respiratory muscles become affected with disease progression. Muscle biopsies of these patients show typical signs of myofibrillar myopathy, including disintegration of myofibrils and aggregation of several proteins into distinct intracellular deposits. Highly similar phenotypes were observed in patients with other mutations in Ig-like domains of FLNc that result in expression of a noxious protein. Biochemical and biophysical studies showed that the mutated domains acquire an abnormal structure causing decreased stability and eventually becoming a seed for abnormal aggregation with other proteins. The disease usually presents only after the fourth decade of life possibly as a result of ageing-related impairments in the machinery that is responsible for disposal of damaged proteins. This is confirmed by mutations in components of this machinery that cause a highly similar phenotype. Transfection studies of cultured muscle cells reflect the events observed in patient muscles and, therefore, may provide a helpful model for testing future dedicated therapeutic strategies. More recently, FLNC mutations were also found in families with a distal myopathy phenotype, caused either by mutations in the actin-binding domain of FLNc that result in increased actin-binding and non-specific myopathic abnormalities without myofibrillar myopathy pathology, or a nonsense mutation in the rod domain that leads to RNA instability, haploinsufficiency with decreased expression levels of FLNc in the muscle fibers and myofibrillar abnormalities, but not to the formation of desmin-positive protein aggregates required for the diagnosis of myofibrillar myopathy.
Keywords: Filamin C, Filaminopathy, Myofibrillar myopathy, Distal myopathy, Limb-girdle muscular dystrophy, Pathomechanism
Introduction
Filaminopathies are recently identified progressive skeletal myopathies manifesting initially by bilateral weakness in either proximal leg muscles or in distal upper limb muscles spreading to other muscle groups and in some forms eventually resulting in tetraparesis and wheelchair dependence [6, 18, 22, 57]. Three distinct types of filaminopathy are recognized. The disease caused by mutations resulting in protein aggregation (so far found at various locations in the FLNc rod domain) presents in the fourth-to-sixth decade of life with slowly progressive predominantly proximal muscle weakness. Associated cardiac and respiratory muscle involvement severely complicate the course of illness [22, 57]. In contrast, mutations in the actin-binding domain (ABD) of FLNc are responsible for the second disease variant that is initially characterized by weakness and wasting of distal muscles, especially intrinsic hand muscles, manifesting in the third decade of life [6]. An intermediate filaminopathy phenotype affecting primarily distal muscles of the upper and lower limbs has recently been described [18]. Muscle biopsies of patients with aggregation-causing FLNC mutations show disintegration of myofibrils and formation of desmin-positive protein aggregates within muscle fibers [30, 39]. These are typical findings in myofibrillar myopathies (MFM), a clinically and genetically diverse group of progressive devastating hereditary skeletal and cardiac myopathies. Thus far MFM has been associated with mutations in seven genes (DES, MYOT, LDB3/ZASP, CRYAB, BAG3, FLNC and FHL1 [33, 40–42]). Muscle biopsies from patients with the second filaminopathy variant show non-specific myopathic abnormalities without MFM pathology [6], while histological evaluation in cases with the intermediate variant indicated disease-associated myofibrillar abnormalities, but desmin-positive protein aggregates required for the diagnosis of MFM were not detected [18].
As a pathological entity, the first variant of filaminopathy related to FLNc rod mutations classifies with a group named protein aggregate myopathy (PAM). PAM is a general term for neuromuscular conditions marked by aggregation of proteins within muscle fibers. This is a diverse group of disorders that in addition to MFM includes among others, nemaline myopathy, myosin storage myopathy, cytoplasmic body myopathies, and reducing body myopathy [15, 16]. The other filaminopathy variants belong to autosomal dominant distal myopathies, of which nine types have been assigned to known genes [50]. The list includes disorders caused by TTID, LDB3, CRYAB, FLNC and DES mutations, genes that have also been associated with MFM, thus indicating a close relationship between these classification units.
This review presents a comparative analysis of the contrasting FLNc mutation-related phenotypes of filaminopathy and their distinct underlying pathomechanisms. It also offers practical considerations regarding diagnostic procedures, severely complicated due to the existence of a FLNC-related pseudogene, as well as health implications and therapeutic strategies.
Clinical aspects
MFM-type FLNc myopathy
Evaluation of about 70 MFM patients with different FLNC mutations has revealed a markedly homogenous clinical phenotype ([2, 22, 23, 28, 44, 48] and our unpublished data). Muscle weakness mostly starts in the fourth or fifth decade of life (range 24–60 years). Proximal muscle weakness leading to difficulty walking uphill and climbing stairs is the initial sign. In the course of illness, most patients develop slowly progressive weakness in both distal and proximal leg and arm muscles (Fig. 1a). Winging of the scapula is a frequent phenomenon. Muscle weakness slowly progresses to the inability to walk. An involvement of respiratory muscles, often requiring nocturnal ventilation, usually occurs with disease progression and contributes to reduced life expectancy.
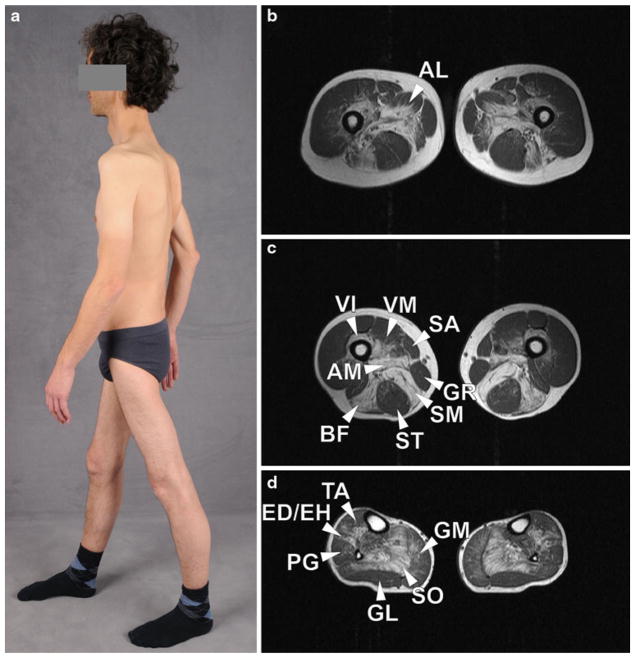
Fig. 1.
Image and transverse T1-weighted muscle MRI of MFM patients harboring mutations in the dimerization domain of FLNc. The patient (a) has predominantly proximal muscle atrophy in the upper and lower limbs and winged scapula. MRI images demonstrate a typical pattern of muscle involvement (hyperintensities reflect lipomatous alterations). On the thigh level (b, c), semimembranosus (SM), adductor magnus (AM) and longus (AL), long head of biceps femoris (BF), vastus intermedius (VI) and medialis (VM) are most affected. Sartorius (SA) and gracilis (GR) appear normal and the semitendinosus (ST) shows only mild lipomatous alterations. In lower legs (d), the soleus muscle (SO) shows pronounced fatty changes. The medial head of the gastrocnemius (GM), the tibialis anterior (TA), extensor digitorum longus, extensor hallucis longus (ED/EH), and peroneal muscles show mild to moderate lipomatous alterations whereas the lateral head of the gastrocnemius (GL) is almost spared
About one-third of the patients showed cardiac abnormalities, including conduction blocks, left ventricular hypertrophy, and diastolic dysfunction. Sudden cardiac arrest as the cause of death was presumed in at least five patients. Creatine kinase (CK) levels were mostly elevated up to tenfold of the upper limit. Electromyography regularly showed typical myopathic changes. Although histological findings indicated neurogenic changes in about one-half of skeletal muscle biopsies [22], clinical examinations and neurophysiological measurements did not reveal a relevant involvement of the peripheral nervous system in any of the FLNc patients. Chronic gastrointestinal complaints were reported by a few patients with p.W2710X and p.K899_V904del/V899_C900ins mutations and may indicate an involvement of smooth muscle [23, 28]. Late-onset cerebellar ataxia with atrophy of cerebellum and vermis was observed in one sporadic patient [48], but the causal relationship with the detected mutation in FLNc Ig-like domain 22 is unclear.
A diagnostic challenge is to discriminate FLNc-based MFM-type myopathy from other myopathies including MFM subtypes, limb-girdle muscular dystrophies (LGMDs), X-chromosomal muscular dystrophy Becker type, myotonic dystrophy type 2 (PROMM), acid maltase deficiency (late-onset Pompe disease), and inclusion body myositis/myopathy. All these diseases are late-onset myopathies that typically present with proximal weakness, slow disease progression and mild to moderate CK elevation. The features of FLNc-based MFM-type myopathy most useful for differential diagnosis appear to be a symmetrical involvement of proximal muscles in the lower extremities, respiratory weakness during the disease course, an autosomal dominant inheritance pattern, MFM-typical histological changes and characteristic muscle imaging findings (see below).
Distal FLNc myopathy
Distal myopathy was associated with missense mutations (p.A193T; p.M251T) located in the N-terminal actin-binding domain of FLNc in families from Australia and Italy [6]. The illness developed in the third decade of life. Intrinsic hand muscles were primarily affected and led to reduced grip strength, followed by leg muscle weakness resulting in difficulties with running and jumping. CK levels were only mildly elevated up to threefold of the normal upper limit. Two patients displayed signs of cardiomyopathy and none had respiratory insufficiency.
In a recently reported Bulgarian family with a frame-shifting deletion mutation in exon 30 of FLNC (p.F1720LfsX63) leading to haploinsufficiency, the disease was associated with distal muscle weakness primarily in the upper limbs with lower limb involvement upon disease progression [18]. It manifests in adulthood between the ages of 20–57 years. Initial symptoms were distal muscle weakness mostly in the upper limbs with subsequent lower limb involvement upon disease progression. CK levels ranged from normal to sixfold elevated. Neurophysiological studies revealed normal motor and sensory nerve conduction velocities. None of the patients had respiratory disturbances and cardiomyopathy was documented in only a single patient. The main clinical differential diagnoses are adult-onset distal myopathies, a group of muscle diseases which share the clinical findings of predominant weakness in lower leg and/or hand muscles. Typical clinical findings characterizing distal FLNc myopathy are weakness in hand and calf muscles with an onset in early adulthood and a family history compatible with an autosomal dominant trait.
All mutations are further specified below in the genetics section (Fig. 3; Table 1).
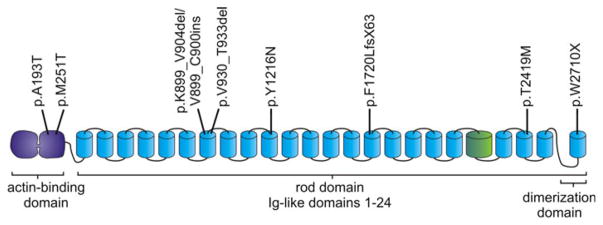
Fig. 3.
Schematic diagram representing the structure of FLNc and the distribution of muscle-disease-associated mutations in FLNc. The aminoterminus consists of two calponin homology domains that together constitute the actin-binding domain (ABD). The ABD is followed by 24 Ig-like domains, the most carboxy terminal of which is responsible for dimerization. Ig-like domain 20 is colored differently since it contains a unique insertion. The positions of the published mutations within FLNc are depicted at the top
Table 1.
Summary of the reported mutations in FLNc
| Mutation | Domain | Type | Disease | References |
|---|---|---|---|---|
| p.A193T | ABD | Point mutation | Distal myopathy | Duff et al. [6] |
| p.M251T | ABD | Point mutation | Distal myopathy | Duff et al. [6] |
| p.K899_V904del/V899_C900ins | Ig-like 7 | Deletion/insertion | MFM, gastrointestinal complaints | Luan et al. [28] |
| p.V930_T933del | Ig-like 7 | Deletion | MFM | Shatunov et al. [44] |
| p.Y1216N | Ig-like 10 | Point mutation | MFM | Avila-Smirnov et al. [2] |
| p.F1720LfsX63 | Ig-like 15 | Frameshift deletion | Distal myopathy | Guergueltcheva et al. [18] |
| p.T2419M | Ig-like 22 | Point mutation | MFM, ataxia | Tasca et al. [48] |
| p.W2710X | Ig-like 24 | Nonsense | MFM | Vorgerd et al. [57] |
Muscle imaging in FLNc myopathy
Magnetic resonance imaging (MRI) is a powerful and non-invasive tool in the diagnostic workup, evaluation of therapeutic efficacy and disease follow-up in neuromuscular disorders. MRI of lower limbs showed a rather homogenous pattern of symmetrical muscle involvement in MFM-type disease caused by FLNC mutations in Ig-like domain 7 and 24 ([10, 22, 23] and our unpublished data). Non fat-saturated T1-weighted images showed a reticular pattern of hyperintensity in less affected patients, whereas homogenous lipomatous alterations were visible in individuals with a more advanced disease. In proximal lower limbs, gluteal muscles, semimembranosus, adductor magnus and longus, long head of biceps femoris, vastus intermedius and vastus medialis were most affected (Fig. 1b, c). The rectus femoris seemed to be more affected in patients carrying the p.V930_T933del mutation than in those with Ig-like domain 24 mutation [23]. The sartorius and gracilis muscles appeared almost normal, even in patients with more advanced clinical course. In lower legs, soleus and the medial head of gastrocnemius were most affected, followed by tibialis anterior, extensor hallucis longus, extensor digitorum longus and peroneal muscles (Fig. 1d). The lateral head of the gastrocnemius was relatively spared. Muscular signal intensities on T2-weighted TIRM images were only mildly elevated, indicating an absence of distinct intramuscular edema. The pattern of muscle involvement in patients with MFM caused by FLNC mutations is similar to that observed in MFM caused by MYOT or ZASP mutations, with only subtle differences detectable by statistical analysis, but is sharply different from that observed in desminopathy or αB-crystallinopathy. Indeed, comparison with other genetically classified MFM subtypes revealed that the combination of the following findings was highly specific for MFM caused by FLNC mutations [10]: (1) semitendinosus and long head of biceps femoris at least equally affected as sartorius, (2) semimembranosus at least equally affected as adductor magnus, (3) medial head of the gastrocnemius more affected than the lateral head. These criteria developed initially in a retrospective study were validated in subsequent MRI analyses of newly identified MFM patients ([23] and our unpublished data).
Muscle imaging data of distal myopathies caused by FLNC mutations are rather limited. In patients with ABD mutations [6], fatty degeneration of semimembranosus and, in contrast to MFM-associated filaminopathy, semitendinosus was described as an early change in thigh muscles, followed by involvement of all hamstring muscles and adductor magnus. Upon further progression of the disease, vastii muscles of the quadriceps showed lipomatous alterations. At the lower leg level, severe fatty degeneration of soleus muscles, asymmetrical involvement of peroneal muscles and a slightly lesser involvement of medial and lateral gastrocnemius muscle were described in a patient with p.M251T mutation. A computer tomography scan in a patient with p.A193T mutation showed severe fatty degeneration of soleus and gastrocnemii muscles and a less severe involvement of peroneal muscles. The anterior compartment and tibialis posterior were spared in both patients. Compared to filaminopathy patients with MFM phenotype, semitendinosus and lateral gastrocnemius seems to be more and tibialis anterior and adductor magnus less affected. Interestingly, the early involvement of the semitendinosus observed in patients with ABD mutations is also typically seen in MFM resulting from mutations in DES or CRYAB [10] and in the recently described hereditary myopathy with early respiratory failure (HMERF) resulting from mutations in the A-band portion of titin [32, 34]. In contrast, the pattern of muscle involvement in the lower legs shows clear differences between these diseases [10, 32, 34].
Muscle MRI of lower limbs in one patient with FLNC haploinsufficiency showed similarities to MFM-associated filaminopathy regarding the most severely affected muscles: gluteus maximus, long head of biceps femoris and semimembranosus at the thigh level and the soleus, medial head of gastrocnemius and the tibialis anterior in the lower legs. In comparison with the MFM subtype of filaminopathy, lipomatous muscle alterations were not only more distinct in lower legs but also markedly more pronounced in distal parts of muscles [18].
Muscle biopsy findings
Muscle biopsy features in filamin C myopathy largely depend on the site of mutation within the FLNc molecule and, more importantly, on the impact that different mutations have on its biophysical and biochemical properties [6, 18, 23]. Affected muscles from patients carrying mutations in the Ig-like domains of FLNc that lead to the expression of a toxic protein show typical features of MFM [23, 26, 28, 44, 48, 57]. General myopathological abnormalities vary from mild variation in fiber size and increased numbers of internal nuclei to more advanced degenerative abnormalities comprising muscle fiber atrophy and hypertrophy, fiber splitting, and fibro-fatty tissue proliferation depending on the stage of illness and the muscle examined (Fig. 2).
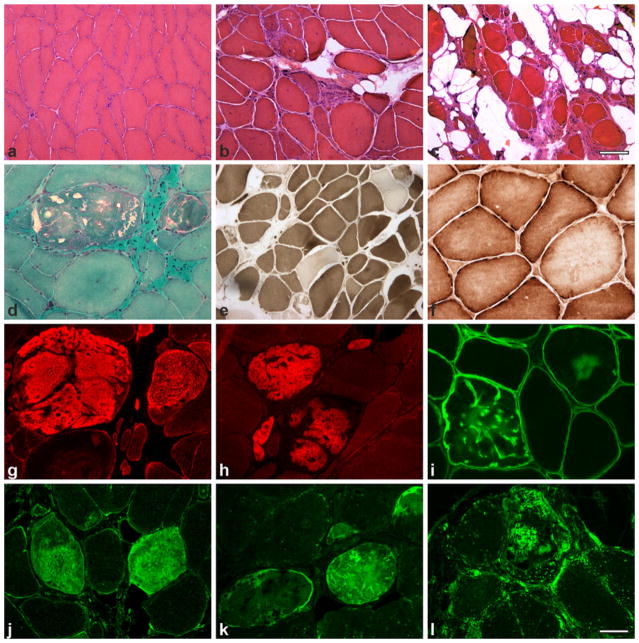
Fig. 2.
Histochemical and immunofluorescence findings in MFM-type FLNc myopathy. Myopathic changes vary from mild variability in fiber size (a), to more pronounced fiber size variation, with atrophic and hypertrophic fibers, and moderate (b) to severe (c) fibro-fatty tissue proliferation. Abnormal fibers show darkly stained areas and small vacuoles (d); ATPase staining reveals type I fiber predominance (e); COX activity is partially reduced in some fiber regions (f). Immunofluorescence analysis showing accumulation of FLNc (g), myotilin (h), δ-sarcoglycan (i), Hsp22 (j), ubiquitin (k) and LAMP-2 (l) in fiber regions corresponding to protein aggregates. a–c HE; d modified trichrome; e ATPase at pH 4,65; f COX. Scale bar in c (also applies to a, b) 100 μm, scale bar in l (also applies to d–k) 50 μm
Muscle fibers undergoing necrosis and phagocytosis are observed, but usually not as a prominent phenomenon. Rimmed and non-rimmed vacuoles are commonly seen. Non-rimmed vacuoles are often marked with strong PAS-positivity. Additionally, increased acid phosphatase activity is observed in some fiber areas. Oxidative activity is partially reduced in some fiber areas resulting in core-like lesions, but rubbed-out fibers are rarely seen. ATPase staining reveals type 1 fiber predominance in the majority of cases. Typically, muscle fibers contain polymorphous cytoplasmic inclusions that correspond to protein aggregates. These are observed as single or multiple plaque-like formations within the cytoplasm, convoluted serpentine inclusions of varying thickness, granular deposits and spheroid bodies. The aggregates are eosinophilic on HE stain, dark-blue to purple on modified trichrome stain and mostly display strong congophilia when congo red stained sections are visualized in rhodamine optics. Oxidative and ATPase activities are partially decreased in fiber regions containing inclusions but oxidative activity is usually enhanced at the periphery. This reflects the absence of mitochondria within the inclusions and increased numbers of them at the periphery. The inclusions can be focal or diffuse occupying the entire cross-section of the fiber; furthermore, abnormal fibers usually show an uneven distribution across the fascicles. Immunohistochemical and immunofluorescence analyses (Fig. 2) show strong positivity for FLNc, myotilin, desmin, the four sarcoglycans, αB-crystallin, BAG3, Xin and multiple other proteins in areas corresponding to protein aggregates [22, 23, 44, 57]. Moreover, proteins involved in protein degradation pathways including heat shock proteins, subunits of the ubiquitin proteasome system and markers of autophagy such as LAMP2 accumulate in areas corresponding to protein deposits (Fig. 2) [23].
Ultrastructural analyses show widespread myofibrillar abnormalities, including Z-disc streaming and accumulation of fine thin filaments that initially emanate at the level of the Z-disc and later coalesce into electron dense inclusions often surrounded by groups of mitochondria. Additionally, nemaline bodies and collections of 15–18 nm tubulofilaments and granulofilamentous material are seen in severely damaged fibers. Autophagic vacuoles containing myelin-like figures and cellular debris are usually present [22, 23, 28, 44].
Two families carrying mutations in the actin-binding domain of FLNc have been reported so far [6]. Muscle biopsies in four affected patients only showed non-specific myopathic features that varied from mild variation of fiber size to more severe dystrophic changes with prominent fibro fatty tissue proliferation. Many fibers showed an uneven distribution of oxidative enzyme activity, but no vacuoles and no protein aggregates were observed. Ultra-structural analysis performed in a single patient revealed no abnormalities [58].
Finally, muscle biopsy from a patient with FLNC haploinsufficiency showed increased variability of fiber size, fiber splitting and pyknotic nuclear clumps. ATPase revealed type I fiber predominance. Oxidative enzyme activity was partially reduced in some fiber areas. Although a few fibers displayed few fine myotilin granular deposits, no definite protein aggregates suggestive of MFM were detected, probably because the truncated mutant protein is not expressed. Ultrastructural analysis revealed some unspecific myofibrillar abnormalities including Z-disc streaming, nemaline bodies, and dappled dense bodies all of which are also observed in patients with the MFM type of filaminopathy [18].
Genetics
Affected gene and its structure
FLNc is a filamin isoform mainly expressed in striated muscles; it contains 2,725 amino acids and has a molecular mass of 291 kDa (GenBank isoform a: NP_001449.3). The FLNC gene is located in 7q32-q35 chromosome band, comprises ~29.5 kb of genomic DNA and contains 49 coding exons [4, 13, 29]. Since the exon encoding the FLNC-specific unique insert in Ig-like domain 20 was numbered 40a, the last FLNC exon carries number 48, not 49 [4]. Conversely, the splice variant that is predominantly expressed in skeletal and cardiac muscles (GenBank isoform b: NP_001120959) lacks exon 32 that encodes the hinge region between Ig-like domains 15 and 16, resulting in a protein of 2,692 amino acids with a molecular mass of 287 kDa [60].
Molecular diagnosis of filaminopathy is hampered by the presence of a pseudogene (pseFLNC) located approximately 53.6 kilobases downstream of the functional FLNC gene in inverted orientation. It is 1,178 base pairs in length and >98 % identical to the functional FLNC exons 46, 47, 48 (including part of the 3′ untranslated region), as well as introns 45 (partly), 46 and 47. In a recent work [31], DNA sequence mismatches between the functional FLNC and pseFLNC have been fully characterized, and an optimized strategy was devised enabling the differentiation of mutations occurring in FLNC from those accumulating in pseFLNC. Reflecting on the difficulty of differentiating between mutations in the functional gene and the pseudogene, some results of FLNC gene studies have been erroneous, as for example a report implicating a c.8107delG variant as the cause of filaminopathy in six patients [24]. The authors tested FLNC exon 48 with primers that amplify both the functional gene and the pseudogene, and the c.8107delG that is present in the pseudogene was misinterpreted as the cause of illness. This mistake could have been avoided, if the functional gene and the pseudogene were sequenced separately [31, 55].
Mutation spectrum
The first FLNc-related disease was described in 2005 when a nonsense mutation (c.G8130A, p.W2710X) in the FLNc dimerization domain was shown to cause skeletal and cardiac myopathy in a large German MFM family [57]. A haplotype-sharing set of further German families also carrying the p.W2710X FLNc mutation was described soon after the first report [22], and the identical mutation was found in three kinships of the Mayo MFM cohort that were not described in detail [41], as well as in two further families from Macedonia and China [23]. These observations established that the p.W2710X mutation is the cause of filaminopathy in genetically unrelated families originating from different ethnic groups, implying that FLNC codon 2710 is a mutational hotspot.
Two families with filamin C myopathy harboring mutations in FLNc Ig-like domain 7 of rod 1 segment have been reported: one harboring an internal 12-nucleotide deletion (c.2997_3008del, p.V930_T933del) [44] and a second exhibiting an 18-nucleotide deletion/6 nucleotide insertion (c.2695–2712del/GTTTGT ins, p.K899_V904del/V899_C900ins) [28]. In addition, an MFM family with a p.Y1216N mutation in Ig-like domain 10 and a single patient with proximal weakness at presentation and MFM-type pathology harboring a c.C7256T, p.T2419M mutation located in FLNc Ig-like domain 22 were recently described [2, 48].
In three distantly related Bulgarian distal myopathy families, a deletion (c.5160delC, p.F1720LfsX63) in exon 30 encoding FLNc Ig-like domain 15 triggers a frameshift, nonsense-mediated decay and haploinsufficiency [18]. Finally, a disorder caused by two different point mutations located in the ABD domain of FLNc (c.577G>A, p.A193T and c.752T>C, p.M251T) has also been associated with a distal myopathy with non-specific myopathic abnormalities on muscle biopsy [6]. Currently known FLNc mutations are shown on mutation chart of Fig. 3 and in Table 1.
This clearly differentiates two FLNc associated phenotypes, one with involvement of predominantly limb-girdle muscles, cardiomyopathy, respiratory failure, and MFM-type pathologies caused by mutations occurring in FLNc Ig-like domains 7, 10, 22 and 24; the other characterized by myopathy seen in distal muscles, no cardiomyopathy or respiratory disturbances, and no typical MFM-type pathology with mutations in the ABD or Ig-like domain 15. This indicates that, as has previously been shown for FLNa and FLNb [3, 7, 37], mutations in different functional domains of filamins can lead to distinct disease phenotypes.
Molecular diagnostics
Timely molecular diagnosis of filaminopathy is important for the prediction and prevention of life-threatening cardiac arrhythmias and respiratory failure that may occur in these patients. A precise diagnosis is also crucial for appropriate counseling. Routine testing of patients for FLNC mutations should be recommended in cases showing limb-girdle distribution of weakness and MFM-type pathological phenomena. Since exon 48 is a hot spot for mutations causing this type of disease, it should be analyzed first. A mutation in the ABD domain of FLNc (exons 1–3 and part of 4) needs to be considered in patients with distal myopathy, especially if thenar and intrinsic hand muscle atrophy are the first clinical symptoms and the family history is consistent with an autosomal dominant pattern of inheritance. This will have to be followed by a full FLNC sequencing in case of a negative result.
Pathophysiology
Protein expression and function
The mammalian filamin family includes three members, A, B and C (FLNa, FLNb and FLNc). They exhibit about 70 % amino acid identity [35]. Whereas Northern blots only detect FLNC mRNA in striated muscles [29], the more sensitive RT-PCR analysis reveal low levels of FLNC expression in multiple other tissues [60]. The lack of FLNc-specific antibodies hampered analysis at the protein level. Staining of multi-tissue slides with an antibody against the carboxy-terminus of FLNc by “the Human Protein Atlas” (http://www.proteinatlas.org/) [51] showed that apart from skeletal and cardiac myocytes, smooth muscle cells, glandular cells and neuronal cells in several tissues are also stained.
Filamins are large proteins that bind to actin and many other proteins (Fig. 4) having diverse physiological functions. Through these connections, filamins stabilize delicate three-dimensional actin filament networks and link it to cellular membranes, thus integrating cell architectural and signaling functions. All three filamin variants bind a plethora of proteins, in particular via their carboxy-terminal Ig-like domains [9, 35, 46, 52]. Some of these interactions may be irrelevant in vivo because of differential expression patterns or significantly differing binding affinities. A prediction of interactions based on simple extrapolation is, therefore, highly questionable.
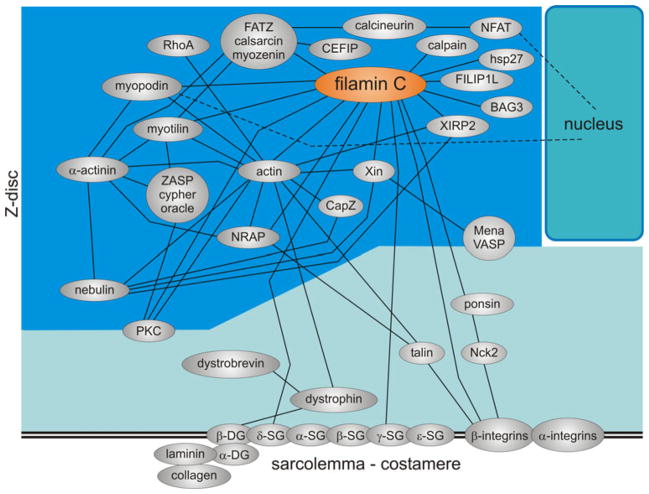
Fig. 4.
Schematic diagram illustrating the complex interactome of FLNc within the Z-disc and at the sarcolemma. Each protein is depicted as an ellipse and direct protein interactions are depicted as connecting lines
FLNc binds essentially two groups of ligands: (1) proteins of the Z-disc including myotilin [17, 56], myopodin [25], the calsarcins [8, 11, 47] and nebulette [21]; (2) sarcolemma-associated proteins such as dystrophin-associated proteins γ-and δ-sarcoglycan [49], the NRAP-talin complex [27], the ponsin-Nck2 complex [14, 61] and β1A integrin [17]. Although often suggested to occur, a direct interaction with the costameric β1D-integrin isoform has been excluded [17]. This implies that only during early developmental stages, FLNc–β1A-integrin interaction may be involved in membrane anchorage of (pre)myofibrils. In mature muscle cells, FLNc most likely indirectly links myofibrils to sarcolemma-associated integrins via the above-mentioned protein complexes. At the same time, FLNc mediates assembly of Z-discs through its interaction with several Z-disc components. Early expression of FLNc during myofibril assembly and its localization to Z-bodies are in line with the proposed role for FLNc in this process [54]. It was also suggested that by shuttling between the sarcolemma and the Z-disc FLNc is involved in signal transduction processes [49, 56].
Protein structure
Filamins consist of an aminoterminal actin-binding domain composed of two calponin homology (CH) domains followed by 24 immunoglobulin-like (Ig-like) domains of 93–103 amino acid residues each (Fig. 3). Two filamin molecules form a homodimer via self-association of their Ig-like domains 24, thus giving rise to large, elongated, Y-shaped molecules that form flexible bridges between two actin filaments [20, 36, 45]. Ig-like domains form an extended rod separated into two segments by hinge regions located between Ig-like domains 15 and 16 and Ig-like domains 23 and 24. However, the FLNc variant that is predominantly expressed in cross-striated muscles lacks the first hinge, implying that this isoform is less flexible than FLNa, and FLNb and FLNc isoforms containing this hinge region [60].
A conspicuous difference between FLNc and the other filamins is a unique insertion in Ig-like domain 20 that is involved in interaction with the Xin-repeat proteins Xin [53] and XIRP2 (unpublished data).
Biochemical and biophysical analysis of mutant proteins
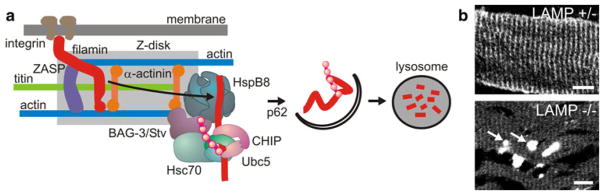
Studies performed in animal models and the data obtained from human patients with haploinsufficiency for FLNc (see below) have demonstrated that the precise stoichiometry of FLNc is of critical importance for muscle function and maintenance. Of particular interest is, therefore, a study that has unraveled a new pathway essential for muscle maintenance that was termed ‘chaperone-assisted selective autophagy’ (CASA). This pathway, which includes the co-chaperone BAG3, the ubiquitin ligase CHIP, the autophagy adaptor p62 and DNAJB6 [1, 38], is essential for the homeostasis of certain proteins, including FLNc [1]. CASA continuously operates at the Z-disc to dispose of mechanically damaged proteins, which distinguishes it from the atrophy-driven degradation pathways [59]. Impairment of this pathway leads to the formation of FLNc-containing protein aggregates, Z-disc disintegration and progressive muscle weakness [1, 19, 38, 43]. In the case of FLNc mutations causing partial protein destabilization, incorrectly folded and damaged mutant FLNc directly drives protein aggregation, thus aggravating CASA (Fig. 5). Indeed, molecular components of CASA were found to be increased in biopsies from such patients (Fig. 2) [23]. Recently, mutations in the DNAJB6 gene have been identified as a cause of limb-girdle muscular dystrophy (LGMD1D) [19, 38].
Fig. 5.
FLNc protein homeostasis. Upon increased muscle activity, damaged FLNc is degraded by BAG3 and CHIP-regulated chaperone-assisted selective autophagy (CASA). a The scheme summarizes the mechanism of selective FLNc release from the Z-disc by BAG3, resulting in ubiquitination, subsequent autophagosome formation and lysosomal degradation. b In LAMP2−/− mice autophagy is blocked and FLNc no longer localizes at Z-discs but instead forms massive aggregates (arrows). Reproduced with permission from [1]
Pathomechanisms
Since the first description of a family with a FLNC mutation causative for a muscle disease, several additional mutations have been found in different parts of the gene. Grossly these mutations can be subdivided in three classes (Fig. 6):
Fig. 6.
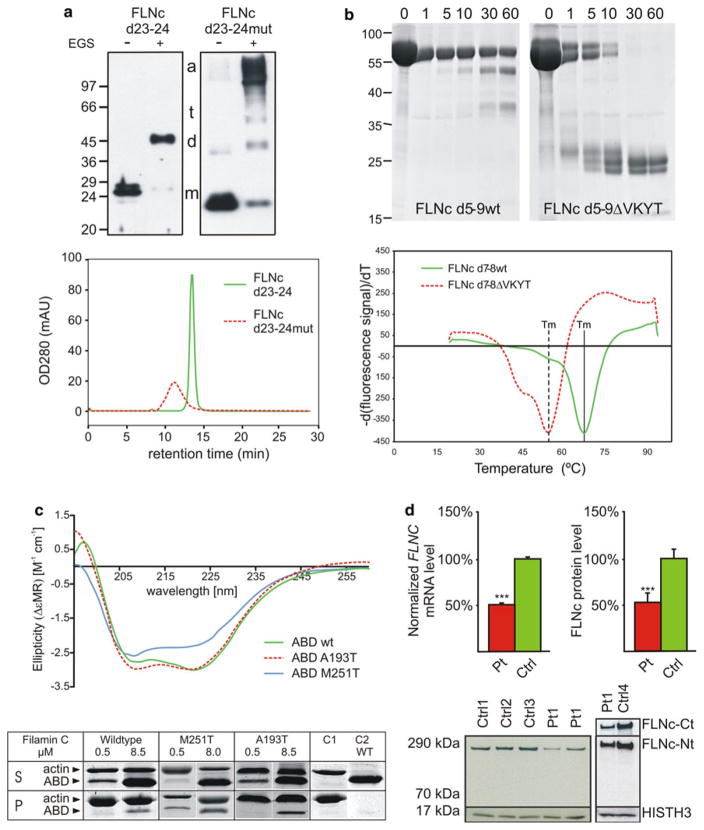
Pathomechanisms of FLNc mutations. a Top panel disturbed dimerization of pW2710X FLNc (top) revealed by chemical cross-linking experiments using wild type (d23–24) and mutant (d23–24mut) filamin constructs. In the presence (+) of the cross-linker EGS, the wild type protein was mainly found in dimer form (d), while the mutant construct was detected in higher molecular mass complexes representing trimers (t) and aggregated (a) oligomers. a Bottom panel analytical gel chromatography shows greatly decreased retention times for the mutant FLNc d23–24 protein indicating aggregation of the mutant but not the wild-type protein. b Decreased stability and increased protease-sensitivity of the p.V930_T933del mutant FLNc. The top panel shows the digestion of wild type and mutant FLNc d5-9ΔVKYT with the protease thermolysin, resulting in complete digestion of the mutant protein after 30 min, while a significant portion of the wild type variant was still intact after 60 min of incubation, indicating less stable folding of the mutant protein. The bottom panel shows temperature denaturation experiments for FLNc d7-8 and deletion mutant FLNc d7-8ΔVKYT. The melting temperature Tm is determined at the inflection point of the fluorescence signal, the first derivative of which is reported here. Solid line represents the wild type, dashed line the deletion mutant, scaled to the wild-type. Note that the mutant protein has a significantly reduced melting temperature in comparison to wild type protein. c Mutant FLNc ABDs have similar structure but stronger actin-binding affinity. The top panel gives circular dichroism spectra obtained with wild-type and mutant filamin ABD constructs, demonstrating that wild-type and p.A193T mutant spectra are almost identical and only minor changes for the p.M251T mutant, indicating only minor structural effects of these mutations. The bottom panel gives high-speed F-actin cosedimentation assays with FLNc wild-type and mutant ABDs, showing increased actin binding activity of the mutant proteins. d Decreased FLNC mRNA and protein levels in patients with the p.F1720LfsX63 mutation. Quantitative FLNC transcript and protein analysis in muscle tissue from patients (Pt) and controls (Ctrl) reveals an approximately 50 % reduction in FLNC mRNA and protein levels in muscle samples from patients. An antibody directed against N-terminal FLNc did not detect expression of a truncated FLNc protein (~190 kDa). Reproduced with permission or adapted from a [26], b [23], c [6] and d [18]
Mutations that lead to the expression of misfolded FLNc, thereby overstraining the ubiquitin proteasome and autophagy pathways in the long run;
Mutations that do not affect protein solubility properties but give rise to a toxic gain of function by altering ligand binding properties;
Mutations causing a premature stop codon and concomitant nonsense-mediated decay, resulting in haploinsufficiency.
While the first type of mutations results in protein aggregation and subsequent impairment of protein homeostasis, giving rise to the typical MFM phenotype, the other two types of mutations result in distal myopathy with no protein aggregates. Although until now aggregation-causing mutations have only been found in Ig-like domains, such mutations may also occur in other portions of the molecule.
Thus far, MFM-causing mutations in Ig-like domains 7 (p.V930_T933del) and 24 (p.W2710X) have been analyzed at the biochemical and cellular level [23, 26, 57]. Since the latter mutation is localized in the last exon (exon 48) of FLNC [57], the mutant mRNA is stable and not prone to degradation by nonsense-mediated decay. The affected part of FLNc is its dimerization domain that is truncated and lacks the carboxyterminal 16 amino acids. Circular dichroism spectroscopy showed that the mutant domain is improperly folded, making it less stable and more susceptible to proteolysis. Hence, the p.W2710X mutation in FLNc impedes its ability to dimerize [26, 57] and instead, the mutant protein acquires a strong tendency for uncontrolled aggregation (Fig. 6a). This results in the deposition of massive protein aggregates that attract multiple other proteins including desmin and other Z-disc-associated proteins. These events ultimately lead to disintegration of myofibrils [26, 57].
The deletion of four amino acids in the β-strand of Ig-like domain 7 that is involved in interactions stabilizing the fold, also causes significant changes in the three-dimensional structure of the mutant domain, as illustrated by a higher proportion of unfolded or disordered structures, reduced stability and increased protease sensitivity (Fig. 6b) [23].
The mutations in the ABD of FLNc that were found in two distal myopathy families apparently cause only minimal structural alterations. Amino acid substitutions, however, are predicted to alter intradomain interactions, thereby facilitating binding to actin and increasing its binding constant (Fig. 6c) [6].
In contrast to the nonsense mutation in Ig-like domain 24, the frameshift deletion mutation in Ig-like domain 15 (c.5160delC, p.F1720LfsX63) does not occur in the last exon and thus activates nonsense-mediated decay of the mutant mRNA [18]. Since no truncated protein could be detected in the patients’ muscles, the 50 % reduction of FLNC mRNA and protein levels is the most probable reason for the disease phenotype (Fig. 6d). The lack of expression of mutant protein precludes the development of major protein aggregates that are a hallmark of MFM.
Cell and animal models
Cell models
The effects of the expression of truncated and full length mutated FLNc constructs were analyzed in tissue culture. Initially, the expression of mutant p.W2710X “mini-filamins” consisting of the ABD and Ig-like domains 15–24 was shown to be sufficient for spontaneous aggregation of the mutant protein in cultured cells [26]. The same effect was found upon transfection of full-length p.W2710X and p.V930_T933del FLNc in C2C12 mouse myoblasts [23]. The up to ten times higher number of transfected cells showing mutant FLNc aggregates in cells transfected with p.W2710X protein indicated that this mutant makes the cells more vulnerable to spontaneous aggregation. Transfection of the FLNc variants p.M251T and p.A193T also resulted in the development of protein aggregates in transfected non-muscle and muscle cells. Many of these aggregates also contained F-actin [6]. These cell models might become valuable tools to study the mechanisms of protein aggregation and evaluate treatments that prevent or reverse this phenomenon.
shRNA constructs in lentiviral vectors were used to generate a C2C12 cell line with a reduction in the level of Flnc mRNA of 93 % [5]. These cells, that expressed only very low levels of FLNc protein, proliferate and fuse normally, but instead of developing long myotubes, cells round up and form multinucleate myoballs upon fusion. This process is associated with a decrease in the expression of myogenin and muscle-specific genes, indicating a direct effect on myogenesis. For these experiments, a cell line showing the highest reduction in Flnc expression was selected. A variant with a more modest knock-down efficiency of Flnc expression could provide a model system for human diseases associated with Flnc haploinsufficiency.
Animal models
Mouse model
The only Flnc mouse model that has been created thus far is B6;129-Flnctm1Lmk/J. In these mice, the last eight exons (exons 41–48) of the Flnc gene were deleted by targeted mutation [5]. This mutation results in the expression of reduced levels of truncated mRNA and very low levels of truncated FLNc protein consisting of the ABD and Ig-like domains 1–19 and part of Ig-like domain 20 that is truncated after the FLNc-unique insertion. Mice homozygous for the knockout allele die at birth due to the inability to breath caused by severe abnormalities of their skeletal muscles. Whereas the heart has a normal appearance, the development of skeletal muscles is grossly disturbed, leading to reduced numbers of muscle fibers, often containing centrally located nuclei. Specifically, intercostal muscles and the diaphragm showed infiltration of connective tissue. Heterozygous mice were viable and fertile, and no abnormalities were reported, indicating that neither the low level of truncated FLNc nor the reduction of the level of wildtype FLNc results in an obvious phenotype [5]. Unfortunately, these mice apparently were not analyzed at older age. Since in man FLNC haploinsufficiency leads to distal myopathy at an average age of approximately 40 years [18], these mice might be a valuable model for this disease.
Medaka
A mutation in one of two flnc genes of medaka (Oryzias latipes, a teleost fish) was identified in zacro (zac) mutants [12]. This strain, which was obtained by N-ethyl-N-nitrosourea treatment, is characterized by disorganization of skeletal muscle fibers and abnormal development of the heart associated with a rupture of the myocardial layer. The causative mutation was found to be a nucleotide substitution in one of the flnc genes, leading to the introduction of a stop codon that would result in the expression of an FLNc variant that is truncated in Ig-like domain 15. Fish heterozygous for the mutation developed normally. All embryos showing the zac phenotype were homozygous for the mutant allele. Instability of the mutant mRNA most likely leads to nonsense-mediated decay and significantly lower the level of flnc mRNA. Although expression of a truncated protein was not analyzed, the lack of FLNc and not the expression of a toxic protein seems to be the most probable explanation for the observed phenotype. This was supported by morpholino-based antisense RNA experiments that resulted in a similar phenotype, at least in the heart [12] which explains why no alterations typical for MFM were found in the muscle of the zacro mutant fish. Heterozygous fish that are expected to be haploinsufficient for flnc should be studied at late adult age to conform to a model for distal myopathy caused by haploinsufficiency.
Future perspectives for research and therapy
The continuing search for mutations in FLNC will certainly result in the identification of more and more disease-associated mutations. Whereas the pathomechanisms of the mutations described thus far are roughly explained, it will be interesting to learn whether mutations in the rod2 segment have an impact on the association of FLNc with its many ligands, and how such a defect would influence the disease phenotype.
The main goal of future research should be the search for approaches to prevent the formation of aggregates in the muscle fibers of filaminopathy (and other MFM) patients. A promising strategy may be the induction of chaperones. For this purpose, appropriate cell and animal models are needed. Preliminary studies in cultured muscle cells indicate that transfection with mutant FLNc leads to aggregate formation. Cell lines stably transfected with constructs expressing mutant FLNc might, therefore, be an invaluable tool. Alternatively, skeletal muscle satellite cells from filaminopathy patients could be immortalized for such studies. Finally, patient fibroblasts could be converted to embryonic stem cells and be forced to develop into skeletal muscle cells.
The currently existing FLNC-related animal models only represent the type of filaminopathy associated with reduced expression levels but not the protein aggregation phenotype. To allow for testing therapeutic interventions in patients with the MFM type of filaminopathy, the development of an animal model would be of great value. A prime candidate would be a knock-in mouse carrying, e.g., the c.G8130A, p.W2710X mutation in one allele, since this represents the most frequent type of human filaminopathy.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, NIH [L.G.G], the German Research foundation [KL 2487/1-1 to R.A.K., FOR1228 to M.V., D.O.F., FOR1352 to D.O.F.], the German Ministry of Education and Research [01GM0887 to R.A.K., P.F.M.v.d.V., M.V., D.O.F.], the Ruhr-University Bochum [FoRUM K042-09 to R.A.K.], and the Spanish Instituto de Salud Carlos III [PI08-574 to M.O.].
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Contributor Information
Dieter O. Fürst, Email: dfuerst@uni-bonn.de, Institute for Cell Biology, University of Bonn, Ulrich-Haberland-Str. 61a, 53121 Bonn, Germany
Lev G. Goldfarb, Email: GoldfarbL@ninds.nih.gov, Clinical Neurogenetics, National Institutes of Health, Bethesda, MD, USA
Rudolf A. Kley, Email: rudolf.kley@rub.de, Department of Neurology, Neuromuscular Center Ruhrgebiet, University Hospital Bergmannsheil, Ruhr-University Bochum, Bochum, Germany
Matthias Vorgerd, Email: matthias.vorgerd@bergmannsheil.de, Department of Neurology, Neuromuscular Center Ruhrgebiet, University Hospital Bergmannsheil, Ruhr-University Bochum, Bochum, Germany.
Montse Olivé, Email: 25169mop@comb.cat, Institute of Neuropathology, Department of Pathology, and Neuromuscular Unit, Department of Neurology, IDIBELL-Hospital Universitari de Bellvitge, Hospitalet de Llobregat, Barcelona, Spain.
Peter F. M. van der Ven, Email: pvdven@uni-bonn.de, Institute for Cell Biology, University of Bonn, Ulrich-Haberland-Str. 61a, 53121 Bonn, Germany
References
- 1.Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Fürst DO, Saftig P, Saint R, Fleischmann BK, Hoch M, Höhfeld J. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 2.Avila-Smirnov D, Béhin A, Gueneau L, Claeys K, Beuvin M, Goudeau B, Richard P, Ben Yaou R, Romero NB, Mathis S, Voit T, Eymard B, Gil R, Fardeau M, Bonne G. A novel missense FLNC mutation causes arrhythmia and late onset myofibrillar myopathy with particular histopathology features. Neuromuscul Disord. 2010;20:623–624. [Google Scholar]
- 3.Bicknell LS, Farrington-Rock C, Shafeghati Y, Rump P, Alanay Y, Alembik Y, Al-Madani N, Firth H, Karimi-Nejad MH, Kim CA, Leask K, Maisenbacher M, Moran E, Pappas JG, Prontera P, de Ravel T, Fryns JP, Sweeney E, Fryer A, Unger S, Wilson LC, Lachman RS, Rimoin DL, Cohn DH, Krakow D, Robertson SP. A molecular and clinical study of Larsen syndrome caused by mutations in FLNB. J Med Genet. 2007;44:89–98. doi: 10.1136/jmg.2006.043687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakarova C, Wehnert MS, Uhl K, Sakthivel S, Vosberg HP, van der Ven PFM, Fürst DO. Genomic structure and fine mapping of the two human filamin gene paralogues FLNB and FLNC and comparative analysis of the filamin gene family. Hum Genet. 2000;107:597–611. doi: 10.1007/s004390000414. [DOI] [PubMed] [Google Scholar]
- 5.Dalkilic I, Schienda J, Thompson TG, Kunkel LM. Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol. 2006;26:6522–6534. doi: 10.1128/MCB.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duff RM, Tay V, Hackman P, Ravenscroft G, McLean C, Kennedy P, Steinbach A, Schöffler W, van der Ven PFM, Fürst DO, Song J, Djinović-Carugo K, Penttilä S, Raheem O, Reardon K, Malandrini A, Gambelli S, Villanova M, Nowak KJ, Williams DR, Landers JE, Brown RH, Jr, Udd B, Laing NG. Mutations in the N-terminal actin-binding domain of filamin C cause a distal myopathy. Am J Hum Genet. 2011;88:729–740. doi: 10.1016/j.ajhg.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrington-Rock C, Firestein MH, Bicknell LS, Superti-Furga A, Bacino CA, Cormier-Daire V, Le Merrer M, Baumann C, Roume J, Rump P, Verheij JB, Sweeney E, Rimoin DL, Lachman RS, Robertson SP, Cohn DH, Krakow D. Mutations in two regions of FLNB result in atelosteogenesis I and III. Hum Mutat. 2006;27:705–710. doi: 10.1002/humu.20348. [DOI] [PubMed] [Google Scholar]
- 8.Faulkner G, Pallavicini A, Comelli A, Salamon M, Bortoletto G, Ievolella C, Trevisan S, Kojic S, Dalla VF, Laveder P, Valle G, Lanfranchi G. FATZ: a filamin, actinin, and telethonin binding protein of the Z-disk of skeletal muscle. J Biol Chem. 2000;275:41234–41242. doi: 10.1074/jbc.M007493200. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 10.Fischer D, Kley RA, Strach K, Meyer C, Sommer T, Eger K, Rolfs A, Meyer W, Pou A, Pradas J, Heyer CM, Grossmann A, Huebner A, Kress W, Reimann J, Schröder R, Eymard B, Fardeau M, Udd B, Goldfarb L, Vorgerd M, Olivé M. Distinct muscle imaging patterns in myofibrillar myopathies. Neurology. 2008;71:758–765. doi: 10.1212/01.wnl.0000324927.28817.9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey N, Olson EN. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J Biol Chem. 2002;277:13998–14004. doi: 10.1074/jbc.M200712200. [DOI] [PubMed] [Google Scholar]
- 12.Fujita M, Mitsuhashi H, Isogai S, Nakata T, Kawakami A, Nonaka I, Noguchi S, Hayashi YK, Nishino I, Kudo A. Filamin C plays an essential role in the maintenance of the structural integrity of cardiac and skeletal muscles, revealed by the medaka mutant zacro. Dev Biol. 2012;361:79–89. doi: 10.1016/j.ydbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Gariboldi M, Maestrini E, Canzian F, Manenti G, De Gregorio L, Rivella S, Chatterjee A, Herman GE, Archidiacono N, Antonacci R. Comparative mapping of the actin-binding protein 280 genes in human and mouse. Genomics. 1994;21:428–430. doi: 10.1006/geno.1994.1288. [DOI] [PubMed] [Google Scholar]
- 14.Gehmlich K, Hayeß K, Legler C, Haebel S, van der Ven PFM, Ehler E, Fürst DO. Ponsin interacts with Nck adapter proteins: implications for a role in cytoskeletal remodelling during differentiation of skeletal muscle cells. Eur J Cell Biol. 2010;89:351–364. doi: 10.1016/j.ejcb.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Goebel HH. Congenital myopathies at their molecular dawning. Muscle Nerve. 2003;27:527–548. doi: 10.1002/mus.10322. [DOI] [PubMed] [Google Scholar]
- 16.Goebel HH. Protein aggregate myopathies. Introduction. Brain Pathol. 2009;19:480–482. doi: 10.1111/j.1750-3639.2009.00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gontier Y, Taivainen A, Fontao L, Sonnenberg A, van der Flier A, Carpén O, Faulkner G, Borradori L. The Z-disc proteins myotilin and FATZ-1 interact with each other and are connected to the sarcolemma via muscle-specific filamins. J Cell Sci. 2005;118:3739–3749. doi: 10.1242/jcs.02484. [DOI] [PubMed] [Google Scholar]
- 18.Guergueltcheva V, Peeters K, Baets J, Ceuterick-de Groote C, Martin JJ, Suls A, De Vriendt E, Mihaylova V, Chamova T, Almeida-Souza L, Ydens E, Tzekov C, Hadjidekov G, Gospodinova M, Storm K, Reyniers E, Bichev S, van der Ven PFM, Fürst DO, Mitev V, Lochmüller H, Timmerman V, Tournev I, De Jonghe P, Jordanova A. Distal myopathy with upper limb predominance caused by filamin C haploinsufficiency. Neurology. 2011;77:2105–2114. doi: 10.1212/WNL.0b013e31823dc51e. [DOI] [PubMed] [Google Scholar]
- 19.Harms MB, Sommerville RB, Allred P, Bell S, Ma D, Cooper P, Lopate G, Pestronk A, Weihl CC, Baloh RH. Exome sequencing reveals DNAJB6 mutations in dominantly-inherited myopathy. Ann Neurol. 2012;71:407–416. doi: 10.1002/ana.22683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Himmel M, van der Ven PFM, Stöcklein W, Fürst DO. The limits of promiscuity: isoform-specific dimerization of filamins. Biochemistry. 2003;42:430–439. doi: 10.1021/bi026501+. [DOI] [PubMed] [Google Scholar]
- 21.Holmes WB, Moncman CL. Nebulette interacts with filamin C. Cell Motil Cytoskeleton. 2008;65:130–142. doi: 10.1002/cm.20249. [DOI] [PubMed] [Google Scholar]
- 22.Kley RA, Hellenbroich Y, van der Ven PFM, Fürst DO, Huebner A, Bruchertseifer V, Peters SA, Heyer CM, Kirschner J, Schröder R, Fischer D, Müller K, Tolksdorf K, Eger K, Germing A, Brodherr T, Reum C, Walter MC, Lochmüller H, Ketelsen UP, Vorgerd M. Clinical and morphological phenotype of the filamin myopathy: a study of 31 German patients. Brain. 2007;130:3250–3264. doi: 10.1093/brain/awm271. [DOI] [PubMed] [Google Scholar]
- 23.Kley RA, Serdaroglu-Oflazer P, Leber Y, Odgerel Z, van der Ven PFM, Olivé M, Ferrer I, Onipe A, Mihaylov M, Bilbao JM, Lee HS, Höhfeld J, Djinović-Carugo K, Kong K, Tegenthoff M, Peters SA, Stenzel W, Vorgerd M, Goldfarb LG, Fürst DO. Pathophysiology of protein aggregation and extended phenotyping in filaminopathy. Brain. 2012;135:2642–2660. doi: 10.1093/brain/aws200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kono S, Nishio T, Takahashi Y, Goto-Inoue N, Kinoshita M, Zaima N, Suzuki H, Fukutoku-Otsuji A, Setou M, Miyajima H. Dominant-negative effects of a novel mutation in the filamin myopathy. Neurology. 2010;75:547–554. doi: 10.1212/WNL.0b013e3181ec7fbd. [DOI] [PubMed] [Google Scholar]
- 25.Linnemann A, van der Ven PFM, Vakeel P, Albinus B, Simonis D, Bendas G, Schenk JA, Micheel B, Kley RA, Fürst DO. The sarcomeric Z-disc component myopodin is a multiadapter protein that interacts with filamin and alpha-actinin. Eur J Cell Biol. 2010;89:681–692. doi: 10.1016/j.ejcb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Löwe T, Kley RA, van der Ven PFM, Himmel M, Huebner A, Vorgerd M, Fürst DO. The pathomechanism of filaminopathy: altered biochemical properties explain the cellular phenotype of a protein aggregation myopathy. Hum Mol Genet. 2007;16:1351–1358. doi: 10.1093/hmg/ddm085. [DOI] [PubMed] [Google Scholar]
- 27.Lu S, Carroll SL, Herrera AH, Ozanne B, Horowits R. New N-RAP-binding partners alpha-actinin, filamin and Krp1 detected by yeast two-hybrid screening: implications for myofibril assembly. J Cell Sci. 2003;116:2169–2178. doi: 10.1242/jcs.00425. [DOI] [PubMed] [Google Scholar]
- 28.Luan X, Hong D, Zhang W, Wang Z, Yuan Y. A novel heterozygous deletion-insertion mutation (2695–2712 del/GTTTGT ins) in exon 18 of the filamin C gene causes filaminopathy in a large Chinese family. Neuromuscul Disord. 2010;20:390–396. doi: 10.1016/j.nmd.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Maestrini E, Patrosso C, Mancini M, Rivella S, Rocchi M, Repetto M, Villa A, Frattini A, Zoppe M, Vezzoni P. Mapping of two genes encoding isoforms of the actin binding protein ABP- 280, a dystrophin like protein, to Xq28 and to chromosome 7. Hum Mol Genet. 1993;2:761–766. doi: 10.1093/hmg/2.6.761. [DOI] [PubMed] [Google Scholar]
- 30.Nakano S, Engel AG, Waclawik AJ, Emslie-Smith AM, Busis NA. Myofibrillar myopathy with abnormal foci of desmin positivity. I. Light and electron microscopy analysis of 10 cases. J Neuropathol Exp Neurol. 1996;55:549–562. doi: 10.1097/00005072-199605000-00008. [DOI] [PubMed] [Google Scholar]
- 31.Odgerel Z, van der Ven PFM, Fürst DO, Goldfarb LG. DNA sequencing errors in molecular diagnostics of filamin myopathy. Clin Chem Lab Med. 2010;48:1409–1414. doi: 10.1515/CCLM.2010.272. [DOI] [PubMed] [Google Scholar]
- 32.Ohlsson M, Hedberg C, Bradvik B, Lindberg C, Tajsharghi H, Danielsson O, Melberg A, Udd B, Martinsson T, Oldfors A. Hereditary myopathy with early respiratory failure associated with a mutation in A-band titin. Brain. 2012;135:1682–1694. doi: 10.1093/brain/aws103. [DOI] [PubMed] [Google Scholar]
- 33.Olivé M, Odgerel Z, Martinez A, Poza JJ, Bragado FG, Zabalza RJ, Jerico I, Gonzalez-Mera L, Shatunov A, Lee HS, Armstrong J, Maravi E, Arroyo MR, Pascual-Calvet J, Navarro C, Paradas C, Huerta M, Marquez F, Rivas EG, Pou A, Ferrer I, Goldfarb LG. Clinical and myopathological evaluation of early- and late-onset subtypes of myofibrillar myopathy. Neuromuscul Disord. 2011;21:533–542. doi: 10.1016/j.nmd.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeffer G, Elliott HR, Griffin H, Barresi R, Miller J, Marsh J, Evila A, Vihola A, Hackman P, Straub V, Dick DJ, Horvath R, Santibanez-Koref M, Udd B, Chinnery PF. Titin mutation segregates with hereditary myopathy with early respiratory failure. Brain. 2012;135:1695–1713. doi: 10.1093/brain/aws102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popowicz GM, Schleicher M, Noegel AA, Holak TA. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem Sci. 2006;31:411–419. doi: 10.1016/j.tibs.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Pudas R, Kiema TR, Butler PJ, Stewart M, Ylänne J. Structural basis for vertebrate filamin dimerization. Structure (Camb) 2005;13:111–119. doi: 10.1016/j.str.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 37.Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R, Orstavik KH, Quarrell OW, Schwartz CE, Shears DJ, Suri M, Kendrick-Jones J, Wilkie AO. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- 38.Sarparanta J, Jonson PH, Golzio C, Sandell S, Luque H, Screen M, McDonald K, Stajich JM, Mahjneh I, Vihola A, Raheem O, Penttilä S, Lehtinen S, Huovinen S, Palmio J, Tasca G, Ricci E, Hackman P, Hauser M, Katsanis N, Udd B. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet. 2012;44:450–452. doi: 10.1038/ng.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schröder R, Schoser B. Myofibrillar myopathies: a clinical and myopathological guide. Brain Pathol. 2009;19:483–492. doi: 10.1111/j.1750-3639.2009.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selcen D. Myofibrillar myopathies. Curr Opin Neurol. 2008;21:585–589. doi: 10.1097/WCO.0b013e32830a752b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selcen D. Myofibrillar myopathies. Neuromuscul Disord. 2011;21:161–171. doi: 10.1016/j.nmd.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selcen D, Bromberg MB, Chin SS, Engel AG. Reducing bodies and myofibrillar myopathy features in FHL1 muscular dystrophy. Neurology. 2011;77:1951–1959. doi: 10.1212/WNL.0b013e31823a0ebe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selcen D, Muntoni F, Burton BK, Pegoraro E, Sewry C, Bite AV, Engel AG. Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann Neurol. 2009;65:83–89. doi: 10.1002/ana.21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shatunov A, Olivé M, Odgerel Z, Stadelmann-Nessler C, Irlbacher K, van Landeghem F, Bayarsaikhan M, Lee HS, Goudeau B, Chinnery PF, Straub V, Hilton-Jones D, Damian MS, Kaminska A, Vicart P, Bushby K, Dalakas MC, Sambuughin N, Ferrer I, Goebel HH, Goldfarb LG. In-frame deletion in the seventh immunoglobulin-like repeat of filamin C in a family with myofibrillar myopathy. Eur J Hum Genet. 2009;17:656–663. doi: 10.1038/ejhg.2008.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sjekloca L, Pudas R, Sjöblom B, Konarev P, Carugo O, Rybin V, Kiema TR, Svergun D, Ylänne J, Djinović-Carugo K. Crystal structure of human filamin C domain 23 and small angle scattering model for filamin C 23–24 dimer. J Mol Biol. 2007;368:1011–1023. doi: 10.1016/j.jmb.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 46.Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 47.Takada F, Vander Woude DL, Tong HQ, Thompson TG, Watkins SC, Beggs AH, Kunkel LM. Myozenin: an α-actinin- and γ-filamin-binding protein of skeletal muscle Z lines. Proc Natl Acad Sci USA. 2001;98:1595–1600. doi: 10.1073/pnas.041609698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tasca G, Odgerel Z, Monforte M, Aurino S, Clarke NF, Waddell LB, Udd B, Ricci E, Goldfarb LG. Novel FLNC mutation in a patient with myofibrillar myopathy in combination with late-onset cerebellar ataxia. Muscle Nerve. 2012;46:275–282. doi: 10.1002/mus.23349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson TG, Chan YM, Hack AA, Brosius M, Rajala M, Lidov HG, McNally EM, Watkins S, Kunkel LM. Filamin 2 (FLN2). A muscle-specific sarcoglycan interacting protein. J Cell Biol. 2000;148:115–126. doi: 10.1083/jcb.148.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Udd B. Distal muscular dystrophies. Handb Clin Neurol. 2011;101:239–262. doi: 10.1016/B978-0-08-045031-5.00016-5. [DOI] [PubMed] [Google Scholar]
- 51.Uhlén M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 52.van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 53.van der Ven PFM, Ehler E, Vakeel P, Eulitz S, Schenk JA, Milting H, Micheel B, Fürst DO. Unusual splicing events result in distinct Xin isoforms that associate differentially with filamin C and Mena/VASP. Exp Cell Res. 2006;312:2154–2167. doi: 10.1016/j.yexcr.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 54.van der Ven PFM, Obermann WMJ, Lemke B, Gautel M, Weber K, Fürst DO. Characterization of muscle filamin isoforms suggests a possible role of γ-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil Cytoskeleton. 2000;45:149–162. doi: 10.1002/(SICI)1097-0169(200002)45:2<149::AID-CM6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 55.van der Ven PFM, Odgerel Z, Fürst DO, Goldfarb LG, Kono S, Miyajima H. Dominant-negative effects of a novel mutation in the filamin myopathy. Neurology. 2010;75:2137–2138. doi: 10.1212/WNL.0b013e3182031bb3. [DOI] [PubMed] [Google Scholar]
- 56.van der Ven PFM, Wiesner S, Salmikangas P, Auerbach D, Himmel M, Kempa S, Hayeß K, Pacholsky D, Taivainen A, Schröder R, Carpén O, Fürst DO. Indications for a novel muscular dystrophy pathway. γ-filamin, the muscle-specific filamin isoform, interacts with myotilin. J Cell Biol. 2000;151:235–248. doi: 10.1083/jcb.151.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vorgerd M, van der Ven PFM, Bruchertseifer V, Löwe T, Kley RA, Schröder R, Lochmüller H, Himmel M, Koehler K, Fürst DO, Huebner A. A mutation in the dimerization domain of filamin c causes a novel type of autosomal-dominant myofibrillar myopathy. Am J Hum Genet. 2005;7:297–304. doi: 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams DR, Reardon K, Roberts L, Dennet X, Duff R, Laing NG, Byrne E. A new dominant distal myopathy affecting posterior leg and anterior upper limb muscles. Neurology. 2005;64:1245–1254. doi: 10.1212/01.WNL.0000156524.95261.B9. [DOI] [PubMed] [Google Scholar]
- 59.Willis MS, Schisler JC, Portbury AL, Patterson C. Build it up-Tear it down: protein quality control in the cardiac sarcomere. Cardiovasc Res. 2009;81:439–448. doi: 10.1093/cvr/cvn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie Z, Xu W, Davie EW, Chung DW. Molecular cloning of human ABPL, an actin-binding protein homologue. Biochem Biophys Res Commun. 1998;251:914–919. doi: 10.1006/bbrc.1998.9506. [DOI] [PubMed] [Google Scholar]
- 61.Zhang M, Liu J, Cheng A, Deyoung SM, Saltiel AR. Identification of CAP as a costameric protein that interacts with filamin C. Mol Biol Cell. 2007;18:4731–4740. doi: 10.1091/mbc.E07-06-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]