The proteasome is one of the major degradation machineries in eukaryotic cells. It terminates the existence of thousands of short-lived, damaged, misfolded or otherwise obsolete proteins and plays pivotal roles in protein quality control and other vital processes in the cell. However, very little has been known about the mechanisms that control and execute the destruction of the proteasome itself. In their recent report in PNAS, Cohen-Kaplan et al. (1) put into place a significant piece of this puzzle by revealing events governing the removal of proteasomes upon amino acid starvation.
The 26S proteasome is a complex proteolytic machine of around 2.5 MDa. It consists of a barrel-shaped protein complex (core particle, CP) that can carry a regulatory lid (the regulatory particle, RP) on one or both ends. To be degraded in the proteasome proteins have to be tagged with Ubiquitin (Ub), in particular with chains of Ub molecules linked through lysine 48 (K48) of Ub (2). Ubiquitinated substrates are recognized by Rpn1, Rpn10, and Rpn13, three subunits of the RP that possess Ub-binding domains (3). Alternatively, substrates are delivered by Ub-binding shuttling proteins (p62, Rad23/HR23, Dsk2/PLIC/Ubiquilin, and Ddi1) that dock at the proteasome via interaction of their Ub-like domain with Rpn1, Rpn10, or Rpn13 (2). After capturing the substrate, the Ub tag is released by an RP-associated deubiquitinating enzyme whereas the substrate is unfolded and threaded through a narrow gate into the interior of the CP, where it is degraded by chymotrypsin-, trypsin-, and caspase-like proteolytic activities (2). This highly controlled process not only eliminates unwanted proteins, terminates or activates signaling pathways, and participates in cell cycle regulation but also provides an important source of amino acids for de novo protein synthesis. Correspondingly, a decline in proteasomal activity is associated with aging, cancer, neurodegenerative diseases, and other late-onset diseases. On the other hand, the strong dependency of highly proliferating cells, such as cancer cells, on an active proteasome is exploited in anticancer therapies by using proteasome inhibitors for inducing cancer cell death (4). Thus, understanding how proteasome abundance and activity are regulated and might be manipulated pharmaceutically is of high interest.
Proteasome abundance is determined by the balance between de novo synthesis/assembly and degradation of proteasome particles. Although the transcriptional regulation of proteasome subunit expression and the assembly into functional proteolytic machines have been dissected in depth, the mechanisms that direct their removal have remained largely elusive. Besides having to digest a multiprotein complex of 2.5 MDa, the major challenge the cell faces during the controlled breakdown of proteasomes concerns the recognition and labeling of nonfunctional, damaged, or superfluous proteasomes. First insights into these processes were gained in 2015 when Marshall et al. observed that proteasomes can be degraded by autophagy in Arabidopsis (5), and in 2016 when two independent reports described the autophagic elimination of proteasomes in Saccharomyces cerevisiae (6, 7). Autophagy is the second major degradation system in eukaryotic cells specialized on long-lived, large, heterogeneous material (8). The hallmark of autophagy is the engulfment of cargo by a double-layered membrane, the phagophore, that closes around the cargo to form the autophagosome. To degrade its content, the autophagosome fuses with the lysosome, whose hydrolytic milieu digests the enclosed material. Bulk autophagy randomly sequesters cytosolic components to provide nutrients and energy in periods of nutrient and/or energy deprivation. However, autophagy can also selectively remove cellular components such as protein aggregates and dysfunctional or superfluous organelles and can be sharply up-regulated in response to various cellular stresses (proteotoxic stress, oxidative stress, organelle damage, proteasome inhibition, etc.) (9). Ub-dependent as well as Ub-independent tags on the cargo serve as recognition signals for selective sequestration by the autophagic machinery (10).
Also, autophagy of proteasomes (proteaphagy) in Arabidopsis can proceed via nonselective and selective routes, depending on the initial trigger. Selective proteaphagy is induced by inhibition of the proteasome and requires the ubiquitination of inactive proteasomes, which is not the case in nonselective proteaphagy occurring upon starvation. Similar to other selective autophagy pathways, the sequestration of ubiquitinated proteasomes into the autophagosome critically depends on autophagy receptors that can simultaneously bind to Ub attached to the cargo and Atg8/LC3, a Ub-like modifier exposed on autophagosomal membranes. Marshall et al. (5) have shown that, in Arabidopsis, Rpn10a fulfills this function. Of note, besides being an integral proteasomal RP subunit, RPN10a can exist also in free (i.e., extraproteasomal) form, and it possesses a newly identified LC3-interacting region (LIR) enabling Rpn10a to take the role of a selective autophagy receptor for ubiquitinated proteasomes. However, neither yeast nor mammalian Rpn10 is able to bind to LC3/Atg8. Instead, Cue5 has been identified as selective autophagy receptor for ubiquitinated proteasomes in yeast. Cue5 and its human homolog Tollip have been implicated only recently in the autophagic clearance of polyQ proteins (11). Moreover, proteaphagy in yeast requires the action of the chaperone Hsp42. Hsp42 delivers the proteasomes to perivacuolar insoluble protein deposit structures that are thought to function as a cytoprotective compartment serving the deposition of potentially toxic damaged or misfolded proteins before their final removal.
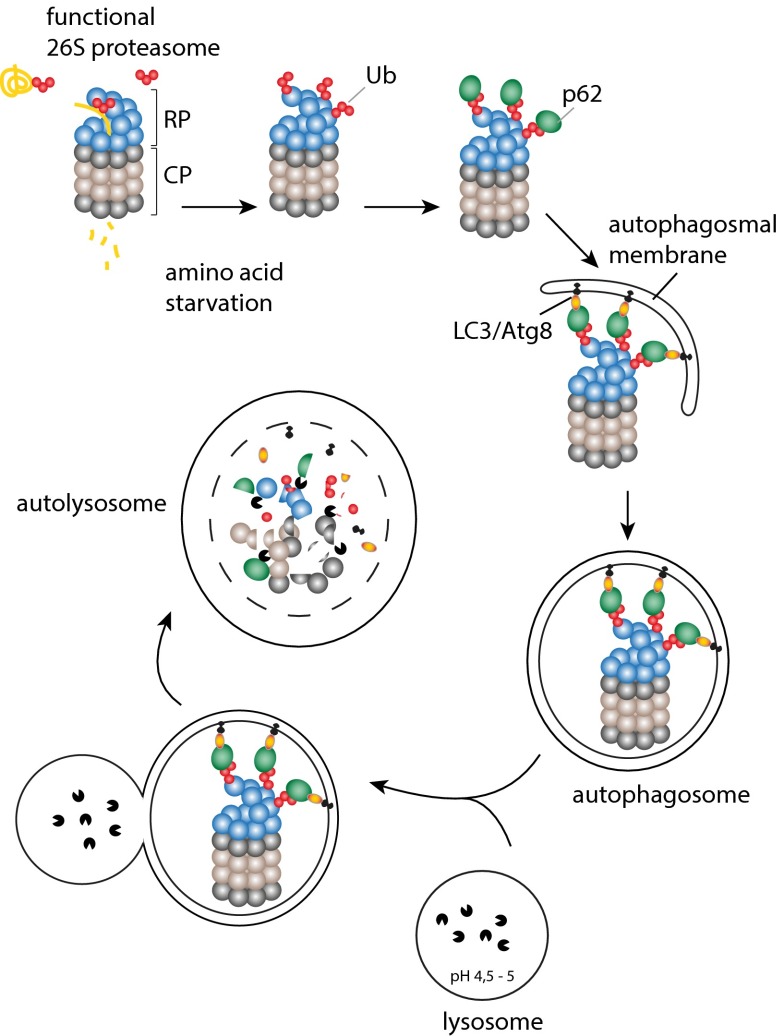
Aaron Ciechanover and his team shed light on the mechanisms governing proteaphagy in mammalian cells that have remained entirely elusive (1). Interestingly, although it could have been assumed that proteaphagy pathways proceed in quite similar manner in different eukaryotes, there are intriguing differences between proteaphagy in Arabidopsis/S. cerevisiae and in mammals. Cohen-Kaplan et al. (1) find that amino acid starvation induces extensive ubiquitination of several RP subunits, in particular, the Ub-receptor subunits Rpn1, Rpn10, and Rpn13. Further, they reveal that the autophagy receptor p62 mediates selective starvation-induced autophagosomal uptake of proteasomes (Fig. 1). This is in contrast to plant cells or yeast that induce Ub-dependent selective autophagy only upon proteasome inhibition but not upon starvation. In fact, Cohen-Kaplan et al. indicate that, in mammalian cells, proteasomes are a specific target of destruction under starvation conditions, whereas other cellular components are randomly taken up, i.e., with significantly lower efficiency. Also, the identification of p62 instead of the human Cue5 homolog Tollip as selective proteaphagy receptor is unexpected; p62 is an intriguing adaptor protein, as it can target Ub-modified proteins to either the proteasome (via its PB1 domain) (12, 13) or the autophagic machinery (via its LIR domain) (14) for degradation, and it is thought that p62 plays a central role in the communication between both degradation systems. Of note, in mammalian cells selective autophagy receptors often work together to facilitate efficient removal of cargo (15), and it remains to be tested whether Tollip or other autophagy receptors team up with p62 in proteaphagy.
Fig. 1.
Under nutrient-rich conditions, the proteasome binds and degrades polyubiquitinated proteins. Amino acid deprivation induces ubiquitination of multiple RP subunits including the Ub receptors Rpn1, Rpn10, and Rpn13. Moreover, mTOR is inhibited, leading to the activation of autophagy core proteins that trigger formation of the nascent autophagosomal membrane; p62 functions as selective autophagy receptor that specifically links polyubiquitinated proteasomes to the growing membrane via interactions with Atg8/LC3. After complete engulfment of the cargo, the autophagosome fuses with the lysosome, whose acidic pH and hydrolases digest the proteasomes.
The findings by Cohen-Kaplan et al. (1) raise several questions: How is the function of p62 switched from a proteasomal shuttling factor delivering ubiquitinated substrates to the proteasome to a
Cohen-Kaplan et al. indicate that, in mammalian cells, proteasomes are a specific target of destruction under starvation conditions, whereas other cellular components are randomly taken up, i.e., with significantly lower efficiency.
selective autophagy receptor that tethers ubiquitinated proteasomes to the autophagosome instead? Obviously, this might be partially achieved by blocking the p62 docking sites on the proteasome by ubiquitination, thereby pushing p62 molecules, including their interaction partners, to the autophagic route. Several studies have shown that already monoubiquitination (i.e., the attachment of a single Ub molecule) impairs the ability of proteasomal Ub-receptors to bind to ubiquitinated substrates and shuttling factors (16–18). However, it is likely that additional players and/or posttranslational modifications help to coordinate the switch from proteasomal degradation toward autophagic removal of the proteasome and likely the accumulating proteasomal substrates as well.
Is p62-dependent selective proteaphagy also relevant upon proteasome inhibition in mammalian cells? Proteasome inhibition is toxic to cells and induces extensive ubiquitination of the proteasome (1, 18). Toxicity is, in part, due to the disturbance of signaling networks, the lack of amino acid recycling, and the accumulation of misfolded aggregation-prone proteins. As a countermeasure, p62 is rapidly induced upon proteasome inhibition to facilitate the clearance of protein aggregates via autophagy. It thereby also contributes to the refeeding of the pool of free amino acids. Thus, the autophagic removal of inactive proteasomes would fit well into its repertoire of functions.
Although the removal of inactive or damaged proteasomal particles is reasonable, an intriguing question is why mammalian cells would actually eliminate functional proteasomes under starvation conditions as shown by Cohen-Kaplan et al. (1). Proteasomal degradation represents a major source of amino acids for de novo protein synthesis, and, therefore, starvation-induced proteaphagy of functional particles seems counterintuitive. It will be interesting to analyze the cellular as well as physiological consequences of blocking selective proteaphagy in different contexts to tackle the specific benefits of the process.
Taken together, the study by Cohen-Kaplan et al. (1) carries on an exciting series of discoveries that shed light on the processes that lead to the destruction of the major degradation machinery in eukaryotic cells. As touched on above, the findings raise several questions that promise further exciting findings and valuable insights relevant not only for basic science but also for the development of treatments for several major diseases, including cancer.
Acknowledgments
I.D. is supported by the Deutsche Forschungsgemeinschaft (DFG)-funded Collaborative Research Centre on Selective Autophagy (SFB 1177), by the DFG-funded Cluster of Excellence “Macromolecular Complexes” (EXC115), by the LandesOffensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) program Ubiquitin Networks (Ub-Net), and by the LOEWE Center for Gene and Cell Therapy Frankfurt (CGT) funded by the State of Hesse/Germany.
Footnotes
The authors declare no conflict of interest.
See companion article on page E7490.
References
- 1.Cohen-Kaplan V, et al. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc Natl Acad Sci USA. 2016;113:E7490–E7499. doi: 10.1073/pnas.1615455113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78(1):477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Husnjak K, Dikic I. Ubiquitin-binding proteins: Decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81(1):291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 4.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458(7237):438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 5.Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell. 2015;58(6):1053–1066. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall RS, McLoughlin F, Vierstra RD. Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Reports. 2016;16(6):1717–1732. doi: 10.1016/j.celrep.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 7.Waite KA, De La Mota-Peynado A, Vontz G, Roelofs J. Starvation induces proteasome autophagy with different pathways for core and regulatory particles. J Biol Chem. 2016;291(7):3239–3253. doi: 10.1074/jbc.M115.699124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: Origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–774. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto K. Organellophagy: Eliminating cellular building blocks via selective autophagy. J Cell Biol. 2014;205(4):435–445. doi: 10.1083/jcb.201402054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26(1):6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell. 2014;158(3):549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 12.Seibenhener ML, et al. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24(18):8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94(1):192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 14.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 15.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16(6):495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 16.Zuin A, et al. Rpn10 monoubiquitination orchestrates the association of the ubiquilin-type DSK2 receptor with the proteasome. Biochem J. 2015;472(3):353–365. doi: 10.1042/BJ20150609. [DOI] [PubMed] [Google Scholar]
- 17.Isasa M, et al. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol Cell. 2010;38(5):733–745. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besche HC, et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 2014;33(10):1159–1176. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]