Significance
S-nitrosylation, addition of an NO group to a cysteine thiol, can regulate protein activity. Aberrant protein S-nitrosylation, however, can disrupt normal enzyme function, as is the case for S-nitrosylated peroxiredoxin (SNO-Prx), which would otherwise catabolize toxic peroxides that occur under neurodegenerative conditions such as Parkinson’s disease. Here, we describe a paradigm of N-phosphorylation–mediated denitrosylation by the enzyme sulfiredoxin that removes NO from Prx. The findings are at the center of redox control of the cell, explaining reactivation by sulfiredoxin of both Prx-SO2H and SNO-Prx and thus describe a master regulator of redox reactions that combats nitrosative and oxidative stress in cells. These results suggest that sulfiredoxin may be an important target for therapeutic intervention in neurodegenerative disorders.
Keywords: sulfiredoxin, peroxiredoxin, denitrosylase, iPSC-derived dopaminergic neuron, S-nitrosylation
Abstract
Recent studies have pointed to protein S-nitrosylation as a critical regulator of cellular redox homeostasis. For example, S-nitrosylation of peroxiredoxin-2 (Prx2), a peroxidase widely expressed in mammalian neurons, inhibits both enzymatic activity and protective function against oxidative stress. Here, using in vitro and in vivo approaches, we identify a role and reaction mechanism of the reductase sulfiredoxin (Srxn1) as an enzyme that denitrosylates (thus removing -SNO) from Prx2 in an ATP-dependent manner. Accordingly, by decreasing S-nitrosylated Prx2 (SNO-Prx2), overexpression of Srxn1 protects dopaminergic neural cells and human-induced pluripotent stem cell (hiPSC)-derived neurons from NO-induced hypersensitivity to oxidative stress. The pathophysiological relevance of this observation is suggested by our finding that SNO-Prx2 is dramatically increased in murine and human Parkinson’s disease (PD) brains. Our findings therefore suggest that Srxn1 may represent a therapeutic target for neurodegenerative disorders such as PD that involve nitrosative/oxidative stress.
Peroxiredoxins (Prxs) belong to an abundant and broadly expressed family of antioxidant enzymes. Prxs catalyze the reduction of H2O2 and thus contribute to intracellular signaling and protection from cytotoxicity. Prx activities are regulated by dimerization, phosphorylation, and redox modification. Among the six Prx isoforms encoded by mammalian genomes, Prx2 is the most abundant in neurons. The various Prx isoforms are classified into two types: 1-Cys and 2-Cys Prxs. Both classes of Prxs contain a conserved cysteine residue that can be peroxidated (thus called the peroxidatic cysteine). 2-Cys Prxs have an additional cysteine, termed the “resolving cysteine residue,” that can react with the sulfenylated peroxidatic cysteine to form a disulfide bond. This disulfide bond can be reduced by a dithiol oxidoreductase. 1-Cys Prxs do not contain a resolving cysteine and therefore require another external electron donor for the reduction of their sulfenic acid intermediate. Prx2 belongs to the 2-Cys Prx class of Prxs.
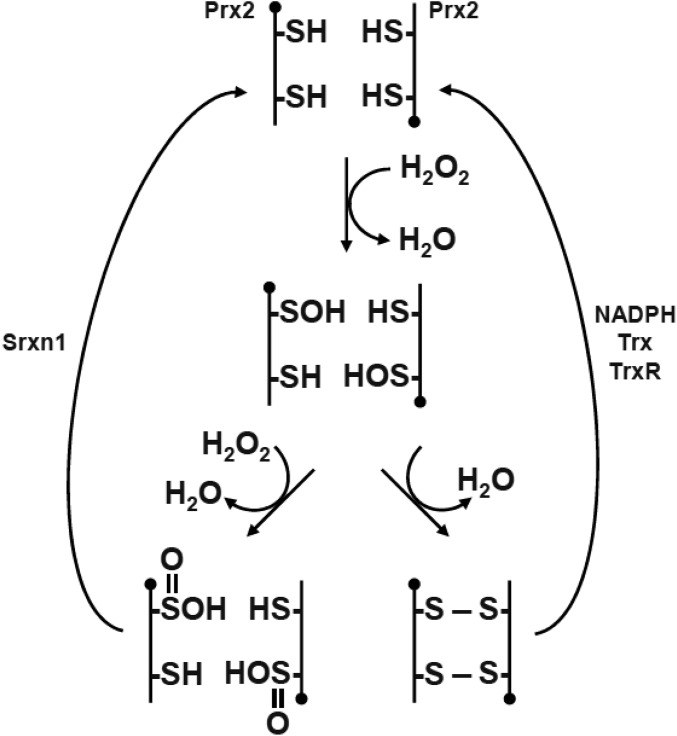
Additionally, the sulfur atom in the side-chain of the peroxidatic cysteine of Prx2 can exist in several different oxidation states (Fig. S1). Peroxides can oxidize the thiol group of the cysteine (Cys-SH) to sulfenic acid (Cys-SOH), which may then react with the resolving Cys-SH to form a disulfide bond (Cys-S-S-Cys). During more severe oxidative stress, however, the sulfenic intermediate can be further oxidized to sulfinic acid (Cys-SO2H) or sulfonic acid derivatives (Cys-SO3H). Oxidization of the cysteine residues of Prx to sulfinic or sulfonic acid leads to the inactivation of the peroxidase activity and an increase in local peroxide levels (1, 2). Importantly, therefore, the cysteine modifications of Prx2 not only reflect the intracellular redox status, but also influence this status. Based on these phenomena, the idea has emerged that the cycle of thiol to sulfinic acid of Prx2 acts as a redox switch that mediates transient cell signaling and cytoprotection (3).
Fig. S1.
Catalytic cycle of Prx2. The peroxidatic cysteine thiol (-SH) of active Prx2 can reduce H2O2 to form a sulfenic acid (-SOH). In turn, this adduct either can be reduced by another active Prx2 to form an intermolecular disulfide bond (S—S) or can be overoxidized to produce sulfinic acid (-SO2H). Both the disulfide bond and overoxidized form can be regenerated to their active states by coordinated action of the thioredoxin (Trx) pathway [NADPH, thioredoxin, and thioredoxin reductase (TrxR)] or by the reductase Srxn1.
Sulfiredoxin (Srxn1) belongs to a family of enzymes involved in the maintenance of cellular redox balance. Srxn1 contains a single conserved cysteine residue, and its expression is induced by hydrogen peroxide (4). One known function of Srxn1 is ATP-dependent reduction of cysteine sulfinic (but not sulfonic) acid derivatives of 2-Cys Prx proteins back to sulfenic acid (4), which leads to reactivation of Prx peroxidase activity (Fig. S1).
Nitric oxide (NO) is a signaling molecule that regulates diverse processes in cells. Protein S-nitrosylation, in which a –NO group is covalently attached to a cysteine thiol or thiolate anion (5–7), constitutes a reversible posttranslational modification that regulates the function of numerous enzymes and transcription factors (8, 9). Previously, we showed that the peroxidase activity of Prx2 can be regulated in vitro and in vivo by S-nitrosylation of its redox-active cysteine residues, forming SNO-Prx2 protein (10). This S-nitrosylation of Prx2, probably representing an aberrant reaction that occurs only in the face of excessive/pathological levels of NO, inhibits the protective function of Prx2 against oxidative stress-mediated neuronal cell death (10). In fact, in Parkinson’s disease (PD), S-nitrosylation of Prx2 has been shown to promote such damage in dopaminergic neurons, the cell type most vulnerable in the disorder (10). Additionally, S-nitrosylation of Prx1, another 2-Cys Prx protein, has been reported to impede its catalytic cycle by directly inhibiting its peroxidase activity and interfering with the recycling of oxidized Prx1 by the thioredoxin system (11). Here, we report that Srxn1 can convert the Cys-SNO of Prx2 to Cys-SH, thus reactivating the enzyme. We further show that this Srxn1-mediated denitrosylation of Prx2 protects dopaminergic neural cells from oxidative stress. Hence, Srxn1 represents a potential drug target for neurodegenerative diseases caused at least in part by oxidative and nitrosative stress.
Results
Denitrosylation of Prx2 in Vitro in the Presence of Srxn1.
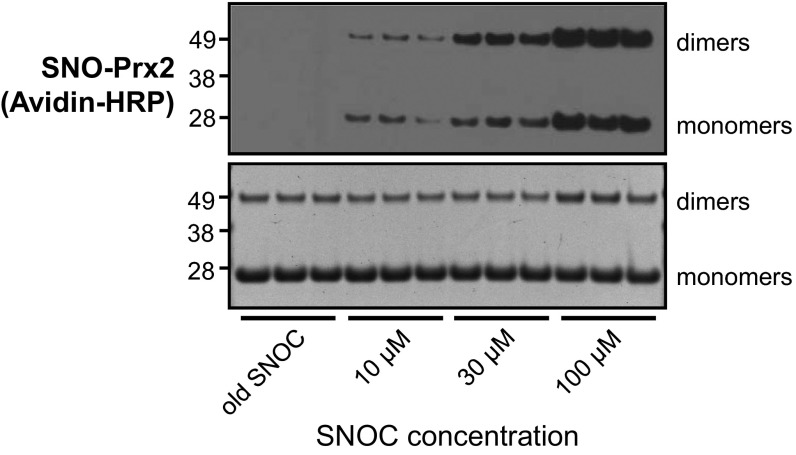
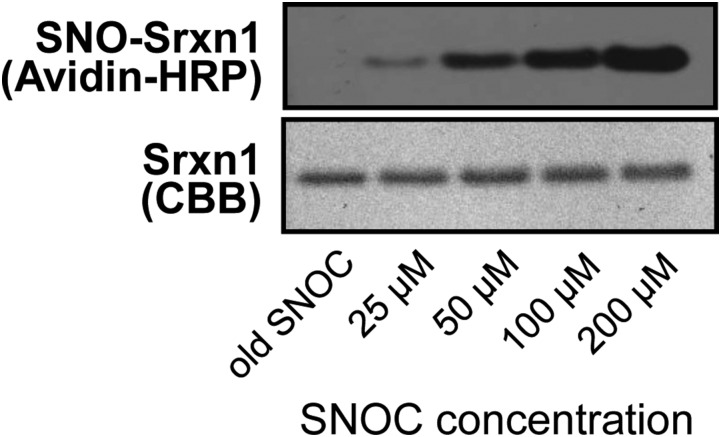
Initially, we tested the ability of recombinant Prx2 and Srxn1 to be S-nitrosylated in vitro by the naturally occurring NO donor, S-nitrosocysteine (SNOC). After exposure to SNOC, recombinant Prx2 and Srxn1 were S-nitrosylated, as detected by a modified biotin-switch assay (6, 8, 12) (Figs. S2 and S3). Coomassie staining revealed an increase in Prx dimers relative to monomers after exposure to 100 μM SNOC, as previously described (13).
Fig. S2.
In vitro S-nitrosylation of recombinant Prx2. (Top) Representative biotin-switch assay showing S-nitrosylation of recombinant Prx2 in the presence of the indicated concentration of SNOC or old SNOC as a negative control. Electrophoresis was performed under nonreducing conditions, and biotinylated proteins were labeled with Avidin-HRP. (Bottom) Corresponding blot stained with Coomassie Brilliant Blue as input control.
Fig. S3.
In vitro S-nitrosylation of recombinant Srxn1. (Top) Representative biotin-switch blot showing S-nitrosylation of recombinant Srxn1 in the presence of increasing concentrations of SNOC or old SNOC as a negative control. Electrophoresis was performed under nonreducing conditions, and biotinylated proteins were labeled with Avidin-HRP. (Bottom) Corresponding blot stained with Coomassie Brilliant Blue as input control.
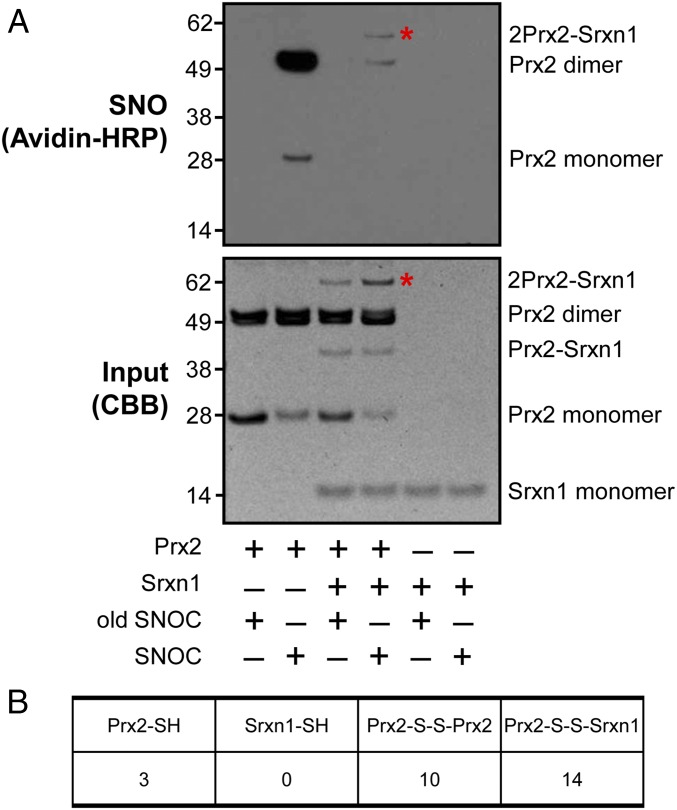
Next, we asked whether Srxn1 could denitrosylate SNO-Prx2 in vitro (Fig. 1A). Again, exposure of recombinant Prx2 to 100 μM SNOC in the absence of Srxn1 led to formation of monomeric and dimeric SNO-Prx2. We then added Srxn1 to the SNO-Prx2 after 1 h. Importantly, when we measured the time course of SNOC action (Fig. S4), we observed that NO had completely dissipated within minutes; thus, no reactive SNOC remained in our reaction mixture when Srxn1 was added to SNO-Prx2. Addition of Srxn1 to the reaction resulted in greatly diminished S-nitrosylation of both monomeric and dimeric Prx2 (Fig. 1 A and B). We also monitored the kinetics of SNO-Prx2 denitrosylation after addition of Srxn1. For this experiment, we incubated Srxn1 with the SNO-Prx2 for varying lengths of time and then performed a biotin-switch assay. We found that Srxn1 completely denitrosylated SNO-Prx2 within 60 min (Fig. S5). Interestingly, however, in these experiments we detected a high-molecular-weight band that was S-nitrosylated (Fig. 1 A and B). By molecular weight, this band appeared to contain two molecules of Prx2 and one of Srxn1.
Fig. 1.
In vitro denitrosylation of Prx2 by Srxn1. (A, Top) Representative biotin-switch blot showing denitrosylation of recombinant Prx2 by recombinant Srxn1 in the presence of 100 μM freshly prepared SNOC or old SNOC (from which NO had been dissipated) as a negative control. Asterisks indicate bands compared in B. (Bottom) Corresponding blot stained with Coomassie Brilliant Blue (CBB) as input. (B) MS analysis to compare the number of free cysteine residues (–SH) and disulfide bonds (–S–S–) between molecules in the bands in A denoted by asterisks. The analysis represents counts of peptides containing these structures as determined by MS (Materials and Methods).
Fig. S4.
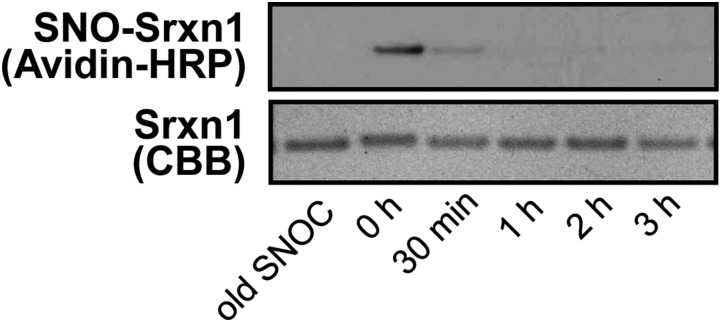
Time course of SNOC action on recombinant Srxn1. (Top) Representative biotin-switch blot showing S-nitrosylation of recombinant Srxn1 in the presence of 100 μM SNOC or old SNOC as a negative control. SNOC was added first, and then recombinant Srxn1 was added to the reaction at various time points, as indicated. Biotinylated proteins were labeled with Avidin-HRP. Because NO is dissipated from SNOC within minutes, substantial SNO-Srxn1 is formed only after exposure to fresh SNOC. (Bottom) Corresponding blot stained with Coomassie Brilliant Blue as input control.
Fig. S5.
Time course of Srxn1 action on SNO-Prx2 mixture. (Top) Representative biotin-switch blot showing denitrosylation of SNO-Prx2 in the presence of Srxn1 incubated for the indicated lengths of time. In this experiment, initially SNOC was added to Prx2 to form SNO-Prx2. Subsequently, after NO had been dissipated from any remaining free SNOC, recombinant Srxn1 was then added to the reaction mixture at various time points, as indicated. (Bottom) Corresponding blot stained with Coomassie Brilliant Blue to show input.
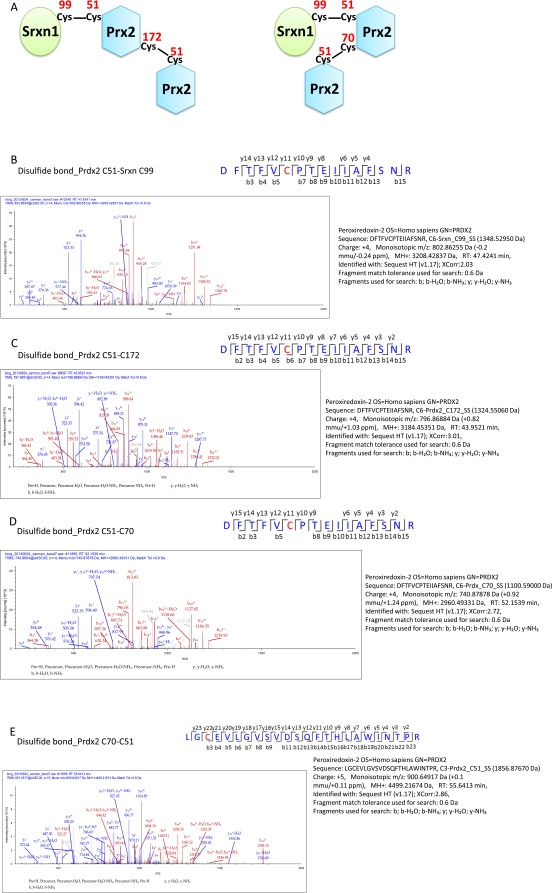
To determine if Prx or Srxn1 was S-nitrosylated in this high-molecular-weight band, we analyzed the band using mass spectrometry (MS). We detected disulfide-bond linkages between Srxn1 and Prx2 as well as between two Prx2 molecules in the band (Fig. 1B). Specifically, we set the delta masses 1348.5295, 1856.8767, 1100.59, and 1324.5506 as variable modifications for cysteines, corresponding to linkages involving the peptide DYFYSFGGCHR of Srxn1 protein as well as peptides DFTFVCPTEIIAFSNR, KLGCEVLGVSV, and DEHGEVCPAGWK of Prx2 protein. These data indicate that disulfide bonds were formed between Cys99 of Srxn1 and Cys51 of Prx2, as well as between Cys51-Cys70 and Cys51-Cys172 of Prx2 (Fig. S6). These data support the presence of a 2Prx2-Srxn1 triple-protein complex. Cys99 is the only cysteine residue in Srxn1, and because we did not detect free cysteines in Srxn1 (see Srxn1-SH in Fig. 1B), our observations are consistent with the notion that Cys99 was used to form a disulfide bond with Prx2.
Fig. S6.
Analysis of 2Prx2-Srxn1 trimer band by mass spectrometry. (A) Schematic of the apparent disulfide linkages in the 2Prx2-Srxn1 band from the mass spec data. (B–E) MS/MS spectra identifying disulfide bonds between Prx2(Cys51)–Srxn1(Cys99), Prx2(Cys51)–Prx2(Cys172), Prx2(Cys51)–Prx2(Cys70), and Prx2(Cys70)–Prx2(Cys51).
In contrast, we found free cysteine residues in Prx2 (i.e., cysteines that were not already involved in disulfide bonds or posttranslational modifications; see Prx2-SH in Fig. 1B). This finding indicates the availability of free thiol groups on Prx2 that could be S-nitrosylated in the large-molecular-weight band. Importantly, under the conditions of this MS experiment, S-nitrosylated thiols would not be observed; rather, free cysteines would be seen, as was the case for Prx2 but not for Srxn1.
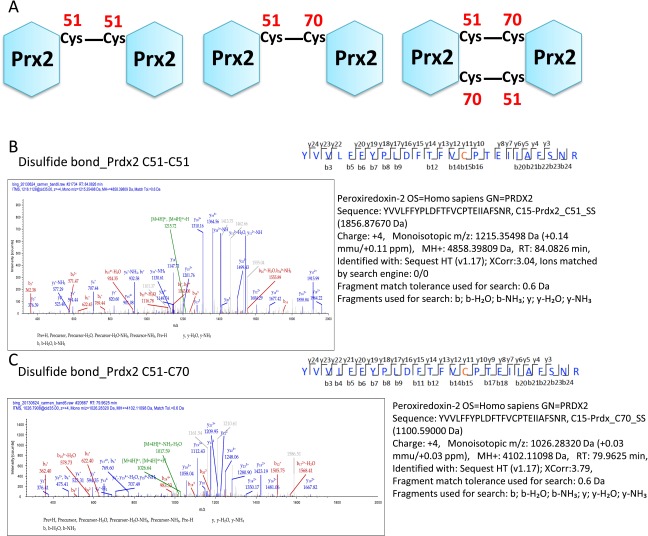
Note also that we observed a double band for Prx2 dimers in Fig. 1A. We therefore analyzed these bands by MS and found that two disulfide bonds, Cys51-Cys51 and Cys51-Cys70, could be formed between two Prx2 molecules (Fig. S7). The appearance of double bands for Prx dimers in nonreducing gel electrophoresis experiments has previously been observed for Prx proteins (14–16). The two bands have been attributed to the simultaneous presence of dimers linked by one (upper band) or two (lower band) disulfide bonds in dimeric Prx proteins (14).
Fig. S7.
Analysis of Prx2-Prx2 dimer band by mass spectrometry. (A) Schematic of the apparent disulfide linkages in the Prx2-Prx2 double band. (B and C) MS/MS spectra identifying the disulfide bonds between Prx2(Cys51)–Prx2(Cys51) and Prx2(Cys51)-Prx2(Cys70).
Dependence of Srxn1-Mediated Prx2 Denitrosylation and Disulfide Formation on Srxn1(Cys99).
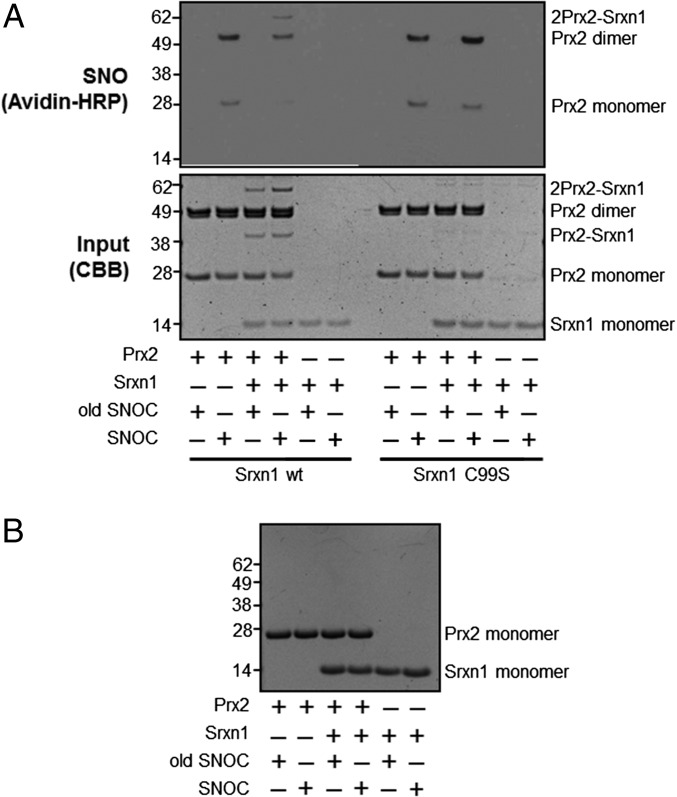
Human Srxn1 has a single Cys residue at position 99 that is known to mediate a transient disulfide linkage that is required for its reductase activity (17). To test whether the Srxn1–Prx2 interaction also involves Srxn1(Cys99), we constructed a mutant human Srxn1 in which Cys99 was replaced by Ser (C99S). Coomassie staining of electrophoresis gels of a mixture of Prx2 and Srxn1 revealed the presence of a complex of appropriate molecular weight for Prx2-Srxn1 (Fig. 2A). In contrast, Srxn1(C99S) mutation abrogated formation of these heterodimers (Fig. 2A), consistent with the notion that Cys99 is involved in the linkage between Prx2 and Srxn1. Additionally, unlike wild-type Srxn1, the Srxn1(C99S) mutant had no effect on SNO-Prx2, suggesting that Srxn1(C99) is necessary for its denitrosylase activity.
Fig. 2.
Denitrosylation of Prx2 by Srxn1 is dependent on Srxn1(Cys99), which is necessary for disulfide bond formation. (A, Top) Representative biotin-switch blot showing denitrosylation of recombinant Prx2 by wild-type (wt) Srxn1 or mutant Srxn1(C99S) in the presence of fresh or old SNOC. (Bottom) Corresponding blot stained with Coomassie Brilliant Blue as input control. (B) Protein gel run with reducing sample buffer containing mercaptoethanol, stained with Coomassie Brilliant Blue.
We also examined the nature of the interaction between Prx2 and Srxn1 by examining heteromer formation under reducing and nonreducing conditions. Under nonreducing conditions, Prx2-Srxn1 heterodimers were present (Fig. 2A), whereas in the presence of the reducing agent β-mercaptoethanol, heterodimers were not detected (Fig. 2B). Collectively, these data are consistent with the notion that Srxn1(Cys99) forms a disulfide bond with Prx2 during the process of Prx2 denitrosylation.
Dependence of Srxn1-Mediated Prx2 Denitrosylation on ATP.
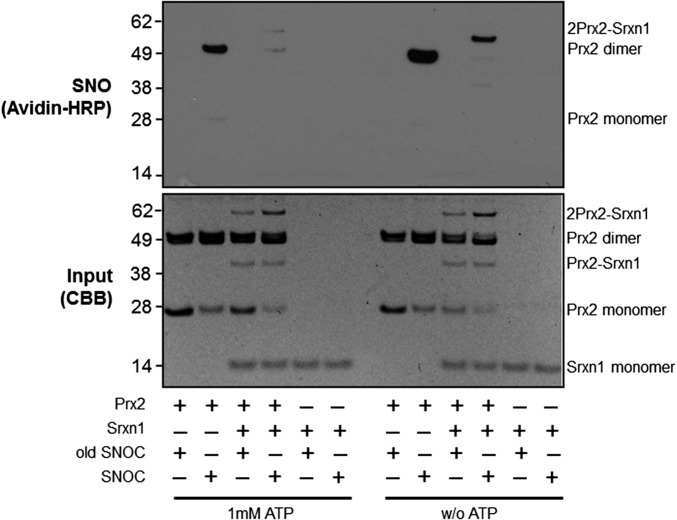
Because the reductase activity of Srxn1 requires ATP hydrolysis (17), we asked whether its denitrosylase activity might also be ATP-dependent. To this end, we incubated SNO-Prx2 with Srxn1 with or without ATP and found that denitrosylation of Prx2 did not occur in the absence of ATP (Fig. 3). Moreover, when Srxn1 was added to SNO-Prx2 in the absence of ATP, SNO-Prx2 remained predominantly in the high-molecular-weight heteromultimeric complex with Srxn1. This finding is consistent with the notion that the SNO-Prx2–Srxn1 complex represents an intermediate for Prx2 denitrosylation by Srxn1.
Fig. 3.
Denitrosylation of Prx2 by Srxn1 is enhanced in the presence of ATP. (Top) Biotin-switch blot in which the denitrosylation of recombinant Prx2 by Srxn1 was compared in the presence and absence of 1 mM ATP. (Bottom) Input control protein blot stained with Coomassie Brilliant Blue.
Transfection with Srxn1 Protects Against H2O2-Induced Cytotoxicity.
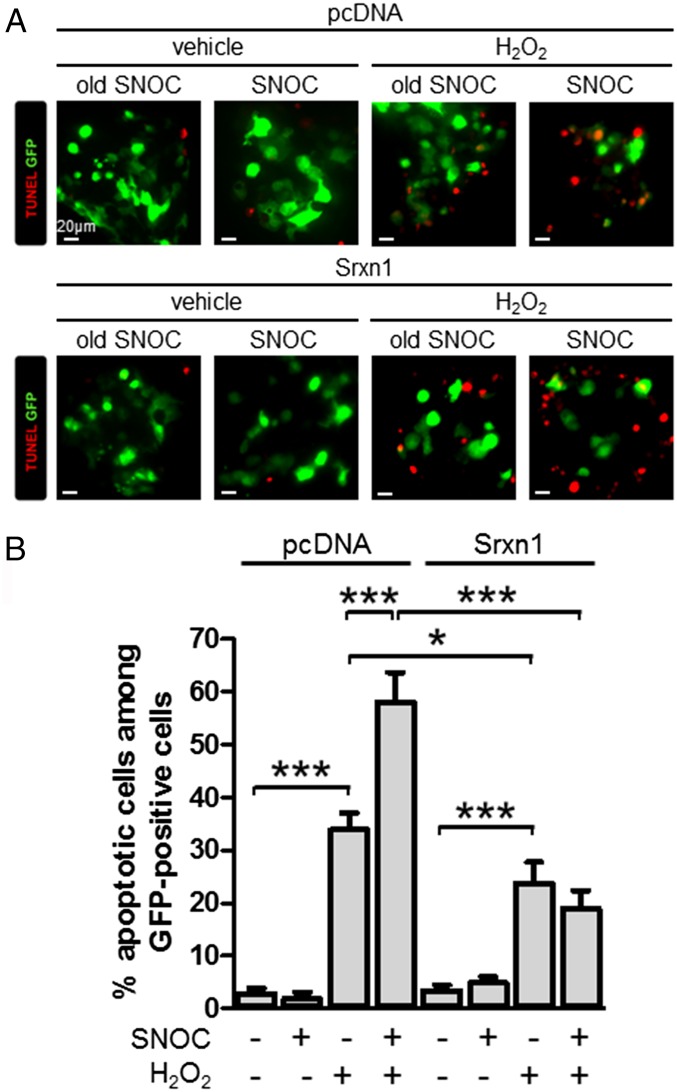
We previously reported that S-nitrosylation of Prx2 decreases its antioxidant and protective activity against H2O2-induced cytotoxicity (10). Empirically, exposing the dopaminergic SH-SY5Y neural cell line to SNOC before H2O2 challenge inhibited the protective activity of Prx2 in these cells. To determine whether Srxn1 activity can alleviate this NO-induced inhibition of Prx2 against oxidative stress, we transiently transfected Srxn1 and evaluated cell survival following preincubation with SNOC and subsequent H2O2 exposure. In agreement with our prior findings (10), SNOC exposure increased cell susceptibility to H2O2-induced cell death, whereas overexpression of Srxn1 counteracted the effect of SNOC (Fig. 4). These data are consistent with the notion that Srxn1 can denitrosylate Prx2 and thus restore its antioxidant activity.
Fig. 4.
Srxn1 transfection can protect against NO-induced hypersensitivity to oxidative stress. (A) Representative deconvolution images of SH-SY5Y cell cultures transfected with Srxn1 or control plasmids coexpressing GFP (green). Cells were exposed to 200 μM SNOC or control media; 30 min later 600 μM H2O2 or control media was substituted. After 24 h, the cultures were assessed for apoptosis with terminal-deoxynucleotidyl–TUNEL (red). (Scale bars, 20 μm.) (B) Quantitative analysis of TUNEL-positive transfected cells (n = 130–622 cells per group). Values are mean + SEM; *P < 0.05, ***P < 0.001.
S-Nitrosylation of Prx2 in Murine and Human Models of Sporadic PD.
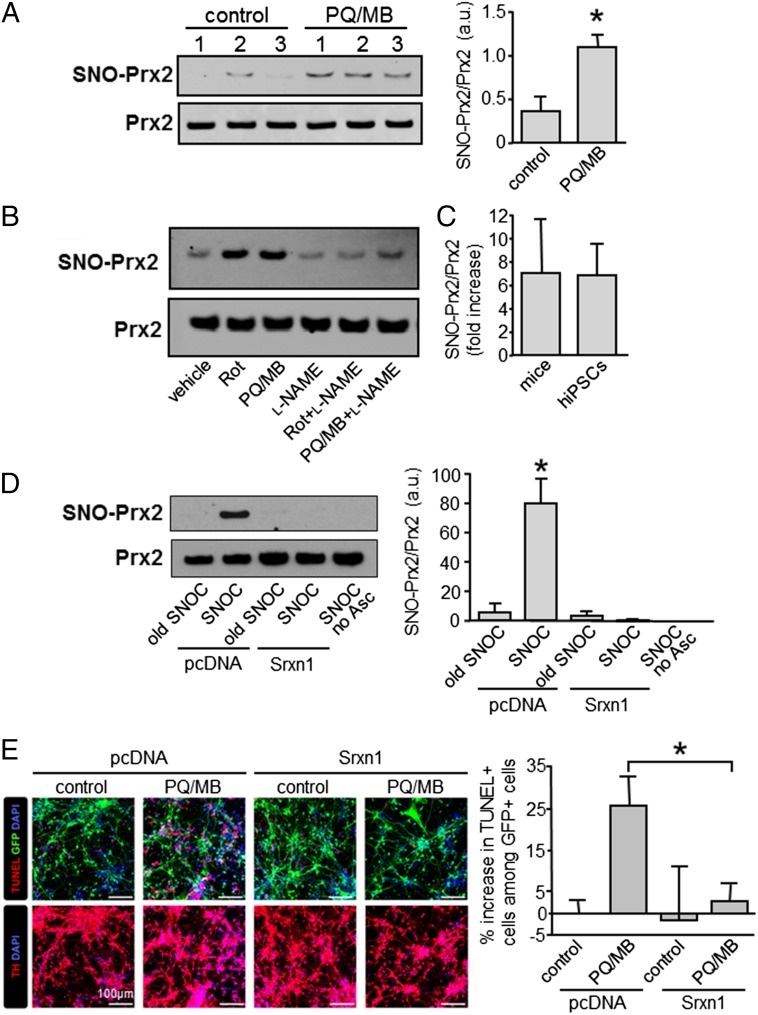
Because nitrosative stress is thought to contribute to a number of neurodegenerative disorders, including PD (8, 9, 12, 18), we asked whether SNO-Prx2 might contribute to the pathogenesis of these disorders. We first asked whether Prx2 is S-nitrosylated in vivo in a mouse model of PD in which selective loss of dopaminergic neurons is induced by exposure to the pesticides paraquat (PQ) and maneb (MB), representing mitochondrial toxins that have been linked epidemiologically to human PD (19–24). In whole-brain lysates of PQ/MB-exposed mice compared with control brains, we found a significant increase in S-nitrosylated Prx2, represented by the ratio of SNO-Prx2 (from the biotin-switch assay) to total Prx2 (from immunoblots) (Fig. 5A).
Fig. 5.
S-nitrosylation of Prx2 in mouse and human PD models. (A, Left) Representative blots showing S-nitrosylation of Prx2 in brain lysates from control wild-type and mice exposed to 28 μM PQ and 5 μM MB. (Right) Quantitative analysis of the blots. Levels of S-nitrosylated (SNO-)Prx2 were normalized to total Prx2 (n = 3 brains per group). Values are mean + SEM; *P < 0.05. (B) Representative biotin-switch blot of S-nitrosylated Prx2 in hiPSC-derived DA neurons exposed to 200 nM Rotenone (Rot) or 28 μM PQ/5 μM MB, alone or combined with 1 mM l-NAME. (C) Ratio of increased SNO-Prx2/total Prx2 in mice and hiPSC-derived DA neurons exposed to PQ/MB (n = 3 blots). (D, Left) Representative biotin-switch assay (one of three such blots) showing that electroporation with Srxn1 decreases SNO-Prx2 in hiPSC-derived DA neurons after exposure to 200 µM SNOC. As controls, control plasmid (pcDNA) was transfected, old SNOC was used instead of freshly prepared SNOC, or ascorbate was omitted during biotin-switch assay. (Right) Quantitative analysis of the blots. Levels of SNO-Prx2 were normalized to total Prx2 (n = 3 independent experiments per group). Values are mean + SEM; *P < 0.01. (E, Left) Representative confocal images of hiPSC-derived DA neurons electroporated with Srxn1 or control plasmids coexpressing GFP (green). Cells were exposed to 28 μM PQ and 1 μM MB for 24 h, fixed, and stained for TUNEL (red, Top panels), tyrosine hydroxylase (TH; red, Bottom panels), and DAPI (blue). (Scale bars, 100 μm.) (Right) Quantitative analysis of the increase in TUNEL-positive transfected cells (n = 3 independent samples with an average of 266 cells scored per group in each experiment). Values are mean + SEM; *P < 0.05.
We next examined whether SNO-Prx2 is increased in PD models in a human context using patient-derived human-induced pluripotent stem cells (hiPSCs). PD initially affects A9-type dopaminergic (DA) neurons of the substantia nigra pars compacta. Hence, we differentiated hiPSCs into A9 DA neurons with high efficiency (>80% of total neurons), as previously described (25, 26). Then, we exposed these DA human neurons to PD-associated pesticides (PQ/MB or rotenone) (22–24). By biotin-switch assay, we found a significant increase in S-nitrosylated Prx2 after exposure to either rotenone or PQ/MB (Fig. 5B). Addition of the broad spectrum NO synthase (NOS) inhibitor Nω-nitro-l-arginine methyl ester (l-NAME) abrogated this increase in SNO-Prx2 (Fig. 5B). Taken together, these results show that SNO-Prx2 is increased in models of both murine and human PD. Moreover, the relative ratio (12) of SNO-Prx2/total Prx2 in these PD models was similar to the ratio that we previously reported in human PD brain (10), indicating that pathophysiologically relevant amounts of SNO-Prx2 are generated in these model systems (Fig. 5C).
Next, to demonstrate causality, we electroporated hiPSC-derived A9 DA neurons with an expression vector encoding Srxn1 or empty vector cDNA and then exposed these cells to the NO donor SNOC. By biotin-switch assay, we found that transfection with Srxn1 abolished S-nitrosylation of Prx2 in these cells (Fig. 5D), consistent with the denitrosylase activity of Srxn1 toward SNO-Prx2.
Finally, to determine whether Srxn1 can alleviate neurotoxic nitrosative stress induced by mitochondrial toxins/pesticides, we evaluated the survival of hiPSC-derived A9 DA neurons after transfection with Srxn1. We found that overexpression of Srxn1 significantly decreased the number of apoptotic neurons after PQ/MB challenge (Fig. 5E). Taken together with our prior observation that S-nitrosylation of Prx2 contributes to neuronal cell death (10), our current data show that Srxn1 rescues human neurons from nitrosative stress by denitrosylating Prx2.
Discussion
Prx2 is an important antioxidant enzyme that limits accumulation of intracellular peroxides in brain cells (26, 27). In a recent study, Prx2 up-regulation by metformin treatment was linked to increased life span in Caenorhabditis elegans (24). Our group had previously shown that S-nitrosylation of Prx2 inhibits its peroxidase activity (10), thus contributing to oxidative stress-induced neuronal cell death in PD models. Cross talk between reactive oxygen and nitrogen (ROS/RNS) metabolism can fundamentally affect various cellular functions, yet molecular insight into mechanisms of this cross talk remain unclear. Our discovery of a role of the reductase Srxn1 as a denitrosylase enzyme for Prx2 advances our understanding in this regard.
Other enzymes have been shown to manifest dual activity as both an l-cystine reductase and a S-denitrosylase. For example, in addition to having broad-spectrum protein disulfide reductase activity, thioredoxin 1 is also a denitrosylase for a large range of nitrosoproteins (28, 29). Additionally, the thioredoxin-related protein of 14 kDa (TRP14) has recently been reported to mediate reduction as well as denitrosylation of specific substrates (30). The enzyme GSNO reductase has also been reported to mediate protein denitrosylation (31, 32), as have carbonyl reductase 1 and protein disulfide isomerase (33). However, the mechanism for Srxn1 reductase/denitrosylase function displays a unique chemical reaction scheme.
Accordingly, the reductase activity of human Srxn1 involves a conserved active-site Cys99 and ATP hydrolysis (34–36). Following the reductase reaction mechanism proposed by Jönsson and colleagues, Cys99 mediates ATP binding and plays a key role in establishing the Srxn1–Prx2 macromolecular interface (34). Interestingly, our findings indicate that the denitrosylase activity of Srxn1 also requires the Cys residue at position 99. Specifically, we found that Srxn1 Cys99 is necessary for binding to Prx2 via disulfide bond formation. Consistent with this observation are reports that the reductive mechanism of 2-Cys Prx2 by Srxn1 also requires this disulfide between both proteins (37, 38). Furthermore, we found that, in the absence of ATP, although the disulfide bond formed between 2-Cys Prx2 and Srxn1, Prx2 denitrosylation did not occur. Hence, the SNO–Prx2/Srxn1 complex appears to represent an intermediate in the denitrosylase reaction catalyzed by Srxn1. Collectively, our data indicate that during Srxn1-mediated denitrosylation of Prx2, the two proteins physically bind via disulfide bond formation, providing a structural basis for the enzymatic reaction that requires ATP hydrolysis.
Prx2 activity is critical for the detoxification of potentially toxic peroxides. Hydrogen peroxide is a signaling molecule that becomes toxic if present in excessive concentration. For example, exposure of SH-SY5Y neural cells to a toxic concentration of H2O2 stimulates apoptotic pathways. We have previously shown that S-nitrosylation of Prx2 enhances this H2O2-mediated neuronal death due to inhibition of Prx2 peroxidase activity (10). Our current data demonstrate that Srxn1 can rescue neurons from this NO-induced increase in vulnerability to H2O2. Our findings are consistent with the notion that Srxn1 acts as a denitrosylase to reactivate the peroxidase activity of Prx2.
Importantly, we had also previously reported that SNO-Prx2 levels are dramatically increased in human PD brains, suggesting the relevance of this finding to the biological process of Prx2 peroxidase activity under pathological conditions (10). In the current study, we observed a similar increase in SNO–Prx2 formation in both human iPSC-derived neurons and in vivo mouse models of PD. Furthermore, Srxn1 levels were concomitantly up-regulated in the mouse model, suggesting a compensatory mechanism for Prx2 inactivation caused by oxidative/nitrosative stressors.
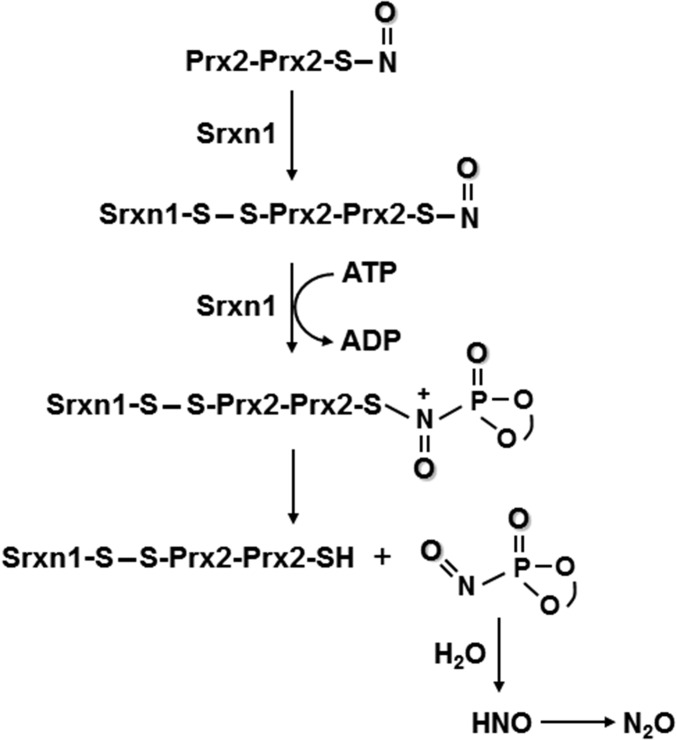
Based on our data, we propose a reaction scheme to explain how the active site Cys99 of Srxn1 catalyzes the denitrosylation of Prx2 using ATP (Fig. 6). Biochemical, MS, and structural analyses support a mechanism whereby thiol groups of Prx2 and Srxn1 form a disulfide bond (38, 39). Then, an ATP molecule, bound tightly to the nucleotide-binding motif of Srxn1, is brought into close proximity to the S-nitrosothiol of Prx2 (35, 37). The direct attack of the ATP phosphate group on Cys-SNO generates a P-nitrosophosphine oxide, a very unstable subproduct of the reaction containing both the phosphate and NO groups (40). Finally, this P-nitrosophosphine oxide hydrolyzes to produce nitroxyl (40), which quickly dimerizes, generating nitrous oxide (N2O). In general, strong nucleophiles will attack the N in S-nitrosothiol compounds. That is the basis for triarylphosphine chemistry and RS-induced transnitrosation, as we and others have previously described. Also, this N-phosphorylation is chemically logical, taking into account that nitrogen is a better nucleophile than oxygen. Note that prior descriptions of the ATP-dependent mechanism for reduction of sulfinic acid to sulfenic acid was proposed to go through oxygen, but this occurred only because oxygen (and not nitrogen) was available to be phosphorylated in those paradigms (17). The novel denitrosylation reaction mechanism of N-phosphorylation described here is therefore more robust. Most importantly, this discovery fits right in the center of redox control of the cell. Because we demonstrate that Srxn1 I reactivates both SO2H-Prx2 and SNO-Prx2, it is a master regulator of the cell redox state.
Fig. 6.
Schematic diagram of proposed mechanism of Srxn1-catalyzed denitrosylation of dimeric Prx2. See text for explanation.
In conclusion, the influence of RNS/ROS on signal transduction and other cellular activities has been shown to be mediated in large part by S-nitrosylation/oxidation of critical cysteine thiol groups found on an ever-increasing number of proteins. As a particular example, S-nitrosylation affects the redox cycle of Prx2, leading to a loss of its peroxidase activity. The diminished activity of SNO-Prx2 interrupts a normal redox cycle and results in accumulation of cellular peroxides, thereby inducing oxidative stress. In the present study, we identify a role for Srxn1 as a denitrosylase for neuronal Prx2. Our finding points to the NO/Srxn1 pair in a previously unrecognized redox cycle that modulates Prx2 peroxidase activity. The discovery of this modulatory system adds another layer of complexity to the regulation of H2O2 in neurons. Additionally, the finding has therapeutic implications for redox homeostasis during nitrosative/oxidative stress that contributes to neurodegenerative diseases such as PD because we show that activation or overexpression of Srxn1 can protect human neurons. Interestingly, the known transcriptional regulators of the Srxn1 gene include AP-1 (41) and Nrf2 (42). We have previously developed small-molecule activators of Nrf2 (43), which could potentially be used to enhance Srxn1 gene expression. Finally, the unique denitrosylase action of Srxn1 yields molecular-level insight into the mechanism underlying the biological processes of peroxide detoxification.
Materials and Methods
Expression and Purification of Recombinant Proteins.
Recombinant DNA experiments were approved by the Scintillon Institutional Biosafety Committee. Prx2, Srxn1, and Srxn1(C99S) expression plasmids were transformed in BL21 CodonPlus (DE3)-RIL competent Escherichia coli cells (Stratagene). Cultures were grown at 37 °C in LB medium to an OD600 of 0.6, induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside, and incubated overnight at 22 °C. Cells were lysed on ice by sonication in the presence of protease inhibitors, and cell debris was separated from the sonicated solution by centrifugation. The supernatant was purified by incubation with nickel-nitrilotriacetic acid agarose beads (Qiagen) and washed with 10 mM imidazole in PBS. The proteins were then eluted using 250 mM imidazole in PBS and analyzed using SDS/PAGE.
Cell Cultures, Transfection, Electroporation, and Exposure to NO or Peroxide Donors.
The use of human cell lines was approved by the Scintillon Institutional Review Board, which determined that the use was “Exempt” and no informed consent is required because all human cell lines were previously established and no personal information is available to the researchers concerning the donors. Dopaminergic SH-SY5Y neural cells were maintained in DMEM (Sigma) supplemented with 10% (vol/vol) heat-inactivated FBS (HyClone), 2 mM l-glutamine (Gibco-Invitrogen), 50 IU/mL penicillin (Omega Scientific), and 50 µg/mL streptomycin (Omega Scientific). Cells were cultured in 100-mm culture plates at 37 °C in a water-saturated atmosphere of 95% air and 5% CO2. Transfections were performed in six-well plates using Lipofectamine LTX and Plus Reagent (Invitrogen) according to the manufacturer’s instructions. Where indicated, cells were exposed for 30 min to a solution containing 200 µM of the NO donor SNOC or a control solution from which NO had been previously dissipated (designated “old” SNOC). In additional experiments, cells were exposed to a solution containing 600 µM H2O2 or its vehicle as a negative control and analyzed after 24 h.
hiPSC-derived A9-type DA neuronal cultures were prepared as previously described (27) and exposed to SNOC or mitochondrial toxins/pesticides, including the herbicide PQ (Fluka), the fungicide MB (Fluka), or the insecticide rotenone (Sigma) (27). Concentrations were chosen as approximately twofold the maximal allowed exposure level by the US Environmental Protection Agency (EPA codes CASRN 1910–42-5 and CASRN 12427–38-2). In some cases, 1 mM of the broad spectrum NOS inhibitor l-NAME (Sigma) was added. Cells were exposed for 6 h at 37 °C in a water-saturated atmosphere of 95% air and 5% CO2. In al experiments, the hiPSC-derived DA neurons were electroporated with a plasmid encoding Srxn1 (107 cells electroporated per group).
Murine Model of Sporadic PD After Pesticide Exposure.
Animal care was conducted in accordance with the US Public Health Service Guide for the Care and Use of the Laboratory Animals, and all experiments were approved by the Scintillon Institutional Animal Care and Use Committee. Exposure to the combination of pesticides PQ and MB has been linked to sporadic cases of human PD in epidemiological studies (22–24) and can produce selective loss of nigrostriatal dopaminergic cell bodies and reduction in dopamine levels in the rodent striatum; the combination of paraquat and maneb has therefore been used as an experimental model of PD induced by pesticides in rodents (19–21, 44, 45). Female mice weighing 20–30 g were injected intraperitoneally with freshly prepared PQ (Fluka; 5 mg/kg) and MB (Fluka; 15 mg/kg) twice a week for 4 wk. Additional groups of animals also received the relatively specific neuronal NOS inhibitor 3-bromo-7-nitroindazole (Enzo Life Sciences; 30 mg/kg) or vehicle [60% (vol/vol) DMSO in PBS] twice a week on alternate days for 4 wk. Animals were killed after treatment, and their brains were frozen at −80 °C.
Protein Extraction and Western Blot Analyses.
Frozen mouse brains were homogenized in HENTS buffer, pH 7.2 (100 mM Hepes, 1 mM EDTA, 0.1 mM neocuproine, 1% Triton X-100, and 0.1% SDS) for biochemical analysis. Protein concentrations were determined using the Bio-Rad protein assay according to the manufacturer’s specifications. For immunoblotting, typically 5–30 µg of proteins were resolved on 4–12% (wt/vol) polyacrylamide NuPAGE-Mes gels (Invitrogen) and then wet-transferred to PVDF membranes (Millipore). Membranes were blocked with 2% (wt/vol) skim milk in Tris-buffered saline (TBS) containing 1% Tween-20 (TBS-T). Immunoblotting was performed using rabbit anti-mouse Srxn1 polyclonal antibody (1:2,000), kindly donated by Sue Goo Rhee (Yonsei University, Seoul, Korea). Immunological complexes were detected with goat anti-rabbit 800CW IR-dye–conjugated secondary antibody (1:15,000; Li-Cor), followed by infrared analysis on an Odyssey instrument (Li-Cor). Densitometric analysis of protein bands was performed using Adobe Photoshop. All values were normalized to their respective loading controls.
Biotin-Switch Assay for Detection of S-Nitrosylated Proteins.
Analysis of SNO-Prx2 with the biotin-switch assay was performed as previously described (6, 8, 12). Briefly, fresh cell or brain tissue samples were homogenized in HEN buffer, pH 7.2 (100 mM Hepes, 1 mM EDTA, 0.1 mM neocuproine) with 1% Triton X-100 and 0.1% SDS. A total of 1 mg of protein per sample was used for the assay. Free thiol groups were blocked by incubation with 20 mM methyl methanethiol-sulfonate (MMTS; Aldrich) for 30 min at 50 °C. Cell extracts were then precipitated with acetone and resuspended in HEN buffer with 1% SDS. S-nitrosothiol groups were then selectively reduced by 50 mM ascorbate to free thiols, which were subsequently biotinylated with 4 mM N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)-propionamide (Soltec Ventures). The biotinylated proteins were pulled down with streptavidin–agarose beads and analyzed by immunoblotting. Total protein as a loading control was quantified by standard immunoblot analysis. Results were expressed as SNO-Prx2 relative to total Prx2 for each sample.
For recombinant Prx2 and Srxn1 proteins, electrophoresis gels were run with nonreducing sample buffer to identify the S-nitrosylation of monomeric and dimeric forms of the proteins. For the visualization of the biotinylated proteins, they were labeled with avidin–peroxidase followed by development with a SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Scientific).
In Vitro Denitrosylation Assay.
To assess denitrosylation of Prx2 by Srxn1, 20 μM Prx2 was S-nitrosylated by preincubation with 100 μM SNOC in a buffer containing 500 mM Tris⋅HCl, pH 7.2, 1 M KCl, 10 mM MgCl2, and 10% (vol/vol) DMSO. After 1 h, 40 μM Srxn1 and 1 mM ATP were added. Following incubation at 30 °C for the indicated length of time, the reaction was stopped with blocking buffer containing 20 mM MMTS, and biotin-switch was performed as described above.
Mass Spectrometry.
Protein bands were excised from Coomassie-stained gels and destained and divided into two aliquots, which were then subjected to an in-gel digestion as described previously (46). Briefly, digestion was performed with 12.5 ng/μL of trypsin and AspN in 50 mM ammonium bicarbonate and incubated overnight at 37 °C. The resultant peptides were extracted with 50% (vol/vol) acetonitrile/5% (vol/vol) formic acid and dried in a vacuum centrifuge. Before measurements, dried peptides were dissolved in 0.1% formic acid.
The peptide mixtures were analyzed by online nanoflow liquid chromatography–tandem mass spectrometry on an Agilent 1200 quaternary HPLC system (Agilent) connected to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) through an in-house built nanoelectrospray ion source. The peptide mixtures were pressure-loaded onto a capillary column (100-µm i.d.) packed with 15 cm of 3-µm Aqua C18 resin (Phenomenex). They were separated with a 90-min gradient from 10 to 50% (vol/vol) acetonitrile in 0.1% formic acid and a flow rate of 220 nL/min (through split). As peptides were eluted from the analytical column, they were electrosprayed (distal 2.5-kV spray voltage) into the mass spectrometer. The MS instrument method consisted of a Fourier Transform full-scan MS analysis (300–1,800 m/z; 60,000 resolution) followed by data-dependent MS/MS scans of the 20 most intense precursors at a 35% normalized collision energy with dynamic exclusion for 60 s. Application of mass spectrometer scan functions and HPLC solvent gradients was controlled by the Xcalibur data system (Thermo Fisher Scientific).
Protein identification was done with Integrated Proteomics Pipeline-IP2 (Integrated Proteomics Applications, Inc., www.integratedproteomics.com/) using ProLuCID (47) and DTASelect (48). MS/MS spectra were extracted using RawXtract (49) (version 1.9.9) and searched with ProLuCID against an EBI IPI Human protein database (version 3.87) concatenated to a decoy database in which the sequence for each entry in the original database was reversed (50). ProLuCID search results were assembled and filtered using DTASelect (version 2) with a false-positive rate below 1% at the peptide level. To identify the disulfide bond linkages between Srxn1 and Prx2 proteins as well as among themselves, the delta masses 1348.5295, 1856.8767, 1100.59, and 1324.5506 were set as variable modifications for cysteine corresponding to the linkage caused by peptide DYFYSFGGCHR of Srxn1 protein and peptides DFTFVCPTEIIAFSNR, KLGCEVLGVSV, and DEHGEVCPAGWK of Prx2 protein. Protein Identification was also performed by Thermo Proteome discoverer (v2.1) with Sequest HT (v1.17) (51). The tandem mass spectra were searched against the UniProt human protein database. The precursor mass tolerance was set as 10 ppm, and the fragment mass tolerance was set as 0.6 Da. The q-values of peptide identification were controlled below 1% using Percolator (52). The variable modification settings were set as described above to identify disulfide bond linkages between Srxn1 and Prx2 proteins as well as among themselves.
Terminal-Deoxynucleotidyl–Transferase dUTP Nick End Labeling Assay and Image Analysis.
SH-SY5Y neural cells and hiPSC-derived DA neurons were grown on coverslips where Transferase dUTP Nick End Labeling (TUNEL) assays were performed. Briefly, cells were fixed with 4% (wt/vol) paraformaldehyde in PBS for 10 min and then permeabilized with 0.1% Triton X-100 in 0.1% sodium citrate for 2 min. Apoptotic cells were labeled using an in situ cell death detection kit (Roche) according to the manufacturer’s instructions, and coverslips were mounted for fluorescence analysis using Fluoro Gel mounting medium (Electron Microscopy Sciences). Apoptotic SH-SY5Y cells and hiPSC-derived neurons were observed and quantified on a deconvolution epifluorescence microscope (Axio, Carl Zeiss), or confocal microscope (Zeiss).
Preparation of SNOC.
SNOC was prepared as described previously (53). Briefly, a mixture of 100 mM l-cysteine and 100 mM sodium nitrite was prepared immediately before use. To this mixture on ice, 10 N HCl was added to a final normality of 0.5. Freshly prepared SNOC was used within 5 min of its preparation. We have previously shown that NO is dissipated from SNOC at room temperature within minutes, so “old SNOC” can be used as a negative control (53).
Statistical Analyses.
Statistical analyses were carried out using GraphPad Prism. Data are presented as mean with error bars representing SEM. Pairwise comparisons of statistical significance were performed using a Student’s t test (two tailed) and, for multiple comparisons, an ANOVA followed by a Newman–Keuls post hoc test.
Acknowledgments
We thank Sue Goo Rhee (Yonsei University, Seoul, Korea) for kindly providing rabbit anti-mouse sulfiredoxin polyclonal antibody; Giles Hardingham (University of Edinburgh) for donating the myc-Srxn1 construct for transfection of human cells; and Mike Shaw (The Scripps Research Institute, La Jolla, CA) for helpful discussions of chemical reaction mechanisms. This work was supported in part by NIH Grants R01 NS086890, DP1 DA041722, and P01 HD29587; La Jolla Interdisciplinary Neuroscience Center Core Grant P30 NS076411; and the Michael J. Fox Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608784113/-/DCSupplemental.
References
- 1.Chae HZ, Kim IH, Kim K, Rhee SG. Cloning, sequencing, and mutation of thiol-specific antioxidant gene of Saccharomyces cerevisiae. J Biol Chem. 1993;268(22):16815–16821. [PubMed] [Google Scholar]
- 2.Yang KS, et al. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem. 2002;277(41):38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 3.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300(5619):650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 4.Woo HA, et al. Reduction of cysteine sulfinic acid by sulfiredoxin is specific to 2-cys peroxiredoxins. J Biol Chem. 2005;280(5):3125–3128. doi: 10.1074/jbc.C400496200. [DOI] [PubMed] [Google Scholar]
- 5.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: Purview and parameters. Nat Rev Mol Cell Biol. 2005;6(2):150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 6.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3(2):193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 7.Hess DT, Matsumoto A, Nudelman R, Stamler JS. S-nitrosylation: Spectrum and specificity. Nat Cell Biol. 2001;3(2):E46–E49. doi: 10.1038/35055152. [DOI] [PubMed] [Google Scholar]
- 8.Yao D, et al. Nitrosative stress linked to sporadic Parkinson’s disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2004;101(29):10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung KK, et al. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304(5675):1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 10.Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci USA. 2007;104(47):18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelman R, et al. Multilevel regulation of 2-Cys peroxiredoxin reaction cycle by S-nitrosylation. J Biol Chem. 2013;288(16):11312–11324. doi: 10.1074/jbc.M112.433755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uehara T, et al. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441(7092):513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- 13.McCarthy SM, Bove PF, Matthews DE, Akaike T, van der Vliet A. Nitric oxide regulation of MMP-9 activation and its relationship to modifications of the cysteine switch. Biochemistry. 2008;47(21):5832–5840. doi: 10.1021/bi702496v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietz KJ, Horling F, König J, Baier M. The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J Exp Bot. 2002;53(372):1321–1329. [PubMed] [Google Scholar]
- 15.Pascual MB, Mata-Cabana A, Florencio FJ, Lindahl M, Cejudo FJ. Overoxidation of 2-Cys peroxiredoxin in prokaryotes: Cyanobacterial 2-Cys peroxiredoxins sensitive to oxidative stress. J Biol Chem. 2010;285(45):34485–34492. doi: 10.1074/jbc.M110.160465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bang YJ, Oh MH, Choi SH. Distinct characteristics of two 2-Cys peroxiredoxins of Vibrio vulnificus suggesting differential roles in detoxifying oxidative stress. J Biol Chem. 2012;287(51):42516–42524. doi: 10.1074/jbc.M112.421214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425(6961):980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 18.Chung KK, Dawson TM, Dawson VL. Nitric oxide, S-nitrosylation and neurodegeneration. Cell Mol Biol (Noisy-le-grand) 2005;51(3):247–254. [PubMed] [Google Scholar]
- 19.Cicchetti F, et al. Systemic exposure to paraquat and maneb models early Parkinson’s disease in young adult rats. Neurobiol Dis. 2005;20(2):360–371. doi: 10.1016/j.nbd.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Thiruchelvam M, Richfield EK, Baggs RB, Tank AW, Cory-Slechta DA. The nigrostriatal dopaminergic system as a preferential target of repeated exposures to combined paraquat and maneb: Implications for Parkinson’s disease. J Neurosci. 2000;20(24):9207–9214. doi: 10.1523/JNEUROSCI.20-24-09207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiruchelvam M, Brockel BJ, Richfield EK, Baggs RB, Cory-Slechta DA. Potentiated and preferential effects of combined paraquat and maneb on nigrostriatal dopamine systems: Environmental risk factors for Parkinson’s disease? Brain Res. 2000;873(2):225–234. doi: 10.1016/s0006-8993(00)02496-3. [DOI] [PubMed] [Google Scholar]
- 22.Costello S, Cockburn M, Bronstein J, Zhang X, Ritz B. Parkinson’s disease and residential exposure to maneb and paraquat from agricultural applications in the central valley of California. Am J Epidemiol. 2009;169(8):919–926. doi: 10.1093/aje/kwp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liou HH, et al. Environmental risk factors and Parkinson’s disease: A case-control study in Taiwan. Neurology. 1997;48(6):1583–1588. doi: 10.1212/wnl.48.6.1583. [DOI] [PubMed] [Google Scholar]
- 24.Meco G, Bonifati V, Vanacore N, Fabrizio E. Parkinsonism after chronic exposure to the fungicide maneb (manganese ethylene-bis-dithiocarbamate) Scand J Work Environ Health. 1994;20(4):301–305. doi: 10.5271/sjweh.1394. [DOI] [PubMed] [Google Scholar]
- 25.Kriks S, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480(7378):547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan SD, et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 2013;155(6):1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Haes W, et al. Metformin promotes lifespan through mitohormesis via the peroxiredoxin PRDX-2. Proc Natl Acad Sci USA. 2014;111(24):E2501–E2509. doi: 10.1073/pnas.1321776111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benhar M, Thompson JW, Moseley MA, Stamler JS. Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry. 2010;49(32):6963–6969. doi: 10.1021/bi100619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sengupta R, et al. Thioredoxin catalyzes the denitrosation of low-molecular mass and protein S-nitrosothiols. Biochemistry. 2007;46(28):8472–8483. doi: 10.1021/bi700449x. [DOI] [PubMed] [Google Scholar]
- 30.Pader I, et al. Thioredoxin-related protein of 14 kDa is an efficient L-cystine reductase and S-denitrosylase. Proc Natl Acad Sci USA. 2014;111(19):6964–6969. doi: 10.1073/pnas.1317320111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, et al. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410(6827):490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 32.Jensen DE, Belka GK, Du Bois GC. S-nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochem J. 1998;331(Pt 2):659–668. doi: 10.1042/bj3310659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bateman RL, Rauh D, Tavshanjian B, Shokat KM. Human carbonyl reductase 1 is an S-nitrosoglutathione reductase. J Biol Chem. 2008;283(51):35756–35762. doi: 10.1074/jbc.M807125200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jönsson TJ, Murray MS, Johnson LC, Poole LB, Lowther WT. Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin. Biochemistry. 2005;44(24):8634–8642. doi: 10.1021/bi050131i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jönsson TJ, Murray MS, Johnson LC, Lowther WT. Reduction of cysteine sulfinic acid in peroxiredoxin by sulfiredoxin proceeds directly through a sulfinic phosphoryl ester intermediate. J Biol Chem. 2008;283(35):23846–23851. doi: 10.1074/jbc.M803244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang TS, et al. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J Biol Chem. 2004;279(49):50994–51001. doi: 10.1074/jbc.M409482200. [DOI] [PubMed] [Google Scholar]
- 37.Jönsson TJ, Johnson LC, Lowther WT. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature. 2008;451(7174):98–101. doi: 10.1038/nature06415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roussel X, et al. Evidence for the formation of a covalent thiosulfinate intermediate with peroxiredoxin in the catalytic mechanism of sulfiredoxin. J Biol Chem. 2008;283(33):22371–22382. doi: 10.1074/jbc.M800493200. [DOI] [PubMed] [Google Scholar]
- 39.Jönsson TJ, Tsang AW, Lowther WT, Furdui CM. Identification of intact protein thiosulfinate intermediate in the reduction of cysteine sulfinic acid in peroxiredoxin by human sulfiredoxin. J Biol Chem. 2008;283(34):22890–22894. doi: 10.1074/jbc.C800124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ware RW, Jr, King SB. P-nitrosophosphate compounds: New N-O heterodienophiles and nitroxyl delivery agents. J Org Chem. 2000;65(25):8725–8729. doi: 10.1021/jo001230z. [DOI] [PubMed] [Google Scholar]
- 41.Glauser DA, Brun T, Gauthier BR, Schlegel W. Transcriptional response of pancreatic beta cells to metabolic stimulation: Large scale identification of immediate-early and secondary response genes. BMC Mol Biol. 2007;8:54. doi: 10.1186/1471-2199-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39(4):199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Satoh T, McKercher SR, Lipton SA. Nrf2/ARE-mediated antioxidant actions of pro-electrophilic drugs. Free Radic Biol Med. 2013;65:645–657. doi: 10.1016/j.freeradbiomed.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norris EH, et al. Pesticide exposure exacerbates alpha-synucleinopathy in an A53T transgenic mouse model. Am J Pathol. 2007;170(2):658–666. doi: 10.2353/ajpath.2007.060359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiruchelvam M, Richfield EK, Goodman BM, Baggs RB, Cory-Slechta DA. Developmental exposure to the pesticides paraquat and maneb and the Parkinson’s disease phenotype. Neurotoxicology. 2002;23(4-5):621–633. doi: 10.1016/s0161-813x(02)00092-x. [DOI] [PubMed] [Google Scholar]
- 46.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 47.Xu T, et al. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics. 2006;5:S174. [Google Scholar]
- 48.Tabb DL, McDonald WH, Yates JR., III, DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald WH, et al. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18(18):2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 50.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: The yeast proteome. J Proteome Res. 2003;2(1):43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 51.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 52.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 53.Lei SZ, et al. Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex. Neuron. 1992;8(6):1087–1099. doi: 10.1016/0896-6273(92)90130-6. [DOI] [PubMed] [Google Scholar]