SUMMARY
MicroRNAs (miRNA) are mostly downregulated in cancer. However, the mechanism underlying this phenomenon and the precise consequence in tumorigenesis remain obscure. Here we show that ERK suppresses pre-miRNA export from the nucleus through phosphorylation of exportin-5 (XPO5) at T345/S416/S497. After phosphorylation by ERK, conformation of XPO5 is altered by prolyl isomerase Pin1, resulting in reduction of pre-miRNA loading. In liver cancer, the ERK-mediated XPO5 suppression reduces miR-122, increases microtubule dynamics, and results in tumor development and drug resistance. Analysis of clinical specimens further showed that XPO5 phosphorylation is associated with poor prognosis for liver cancer patients. Our study reveals a function of ERK in miRNA biogenesis and suggests that modulation of miRNA export has potential clinical implications.
In Brief
Sun et al. find that ERK phosphorylates XPO5, which induces a Pin1-mediated conformational change that inhibits the ability of XPO5 to load and export pre-miRNA from the nucleus. Phosphorylation of XPO5 is associated with global miRNA downregulation and correlates with poor survival in hepatocellular carcinoma.
INTRODUCTION
MicroRNAs (miRNA) are a class of small non-coding RNAs that regulate gene expression through inhibition of translation or stability of target mRNAs. The biogenesis of miRNA involves multiple steps, including transcription of primary miRNA (pri-miRNA) by RNA polymerase II, cleavage of pri-mRNA to precursor miRNA (pre-miRNA) by Drosha, nucleocytoplasmic export of pre-miRNA by exportin-5 (XPO5), processing of pre-miRNA to mature miRNA by Dicer, and formation of functional RNA-induced Silencing Complex containing Argonaute (Krol et al., 2010). Depending on the cellular context, a miRNA can function as an oncogene or a tumor suppressor (Croce, 2009). However, global downregulation of miRNA expression has been observed in many tumors (Lu et al., 2005). Interestingly, systematic evaluation of miRNA levels in cancer cell lines demonstrated that many pre-miRNAs are retained in the nucleus (Lee et al., 2008), implying that function of the nuclear-cytoplasmic export machinery may be compromised in tumors.
XPO5 is currently considered the indispensable transport receptor for pre-miRNA in most organisms examined (Katahira and Yoneda, 2011). Upon binding to Ran-GTP, XPO5 uses a baseball mitt-like structure to shield the pre-miRNA stem region and the tunnel-like structure to recognize the 2-nucleotide 3′-overhang (Okada et al., 2009). Following GTP hydrolysis, XPO5 releases the pre-miRNA into the cytoplasm. It has been proposed that XPO5 is unique among classical haploinsufficient tumor suppressor genes since partial, but not complete, loss promotes tumor development (Melo et al., 2010). However, the exact role of XPO5 in tumor progression remains elusive. XPO5 is downregulated in lung cancer (Chiosea et al., 2007), but upregulated in breast and prostate cancer (Leaderer et al., 2011). Furthermore, the C/C genotype of rs11077 in the XPO5 3′UTR, which decreases XPO5 protein expression, is associated with poorer prognosis of esophageal and renal cancer (Horikawa et al., 2008) but better prognosis of non-small-cell lung cancer, multiple myeloma, and liver cancer (Liu et al., 2014).
Development of hepatocellular carcinoma (HCC) is a multistep sequential process going from chronic inflammation, cirrhosis, primary HCC to metastatic HCC. It has been reported that aberrant miRNA expression drives HCC development and global miRNA downregulation at a later stage promotes metastasis (Wong et al., 2012). Since the reduced miRNA expression in liver cancer is attributed to defective miRNA biogenesis (Lee et al., 2008) and the activity of XPO5 is the rate-limiting step for the production of mature miRNA (Yi et al., 2005), we decided to investigate the role of XPO5 in liver cancer.
RESULTS
ERK Activation Suppresses Nuclear Export Activity of XPO5
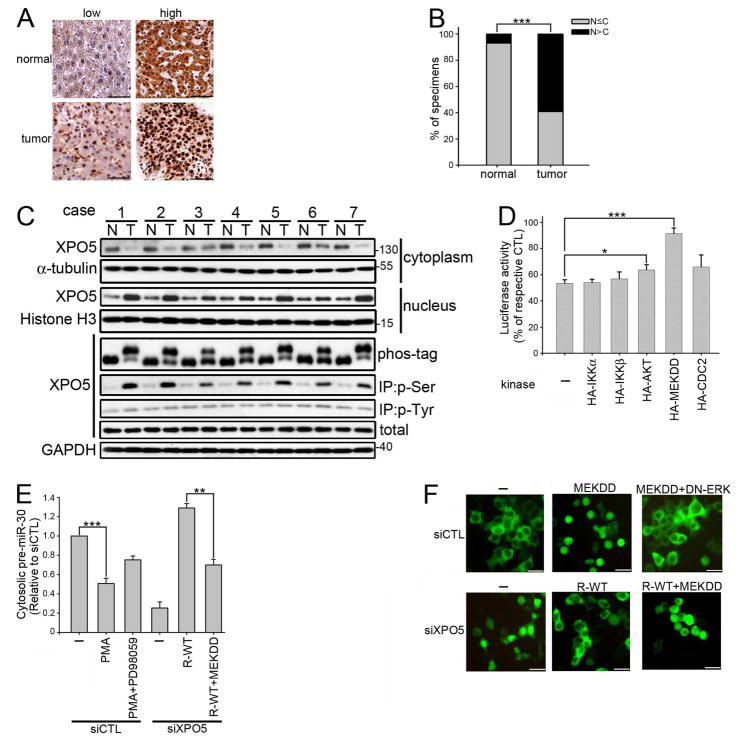
To determine the role of XPO5 in the development of HCC, we first investigated the expression of XPO5 in liver cancer patients. Although the expression level of XPO5 was similar in normal and tumor tissues (Figure S1A), we noticed that more XPO5 was retained in the nucleus of tumor cells (Figures 1A and 1B). Consistent with the findings from IHC staining, Western blot analysis showed nuclear localization of XPO5 in liver tumor lysates (Figure 1C). Because phosphorylation-dependent mobility shift could be detected by phos-tag (Kinoshita et al., 2006) in tumors, we further compared phosphorylation status of XPO5. Interestingly, serine phosphorylation of XPO5 was significantly increased in tumors (Figure 1C).
Figure 1. ERK Activation Suppresses XPO5 Function.
(A) Representative specimens of low and high XPO5 expression in paired human normal and liver tumor tissues on Human HistoArray IMH-342 and IMH-318 by IHC staining of XPO5. Scale bar: 50 μm. (B) Percentage of human normal liver and tumor specimens with the majority of XPO5 localized in the nucleus (N) or cytoplasm (C) was determined. (n=35 per group, p values were calculated by Pearson’s chi-square test). (C) Comparison of localization and phosphorylation of XPO5 in paired human HCC (T) and adjacent benign liver (N). Tissue lysates were subjected to immunoprecipitation (IP) with p-serine or p-tyrosine antibody, or separation of cytoslic and nuclear fraction. Proteins were separated by regular SDS-PAGE or Phos-tag SDS-PAGE then detected by immunoblotting (IB) with XPO5 antibody. (D) 293T cells were transfected with pCMV-Luc-miR-30, pKmyc-XPO5, and the indicated kinase expression vectors for 48 hr before luciferase assay. Data were presented as relative to the cells transfected with the kinase expression vector without XPO5 overexpression. (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (E) Control (siCTL) cells were pre-treated with or without 30 μM PD98059 for 1 hr then treated with 100 nM PMA for 8 hr, while XPO5-depleted (siXPO5) cells were transfected with siRNA-resistant XPO5 (R-WT) with or without MEKDD for 48 hr. All cells were co-transfected with pCMV-miR-30 because 293T cells do not express Northern blot-detectable miR-30 (Zeng et al., 2002). Cytoplasmic RNA was extracted using PARIS kit then detected by qRT-PCR. Data were presented as relative to the cytoplasmic pre-miR-30 expression in siCTL 293T cells transfected with pCMV-miR-30 (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (F) siCTL cells were transfected with MEKDD with or without DN-ERK, while siXPO5 cells were transfected with siRNA-resistant XPO5 (R-WT) with or without MEKDD. All cells were co-transfected with GFP-NLS-eEF1A and observed by florescence microscopy after 48 hr. Scale bars: 50 μm. See also Figure S1.
XPO5 overexpression is known to increase nuclear export of pre-miRNA, leading to the enhanced inhibition of miRNA-targeted gene expression. Therefore, we assessed the effects of XPO5 phosphorylation on its function using an indicator luciferase reporter containing eight perfect miR-30a binding sites. Consistent with previous reports (Yi et al., 2005), overexpression of XPO5 exports the endogenous miR-30 to inhibit luciferase expression (Figure 1D). Interestingly, when we co-transfected five different oncogenic serine/threonine kinases, including IKKα, IKKβ, myristoylated-AKT, MEKDD, or CDC2; only constitutively active MEK (MEKDD), which activates ERK, significantly reversed the effects of XPO5 (Figure 1D). In addition, ERK activation increased the slow-migrating and serine phosphorylated form of XPO5 (Figure S1B). The activities of these five kinases were confirmed using previously reported substrates (Figure S1C). Distribution of pre-miR-30 in the cytoplasm was reduced by knockdown of XPO5 and restored by overexpression of shRNA-resistant XPO5 (R-WT) (Figure 1E). In contrast, ERK activation by phorbol 12-myristate 13-acetate (PMA) or co-transfection with MEKDD abrogated the cytoplasmic accumulation of pre-miR-30 (Figures 1E and S1D). Treatment of cells with MEK1 inhibitor, PD98059, was found to reverse the decrease in pre-miR-30 triggered by PMA. Northern blot analysis confirmed inefficient pre-miRNA export in cells with activated ERK (Figure S1E). Eukaryotic translation elongation factor 1A (eEF1A), a nuclear export substrate of XPO5 (Bohnsack et al., 2002), was also used as a readout of XPO5 function. Consistently, MEK activation, whose effect can be reversed by dominant negative (DN) ERK, also blocks XPO5-mediated eEF1A nuclear export (Figure 1F). In summary, these results suggest that the activation of ERK inhibits nuclear export activity of XPO5.
ERK Phosphorylates XPO5 at T345, S416 and S497
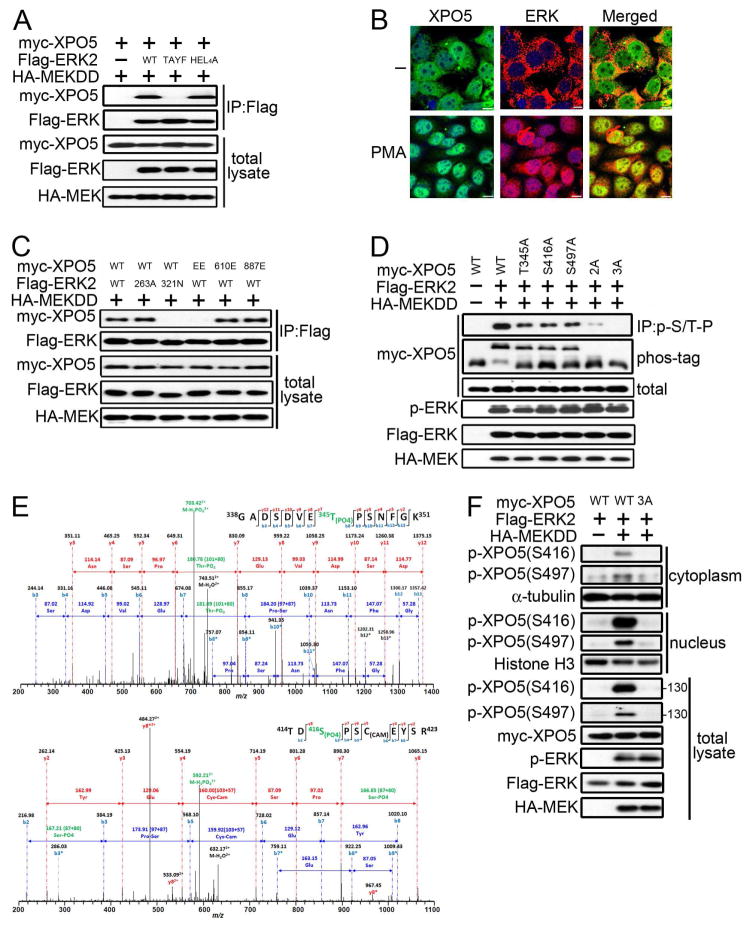
To determine how ERK inhibits XPO5 activity, we first tested whether ERK interacts with XPO5. Co-immunoprecipitation experiments showed that XPO5 was associated with ERKs upon MEK activation (Figure S2A). Although ERK1 and ERK2 are highly similar and possess identical substrate specificity in vitro, we focused on the ERK2 because its expression exceeds that of ERK1 in most cells, and Erk2 knockout results in embryonic lethality while Erk1 knockout does not (Pages et al., 1999; Yao et al., 2003). The interaction between endogenous ERK2 and XPO5 was observed after PMA stimulation (Figure S2B). After MEK stimulation, the phosphorylated ERK translocates into the nucleus to activate nuclear substrates or forms a dimer to activate cytoplasmic substrates (Casar et al., 2009). As shown in Figure 2A, XPO5 does not interact with phosphorylation site mutant TAYF but still binds to dimerization mutant HEL4A of ERK, suggesting that XPO5 could be a nuclear substrate of ERK. Indeed, ERK co-localizes with XPO5 in the nucleus (Figure 2B).
Figure 2. ERK Interacts with and Phosphorylates XPO5.
(A) Lysates of 293T cells transfected as indicated were subjected to IP with anti-Flag antibody and IB. Interaction was assessed between XPO5 and WT ERK2, dimerization mutant ERK2-H178E/L335A/L338A/L343A/L346A (HEL4A), and MEK-phosphorylation site mutant ERK2-T185A/Y187F (TAYF). (B) Immunofluorescence analysis of XPO5 (green) and ERK (red) of 293T cells untreated or treated with 100 nM PMA for 30 min. DAPI (blue) was used to mark the nucleus. Scale bars: 10 μm. (C) Interaction between WT or mutant ERK2 and XPO5 in lysates from MEKDD expressing 293T cells transfected as indicated was examined by co-IP. EE: R284E/K285E. (D) Lysates of 293T cells transfected with WT or non-phosphorylatable alanine mutants of XPO5 were subjected to IP and IB. 2A: T345A/S416A, 3A: T345A/S416A/S497A. (E) Mass spectrometry detected T345 and S416 phosphorylation in GST-XPO5 (251–550) incubated with active recombinant ERK2. (F) Characterization of p-XPO5 (S416) and p-XPO5 (S497) antibodies by IB analysis of cytosolic, nuclear, and total lysates of 293T cells transfected as indicated with MEKDD, ERK2, and WT or non-phosphorylatable alanine mutant (3A) XPO5. See also Figure S2.
ERK displays specificity for phosphorylation at the serine/threonine-proline (S/T-P) motif. Since the S/T-P motif is found in many proteins, ERK uses a docking motif to ensure its substrate specificity. The best characterized docking sites on ERK are the F-site recruitment site (FRS) and common docking (CD) domain, which responds to the F-site (FX-F/Y-P) and D domain (K/R0–2-X1–6-ϕ-X-ϕ) on substrates (Roskoski, 2012). We found that CD mutant (321N), but not FRS mutant (263A) of ERK abolishes interaction with XPO5 (Figure 2C). Mutational analysis of three potential D domains of XPO5, identified by Eukaryotic Linear Motif database, further revealed that XPO5 residues 284–291 are critical for the association (Figures 2C and S2C).
Given the physical interaction between ERK and XPO5 and that calf intestinal alkaline phosphatase can eliminate the mobility shift of XPO5 induced by ERK (Figure S2D), we examined whether XPO5 is a physiological substrate of ERK. Phospho-S/T-P antibody detected XPO5 phosphorylation correlated with ERK activation (Figures S2E and S2F). In vitro kinase assays and mutational analyses suggested that three highly conserved residues T345/S416/S497 of XPO5 are major ERK phosphorylation sites (Figures 2D, S2G and S2H). Mass spectrometry analysis showed that ERK phosphorylated XPO5 at T345/S416 in vitro and S416 in vivo (Figures 2E and S2I). Although phosphorylation at S497 was not detected by mass spectrometry, it was confirmed by a phosphorylation site specific polyclonal antibody (Figure 2F). The original intention was to generate all three phospho-specific antibodies, but only those against p-S416 and p-S497 were successful. Considering the nucleus-enriched specificity and stronger signal intensity (Figure 2F), p-S416 antibody was used as a tool to monitor XPO5 phosphorylation hereafter (Figure S2J). In conclusion, we demonstrate that ERK interacts with XPO5 and phosphorylates it at T345/S416/S497.
XPO5 Phosphorylation Globally Downregulates miRNA Expression
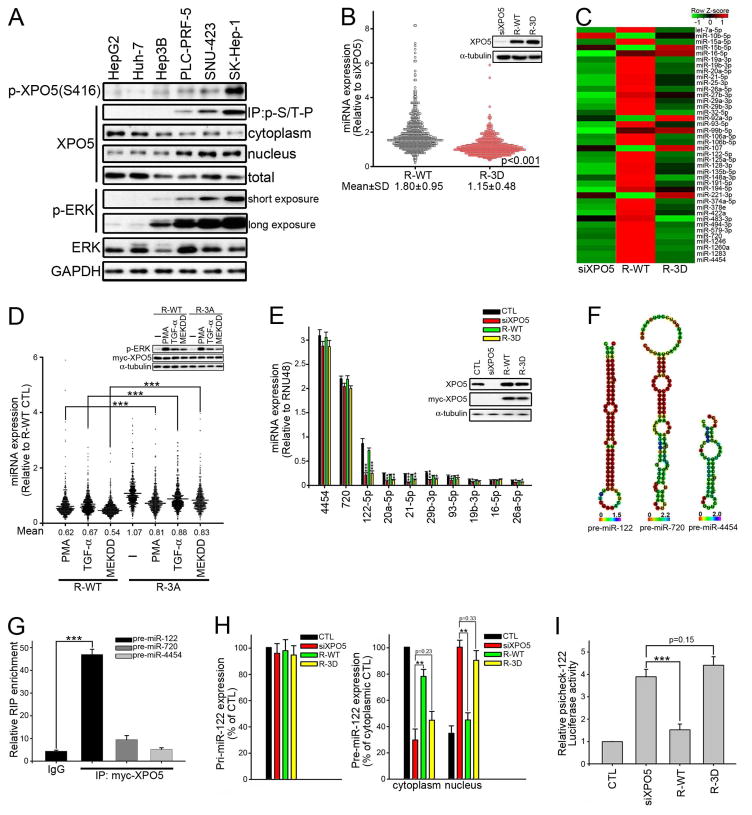
Before investigating how phosphorylation inhibits XPO5 activity, we first examined the functional importance of XPO5 phosphorylation in miRNA regulation. Comparison of the phosphorylation of ERK and XPO5 in a panel of liver cancer cell lines showed that XPO5 phosphorylation correlated positively with ERK phosphorylation (Figure 3A) and negatively with sensitivity to sorafenib (Figure S3A), a RAF inhibitor approved for the treatment of advanced liver cancer (Bai et al., 2009). To show the global trend of miRNA expression, dot density plot was used to compare expression of 798 miRNAs identified by Nanostring nCounter miRNA expression assay. We found that XPO5-knockdown induced global downregulation of miRNA in the low ERK phosphorylated cell lines, Huh-7 and HepG2, which could be rescued by wild-type XPO5, but not a phosphomimetic 3D mutant (Figures 3B and S3B). Moreover, non-phosphorylatable 3A mutant upregulated miRNA expression more than wild-type XPO5 in the high ERK phosphorylated cell lines, SNU-423 and SK-Hep-1 (Figures S3C and S3D). Based on relative expression levels, we classified the miRNA into three sets: downregulated (<0.67-fold), unchanged (0.67- to 1.5-fold), and upregulated (>1.5-fold) (Figure S3E). The 40 most abundantly expressed miRNAs were also selected to generate the heat map showing the trend of miRNA expression (Figures 3C and S3F–H). The Huh-7 cell line was chosen as a model to study the effect of XPO5 phosphorylation hereafter because it exhibits the lowest ERK and XPO5 phosphorylation levels (Figure 3A). ERK activation by PMA, TGF-α, or MEKDD induced global downregulation of miRNA in the presence of wild-type XPO5, but this effect of ERK activation was attenuated in cells expressing 3A XPO5 (Figure 3D). These results support the notion that ERK activation leads to globally downregulated miRNA levels by phosphorylating XPO5 at T345/S416/S497.
Figure 3. Phosphorylation of XPO5 by ERK Globally Downregulates miRNA Expression.
(A) Basal XPO5 expression, localization, and phosphorylation in a panel of human liver cancer cell lines were analyzed by IB. (B) Dot density plot depicting the expression changes of miRNA in siXPO5 Huh-7 cells stably overexpressing siXPO5-resistant WT (R-WT) or phosphomimetic mutant (R-3D) XPO5. Data were presented as relative to the miRNA expression in siXPO5 cells (n=2). Inset: IB of XPO5. (C) Heat map generated using R program shows relative miRNA expression in Huh-7 stable transfectants as indicated by the green to red key bar at the top of the map. NanoString counts >350 are shown. (D) Dot density plot was used to examine the effect of ERK activation on global miRNA expression. WT or non-phosphorylatable 3A XPO5 expressing Huh-7 cells were treated with 100 nM PMA, 100 ng/ml TGF-α or transfected with MEKDD for 48 hr. Data were presented as relative to the miRNA expression in untreated R-WT Huh-7 cells (n=2). Inset: IB of ERK phosphorylation across treatment groups. WT or non-phosphorylatable 3A XPO5 expressing Huh-7 cells were treated with 100 nM PMA, 100 ng/ml TGF-α for 30 min or transfected with MEKDD for 48 hr. (E) Top ten highly expressed miRNAs in Huh-7 stable transfectants were quantified by qRT-PCR. miRNA expression was normalized to RNU48 (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (F) Secondary structure of pre-miR-122, pre-miR-720 and pre-miR-4454. MFE structure drawing encoding positional entropy was predicted by RNAfold. (G) Lysates of Huh-7 R-WT stable transfectant were subjected to RNA-binding protein immunoprecipitation (RIP) analysis. Extracts of Huh-7 cells were subjected to IP with IgG or anti-myc tag antibody. Pull-down RNA was analyzed by qRT-PCR using specific primers for pre-miR-122, pre-miR-720, and pre-miR-4454 (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (H) Comparison of pri-miR-122 and pre-miR-122 expression in Huh-7 stable transfectant (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). Left panel: RNA was isolated by TRIzol for Taqman pri-miR-122 assay. Data were presented as relative to CTL Huh-7 cells. Right panel: Cytoplasmic and nuclear RNA fractions were isolated using PARIS kit then subjected to pre-miR-122 qPCR analysis. Data were presented as relative to the cytoplasmic pre-miR-122 expression in CTL Huh-7 cells. (I) Huh-7 stable transfectants were transfected with a reporter with miR-122-binding sites for 48 hr before luciferase assay (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). See also Figure S3.
Considering that abundance of miRNA is crucial for miRNA function, expression levels of the top ten abundantly expressed miRNAs were validated by qRT-PCR (Figure 3E). MiR-122 is known to dominate the miRNA content in the normal adult liver (Jopling, 2012). In Huh-7 cells, we noticed that miR-4454 and miR-720 are expressed at higher levels than miR-122, but are minimally affected by XPO5. MiRNA precursors fold into stem-loop structures and are recognized by XPO5; however, a large number of similar hairpins that are not pre-miRNAs can also be found in the genome and are called pseudo-hairpins. RNA-fold (Hofacker, 2003) shows these two pre-miRNAs have significantly higher minimum free-energy (Figure 3F), and MiPred (Jiang et al., 2007) does not classify them as pre-miRNA like hairpins (Figure S3I). Consistent with the in silico prediction, RNA-binding protein immunoprecipitation (RIP) and RNA pull-down suggest miR-4454 and miR-720 minimally interact with XPO5, compared to miR-122 (Figures 3G and S3J).
Considering that Huh-7 cells resemble normal hepatocytes in expressing high levels of miR-122 (Chang et al., 2004), miR-122 was chosen hereafter as a functional indicator for XPO5 phosphorylation-induced miRNA downregulation. Defective in pre-miR-122 export in 3D mutant expressing Huh-7 cells (Figures 3H and S3K) resulted in the de-repression of miR-122 targets as measured by miR-122 reporter luciferase or EGFP harboring miR-122 binding sites in the respective 3′-UTR (Figures 3I and S3L), indicating that miR-122 function is inhibited upon XPO5 phosphorylation.
Suppression of miR-122 Confers Taxol Resistance by Upregulating Septin-9
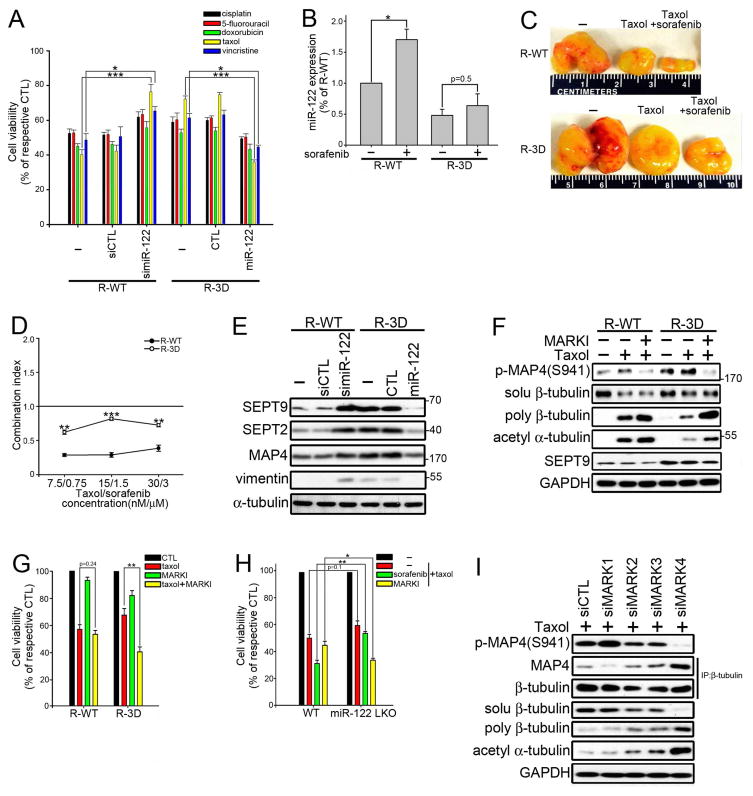
Because XPO5 phosphorylation decreased miR-122 expression and loss of miR-122 reduced chemosensitivity (Xu et al., 2011; Yang et al., 2011; Yin et al., 2011), we examined whether phosphorylation of XPO5 confers chemoresistance. As shown in Figure 4A, taxol resistance was induced by 3D XPO5 and reversed by miR-122 mimetic. Knockdown of miR-122 (simiR-122) also increased taxol resistance, both in vitro and in vivo (Figures 4A and S4A). We then investigated whether inhibition of the ERK pathway could increase miR-122 and attenuate taxol resistance. As expected, sorafenib, an inhibitor to reduce ERK activation, increased miR-122 expression and sensitized tumor cells to taxol (Figures 4B and 4C and S4B–S4E). However, the 3D XPO5 mutant was resistant to sorafenib-induced miR-122 accumulation and taxol sensitization, as confirmed by the upward shift of the combination index (CI) plot (Figure 4D). The importance of XPO5 phosphorylation in the synergism of ERK inhibition and taxol was further substantiated by DN-ERK and other pharmacological inhibitors of the RAF/MEK/ERK pathway (Figure S4F).
Figure 4. Depletion of miR-122 Upregulates SEPT9 to Induce Taxol Resistance.
(A) Vector-transfected and simiR-122-expressing R-WT Huh-7 cells or vector-transfected and miR-122-expressing R-3D Huh-7 cells were treated with 10 μM cisplatin, 50 μM 5-fluorouracil, 1 μM doxorubicin, 0.1 μM taxol, or 0.1 μM vincristine for 48 hr before measuring cell viability by MTT assay. Data were presented as relative to the cells without drug treatment (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (B) Stable Huh-7 transfectants were incubated with 5 μM sorafenib for 24 hr followed by miR-122 quantification by qRT-PCR. Data were presented as relative to the miR-122 expression in the R-WT cells without sorafenib treatment (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (C) Representative photograph of engrafted tumors of Huh-7 xenograft treated with taxol with or without sorafenib. (D) The combination index plot for the Huh-7 stable transfectants exposed to fixed molar ratios, 1:100, of taxol to sorafenib for 48 hr (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test) (E) IB of four proteins identified as miR-122 targets in Huh-7 stable transfectants. (F) After 24 hr treatment with 30 nM taxol with or without 30 μM MARK inhibitor (MARKI), total cell lysates or tubulin fractions (soluble or polymerized tubulin) from Huh-7 stable transfectants were analyzed by IB. (G) Cell viability of Huh-7 stable transfectants after treatment with 30 nM taxol and 30 μM MARKI for 48 hr was measured by MTT assay. Data were presented as relative to the Huh-7 stable transfectants without drug treatment (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (H) Primary hepatocytes isolated from WT or miR-122 liver-specific knockout (LKO) mice were treated with 30 nM taxol, 3 μM sorafenib, and 30 μM MARKI for 48 hr before determining cell viability by MTT assay. Data were presented as relative to the hepatocytes without drug treatment (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (I) Total cell lysates or tubulin fractions of siMARK Huh-7 R-3D stable transfectants treated with 100 nM taxol for 24 hr were analyzed by IB. See also Figure S4.
miR-122 plays a central role in diverse hepatic functions, as highlighted by its direct role in fat metabolism, tumor suppression, inflammation and fibrosis (Hsu et al., 2012). To find a therapeutic target to circumvent loss-of-miR-122-induced taxol resistance, we noticed that 5 of 30 genes identified by Gene ontology and biological association analysis of the miR-122 targets (Boutz et al., 2011) are associated with microtubule dynamics, including septin-2 (SEPT2), septin-9 (SEPT9), MAP1B, MAP4, and vimentin. Western blot analysis showed that SEPT9 was significantly increased in cells depleted of miR-122 (Figure 4E) and luciferase reporter assay indicated that miR-122 negatively regulates SEPT9 expression by interacting with its 3′ UTR (Figure S4G). Because septins may increase microtubule dynamics through scaffolding microtubule-associated proteins (MAP) and microtubule affinity regulating kinase (MARK) (Spiliotis, 2010), we compared MAP4 phosphorylation in XPO5 stable transfectants and examined the role of MARK. MAP4 phosphorylation was higher in 3D XPO5 cells, resulting in susceptibility to MARK inhibitor (MARKI) (Figures 4F, 4G and S4H). Consistently, miR-122 KO hepatocytes that express more SEPT9 are resistant to sorafenib, but sensitive to the combination of MARKI and taxol (Figures 4H and S4I). We further knocked down four isoforms of MARK in 3D XPO5 cells and found MARK4 depletion significantly inhibited MAP4 phosphorylation to restore its association with tubulin, thereby conferring taxol sensitivity (Figure 4I). Combined with the finding that MARK4 is localized to microtubules (Trinczek et al., 2004) and upregulated in liver cancer (Figure S4J) (Kato et al., 2001), our results suggest that MARK4 could be a therapeutic target to overcome taxol resistance caused by depletion of miR-122.
Pin1-Induced Conformational Changes in XPO5 Decrease Pre-miRNA Loading
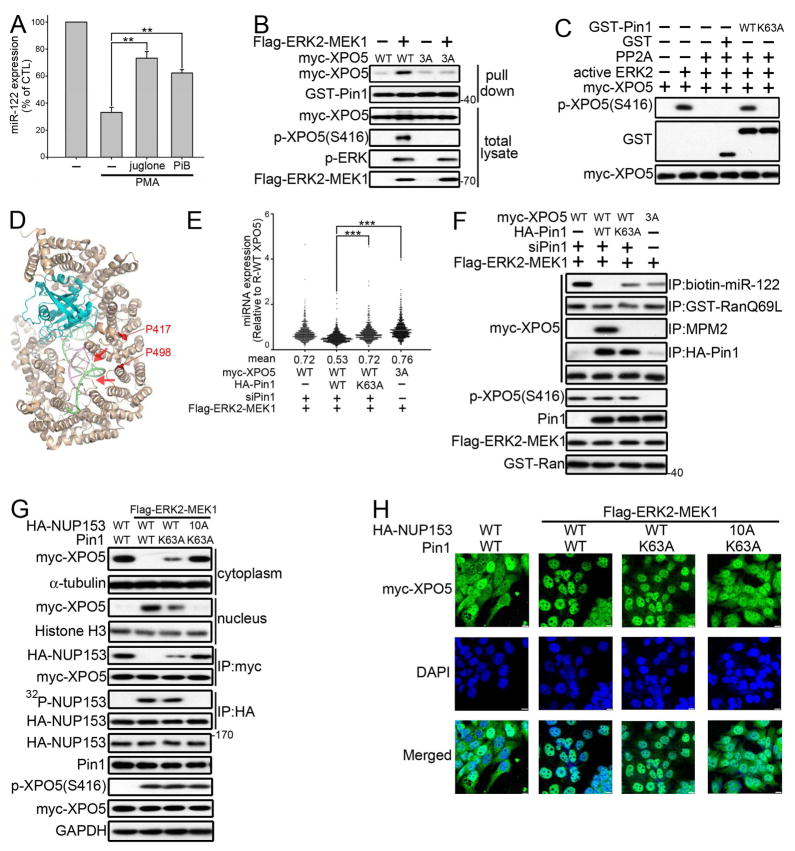
To gain insight on how phosphorylation impairs XPO5 activity, we analyzed the structure of XPO5. Based on WESA (weighted ensemble solvent accessibility) algorithm (Chen and Zhou, 2005) and the secondary structure of XPO5 (Figures S5A and S5B), three ERK phosphorylation sites are fairly accessible and not part of defined helix structures. The structure of XPO5 resembles a flexible wound spring; therefore, small changes in the relative orientation of successive HEAT repeats could cumulatively generate substantial changes in the helicoidal pitch (Okada et al., 2009). It has been shown that prolyl isomerase Pin1 interacts with proteins phosphorylated at S/T-P motifs, thereby controlling their activity by promoting cis-trans isomerization (Liou et al., 2011). Because ERK phosphorylates substrate at S/T-P motifs, we investigated whether Pin1 plays a role in ERK-mediated impairment of XPO5 functions. Treatment of cells with two structurally distinct Pin1 inhibitors, juglone or PiB, was found to reverse the decrease in miR-122 triggered by PMA (Figure 5A). GST pull-down assay shows that ERK activation by ERK2-L4A-MEK1 fusion, a constitutively active and nuclear form of the ERK2, enhances wild-type, but not 3A, XPO5 to associate with Pin-1 (Figure 5B). The interaction between endogenous XPO5 and Pin-1 also increased when ERK was activated (Figure S5C). To investigate if Pin1 changes the conformation of XPO5, we performed partial proteolysis assay with subtilisin, a protease particularly sensitive to substrate conformation (Lu and Zhou, 2007). Incubation with wild-type, but not with catalytically inactive K63A (Zhou et al., 2000) Pin1 mutant protects XPO5 from proteolytic cleavage (Figure S5D). Notably, Pin1 fails to protect 3A XPO5 from proteolysis. Given that Pin1 induces conformational changes in XPO5, we used PP2A, a trans-pSer/Thr-Pro isomer specific phosphatase (Zhou et al., 2000), to examine the cis/trans status of XPO5. Wild-type, but not K63A, Pin1 prevents dephosphorylation of XPO5 by PP2A, indicating that trans-conformation of pSer/Thr-Pro motif on XPO5 was isomerized to cis-conformation in the presence of Pin1 (Figure 5C).
Figure 5. Pin1 Inhibits the Loading of Pre-miRNA to XPO5.
(A) Huh-7 cells were pre-treated with or without 5 μM juglone or 5 μM PiB for 1 hr then treated with 100nM PMA for 24 hr before measuring miR-122 by qRT-PCR. Data were presented as relative to the miR-122 expression in the Huh-7 cells without drug treatment (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (B) Lysates of 293T cells transfected as indicated were incubated with GST-Pin1 then subjected to GST pull-down and IB. (C) Ectopically expressed myc-XPO5 was purified from Pin1 knocked down 293T cells followed by phosphorylation with active ERK2. Purified XPO5 was then incubated with GST, GST-Pin1, or GST-Pin1 (K63A) for 30 min before PP2A treatment for 5 min. (D) Relative position of P417 and P498 (red) in the crystal structure of XPO5 (gray, PDB 3A6P) in complex with RanGTP (cyan) and pre-miR30a (green and pink). Possible movements of helices 9 and 10 when changed to cis form are shown as red arrows. (E) miRNA of Huh-7 Flag-ERK2-MEK1 stable transfectant with Pin1-depleted (siPin1), siRNA-resistant Pin1 (R-WT), catalytic inactive K63A Pin1, or Pin1-binding-defective 3A XPO5 were profiled by nanostring assay. Data were presented as relative to the miRNA expression in R-WT XPO5 Huh-7 cells though dot density plot (n=2) (F) Lysates of Huh-7 stable transfectants expressing Flag-ERK2-MEK1 were incubated with 3′biotin-labeled pre-miR-122, GST-RanQ69L, MPM2 antibody, or HA antibody. Pull-down proteins were subjected to IB. (G) Lysates of Huh-7 stable transfectants were subjected to separation of cytoslic and nuclear fraction or IP with myc or HA antibody before IB. For detection of the NUP153 phosphorylation, cells were labeled with 32P orthophosphate. (H) Immunofluorescence analysis of XPO5 (green) in Huh-7 stable transfectants. DAPI (blue) was used to mark the nucleus. Scale bars: 10 μm. See also Figure S5
EMBOSS analysis (Jones et al., 2002) with WATER algorithm revealed that 416S at HEAT9 and 497S at HEAT10 of XPO5 aligned better with WFYS*PR, an optimal binding sequence for Pin1 (Yaffe et al., 1997) (Figure S5E). Considering the inner helix of HEAT9 provides the closest contact with the stem of pre-miRNA (Okada et al., 2009), we asked whether Pin1-induced conformational changes interfere pre-miRNA binding. Despite the availability of the crystal structure of XPO5, the disorder region in the helix of HEAT10 hampered accurate in silico simulation (Figure S5B). The structure, however, indicates that the possible rearrangement of helices 9 and 10 upon trans-to-cis conversion of P417 and P498, will obstruct pre-miRNA loading (Figure 5D). We therefore investigated whether the Pin1-induced XPO5 conformational changes contribute to ERK-mediated global miRNA downregulation. As shown in Figure 5E, miRNA expression was globally downregulated in cells expressing wild-type Pin1, but this effect was attenuated in the presence of K63A Pin1 or Pin1-binding-defective 3A XPO5. The assembly of the pre-miRNA/XPO5/Ran-GTP complex after Pin1-mediated isomerization was further examined. Because changes in accessibility to the MPM2 antibody have been suggested to reflect conformational changes induced by Pin1 (Stukenberg and Kirschner, 2001), we used it as a tool to evaluate the conformation of XPO5. Consistent with the in silico predictions, the conformation-altered XPO5 was not able to load pre-miR-122 (Figures 5F and S5F), resulting in the inhibition of pre-miR-122 export (Figure S5G), de-repression of miR-122 targets, and resistance to taxol-induced tubulin polymerization (Figures S5H–J). The involvement of Pin1-mediated conformational changes in pre-miRNA loading can also be observed in HepG2 and SK-Hep-1 cells (Figure S5K). Considering that pancreatic cancer exhibits the highest incidence of KRAS mutations and that melanoma has the highest rate of BRAF mutations (Cheng et al., 2013; Neuzillet et al., 2013), we investigated whether ERK-mediated impairment of XPO5 functions also occurs in these two types of cancer. Our results demonstrated that Pin1-induced XPO5 conformational changes contribute to KRASG12V and BRAFV600E-induced impairment of XPO5 function in pancreatic cancer and melanoma, respectively (Figure S5L and S5M).
Because phosphorylated XPO5 is enriched in the nucleus (Figure 2F), we also examined the role of Pin1 in XPO5 localization. As shown in Figure 5G, Pin1 loss-of-function partially restored nuclear export of XPO5 and interaction with NUP153, which is essential in the nuclear export of the pre-miRNA-XPO5 complex (Wan et al., 2013). Interestingly, it has been reported that ERK is a physiological nucleoporin kinase that targets the FG repeat regions to disrupt karopherin-nucleoporin interactions. ERK phosphorylates nucleoporin NUP153 at Ser257, Ser320, Ser334, Ser338, Thr369, Thr388, Thr413, Ser516, Ser522 and Ser529. Through generating phospho-specific antibodies, it was shown that ERK directly phosphorylates NUP153 at least at Ser338 and Ser529 (Kosako et al., 2009). NUP153 was not found to directly bind Pin1 (Figure S5N). We found non-phosphorylatable S257A/S320A/S334A/S338A/T369A/T388A/T413A/S516A/S522A/S529A (10A) NUP153 further restored the interaction with XPO5 in cells expressing catalytically inactive Pin1 with activated ERK, resulting in the export of XPO5 and pre-miR-122 (Figures 5G, 5H and S5O). These results support the conclusion that ERK phosphorylates both XPO5 and NUP153 to trap XPO5 and pre-miRNA in the nucleus.
XPO5 Phosphorylation Promotes Tumor Development and Associates with Poor Prognosis
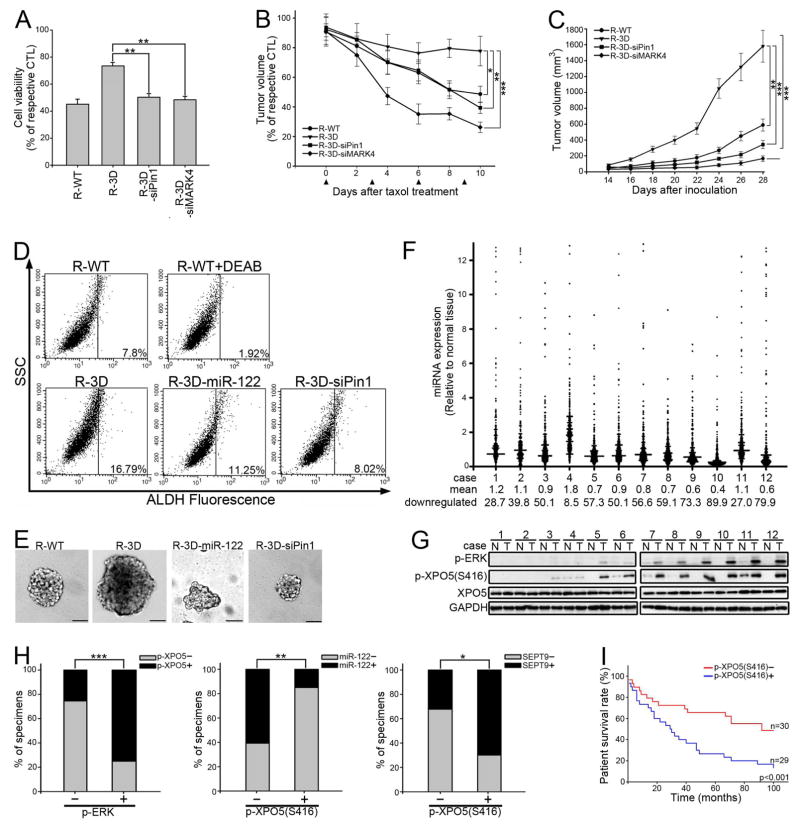
We showed above that depleting Pin1 or MARK4 may increase taxol sensitivity in XPO5-phosphorylated cells (Figures 4I and S5J). Indeed, inhibition of Pin1 or MARK4 restored taxol-induced tubulin polymerization (Figure S6A), reduced cell viability (Figure 6A), and decreased tumor burden (Figures 6B and S6B). These data support the idea that Pin1 and MARK4 could be therapeutic targets in XPO5 phosphorylation-associated drug resistance. Although the XPO5 phosphomimetic did not significantly increase Huh-7 cell proliferation in vitro (Figure S6C) in 2 days, we noticed that in vivo tumor growth was increased when 3D XPO5 was expressed (Figures 6C and S6D). Interestingly, the cancer stem cell marker CD133 and EPCAM is detected in the liver of young miR-122 KO mice, followed by further increase in the tumors (Hsu et al., 2012; Tsai et al., 2012). Consistently, we found cell surface markers CD133 and EPCAM, functional marker aldehyde dehydrogenase (ALDH) activity, and tumor sphere formation was increased in 3D XPO5 cells, but attenuated when miR-122 was overexpressed or Pin1 was knocked down (Figures 6D, 6E, S6E and S6F). Taken together, our data supports the conclusion that XPO5-dependent downregulation of miRNA contributes to tumor development through increasing cancer stem cell formation and chemoresistance.
Figure 6. XPO5 phosphorylation promotes tumor development and associates with poor prognosis.
(A) Huh-7 stable transfectants were treated with 100 nM taxol for 48 hr before MTT assay. Data were presented as relative to the cells without taxol treatment (n=3 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (B) Taxol (20 mg/kg once every 3 days, i.v.) was injected into mice for 4 cycles when tumors volume reached 150 mm3. Data shown represent percentage tumor volume relative to xenograft without taxol treatment (n=5 per group, data represents mean ± SEM, p values were calculated by Student’s t test). Arrowheads indicate time points of taxol injection. (C) Tumor volume of engrafted Huh-7 stable transfectants as indicated over time (n=5 per group, data represents mean ± SEM, p values were calculated by Student’s t test). (D) Representative flow cytometry analyses of Huh-7 stable transfectants for ALDH activity using an Aldefluor kit. WT cells treated with the ALDH inhibitor diethylaminobenzaldehyde (DEAB) served as the negative control. The gated cells represent the subpopulation of cells that are positive for ALDH activity. (E) Representative tumor spheres of Huh-7 stable transfectants were imaged under a phase contrast microscope. Scale bars: 50 μm. (F) Dot density plot illustrates the miRNA level in tumor relative to adjacent normal tissue (n=2). The overall average of miRNA levels is marked as mean. Expression levels lower than 0.67 were classified as downregulated miRNA. (G) Paired human normal (N) and liver tumor (T) tissues were analyzed by IB. (H) Correlation between p-ERK, p-XPO5, miR-122, and SEPT9 in 59 human tumor specimens on Human HistoArray IMH-318. p values were calculated by Pearson’s chi-square test. (I) Kaplan-Meier survival curves of liver cancer patients on Human HistoArray IMH-318 grouped by p-XPO5 expression. See also Figure S6.
To examine whether these conclusions are in accord with findings in human tumors, we performed nanostring miRNA profiling and immunoblotting using 12 paired HCC tumor specimens and adjacent normal tissues. XPO5 phosphorylation was found to be associated with global miRNA downregulation (Figures 6F, 6G and S6G). Because miR-122 dominates the miRNA content in the normal liver, we choose it as an indicator and studied its expression using liver tumor tissue array that contained 59 HCC specimens. Consistent with our findings, XPO5 phosphorylation was associated with ERK phosphorylation, miR-122 downregulation, and SEPT9 expression (Figures 6H and S6H). We next correlated our findings with patient survival. The Kaplan-Meier survival curves showed that high p-XPO5 (S416) were associated with poor survival (Figure 6I). Moreover, p-XPO5/p-ERK double positive patients had reduced overall survival relative to the p-XPO5 negative/p-ERK positive group (Figure S6I), supporting that ERK-mediated phosphorylation of XPO5 is associated with worse prognosis. Consistent with the discovery cohort, we also observed that XPO5 phosphorylation correlated with poor survival in the validation cohort (Figure S6J). A sex, age, stage, and adjacent liver status (normal, viral hepatitis, viral hepatitis related cirrhosis)-adjusted multivariate regression analysis suggested that p-XPO5 was independently associated with patient survival (Figure S6K). Taken together, analyses of tumor specimens further strengthened the notion that phosphorylation of XPO5 by ERK downregulates miRNA expression and is associated with poor clinical outcome of liver cancer patients.
DISCUSSION
Hepatocellular carcinoma is often diagnosed at advanced stages when surgery is not feasible and is also highly resistant to conventional chemotherapy. Therefore, identification of signaling pathways regulating liver carcinogenesis is important in developing targeted therapy. In this study, we demonstrated that ERK phosphorylates XPO5 to impair its ability to export pre-miRNA. However, ERK is also known to increase Drosha and Dicer by phosphorylating DGCR8 and TRBP, a consequence of increasing pro-growth miRNA levels (Herbert et al., 2013; Paroo et al., 2009). Further investigation is therefore needed to establish how ERK affects miRNA biogenesis. It is important to mention that nuclear export by XPO5 has been suggested as a rate-limiting step in miRNA biogenesis. Only XPO5, but not Drosha or Dicer, overexpression enhances inhibition of gene expression by miRNA (Yi et al., 2005). Concordant with our results, most miRNAs were downregulated in PMA-treated myeloid leukemia (Wang et al., 2013) and upregulated in sorafenib-treated HCC (Zhou et al., 2011). But in colon cancer, which has been reported to express an XPO5 frameshift mutant (Melo et al., 2010), expression of miRNAs was upregulated by KRAS and inhibited by MEK/ERK inhibitors (Ragusa et al., 2012). Based on our results and the published literature, an interesting model can be proposed: in cells with wild-type XPO5, activated ERK phosphorylates XPO5, which results in decreased miRNA production. However, if XPO5 is mutated, the effect of ERK on Drosha and Dicer becomes apparent, meaning ERK can globally increase miRNA expression. This model suggests that XPO5 plays a critical role in deciding how ERK reprograms miRNA biogenesis.
Shuttling of RNAs and proteins out of the nucleus is essential in maintaining normal cellular function. Cancer cells can usurp this process to stimulate tumor growth and evade apoptosis (Gravina et al., 2014). However, among all of the potential targets in nucleo-cytoplasmic transport, only XPO1 is better understood and is inhibited by selective inhibitor of nuclear export (SINE), which is currently in phase 2 clinical trial (Gerecitano, 2014). An in-depth understanding of miRNA export machinery is needed for the successful design of clinical therapeutics. Our study showed that ERK phosphorylation followed by Pin1-mediated isomerization impairs XPO5 activity. These findings suggest that XPO5 and global miRNA downregulation may become druggable targets, although systems biology and network analysis are necessary to determine the effect of XPO5 restoration in a holistic manner. Pin1 is an attractive target for designing small molecule inhibitors because of its well-defined active site. However, the available Pin1 inhibitors still lack the required specificity and potency (Moore and Potter, 2013). Interestingly, a recent report showed that all-trans retinoic acid (ATRA) directly binds and degrades the active form of Pin1 that is overexpressed in cancer (Wei et al., 2015). Thus, development of more potent and specific Pin1-targeted ATRA derivatives may be a strategy to restore XPO5 function.
XPO5 is the only known transporter to export pre-miRNA to the cytoplasm. In our study, we found that cells with XPO5 knockdown or phosphomimetic XPO5 were characterized as having global down-regulation of miRNA expression. Nonetheless, nanostring nCounter analysis revealed that some miRNAs levels were not altered by the impairment of the XPO5. Two splicing-independent mirtrons, miR-1225 and 1228, are also known to be independent of XPO5 for their biogenesis (Havens et al., 2012). We plan to use affected and unaffected pre-miRNA, such as pre-miR-122 and pre-miR-221, as probes to seek additional nuclear export pathways. Among the miRNAs downregulated after XPO5 phosphorylation, we further explored the consequence of miR-122 reduction because it is an abundant liver-specific miRNA and is essential for the maintenance of liver homeostasis. Previous reports suggested that restoring miR-122 may be a strategy to limit HBV/leishmania infection, suppress HCV negative HCC, and prevent the development of nonalcoholic steatohepatitis and cirrhosis (Thakral and Ghoshal, 2015). However, the efficacy of the miRNA mimetic and delivery method still needs to be resolved before advancing to clinical trial. An alternative method to restore miRNA function is to attack downstream targets of the miRNA. Our results suggest that MARK4 may be a therapeutic target to overcome miR-122 loss-induced tumorigenesis and drug resistance. Although the crystal structure of MARK4 is still unknown, combined molecular dynamic simulation and pharmacophore-based virtual screening recently identified six potent compounds specific for MARK4 (Jenardhanan et al., 2014). These lead compounds deserve further testing as potential candidates for miR-122-related diseases.
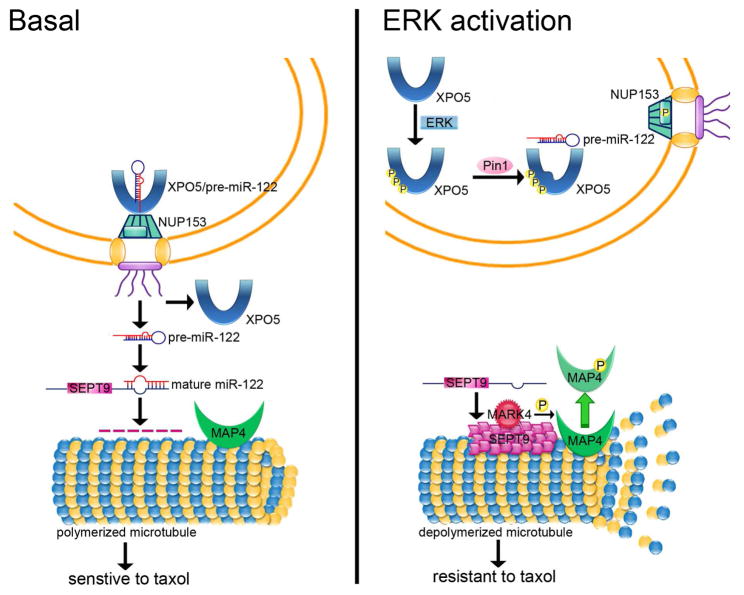
In summary, we propose a model based on the current findings for ERK-induced tumorigenesis in liver cancer cells (Figure 7). In the absence of ERK activation, pre-miR-122 is exported by XPO5 from nucleus to cytoplasm through the nuclear pore complex to produce mature miR-122 to inhibit SEPT9 expression. Un-phosphorylated MAP4 binds to tubulin, causing cells to respond to taxol (left panel). When ERK is activated, XPO5 is phosphorylated at T345/S416/S497, followed by isomerization by Pin1, which impairs its ability to load pre-miR-122. ERK also phosphorylates NUP153 to further inhibit pre-miRNA-XPO5 complex export. SEPT9 then acts as a scaffold for MAP4 and MARK4 and causes phosphorylated MAP4 to detach from microtubules to impart taxol resistance (right panel). The identification of XPO5 as a downstream substrate for ERK links the mitogenic signaling and miRNA nuclear export machinery, and suggests that strategies toward restoring the activity of XPO5 may have therapeutic value.
Figure 7. XPO5 Phosphorylation by ERK Leads to Taxol Resistance in Liver Cancer by Decreasing miR-122 Export.
Under basal condition, pre-miR-122 is exported by XPO5 to produce mature miR-122 to inhibit SEPT9 expression. Un-phosphorylated MAP4 binds to tubulin to maintain taxol sensitivity. When ERK is activated, XPO5 is phosphorylated followed by isomerization by Pin1, which impairs its ability to load pre-miR-122. SEPT9 then acts as a scaffold for MAP4 and MARK4 to cause phosphorylated MAP4 to detach from microtubules therefore conferring taxol resistance.
EXPERIMENTAL PROCEDURES
Mouse model for tumorigenesis
All mice were handled according to the procedures approved by Ohio State University Institutional Laboratory Animal Care and Use Committee.
Human liver tumor samples
The Ohio State University Institutional Review Board approved this study. Primary human liver cancer and adjacent normal tissues were obtained from the Cooperative Human Tissue Network at the Ohio State University James Cancer Hospital. Informed consent was obtained from all patients.
Statistical analysis
Each experiment was performed at least three times, and representative data are shown. Data in bar graph are given as the mean ± SEM. Means were checked for statistical difference using Student’s t test or Pearson’s chi-square test, and p values less than 0.05 were considered significant (* p<0.05, ** p<0.01, *** p<0.001). For dot density plot, Mann-Whitney rank sum test was applied. For survival analysis, Kalpan-Meier analysis and log-rank test were applied.
Supplementary Material
Significance.
Systemic analysis of miRNAs has revealed that many pre-miRNA are retained in the nucleus of cancer cells. In this study, we have identified that ERK phosphorylation coupled with Pin1-mediated conformational changes in XPO5 inhibits miRNA export. Global downregulation of miRNA expression including that of miR-122 is observed in liver cancer when XPO5 is phosphorylated. Depletion of miR-122 activates MARK4 to increase microtubule dynamics, thereby inducing drug resistance and tumorigenesis. Furthermore, XPO5 phosphorylation correlates with poor prognosis in liver cancer patients. Our findings identified a mechanism of global downregulation of miRNA in liver cancer and targets, Pin1 and MARK4, for therapeutic intervention.
Highlights.
ERK phosphorylation followed by Pin1-mediated isomerization impairs XPO5 activity.
Down-regulation of miR-122 leads to taxol resistance through septin-9 and MARK4.
XPO5 phosphorylation correlates with poor prognosis in HCC patients.
Pin1 and MARK4 are potential targets for clinical intervention in liver cancer.
Acknowledgments
We thank the Ohio State University Comprehensive Cancer Center microscopy, genomics, nucleic acids, flow cytometry, mass spectrometry, and comparative pathology core facility. We thank Dr. M.-C. Hung, J. Dahlberg, P. Sarnow, A. Deiters, Y. Zheng, C.-S. Chen, T. Williams for providing reagents; Dr. K. Huebner, W.-L. Shih, T. Banh for editing; Dr. P. Fadda for nanostring profiling technical support; Dr. S. Palko and Dr. D. Wernicke-Jameson for administrative support. This study was supported by the US National Institutes of Health (R35CA197706 to C.-M. Croce; R01CA193244 to K. Ghoshal); Taiwan Ministry of Science and Technology (NSC99-2320-B400-008-MY3 and NSC 99-2628-B002-024-MY3 to C.-M. Teng; 101-2917-I-564-052 to H.-L. Sun; MOST 105-2632-B-715-001 and MOST 104-2320-B-715-002 to S.-W. Wang); Pelotonia Graduate Student Fellowship (to K.-Y. T); National Key Research & Development Program of China (2016YFA0502204) and National Natural Science Foundation of China (81572739) to Y. Peng.
Footnotes
Supplemental information, including supplemental experimental procedures and six figures can be found with this article online.
AUTHOR CONTRIBUTIONS
C. M. C, Y. P, C.-M. T and M.-C. H conceived the project. H.-L. S, R. C., and Y. P. designed experiment. R. C, Y.-H. H, Z. L, N. Z. performed in vivo experiments. K.-Y. T, H. K, and K. G. provided RNA and tissue extracts from HCC patients’ specimens and miR-122 KO mouse livers and hepatocytes. K. N. and G. S. analyzed the crystal structure of XPO5. M. F. and C. V. performed miRNA in situ hybridization and pathology study. J. Z, H.-L. H., T. K., T. J. L, and Y.-J. J provided technical assistance. K.N., E.T., F. L., S.-W. W, J.-H. G, M.-C. H, C.-M. T., Y.P., K.G., and C. M. C edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 2002;21:6205–6215. doi: 10.1093/emboj/cdf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz DR, Collins PJ, Suresh U, Lu M, Ramirez CM, Fernandez-Hernando C, Huang Y, de Abreu RS, Le SY, Shapiro BA, et al. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J Biol Chem. 2011;286:18066–18078. doi: 10.1074/jbc.M110.196451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casar B, Pinto A, Crespo P. ERK dimers and scaffold proteins: unexpected partners for a forgotten (cytoplasmic) task. Cell Cycle. 2009;8:1007–1013. doi: 10.4161/cc.8.7.8078. [DOI] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhou HX. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res. 2005;33:3193–3199. doi: 10.1093/nar/gki633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhang G, Li G. Targeting MAPK pathway in melanoma therapy. Cancer Metastasis Rev. 2013;32:567–584. doi: 10.1007/s10555-013-9433-9. [DOI] [PubMed] [Google Scholar]
- Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, Dacic S. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecitano J. SINE (selective inhibitor of nuclear export)--translational science in a new class of anti-cancer agents. J Hematol Oncol. 2014;7:67. doi: 10.1186/s13045-014-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravina GL, Senapedis W, McCauley D, Baloglu E, Shacham S, Festuccia C. Nucleo-cytoplasmic transport as a therapeutic target of cancer. J Hematol Oncol. 2014;7:85. doi: 10.1186/s13045-014-0085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012;40:4626–4640. doi: 10.1093/nar/gks026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Pimienta G, DeGregorio SJ, Alexandrov A, Steitz JA. Phosphorylation of DGCR8 increases its intracellular stability and induces a progrowth miRNA profile. Cell Rep. 2013;5:1070–1081. doi: 10.1016/j.celrep.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, Gu J, Lin J, Habuchi T, Wu X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res. 2008;14:7956–7962. doi: 10.1158/1078-0432.CCR-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenardhanan P, Mannu J, Mathur PP. The structural analysis of MARK4 and the exploration of specific inhibitors for the MARK family: a computational approach to obstruct the role of MARK4 in prostate cancer progression. Mol Biosyst. 2014;10:1845–1868. doi: 10.1039/c3mb70591a. [DOI] [PubMed] [Google Scholar]
- Jiang P, Wu H, Wang W, Ma W, Sun X, Lu Z. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:W339–344. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J, Field JK, Risk JM. A comparative guide to gene prediction tools for the bioinformatics amateur. Int J Oncol. 2002;20:697–705. doi: 10.3892/ijo.20.4.697. [DOI] [PubMed] [Google Scholar]
- Jopling C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012;9:137–142. doi: 10.4161/rna.18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Yoneda Y. Nucleocytoplasmic Transport of MicroRNAs and Related Small RNAs. Traffic. 2011 doi: 10.1111/j.1600-0854.2011.01211.x. [DOI] [PubMed] [Google Scholar]
- Kato T, Satoh S, Okabe H, Kitahara O, Ono K, Kihara C, Tanaka T, Tsunoda T, Yamaoka Y, Nakamura Y, et al. Isolation of a novel human gene, MARKL1, homologous to MARK3 and its involvement in hepatocellular carcinogenesis. Neoplasia. 2001;3:4–9. doi: 10.1038/sj.neo.7900132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Kosako H, Yamaguchi N, Aranami C, Ushiyama M, Kose S, Imamoto N, Taniguchi H, Nishida E, Hattori S. Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat Struct Mol Biol. 2009;16:1026–1035. doi: 10.1038/nsmb.1656. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Leaderer D, Hoffman AE, Zheng T, Fu A, Weidhaas J, Paranjape T, Zhu Y. Genetic and epigenetic association studies suggest a role of microRNA biogenesis gene exportin-5 (XPO5) in breast tumorigenesis. Int J Mol Epidemiol Genet. 2011;2:9–18. [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36:501–514. doi: 10.1016/j.tibs.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, An J, Lin J, Liu Y, Bao L, Zhang W, Zhao JJ. Single Nucleotide Polymorphisms of MicroRNA Processing Machinery Genes and Outcome of Hepatocellular Carcinoma. PLoS One. 2014;9:e92791. doi: 10.1371/journal.pone.0092791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nature reviews Molecular cell biology. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Moore JD, Potter A. Pin1 inhibitors: Pitfalls, progress and cellular pharmacology. Bioorg Med Chem Lett. 2013;23:4283–4291. doi: 10.1016/j.bmcl.2013.05.088. [DOI] [PubMed] [Google Scholar]
- Neuzillet C, Hammel P, Tijeras-Raballand A, Couvelard A, Raymond E. Targeting the Ras-ERK pathway in pancreatic adenocarcinoma. Cancer Metastasis Rev. 2013;32:147–162. doi: 10.1007/s10555-012-9396-2. [DOI] [PubMed] [Google Scholar]
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- Pages G, Guerin S, Grall D, Bonino F, Smith A, Anjuere F, Auberger P, Pouyssegur J. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 1999;286:1374–1377. doi: 10.1126/science.286.5443.1374. [DOI] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa M, Statello L, Maugeri M, Majorana A, Barbagallo D, Salito L, Sammito M, Santonocito M, Angelica R, Cavallaro A, et al. Specific alterations of the microRNA transcriptome and global network structure in colorectal cancer after treatment with MAPK/ERK inhibitors. J Mol Med (Berl) 2012;90:1421–1438. doi: 10.1007/s00109-012-0918-8. [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Spiliotis ET. Regulation of microtubule organization and functions by septin GTPases. Cytoskeleton (Hoboken) 2010;67:339–345. doi: 10.1002/cm.20448. [DOI] [PubMed] [Google Scholar]
- Stukenberg PT, Kirschner MW. Pin1 acts catalytically to promote a conformational change in Cdc25. Mol Cell. 2001;7:1071–1083. doi: 10.1016/s1097-2765(01)00245-3. [DOI] [PubMed] [Google Scholar]
- Thakral S, Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr Gene Ther. 2015;15:142–150. doi: 10.2174/1566523214666141224095610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinczek B, Brajenovic M, Ebneth A, Drewes G. MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes. J Biol Chem. 2004;279:5915–5923. doi: 10.1074/jbc.M304528200. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Zhang X, Langley RR, Liu Y, Hu X, Han C, Peng G, Ellis LM, Jones SN, Lu X. DNA-damage-induced nuclear export of precursor microRNAs is regulated by the ATM-AKT pathway. Cell Rep. 2013;3:2100–2112. doi: 10.1016/j.celrep.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu L, Xie L, Xiang G, Zhou Y. Induction of differentiation-specific miRNAs in TPA-induced myeloid leukemia cells through MEK/ERK activation. Int J Mol Med. 2013;31:59–66. doi: 10.3892/ijmm.2012.1191. [DOI] [PubMed] [Google Scholar]
- Wei S, Kozono S, Kats L, Nechama M, Li W, Guarnerio J, Luo M, You MH, Yao Y, Kondo A, et al. Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat Med. 2015;21:457–466. doi: 10.1038/nm.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CM, Wong CC, Lee JM, Fan DN, Au SL, Ng IO. Sequential alterations of microRNA expression in hepatocellular carcinoma development and venous metastasis. Hepatology. 2012;55:1453–1461. doi: 10.1002/hep.25512. [DOI] [PubMed] [Google Scholar]
- Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, Liu J, Cui Y, Bian X, Bie P, et al. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011;310:160–169. doi: 10.1016/j.canlet.2011.06.027. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, et al. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- Yang F, Zhang L, Wang F, Wang Y, Huo XS, Yin YX, Wang YQ, Sun SH. Modulation of the unfolded protein response is the core of microRNA-122-involved sensitivity to chemotherapy in hepatocellular carcinoma. Neoplasia. 2011;13:590–600. doi: 10.1593/neo.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Li W, Wu J, Germann UA, Su MS, Kuida K, Boucher DM. Extracellular signal-regulated kinase 2 is necessary for mesoderm differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12759–12764. doi: 10.1073/pnas.2134254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11:220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Tang HF, Xiang Q, Yu J, Yang XY, Hu N, Lei XY. MiR-122 increases sensitivity of drug-resistant BEL-7402/5-FU cells to 5-fluorouracil via down-regulation of bcl-2 family proteins. Pharmazie. 2011;66:975–981. [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- Zhou C, Liu J, Li Y, Liu L, Zhang X, Ma CY, Hua SC, Yang M, Yuan Q. microRNA-1274a, a modulator of sorafenib induced a disintegrin and metalloproteinase 9 (ADAM9) down-regulation in hepatocellular carcinoma. FEBS Lett. 2011;585:1828–1834. doi: 10.1016/j.febslet.2011.04.040. [DOI] [PubMed] [Google Scholar]
- Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Kullertz G, Stark M, Fischer G, Lu KP. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Molecular cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.