Abstract
Simple and efficient methods are presented for creating precise modifications of the zebrafish genome. Edited alleles are generated by homologous recombination between the host genome and double-stranded DNA (dsDNA) donor molecules, stimulated by the induction of double-strand breaks at targeted loci in the host genome. Because several kilobase-long tracts of sequence can be exchanged, multiple genome modifications can be generated simultaneously at a single locus. Methods are described for creating: (1) alleles with simple sequence changes or in-frame additions, (2) knockin/knockout alleles that express a reporter protein from an endogenous locus, and (3) conditional alleles in which exons are flanked by recombinogenic loxP sites. Significantly, our approach to genome editing allows the incorporation of a linked reporter gene into the donor sequences so that successfully edited alleles can be identified by virtue of expression of the reporter. Factors affecting the efficiency of genome editing are discussed, including the finding that dsDNA products of I-SceI meganuclease enzyme digestion are particularly effective as donor molecules for gene-editing events. Reagents and procedures are described for accomplishing efficient genome editing in the zebrafish.
INTRODUCTION
Studies in zebrafish have made substantial contributions to our understanding of gene function in vertebrates. The ability to conduct forward genetic screens combined with the ability often to detect phenotypic consequences that result immediately and directly from aberrant gene activity have contributed to use of the zebrafish as a platform for discovering gene functions and for obtaining novel insights into previously characterized genes. Implementation of antisense gene knockdown, targeted screening of randomly mutagenized genomes, and gene knockout methods that utilize targeted mutagenesis with programmable nucleases has expanded uses of the zebrafish by creating the opportunity to apply the power of phenotypic analyses in the zebrafish to selected genes of interest (Lawson & Wolfe, 2011). Hence genes initially discovered experimentally in other species (Giraldez et al., 2005), by virtue of their association with human diseases (Jurynec et al., 2008; Phillips & Westerfield, 2014), or predicted as a consequence of genome and expression analyses (Ulitsky, Shkumatava, Jan, Sive, & Bartel, 2011) have been studied through loss-of-function analysis in the zebrafish. Analysis of loss-of-function conditions is a powerful approach for uncovering the earliest acting functions of a gene and has particular usefulness for identifying genes that may contribute to a shared molecular or developmental pathway (Gritsman et al., 1999; Jurynec et al., 2008; Langdon & Mullins, 2011). Nevertheless, as discussed below, these methods of analysis do not give access to the full range of functions of a gene. New techniques for directed modification of the genome are being developed to expand the kinds of inquiry that can be conducted in the zebrafish.
Null alleles of essential genes are most useful for uncovering the first stage at which a gene is required. However, as many genes governing signal transduction pathways, tissue patterning, growth regulation, etc. are used in multiple contexts during development and tissue homeostasis, we need tools to control tissue-specific and temporal gene expression in order to study the context-specific functions of any gene. For example, as the pathways governing tissue turnover and regeneration often have essential roles also during embryogenesis (Beachy, Karhadkar, & Berman, 2004), the ability to generate and utilize conditional alleles will have dramatic impact on the ability to analyze genes governing tissue maintenance in adults.
Constitutive loss-of-function mutations also have limitations with respect to their ability to recapitulate disease states or provide insights into a gene’s role in disease or its range of developmental activities. Many alleles associated with developmental, physiological, or behavioral disorders produce altered gene products or affect gene expression levels. Similarly, a main source of phenotypic variation among members of a species is likely due to sequence variants that do not eliminate gene function (Wray, 2007). Hence, tools for generating precise sequence modifications of the zebrafish genome are needed to more fully exploit this organism for purposes such as modeling disease states and understanding the roles of naturally occurring gene variants.
In addition, tools for modifying the zebrafish genome will allow study of the consequences of mutations that arise mosaically in the soma. Sporadically arising gain- or loss-of-function mutations are known to contribute to the origin of cancers and additional conditions including neurological disorders (Lupski, 2013; Poduri, Evrony, Cai, & Walsh, 2013). In sum, techniques to precisely modify the zebrafish genome will promote new approaches to the study of gene function that have the potential to render genuinely new insights into the phenotypic consequences of mutations.
Finally, the ability to precisely modify the genome may revolutionize developmental and cell biological studies in the zebrafish. Gene editing will allow us to generate modified proteins or introduce completely novel products that are expressed under conditions that precisely mimic an endogenous gene product. We anticipate these types of modifications will allow fate tracing or ablation of cells in developmental studies, visualization of the dynamic utilization of subcellular components, and identification of molecules that physically interact with the modified proteins of interest.
1. OVERVIEW OF CONTEMPORARY APPROACHES TO GENOME EDITING
The potential of genome-editing applications has spawned many efforts to develop flexible and reliable methods for modifying the zebrafish genome (Auer, Duroure, Concordet, et al., 2014; Auer, Duroure, De Cian, Concordet, & Del Bene, 2014; Bedell et al., 2012; Chang et al., 2013; Hisano et al., 2015; Hoshijima, Jurynec, & Grunwald, 2016; Hruscha et al., 2013; Irion, Krauss, & Nusslein-Volhard, 2014; Kimura, Hisano, Kawahara, & Higashijima, 2014; Li et al., 2015; Shin, Chen, & Solnica-Krezel, 2014; Zu et al., 2013). All current approaches to genome editing utilize the host cellular pathways that are normally responsible for repairing DNA damage (Fig. 1). The major advance that sets the groundwork for these methods was the development of sequence-specific programmable nucleases, which are used to initiate double-strand breaks (DSBs) at targeted loci and thus trigger repair (Carroll, 2014; Hsu, Lander, & Zhang, 2014; Kim & Kim, 2014). As the technology for efficiently inducing targeted DSBs is relatively young, the current approaches for modifying the genome are still at nascent stages.
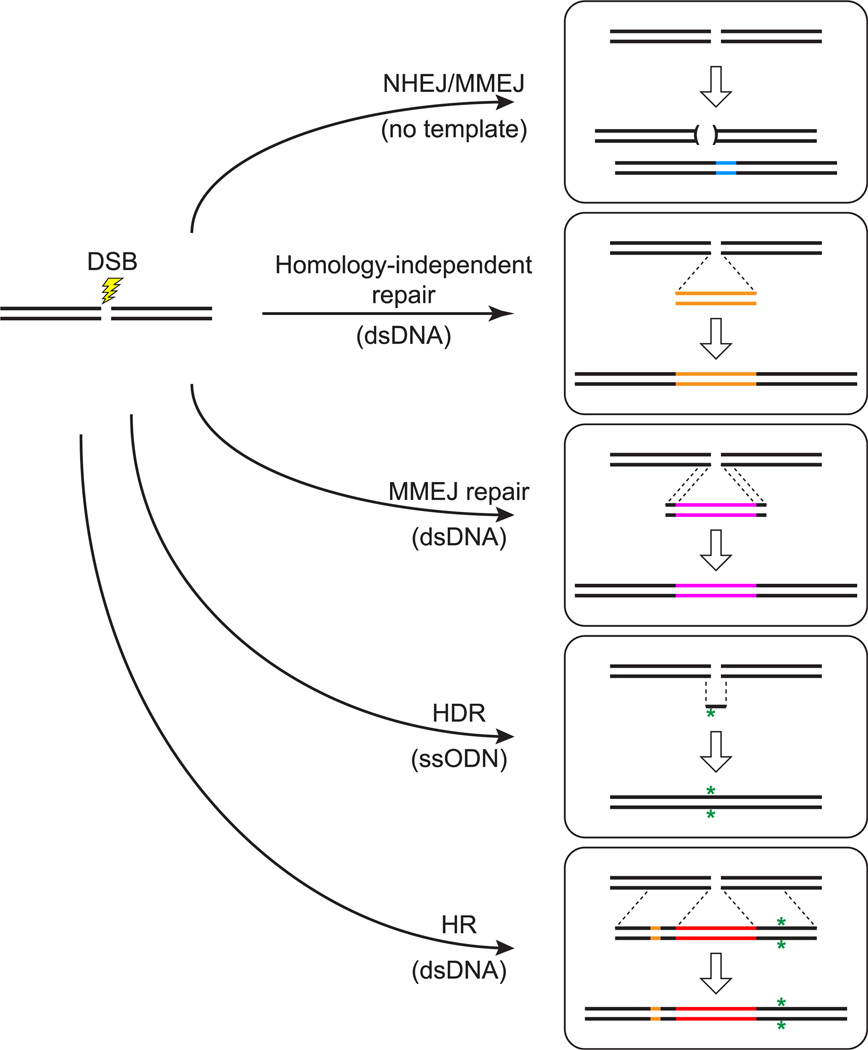
FIGURE 1. Approaches to genome editing.
Repair and recombination events are stimulated by double-strand breaks (DSBs) induced by targeted cleavage (lightning bolt) of the genome with programmable nucleases. In the absence of template DNA to guide repair, the nonhomologous end-joining (NHEJ) or microhomology-mediated end-joining (MMEJ) pathways heal broken ends of chromosomes in a process that often generates small deletions and/or insertions at the site of the lesion. These repair pathways can also facilitate integration of exogenously supplied DNA sequences at the lesion site. In the presence of linear double-stranded DNA (dsDNA) donor molecules that bear no homology to the targeted locus, the NHEJ pathway may join donor DNA sequences to the broken chromosome ends. End-joining occurs in a homology-independent, imprecise manner, and foreign sequences may be integrated in either orientation. If the borders of the donor DNA contain short sequences homologous to the regions immediately flanking the DSB site, end resection may uncover short stretches of complementarity that guide repair and result in incorporation of the donor sequences at the lesion. Two additional methods have been employed with the purpose of precisely modifying the host genome. When single-stranded oligodeoxynucleotides (ssODN) with close homology to the site of the DSB are available, the donor sequences may guide homology-directed repair (HDR) resulting in the modification of host sequences. Finally, when DSBs are induced in the presence of dsDNA molecules with extensive homology to the targeted region, true crossover events mediated by homologous recombination (HR) may occur, resulting in exchange of several kilobase-long regions between the host genome and the exogenously supplied DNA. This approach can be used to produce several closely linked genome modifications simultaneously. (See color plate)
Because chromosome breaks stimulate repair and recombination pathways at the site of the lesion, synthetic sequence-specific nucleases can be used to direct repair-dependent modifications of the zebrafish genome to a particular locus of interest (Fig. 1) (reviewed in Auer & Del Bene, 2014). Zinc-finger nucleases, TALENs, and RNA-guided nucleases of the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system have all been demonstrated capable of inducing DSBs at targeted sites in the zebrafish genome and triggering repair. In the absence of a DNA template to direct repair, DSB lesions can be healed by the nonhomologous end-joining (NHEJ) or the microhomology-mediated end-joining (MMEJ) pathway, both of which often result in small sequence modifications at the lesion site. This approach has become widely applied for the purposes of generating frame-shift or deficiency mutations at a targeted locus.
In addition, both repair pathways have been exploited as means for inserting exogenously provided donor sequences into a targeted zebrafish locus (Auer, Duroure, De Cian et al., 2014; Hisano et al., 2015; Kimura et al., 2014; Li et al., 2015). In the absence of homology between donor and targeted locus, donor sequences can be inserted imprecisely via NHEJ, with sequence alterations arising at the junction sites. When the ends of linear donor molecules are flanked by short sequences homologous to those flanking the DSB in the host, the homologies may facilitate MMEJ repair leading to intact incorporation of all novel sequences at the lesion site. These approaches have been used to insert protein-coding sequences in a manner that allows their expression to be regulated by the targeted locus.
In contrast to the editing methods described in the preceding paragraphs, two additional methods have been introduced with the express purpose of precisely revising the host genome. The exact molecular mechanisms mediating these repair/recombination events are not clear, and therefore, the factors that contribute to the efficiency of each type of editing event are not yet well defined.
In the first method, designated homology-directed repair (HDR), single-stranded oligodeoxynucleotides (ssODN) sharing homology with the lesion site are supplied as donor sequences to guide repair of the broken chromosome (Bedell et al., 2012; Chang et al., 2013; Hruscha et al., 2013). The repair process drives replacement of host sequences with donor sequences, allowing both the precise modification of host sequences and the insertion of short stretches of novel sequence into the genome. One strength of the approach is the ease with which donor templates can be designed and generated. However, as currently implemented, there are three substantial limitations to the use of HDR with ssODN donor molecules. First, because the donor templates are short, generally less than 100 nt, only small sequence modifications are produced by this method. Second, the recovery of edited alleles usually requires laborious DNA-based screening. Third, at least half the repair events are imprecise, accompanied by unintended changes to the genome. We expect the fidelity of this approach will improve with further study.
An alternative approach utilizes homologous recombination (HR) between genomic sequences flanking the DSB lesion site and double-stranded DNA (dsDNA) donor molecules, allowing the precise replacement of several kilobase-long stretches of host sequence with exogenously provided sequences (Hoshijima et al., 2016; Irion et al., 2014; Shin et al., 2014; Zu et al., 2013). Donor templates are supplied as dsDNA molecules that generally contain homology arms of 0.5–2 kbp, which may or may not border regions of nonhomology. Because this approach utilizes true crossover events involving the homology arms, it can be used simply to edit host sequences or to accomplish more complex genome modifications. As illustrated in Fig. 2A, one simple application is to produce the precise in-frame insertion of entire protein-coding sequences, leading to expression of a novel protein such as green fluorescent protein (GFP) from an endogenous locus. Significantly, the approach can also be used to produce multiple modifications distributed over several kilobases of a targeted locus (Hoshijima et al., 2016). As illustrated in Fig. 2B, in addition to critical sequence modifications of interest, we often include a reporter gene within the donor sequences. Recombination events that utilize both homology arms will produce the intended editing changes and also lead to incorporation of the linked reporter gene, which can be used initially to help identify an edited allele and subsequently to track its inheritance. Use of the linked reporter gene makes it easy to identify and recover edited alleles, such as loxP-flanked conditional mutations, that may not confer an overt phenotype on the carrier. In practice, inclusion of the reporter within the donor sequences greatly streamlines the recovery of edited alleles: we find 30–50% of the reporter-expressing genomes recovered from treated animals harbor precisely edited alleles that include all intended modifications. Moreover, when the reporter gene is bordered by FRT sites, it can be readily excised upon expression of FLP recombinase (Hoshijima et al., 2016).
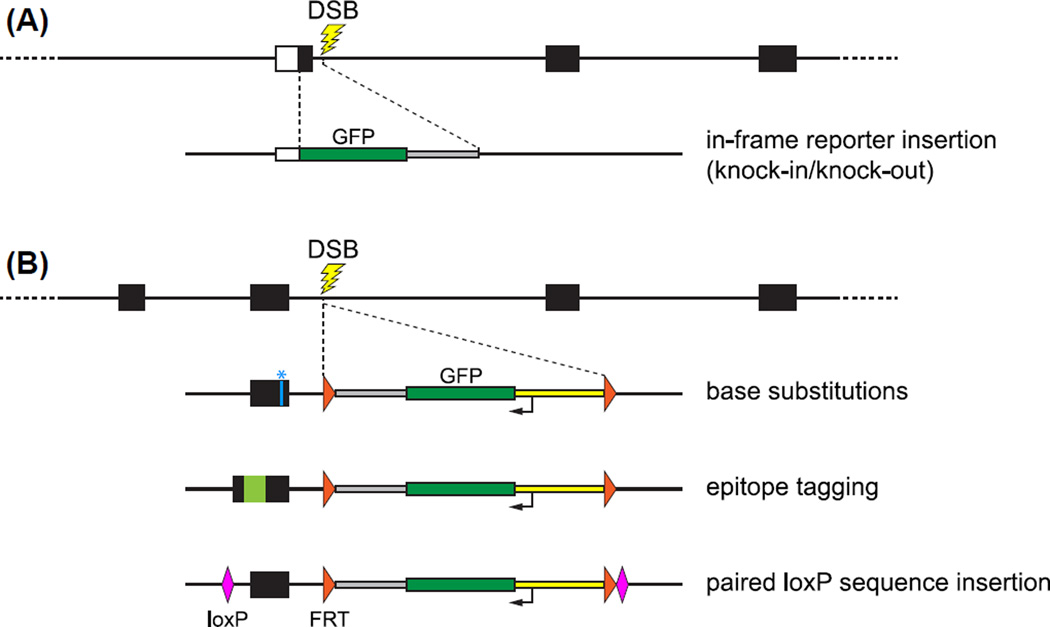
FIGURE 2. Examples of genome modifications produced by DSB-stimulated homologous recombination (HR) using dsDNA as donor template molecules.
Recombination events are stimulated by targeted cleavage (lightning bolts) of the genome with programmable nucleases. The top line of each panel represents the host genome, with boxes indicating exons (white, 5′ untranslated region; black, coding sequences). Below the genome cartoons are examples of dsDNA donor templates that have been used to generate the indicated types of edited alleles. Except as indicated, donor sequences are identical to those of the genome: green boxes indicate GFP coding sequences, gray boxes indicate transcription termination sequences, yellow boxes indicate α-crystallin promoter sequences, orange arrowheads are FRT sites, and pink diamonds are loxP sites. (A) Generation of a knockin/knockout allele. The DSB was induced in intron sequences just 3′ to the first exon. Recombination with donor sequences introduced GFP reporter sequences in-frame just downstream of the endogenous translation initiation codon. Animals inheriting this kind of edited allele can be recognized by virtue of expression of GFP under control of the endogenous promoter. (B) Use of a linked reporter gene to tag genomic modifications. In addition to the desired editing events, donor DNAs can be designed so they introduce into the host genome a small reporter gene (here α-crystallin::GFP flanked by directly repeated FRT sites). Edited alleles that incorporate precise alteration of coding sequences (blue *–base substitutions), in-frame introduction of epitope sequences (pale green–epitope tagging), or introduction of a pair of loxP recombination sites flanking an exon can be identified by expression of the linked tissue–specific reporter gene. Screening for GFP fluorescence allows for the efficient recovery of phenotypically silent modification events and obviates the need for laborious DNA-based screening of F1 individuals. Following identification of successful editing events, the reporter gene can be excised using FLP-mediated recombination. (See color plate)
Each of the approaches to genome editing described in the preceding paragraphs is sufficiently efficient to be practicable. Because of the range of types of genome modifications that can be produced, the precision with which edited alleles may be generated, and the ease with which reporter-marked alleles can be recovered, our focus has been on developing genome-editing methods that utilize DSB-stimulated HR with dsDNA donor molecules. Several groups using this approach have found between five percent and 15% of the animals that arise from zygotes injected with a combination of programmable nuclease and dsDNA donor molecules transmit precisely edited alleles to their progeny (Hoshijima et al., 2016; Irion et al., 2014; Shin et al., 2014). In our experience, about 6% (range 0.6–23) of the gametes of a founder with an edited germ line will transmit the modified allele of interest (Hoshijima et al., 2016). Thus marking donor sequences with a linked reporter gene can greatly facilitate the identification and recovery of edited alleles.
2. SUMMARY OF WORKFLOW
Our current focus is to edit genomes at early stages during the embryonic growth of a founder, with the expectation that a subset of the edited genomes will enter the germ line and become heritable. In experiments aimed at optimizing the induction of heritable modified alleles, we initially measured the production of edited alleles in the somatic tissues of 1–2-day-old founder embryos, assuming these events are representative of the germ-line genomes (Hoshijima et al., 2016). Our method of approach to gene editing has been informed by such experiments, some of which are described below.
Briefly, to stimulate HR events, just-fertilized zebrafish zygotes are injected with a mixture of synthetic nuclease targeting a unique locus and donor dsDNA consisting of novel sequences flanked by 1-kbp left and right homology arms (Fig. 3). At 2-day post-fertilization (dpf), the genomes of 8–12 injected embryos are individually analyzed for the presence of correctly edited alleles. Polymerase chain reaction (PCR) analysis functions as a preliminary test to determine whether the targeted locus has acquired donor sequences in the expected configuration, using one primer specific to novel donor sequences and the second primer specific to host sequences distal to the homology regions. As shown in Fig. 3, nuclease activity has a profound effect stimulating HR. Under typical conditions, we cannot detect edited alleles in embryos injected only with donor DNA, but all embryos injected with nuclease and donor DNA have edited genomes. When injection leads to detectable genome editing in virtually all (>95%) founder embryos, sibling injected embryos are grown to adulthood and examined for the ability to transmit modified alleles to offspring. Depending on the type of genome modification that has been designed, transmission of edited alleles is detected in F1 offspring either by the expression of newly acquired coding sequences, such as a reporter protein, or by the analyses of genomic DNA.
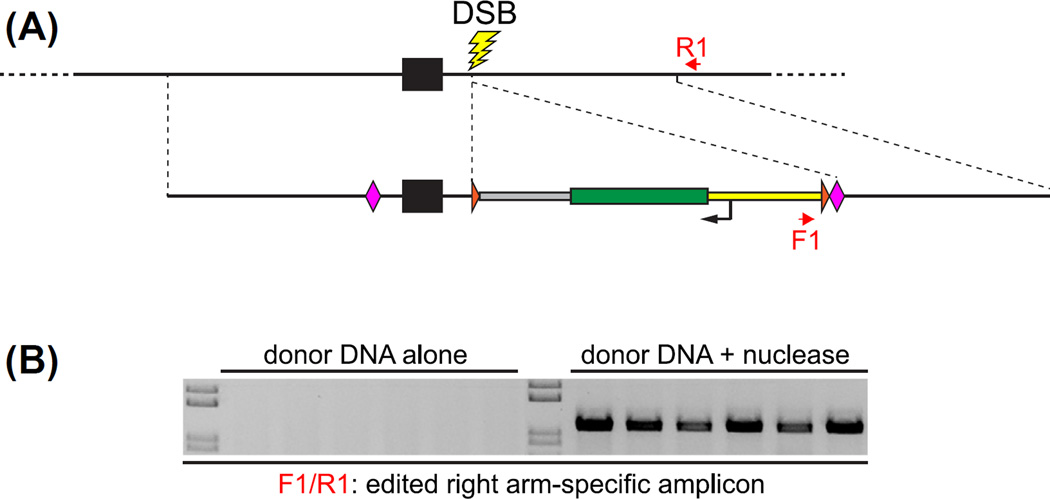
FIGURE 3. PCR-based screening to identify edited alleles in F0 embryos.
(A) Schematic representation (as in Fig. 2) of a genome-editing event in which HR with the dsDNA donor molecule introduces a pair of loxP recombination sites flanking an exon as well as the α-crystallin::GFP reporter gene. To detect the presence of correctly edited alleles in F0 embryos that had been injected with nuclease and donor molecules, genomic DNA is isolated from individual 1 dpf F0 embryos, and PCR analysis is performed using one primer (F1) specific to novel donor sequences and a second primer (R1) specific to endogenous host sequences distal to the homology region. (B) HR is dependent on nuclease activity. When donor DNA is injected without nuclease, edited alleles are not detected in F0 embryos. However, under typical efficient editing conditions, following injection of donor DNA with nuclease, every F0 embryo harbors precisely edited alleles.
3. FACTORS TO CONSIDER FOR GENOME EDITING VIA HOMOLOGOUS RECOMBINATION
Although the approach described here has produced precisely modified alleles of many loci (Hoshijima et al., 2016), we realize the methods are not yet perfected and many parameters that might affect the efficiency of editing have yet to be tested. In this section, we discuss factors that we consider in the design of a genome-editing experiment, highlighting aspects of the technology that need to be further investigated and optimized.
3.1 CHOICE OF NUCLEASE TARGET SITE
The induction of DSBs in the host genome is responsible for recruiting repair machinery and stimulating HR events between the affected locus and donor molecules harboring homologous sequences. However, DSBs also stimulate the NHEJ and MMEJ repair pathways, which simply rejoin the chromosome arms, often producing indel mutations at the DSB site (Fig. 1). Hence, there is potential for a conflict, for as the frequency of DSBs is increased to maximize the occurrence of HR, there will likely be a concomitant rise in the induction of indel mutations at targeted loci. For this reason, especially when we attempt to modify an essential gene, we prefer to design and utilize programmable nucleases that generate DSBs in intron regions devoid of conserved sequences, with the intent to minimize the production of mutant cells in the soma of injected embryos. In the future, the development of tools that stimulate gene editing specifically in the germ line may obviate this concern.
A second consideration is the distance between the DSB site and the closest position at which nonhomologous sequences are to be introduced. Indeed, there are no published empirical data addressing this issue in the zebrafish. Published studies have tended to induce DSBs at genomic sites very close to the sequences that are to be modified by the donor. Similarly, we do not know how far recombination events initiated at a DSB will travel, and thus how much donor sequence can be incorporated efficiently into the genome. Several reports indicate that tracts of about 2-kbp of sequence can be readily introduced into the host genome (Hoshijima et al., 2016; Shin et al., 2014; Zu et al., 2013).
Third, we found that DSBs with 5′ overhangs, produced by TALEN-mediated cleavage, and DSBs initiated by the blunt-cutting CRISPR/Cas9 system can each be used promote HR efficiently in the zebrafish. Either type of target site can be used to stimulate gene editing.
For several reasons, we attempt to minimize sequence heterogeneity at the target locus among the genomes that are subjected to editing. First, polymorphisms within the nuclease recognition site may reduce the induction of DSBs. Second, polymorphisms close to the targeted site may affect the ability to measure nuclease activity, because the induction of DSBs is often measured indirectly as the occurrence of repair-induced sequence heterogeneity close to the DSB site (Dahlem et al., 2012; Hwang et al., 2013; Jao,Wente, & Chen, 2013). Third, it is unclear how mismatches between the host locus and donor sequences affect recombination efficiency, although we note that donor sequences are generally designed so that their integration will destroy the nuclease recognition site and render the edited locus immune to further nuclease activity. For these reasons, we utilize a selected breeding population that lacks polymorphism near the DSB target site to produce the genomes to be edited.
3.2 INDUCTION OF TARGETED DSBs
To induce DSBs at a selected site in the host genome, we utilize TALENs or components of the CRISPR/Cas9 with equal abandon. As TALENs are delivered in the form of injected mRNA that needs to be translated prior to the onset of enzyme activity, our bias is to initiate DSBs by injecting a complex of sgRNA (single guide RNA) and Cas9 protein (CRISPR-associated protein 9) (Gagnon et al., 2014), components that should be capable of catalyzing DSBs soon after injection.
To achieve high rates of HR in the embryo, it is critical to identify nucleases that efficiently generate DSBs at the target site. To determine the relative cleavage activity of TALENs or sgRNAs in vivo, we inject candidate nucleases without donor DNA into zygotes and analyze the presence of mis-repaired alleles in the genomes of 8–12 individual 1 dpf embryos (Fig. 4) (Dahlem et al., 2012; Hoshijima et al., 2016). As the starting population of targeted genomes had been selected to lack polymorphisms in the region bordering the nuclease target site, the relative abundance of newly induced mutations reflects DSB activity. Newly arising sequence polymorphisms can be detected by any of a number of methods, including direct sequencing, high-resolution melt analysis (HRMA) (Dahlem et al., 2012), capillary electrophoresis–based fragment analysis (Carrington, Varshney, Burgess, & Sood, 2015), or the Surveyor or T7 endonuclease mismatch detection assays (Qiu et al., 2004; Reyon et al., 2012). We attempt genome editing only with programmable nucleases that are shown to induce somatic mutations in every treated embryo. In experiments that have led to successful gene editing, we have used nucleases that induce mutations in 30–70% of the host genomes.
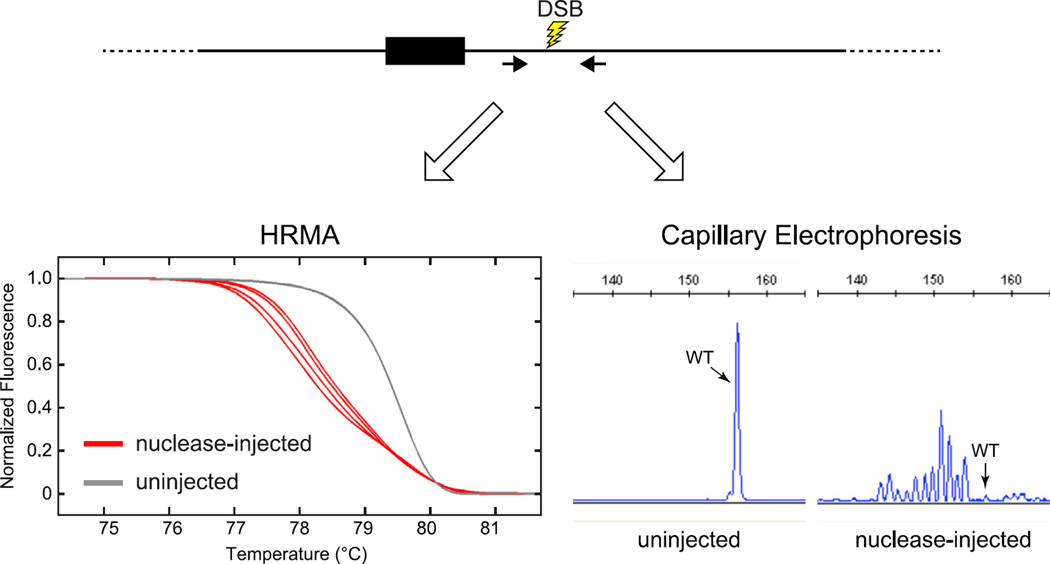
FIGURE 4. PCR-based methods to detect DSB repair-induced mutations.
Prior to conducting a genome-editing experiment, the in vivo activity of the programmable nuclease used to induce DSBs is assayed by the induction of small indels at the nuclease target site. Several methods are available for detecting repair-induced mutations. We use HRMA (Dahlem et al., 2012) or capillary electrophoresis (Carrington et al., 2015) for this purpose. Genomic DNA is isolated from individual 1 dpf embryos that had been injected at the one-cell stage with nuclease. PCR primers (black arrows) are used to amplify a 90–150-bp product that is centered on the nuclease target site. DNA from individual uninjected or nuclease-injected embryos is amplified in the presence of a dsDNA-binding dye (HRMA) or with fluorescently labeled primers (capillary electrophoresis). For HRMA, PCR products are denatured and renatured, and duplexes with mismatches are detected by their altered thermal denaturation profile (red curves (black in print versions)). For capillary electrophoresis analysis, products are resolved according to size, and the fraction of amplicons with altered size can be determined. Representative HRMA and capillary electrophoresis traces indicate a highly active nuclease.
3.3 DESIGN OF DONOR SEQUENCES
Experiments have not yet been reported that determine requirements for homology between the donor and the sequences immediately surrounding the site of the induced lesion. For this reason, we usually generate donor molecules that carry sequences bridging the cleavage site. Furthermore, to prevent the programmed nuclease from cleaving a successfully edited allele, donor sequences must not contain an intact nuclease recognition site. If the nuclease target site lies within host coding sequences that are to be retained in the edited allele, silent mutation changes can be introduced into the donor that destroy the target site but maintain coding capacity.
Donor sequences sharing perfect homology with the targeted locus should flank the nonhomologous sequences to be introduced into the genome. We use 1-kbp homology arms because they are sufficient to achieve HR in zebrafish embryos with high efficiency, and yet they allow for the detection of precise integration events by PCR amplification (Fig. 3). However, we note the optimal extent of homology arms has yet to be determined, and some reports have found that longer homology arms in donor molecules enhance the frequency of HR (Shin et al., 2014). To create homology arms harboring sequences identical with the genomes to be targeted, we first identify a selected breeding population with absent/reduced polymorphism bordering the nuclease target site and then PCR amplify sequences from founder genomes or their offspring.
3.4 CONFIGURATION OF DONOR MOLECULES
As important as the induction of DSBs is for the stimulation of repair/recombination machinery, so too, is the availability of donor template for participation in HR with genomic sequences at the targeted site. We do not understand how injected donor DNA comes to be recruited to a recombination complex in zebrafish cleavage stage embryos. Three factors potentially affect the accessibility of donor DNA (Stuart, McMurray, & Westerfield, 1988; Udvadia & Linney, 2003). First, injected plasmid DNA does not readily diffuse or disperse throughout the cytoplasm of cleavage stage embryos. Second, injected circular or linear plasmid DNA is often rearranged into long molecules consisting of imperfectly repeated units. Third, exogenously supplied DNA may be subjected to degradation. In truth, there is little experimental evidence that informs us as to how best to deliver exogenously supplied donor DNA so as to maximize its ability to participate in HR events with the host genome.
As a result, different investigators have tested the effect of altering donor DNA configuration on the efficiency of gene editing. Although little consensus can be derived from these preliminary studies, we, and others, found injection of circular donor DNA is better than injection of linear molecules for producing edited alleles (Hoshijima et al., 2016; Irion et al., 2014).
In our efforts to identify factors that could improve the ability of donor DNA to participate in HR-mediated genome editing, we generated linear donor fragments produced by cleavage with I-SceI endonuclease, whose recognition sites were arranged in a head-to-head orientation bordering the donor sequences. I-SceI enzyme cleaves its 18-bp recognition site asymmetrically, producing a longer “head” portion to which the I-SceI protein remains associated (Perrin, Buckle, & Dujon, 1993).We reasoned that linear fragments associated with end-capped protein might provide good substrate for HR.
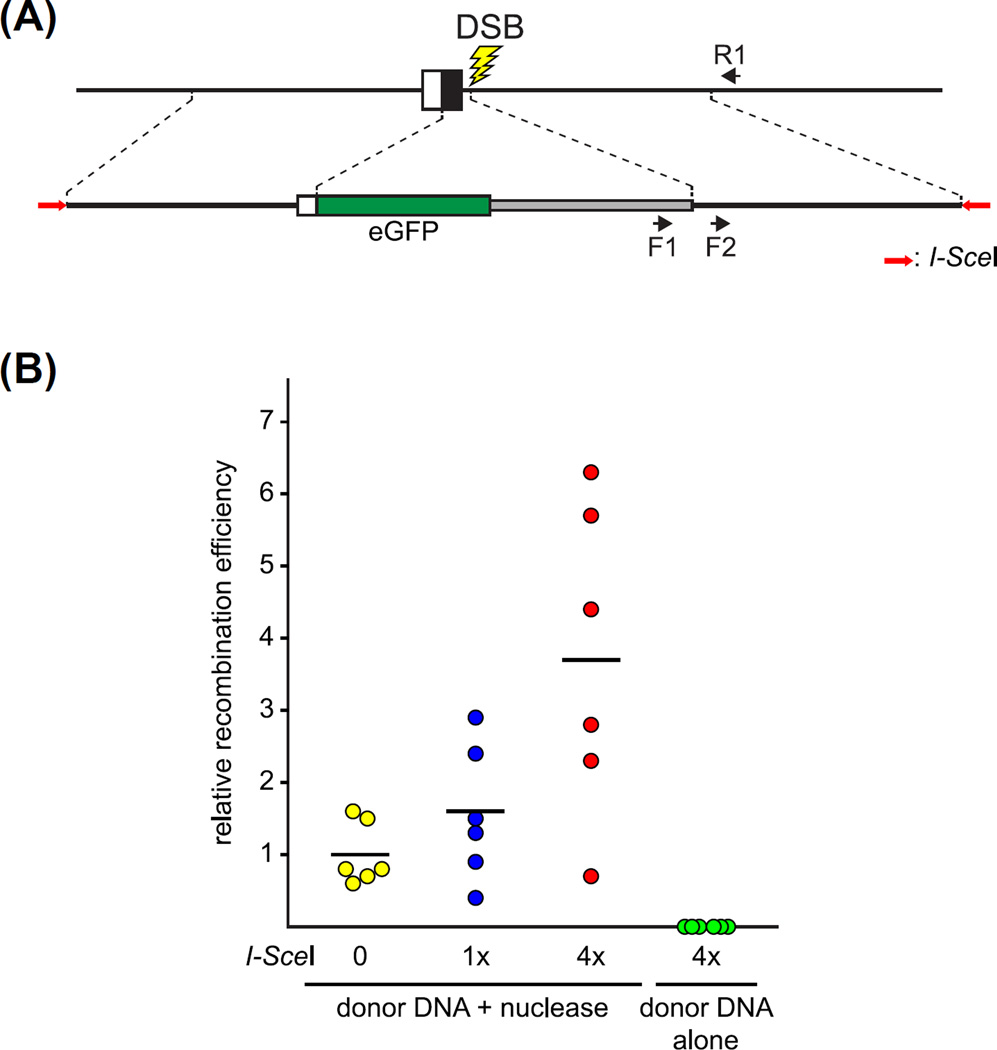
Cleavage of donor DNA molecules with I-SceI enzyme stimulates the ability of donor molecules to participate in HR and produce edited alleles. Fig. 5A illustrates an experiment that produced a knockin/knockout allele at the kcnh6a locus of zebrafish (Hoshijima et al., 2016). Donor sequences flanked by head-to-head I-SceI recognition sites were cloned into the pKHR4 vector (Fig. 6). Donor plasmid DNA was injected with or without targeting nuclease into one-cell embryos, and the generation of edited alleles in individual 2 dpf embryos was measured in quantitative PCR assays (Fig. 5B). Digestion of the circular plasmid donor DNA with I-SceI enzyme in vitro prior to being mixed with programmable nuclease and injected into zygotes stimulated the formation of edited kcnh6a alleles in a dose-dependent manner. We obtained similar results at additional loci. Hence, we developed a series of donor pKHR vectors (Fig. 6) that allow ready construction of donor sequences bordered by head-to-head-oriented I-SceI recognition sites.
FIGURE 5. Enhancement of genome editing using donor molecules predigested with I-SceI meganuclease.
(A) Schematic representation of a genome-editing event to produce a reporter knockin/knockout allele, as in Fig. 2A. Donor DNA sequences were flanked by a pair of head-to-head oriented I-SceI sites (red arrows) present within the pKHR4 plasmid vector (Fig. 6). The relative abundance of edited alleles within the genomes of injected F0 embryos was determined following quantitative PCR (qPCR). Primers used for qPCR are depicted as black arrows: the F1/R1 pair specifically amplifies the edited allele, whereas the F2/R1 pair amplifies edited as well as unedited forms of the endogenous locus. (B) Genomic editing is enhanced by I-SceI digestion of a donor plasmid prior to injection. The donor plasmid was digested with increasing amounts of I-SceI enzyme in vitro and subsequently injected with programmable nuclease into zygotes. As a control, I-SceI-digested donor plasmid was injected alone, without added nuclease to target the host genome. The fraction of edited alleles (detected with the F1/R1 primer pair) relative to total targeted loci (detected with the F2/R1 primer pair) present in injected 2 dpf embryos was determined by qPCR. The relative recombination efficiency was determined by normalizing the mean fraction of edited alleles following injection of nuclease and undigested donor plasmid DNA to 1.0. For each condition, six individual embryos were analyzed (circles) and the mean relative recombination efficiency is indicated (horizontal dash). Unpaired t-test analysis indicated that digestion of donor DNA with 4× enzyme significantly stimulated the production of edited alleles as compared with untreated donor DNA (p < 0.01). Digestion with 1× enzyme did not yield a significant increase in recombination efficiency as compared with untreated DNA. Injection of I-SceI-digested DNA without programmable nuclease failed to produce a detectable level of edited target alleles. (See color plate)
FIGURE 6. pKHR plasmid vectors for genome editing.
pKHR4: pKHR4 is an ampR plasmid built from Bluescript SK(+) with its multiple cloning site (mcs) flanked by inverted head-to-head oriented I-SceI sites. pKHR5: pKHR5 is derived from pKHR4 by inserting the CV reporter gene cassette, FRT-CV-FRT-loxP, between the EcoRI and EcoRV sites. The cassette consists of the α-crystallin promoter driving expression of the Venus version of the GFP protein in the lens (Hesselson, Anderson, Beinat, & Stainier, 2009) flanked by FRT sites and bordered on the 3′ end with a single loxP site. pKHR5 contains two mcs for independently introducing left and right homology arms to flank the reporter cassette. pKHR7: pKHR7 contains a simple modification of pKHR4 that allows detection of imprecise or random insertion events. pKHR7 contains a cmlc2::mCherry (red heart) reporter gene that resides within the donor fragment that is produced by I-SceI digestion, but outside any homology region. Incorporation of cmlc2::mCherry with donor sequences can only occur as a consequence of imprecise recombination or random integration but never as a result of precise HR. pKHR8: pKHR8 contains a simple modification of pKHR5 that allows detection of imprecise or random insertion events. pKHR8 carries the CV reporter gene cassette, as in pKHR5, but also has a cmlc2::mCherry (red heart) reporter gene, which resides within the donor fragment produced by I-SceI digestion, but outside any homology region. Successful homologous recombination events of interest should acquire the green lens reporter but not the red heart reporter. Sequences for the donor vectors pKHR4, 5, 7, 8 have been deposited in GenBank (KU144822-KU144825). The plasmids are available through Addgene. (See color plate)
4. METHOD OF APPROACH
4.1 DESIGN AND PREPARATION OF PROGRAMMED NUCLEASES
4.1.1 Design of nucleases
We utilize the TALEN or CRISPR/Cas9 nuclease systems to initiate targeted DSBs. Once a target region of interest has been selected, we retrieve 400–600 bp of genomic sequence covering the region from Ensembl and search for potential TALEN target sites using TALEN Targeter (https://tale-nt.cac.cornell.edu/) or potential sgRNA target sequences using web resources such as CRISPR Design (crispr.mit.edu) or CHOPCHOP (https://chopchop.rc.fas.harvard.edu/).
4.1.2 Target sequence confirmation and selection of a breeding population
Prior to the generation of nucleases, the actual fish genomes to be targeted should be analyzed to confirm they carry the target sequences as described in the reference sequence database. The purpose of this step is to establish a small breeding population that harbors the nuclease target and exhibits minimal sequence heterogeneity around the target site, a concern especially relevant when designing nucleases to cleave within an intron. A 400–600-bp genomic region surrounding the nuclease recognition site should be amplified from genomic DNAs prepared from eight or more adult fish. Amplicons should be sequenced and the existence of target sequences verified. Simultaneously, sequence chromatographs are analyzed to determine the presence of polymorphic sequences. Adults that carry the exact target sequence and exhibit minimal additional polymorphisms should be selected as a breeding population. In our experience, wild-type strains differ greatly in the distributions of polymorphic sequences. While one strain (eg, AB) may be rich in sequence variation at a particular gene, another strain (eg, TU) may be completely devoid of such polymorphisms.
4.1.3 Synthesis of programmable nucleases
Programmable nucleases are prepared according to standard published methods. TALEN plasmids are constructed using the Golden Gate System using DDD/RRR FokI nuclease domains for obligate heterodimerization between left and right TALEN monomers (Dahlem et al., 2012). TALEN mRNAs are prepared from these templates by in vitro transcription. Target-specific sgRNAs for the CRISPR/Cas9 system are prepared by in vitro transcription from an oligonucleotide template DNA (Gagnon et al., 2014).
4.1.4 Determination of nuclease cleavage activity
Cleavage activity of the targeting nucleases must be analyzed in zebrafish embryos prior to performing gene editing with donor molecules. For TALENs, 50 pg left and 50 pg right TALEN mRNA are injected with 0.05% phenol red into the cytoplasm of one-cell stage of zygotes generated from the breeding population identified above. For the CRISPR/Cas9 system, 200–300 ng sgRNA is coinjected with 600 pg Cas9 protein (PNA BIO) and 0.05% phenol red. Under these conditions, most of the RNA-injected embryos should develop normally (80–90%). If a large fraction of injected embryos display developmental abnormalities, the amount of injected RNA should be reduced.
As described in Section 3.2, cleavage activity is actually measured as the induction of novel sequence changes at the nuclease target site in the somatic genomes of 1 dpf injected embryos. Genomic DNA is extracted from 8 to 12 individual embryos using the following protocol:
Place individual dechorionated embryos in 50 µL of 50 µM NaOH.
Incubate 95°C, 20 min.
Transfer onto ice, 4°C.
Add 5 µL 1 M Tris–HCl (pH 8.0).
-
Mix well and spin down.
Store at −20°C.
To analyze the induction of polymorphisms, 90–150 bp bordering the targeted region is amplified from genomic and analyzed by HRMA, Surveyor/T7 Endonuclease activity, or capillary electrophoresis. Strong cleavage activity should be detected in every embryo before proceeding.
4.2 DESIGN AND PREPARATION OF DONOR MOLECULES USING pKHR VECTORS
A set of pKHR vectors (Fig. 6) was designed to allow ready construction of donor sequences bordered by head-to-head-oriented I-SceI recognition sites. pKHR4 simply has a multiple cloning site between I-SceI cleavage sites. pKHR5, pKHR7, and pKHR8 provide additional sequences including a reporter gene placed between the homology arms to track acquisition of donor sequences via HR and/or a reporter gene residing distal to the homology arms to track imperfect or random insertion events.
4.2.1 Preparation of homologous arms and novel donor sequences
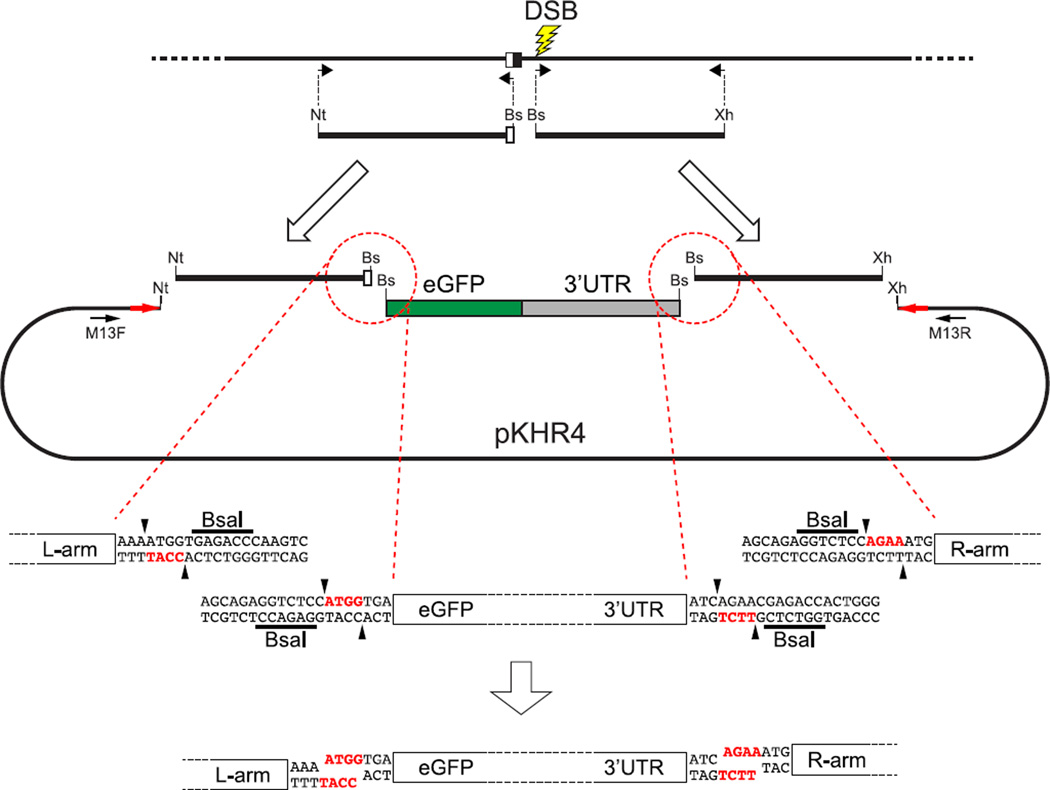
Genomic DNA to be used as template to generate homology arms should be extracted from breeding population or F0 animals that are likely to have uniform sequence in the region of the target site. As illustrated in Figs. 7 and 8, typically three PCR-amplified fragments are initially generated and joined to vector sequences by classical ligation methods in the preparation of a donor plasmid. Each homology arm (about 1 kbp) is prepared by PCR amplification using high fidelity DNA polymerase with primers that introduce at the ends of the arms appropriate restriction enzyme recognition sequences. Nonhomologous sequences to be introduced, for example GFP coding sequences with 3′ UTR sequences, are also prepared by PCR amplification from source plasmids with primers that provide restriction enzyme sequences. As denoted in Fig. 7, a type-II restriction enzyme, such as BsaI, is convenient for generating amplicons with unique and complementary protruding ends.
FIGURE 7. Preparation of a donor plasmid for targeted reporter integration.
To integrate a reporter gene consisting of eGFP coding sequences (green (dark gray in print versions)) and translation/transcription termination sequences (gray) at a specific target site, about 1 kbp of genomic sequences upstream and downstream of the reporter integration site are prepared by PCR amplification for left and right homology arms, respectively. The homology arms should include a mutated nuclease target sequence, so that integration of donor sequences will produce an edited allele that cannot be cleaved by the nuclease. Each amplified fragment is bordered by unique restriction enzyme recognition sequences derived from PCR primers. In this case, the left-arm fragment is bordered by NotI (Nt) and BsaI (Bs) sites, and the right arm has BsaI and XhoI (Xh) sites at its ends. BsaI is a type-II restriction enzyme that produces a staggered cut next to the enzyme recognition site. As a result, it can be used to generate unique, complementary protruding ends (red letters (black in print versions)) independent of the enzyme recognition sequence (underlined). The reporter sequence is prepared as a middle fragment by PCR amplification with ends containing BsaI recognition sites whose digestion would yield protruding ends complementary to the digested left- and right-arm fragments. Restriction enzyme-digested amplicons are individually purified. Because each digested end is complementary to a unique partner fragment, ligation of the digested fragments leads to ordered assembly, and a single cloning step is used to assemble the homology arms and the reporter middle fragment in correct sequence into the pKHR4 backbone, which had been predigested with NotI and XhoI.
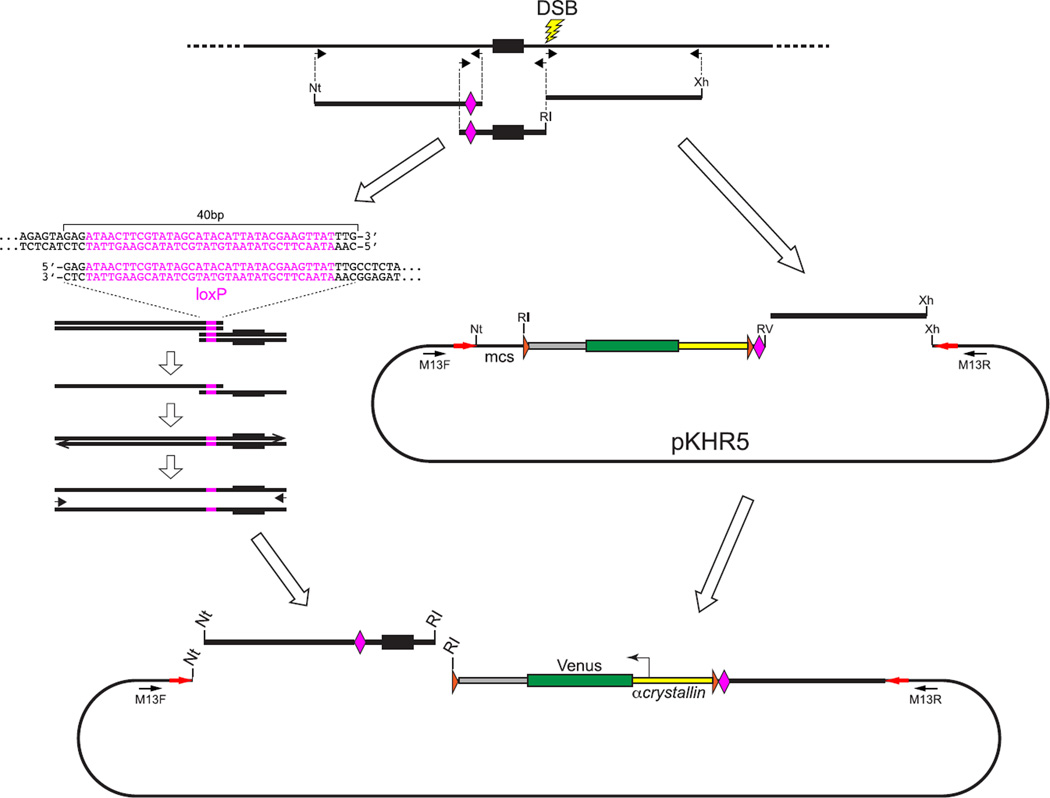
FIGURE 8. Preparation of a donor plasmid to create a loxP-flanked conditional allele marked by a linked reporter gene.
A donor plasmid to create a conditional allele in which loxP sites flank an exon can be constructed using the pKHR5 or pKHR8 vectors (Fig. 6). These vectors provide the CV reporter gene cassette, which consists of the α-crystallin::Venus reporter gene flanked by FRT sites (orange arrowheads) with a single loxP site (pink diamond) located at the border between the reporter and the right homology arm. The right homology arm, consisting of about 1-kbp sequence extending 3′ from the lesion site, is amplified with primers to produce a blunt phosphorylated 5′ end and a 3′ end with an XhoI (Xh) site. The right arm is then cloned into an EcoRV + XhoI-digested vector. As illustrated, the vector-provided loxP site will be inserted into intron sequences downstream (3′) of the targeted exon, and so a second loxP site needs to be introduced into an extended left homology arm, 5′ to the exon. The left arm containing the loxP site is prepared by overlapping PCR amplification to create a fused template from two fragments initially generated from the targeted locus. One fragment contains the exon, extending from the DSB lesion site to the intended position of the second loxP site. Primers used to generate this fragment introduce novel end sequences, an EcoRI (RI) recognition sequence at the lesion site and a 40-bp sequence that includes the loxP site at the end terminating 5′ of the exon. The second fragment extends about 1 kbp further upstream from the position of the loxP site. Primers used to generate this amplicon produce a fragment bordered 5′ by a NotI (Nt) site and 3′ by the 40-bp sequence that includes the loxP site. Overlapping PCR is used to generate a single extended left-arm fragment that is bordered by NotI and EcoRI sites and extends from the lesion site into the upstream intron. The left arm is cloned into a NotI + EcoRI digested vector that already contains the right arm homology. (See color plate)
4.2.2 Generating a loxP site within a homology arm
As illustrated in Fig. 8, to introduce the 34-bp loxP sequence (or other similar small sequence addition) into the host genome, an extended homology arm is created in the donor vector by overlapping PCR amplification. Using primers that introduce loxP sequences at the desired point of insertion, two overlapping amplicon fragments are initially generated from genomic template, one extending 5′ and the other extending 3′ from the point of insertion. Consequently, as shown in Fig. 8, each amplified fragment will have overlapping sequence that includes the loxP sequence. To create the extended homology arm, overlapping PCR amplification is carried out in two steps as follows:
-
1Annealing and extension reaction to create a fused template:
25 µL 2× KAPA HiFi HotStart Ready Mix (KAPA) 4 µL 10 ng/µL gel-purified amplicon#1 4 µL 10 ng/µL gel-purified amplicon#2 17 µL Nuclease-free water
Cycle conditions:
Denature: 95°C, 5 min
Ten cycles: [98°C, 20 s; 60°C, 15 s; 72°C, 30 s]
Final extension: 72°C, 1 min
Store: 10°C
Purify the fused template using QIAquick PCR purification Kit (QIAGEN).
To create the extended homology arm ready for cloning into a pKHR vector, the fused template is further amplified using forward and reverse primers that provide appropriate restriction enzyme sequences for subsequent cloning.
-
2PCR amplification to generate extended homology arm:
25 µL 2× KAPA HiFi HotStart Ready Mix (KAPA) 7.5 µL 2 µM forward primer 7.5 µL 2 µM reverse primer 10 µL 10 ng/µL purified fused template
Cycle conditions (annealing temperature* is dependent on primer Tm):
Denature: 95°C, 5 min
Fifteen cycles: [98°C, 20 s; 66°C*, 15 s; 72°C, 30 s]
Final extension: 72°C, 1 min
Store: 10°C
Purify the amplicon using the QIAquick PCR purification Kit (QIAGEN).
4.2.3 Assembling donor sequences into pKHR vectors
As illustrated in Fig. 7, a single cloning step can be used to introduce donor sequences into the pKHR4 or pKHR7 vectors. These vectors do not themselves provide sequences to be incorporated into the genome. Hence, a single continuous assembly of donor sequences, generated from amplicons representing the left and right homology arms and a nonhomologous middle fragment is introduced into the vector. To ligate these fragments in correct orientation, each fragment should have unique protruding sequences that direct assembly in the desired order.
In contrast, the purpose of vectors pKHR5 and pKHR8 is to provide an αcrystallin- Venus reporter gene and associated recombination sites that will be incorporated into the host genome upon HR. The reporter gene can be used to identify inheritance of an edited allele, and subsequently it can be readily excised by FLP-mediated recombination (Hoshijima et al., 2016). Assembly of donor sequences in these vectors requires independent insertion of left and right homology arms so that they flank the reporter gene. As illustrated in Fig. 8, to generate a Cre-dependent, loxP-mediated conditional allele, left and right homology arm fragments are inserted sequentially into the pKHR vector.
4.2.4 Sequence verification of donor plasmids
The entire donor region of constructed plasmids should be sequenced to ensure that there have been no unintended amplification errors. Sequencing of several independent donor plasmids can indicate if there is extensive sequence polymorphism in the homology regions. If this occurs, one can identify a predominant haplotype and reselect breeder fish with the desired haplotype for subsequent donor injection.
4.3 MICROINJECTION OF PROGRAMMED NUCLEASE AND I-SceI-DIGESTED DONOR DNA
Column-purified donor plasmid DNA is further purified to remove all traces of RNase activity by phenol/chloroform extraction, chloroform extraction, and ethanol precipitation with sodium acetate. Plasmid DNA is dissolved in nuclease-free water, quantified, diluted to 500 ng/µL, and stored at −20°C. Prior to injection into zebrafish embryos, 500 ng of donor plasmid DNA is digested with I-SceI enzyme in 5 µL 1× I-SceI buffer:
| I-SceI Digestion: | |
| 0.5 µL | 10× I-SceI buffer (NEB) |
| 1 µL | 500 ng/µL donor plasmid DNA |
| 2 µL | 5 U/µL I-SceI (NEB) |
| 1.5 µL | Nuclease-free water |
| Incubate 37°C, 1 h | |
| Store on ice, 4°C |
Meanwhile prepare sgRNA and Cas9 protein mixture or TALEN mRNA solution on ice.
| sgRNA with Cas9 protein: | |
| 2 µL | 1 µg/µL sgRNA |
| 1.2 µL | 5 µg/µL Cas9 protein (PNA BIO) |
| 1 µL | 0.5% phenol red |
| 0.8 µL | Nuclease-free water |
| TALEN mRNA solution: | |
| 1 µL | 500 ng/µL left TALEN mRNA |
| 1 µL | 500 ng/µL right TALEN mRNA |
| 1 µL | 0.5% phenol red |
| 2 µL | Nuclease-free water |
After I-SceI digestion, mix 5 µL donor DNA solution with 5 µL sgRNA/Cas9 protein mixture or 5 µL TALEN mRNA solution on ice and inject 1 nL of the cocktail into the cytoplasm of just-fertilized zebrafish zygotes. About 50–70% of embryos injected with cocktail mix should develop normally. The amount of donor DNA and nuclease should be adjusted if injection leads to excessive lethality.
4.4 DETECTION OF RECOMBINATION EVENTS
HR-mediated integration of donor sequences into the target site should be observed in almost every embryo injected with nuclease and donor DNA. It is important, and reassuring, to confirm the production of correctly edited alleles in treated embryos before growing them to adulthood. At 2 dpf, genomic DNA is extracted from 8 to 12 individual donor-injected embryos by the method described in Section 4.1.4 and analyzed as illustrated in Fig. 3 by PCR amplification with a donor sequence–specific primer and a host genome–specific primer. As a control, confirm that the edited allele–specific fragment can be amplified from embryos injected with donor DNA and nuclease, but not from embryos injected only with donor DNA. We note that expression of the reporter genes present in pKHR vectors cannot be used as an indicator of integration, as these genes can be expressed in transient fashion without integration into the genome.
4.5 ISOLATION OF FOUNDER FISH THAT TRANSMIT PRECISELY EDITED GENOMES
Once it is confirmed that injected F0 embryos harbor correctly edited alleles, about 100 sibling injected F0 founders should be raised to adulthood. Analysis of F1 progeny is used to identify founders that have acquired and transmit donor plasmid-derived sequences.
4.5.1 Identification of F0 founders that transmit donor DNA sequences to progeny
In cases where acquisition of the α-crystallin-Venus reporter gene was used to indicate incorporation of donor sequences, about 100 F1 embryos should be generated from each F0 founder and analyzed at 2 dpf for expression of GFP in the lens. To expedite the screening process, we incross F0 adults. If GFP-positive embryos are identified among the progeny of an incross, then F0’s can be outcrossed with a WT animal to identify the carrier. In cases where donor plasmids carried the cmlc2:: mCherry reporter outside of the homology regions, expression of mCherry in the heart indicates imprecise integration of donor sequences. F1 embryos that express GFP but not mCherry are likely to harbor precisely edited alleles. The genomic DNA of a few of these should be characterized in detail (below) to confirm the presence of precise editing events, and then sibling embryos expressing appropriate reporter genes should be raised to adulthood.
In cases where integration of donor sequences does not lead to the expression of any visible marker, the genomic DNA of about 50 F1 embryos from each F0 founder should be analyzed for the inheritance of donor DNA. Typically, genomic DNA is prepared from 8 or more pools of 6 F1 embryos and analyzed by PCR amplification with primers that specifically detect donor sequences. Once transmission of donor DNA is detected, detailed analyses are performed to determine whether F1 individuals have inherited precisely modified alleles.
4.5.2 Identification of founder fish carrying precisely edited genomes
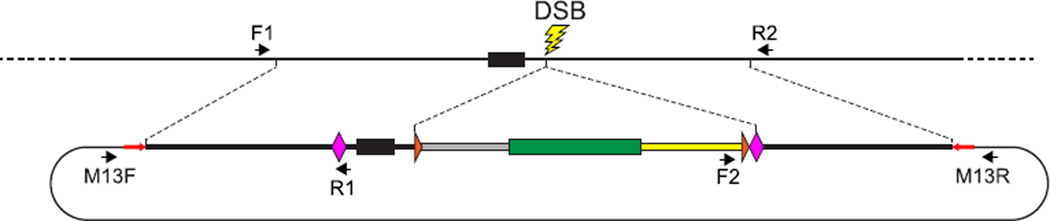
Integration of donor sequences into the host genome can arise as a consequence of: (1) precise integration via HR, (2) imprecise or partial integration of donor sequences at the target locus, or (3) nontargeted random insertion. In the case of precise target integration, novel donor sequences should be integrated as a single copy without additional indel mutations. As described in Fig. 9, preliminary analyses should be performed to detect (1) junction fragments that would be created upon precise integration (Fig. 3) and (2) donor–vector junction sequences whose presence would indicate imprecise integration events.
FIGURE 9. Diagnostic amplification to identify a precisely edited allele.
Once a candidate edited allele has been detected either by expression of the linked reporter gene or by preliminary genomic analysis, detailed PCR and sequence analysis of the targeted locus is performed to confirm that a precisely edited allele has been generated. Recombination events that correctly modify the targeted locus can be detected with primer pairs such as F1/R1 and F2/R2, which specifically amplify host–donor junction fragments and are expected to be present only in genomes harboring a precisely edited allele. In addition, to detect imprecise integration events, the edited genome is probed with primer pairs that amplify donor–vector backbone junction fragments, such as M13F/R1 and M13R/ R2. Genomes that likely harbor only precisely edited alleles are finally analyzed by amplification of the entire edited region, using the F1 and R2 primers, which are complementary to host genomic sequences and not present in the donor DNA. The size of the F1/R2 amplicon should be consistent with that expected from a simple HR-mediated gene-editing event, and the accuracy of the editing event should be verified by sequencing the entire amplicon.
The entire sequence of an edited allele should be amplified from an F1 genome as a single intact amplicon and sequenced to confirm the accuracy of the edited allele. To accomplish this analysis, genomic DNA gently extracted from a pool of candidate embryos carrying precisely edited genomes should be amplified using primers that recognize genomic sequences outside of the homology regions. The size of the amplicon should be consistent with that of the expected edited allele, confirming single-copy integration. Once a founder carrying a precisely edited genome is isolated, carriers can be easily identified by expression of the GFP reporter or by DNA analysis from fin biopsies.
CONCLUSIONS
Although the methods presented here represent an early stage in the evolution of genome editing in the zebrafish, the approach described in this Chapter already enables precise modification of the zebrafish genome. Changes ranging from single base pair substitutions to additions and/or deletions of several kilobase-long stretches of sequence can be generated efficiently. Significantly, genome editing accomplished by DSB-stimulated HR can be used to produce several closely linked genome modifications simultaneously. Thus incorporation of a linked reporter gene can be used to tag edited alleles, and the expression of the linked reporter makes it very simple to identify and recover precisely edited alleles. The ease with which genome editing can be accomplished will revolutionize use of the zebrafish allowing new types of studies of cell biological processes, development and tissue homeostasis, and disease modeling.
Acknowledgments
Work reported here utilized University of Utah Cores for sequencing, HRMA, and oligonucleotide synthesis. These studies were supported by grants to D.J.G. from the University of Utah and the National Institutes of Health (5R21HD073847, 1R21OD018323, and 1R01HD081950) and by a subaward from 5P30CA042014.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- Auer TO, Del Bene F. CRISPR/Cas9 and TALEN-mediated knock-in approaches in zebrafish. Methods. 2014;69:142–150. doi: 10.1016/j.ymeth.2014.03.027. [DOI] [PubMed] [Google Scholar]
- Auer TO, Duroure K, Concordet JP, Del Bene F. CRISPR/Cas9-mediated conversion of eGFP- into Gal4-transgenic lines in zebrafish. Nature Protocols. 2014;9:2823–2840. doi: 10.1038/nprot.2014.187. [DOI] [PubMed] [Google Scholar]
- Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Research. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington B, Varshney GK, Burgess SM, Sood R. CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Research. 2015;43:e157. doi: 10.1093/nar/gkv802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with targetable nucleases. Annual Review of Biochemistry. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Research. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genetics. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Schier AF. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–132. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14896–14901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano Y, Sakuma T, Nakade S, Ohga R, Ota S, Okamoto H, Kawahara A. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Scientific Reports. 2015;5:8841. doi: 10.1038/srep08841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima K, Jurynec MJ, Grunwald DJ. Precise editing of the zebrafish genome made simple and efficient. Developmental Cell. 2016;36:654–667. doi: 10.1016/j.devcel.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature Biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U, Krauss J, Nusslein-Volhard C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development. 2014;141:4827–4830. doi: 10.1242/dev.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurynec MJ, Xia R, Mackrill JJ, Gunther D, Crawford T, Flanigan KM, Grunwald DJ. Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12485–12490. doi: 10.1073/pnas.0806015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nature Reviews Genetics. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Scientific Reports. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon YG, Mullins MC. Maternal and zygotic control of zebrafish dorsoventral axial patterning. Annual Review of Genetics. 2011;45:357–377. doi: 10.1146/annurev-genet-110410-132517. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Wolfe SA. Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Developmental Cell. 2011;21:48–64. doi: 10.1016/j.devcel.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang BB, Ren YG, Gu SY, Xiang YH, Du JL. Intron targeting-mediated and endogenous gene integrity-maintaining knockin in zebrafish using the CRISPR/Cas9 system. Cell Research. 2015;25:634–637. doi: 10.1038/cr.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR. Genetics. Genome mosaicisme–one human, multiple genomes. Science. 2013;341:358–359. doi: 10.1126/science.1239503. [DOI] [PubMed] [Google Scholar]
- Perrin A, Buckle M, Dujon B. Asymmetrical recognition and activity of the I-SceI endonuclease on its site and on intron-exon junctions. EMBO Journal. 1993;12:2939–2947. doi: 10.1002/j.1460-2075.1993.tb05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JB, Westerfield M. Zebrafish models in translational research: tipping the scales toward advancements in human health. Disease Models & Mechanisms. 2014;7:739–743. doi: 10.1242/dmm.015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Shandilya H, D’Alessio JM, O’Connor K, Durocher J, Gerard GF. Mutation detection using surveyor nuclease. Biotechniques. 2004;36:702–707. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nature Biotechnology. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Chen J, Solnica-Krezel L. Efficient homologous recombination-mediated genome engineering in zebrafish using TALE nucleases. Development. 2014;141:3807–3818. doi: 10.1242/dev.108019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart GW, McMurray JV, Westerfield M. Replication, integration and stable germ-line transmission of foreign sequences injected into early zebrafish embryos. Development. 1988;103:403–412. doi: 10.1242/dev.103.2.403. [DOI] [PubMed] [Google Scholar]
- Udvadia AJ, Linney E. Windows into development: historic, current, and future perspectives on transgenic zebrafish. Developmental Biology. 2003;256:1–17. doi: 10.1016/s0012-1606(02)00083-0. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Lin S. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nature Methods. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]