Significance
Gram-negative pathogens represent a major public health issue. With increases in antibiotic resistance and the limited availability of preventive strategies, new ways are needed to target these bacteria. Here, we propose a narrow-spectrum immunization strategy to block the ability of a gastrointestinal pathogen to acquire the essential metal nutrient iron. This approach involves the chemical synthesis of bacterial iron chelators, known as siderophores, and their conjugation to immunogenic carrier proteins. Mice immunized with these compounds developed antibodies against siderophores in the intestine, which hindered proliferation of the gut pathogen Salmonella. Because similar but distinct molecules are secreted by many bacterial and fungal pathogens, this synthesis and immunization strategy could be applied to a broad range of infectious agents.
Keywords: siderophore, immunization, iron, Salmonella, microbiota
Abstract
Infections with Gram-negative pathogens pose a serious threat to public health. This scenario is exacerbated by increases in antibiotic resistance and the limited availability of vaccines and therapeutic tools to combat these infections. Here, we report an immunization approach that targets siderophores, which are small molecules exported by enteric Gram-negative pathogens to acquire iron, an essential nutrient, in the host. Because siderophores are nonimmunogenic, we designed and synthesized conjugates of a native siderophore and the immunogenic carrier protein cholera toxin subunit B (CTB). Mice immunized with the CTB–siderophore conjugate developed anti-siderophore antibodies in the gut mucosa, and when mice were infected with the enteric pathogen Salmonella, they exhibited reduced intestinal colonization and reduced systemic dissemination of the pathogen. Moreover, analysis of the gut microbiota revealed that reduction of Salmonella colonization in the inflamed gut was accompanied by expansion of Lactobacillus spp., which are beneficial commensal organisms that thrive in similar locales as Enterobacteriaceae. Collectively, our results demonstrate that anti-siderophore antibodies inhibit Salmonella colonization. Because siderophore-mediated iron acquisition is a virulence trait shared by many bacterial and fungal pathogens, blocking microbial iron acquisition by siderophore-based immunization or other siderophore-targeted approaches may represent a novel strategy to prevent and ameliorate a broad range of infections.
Gram-negative pathogens cause a range of human diseases, including foodborne illness, urinary tract infections, and sepsis. Infections with these organisms pose a serious global public health threat that is exacerbated by increasing antibiotic resistance, the dearth of new antibiotics in the drug pipeline, and the limited availability of vaccines (1). To slow the emergence of antibiotic resistance and to reduce the incidence of secondary infections, narrow-spectrum therapeutic strategies are needed, specifically ones that limit disruption of the gut microbiota, which comprises beneficial microbes that provide colonization resistance to pathogens (2). Many studies have elucidated molecular mechanisms by which pathogens thrive in the host, thus indicating potential targets for the prevention and treatment of infection. In recent years, bacterial metabolism has been proposed to be a key factor in promoting pathogenicity (3).
The vast majority of bacterial pathogens have a metabolic requirement for iron. Because the vertebrate host tightly controls the concentration of free iron (e.g., ∼10−24 M in serum), many microbes biosynthesize and export secondary metabolites called siderophores to scavenge iron from the host (4). These small molecules chelate ferric iron (Fe3+) with high affinity [e.g., enterobactin (Ent)] (5–8) (Kd ∼10−25 M at neutral pH; Fig. 1A). Once a siderophore coordinates iron in the extracellular space, the iron-bound siderophore is recognized and transported into a microbial cell by a dedicated membrane receptor. Following cellular uptake, the iron is released from the siderophore to support microbial metabolism and replication. Siderophores are regarded as major virulence factors during infection with bacterial and fungal pathogens, examples of which include Salmonella enterica, uropathogenic Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Mycobacterium tuberculosis, Staphylococcus aureus, and Aspergillus fumigatus (9).
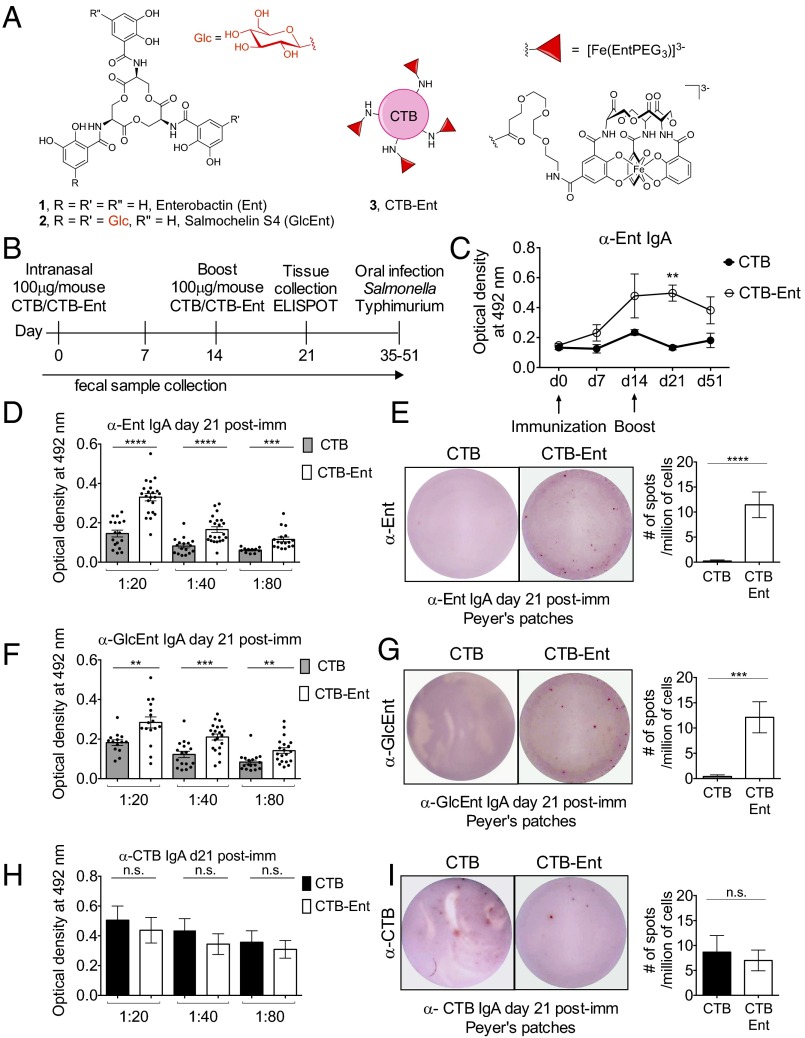
Fig. 1.
Intranasal immunization with CTB-Ent induces the development of anti-Ent and antisalmochelin IgA antibodies in the gut mucosa. (A) Chemical structures of Ent (1) and salmochelin S4 (GlcEnt; 2) and a cartoon depicting the CTB-Ent (3) conjugate. Ent and GlcEnt are cyclic trimers of N-2,3-dihydroxylbenzoyl-l-serine. (B) Description and time line of the immunization protocol. The timing of intranasal immunization (day 0 and day 14) and the dose of antigen (100 μg/mL) are indicated. Feces were collected weekly. An ELISPOT assay was carried out at day 21 postimmunization. Mice were infected with S. enterica serovar Typhimurium per os between day 35 and day 51 postimmunization. (C) Anti-Ent IgA antibodies were quantified by using an in-house ELISA in fecal samples from mice immunized with either CTB (n = 5) or CTB-Ent (n = 7) during the indicated time course. Day 0 represents antibodies from naive mice before immunization. A twofold dilution was used for detection of fecal anti-Ent IgA (D) or anti-GlcEnt IgA (F) by ELISA in mice immunized with either CTB (n = 15–20) or CTB-Ent (n = 15–20) at day 21 postimmunization. Representative ELISPOT images of supernatant from B cells producing anti-Ent IgA (E) or anti-GlcEnt IgA (G) isolated from Peyer’s patches of mice immunized with either CTB or CTB-Ent at day 21 postimmunization are shown. The average number of spots detected in CTB- or CTB-Ent–immunized mice per 1 million Peyer’s patches cells (n = 5–7) is shown. (H) Twofold dilution for detection of fecal anti-CTB IgA in CTB (n = 6) and CTB-Ent (n = 6) by in-house ELlSA. (I) Representative ELISPOT images of CTB IgA from Peyer’s patches of mice immunized with either CTB or CTB-Ent at day 21 postimmunization. The average number of spots detected in the CTB or CTB-Ent–immunized mice per 1 million Peyer’s patches cells (n = 3) is shown. In C–I, bars represent the mean ± SE. Immunization experiments were repeated at least three times with different batches of CTB/CTB-Ent. ****P < 0.0001; ***P < 0.001;**P < 0.01. n.s., not significant; post-imm, postimmunization.
Ent (Fig. 1A) is a siderophore biosynthesized and deployed by commensal and pathogenic Enterobacteriaceae (10). In a process known as “nutritional immunity,” the host responds to microbial infection and inflammation by limiting the availability of essential nutrient metals, including iron (11). To prevent Ent-mediated microbial iron acquisition, epithelial cells and neutrophils secrete the host-defense protein lipocalin-2 (LCN2). This protein inhibits siderophore-mediated iron uptake by capturing ferric Ent in the extracellular milieu (12). By inhibiting the growth of Enterobacteriaceae that rely on Ent-mediated iron acquisition, LCN2 plays an essential role in preventing lethal infection by these organisms (12, 13). Nevertheless, various Gram-negative pathogens evade LCN2 by producing and using “stealth siderophores” that cannot be captured by this host-defense protein. For example, Salmonella spp. and strains of pathogenic E. coli overcome LCN2 by biosynthesizing a family of C-glucosylated Ent (GlcEnt) derivatives named salmochelins (14, 15) (Fig. 1A). These siderophores allow the pathogen to thrive in the inflamed gut in the presence of LCN2 and outcompete the microbiota (14–16). Indeed, Salmonella mutants that lack the GlcEnt receptor IroN are susceptible to the LCN2-mediated host response; these mutants exhibit reduced colonization in the inflamed intestine and cannot outcompete the microbiota (17, 18). In the absence of intestinal inflammation or LCN2 expression, iron is more readily available and mutants in siderophore receptors are not attenuated (19).
Inspired by seminal studies exemplifying the importance of siderophore-mediated iron acquisition during infection with Salmonella as well as other pathogens (18, 20), we hypothesized that boosting host nutritional immunity by blocking siderophore-based iron acquisition would reduce microbial burden and improve the outcome of infection. To address this hypothesis, we designed and synthesized conjugates of a native siderophore used by Salmonella and an immunogenic carrier protein, and we immunized mice with the compounds to induce an antibody response against siderophores. Herein, we show that immunization of mice with the siderophore-carrier protein conjugate elicited an antibody response to siderophores in the intestinal mucosa and significantly reduced Salmonella colonization of the inflamed gut.
Results and Discussion
Immunization with Siderophore Conjugates Elicits a Mucosal Antibody Response.
To induce the development of an antibody response against siderophores, we selected cholera toxin subunit B (CTB) as the immunogenic carrier protein because it induces a strong mucosal immune response (21). We reasoned that immunizing mice with a CTB–siderophore conjugate should result in the production of anti-siderophore antibodies in the gut mucosa, which is the primary site of Salmonella infection. Because Ent and GlcEnt (Fig. 1A; 1 and 2, respectively) share several structural features, including the catechol moieties and trilactone ring, and GlcEnt is a derivative of Ent, we hypothesized that immunization with CTB-Ent would elicit an antibody response against both Ent and GlcEnt. In addition, it could be difficult to elicit a specific antibody response to only one of these siderophores given the shared structural attributes. Although Ent is captured by LCN2 in the inflamed gut, we also reasoned that the development of antibodies that capture both Ent and GlcEnt would be advantageous, especially if LCN2 levels are low in response to infection, for instance, in immunocompromised patients (22).
We thus designed and prepared a CTB-Ent conjugate that harbors the native Ent scaffold. We assembled CTB-Ent (Fig. 1A, 3) from CTB and intermediate 8 (SI Appendix, Fig. S1), an Ent derivative monofunctionalized with a polyethylene glycol (PEG3) linker using standard amide coupling methods (Fig. 1A and SI Appendix, Figs. S1–S3 and Supplementary Discussion). This procedure results in the Ent haptens covalently attached to surface-exposed lysine residues of CTB via the PEG3 linker (Fig. 1A). The average number of Ent-labeled lysine residues is approximately equal to four out of nine surface-exposed lysine residues in one subunit of CTB (SI Appendix, Supplementary Discussion). We selected a PEG3 linker because it is stable and affords both flexibility and enhanced solubility in aqueous solution (23). In prior work, we modified the native Ent scaffold with a PEG3 linker and established that these molecules coordinate Fe3+ and are recognized and transported by the Ent receptor FepA (23, 24). From these prior studies, we reasoned that antibodies that bind the Ent moiety of CTB-Ent will also recognize and bind unmodified Ent.
We immunized mice at day 0 and boosted at day 14 (Fig. 1B) by intranasal administration of either unmodified CTB (100 μg per mouse, mock immunization) or CTB-Ent (100 μg per mouse). To determine whether mice immunized with CTB-Ent developed a specific antibody response against Ent, blood and fecal samples were collected weekly for detection of anti-Ent antibodies by using an in-house ELISA and dot-blot. As expected, the amount of total fecal IgA was similar between the two groups of mice (SI Appendix, Fig. S4A). However, only mice immunized with CTB-Ent developed specific fecal IgA antibodies that recognized Ent (Fig. 1 C and D and SI Appendix, Fig. S4B), which were undetectable before immunization (day 0; Fig. 1C). The production of fecal anti-Ent IgA significantly increased at day 21 postimmunization in mice immunized with CTB-Ent (Fig. 1 C and D). An enzyme-linked immunospot (ELISPOT) assay confirmed that mice immunized with CTB-Ent developed B cells secreting antigen-specific antibodies against Ent in the Peyer’s patches (Fig. 1E).
We next investigated whether the mucosal IgA elicited by CTB-Ent immunization would also recognize GlcEnt. Consistent with our expectation based on the structural attributes shared by Ent and GlcEnt (Fig. 1A), fecal IgA from mice immunized with CTB-Ent also recognized GlcEnt (Fig. 1 F and G and SI Appendix, Fig. S4B). This immunization appeared to trigger mucosal immunity to Ent and GlcEnt specifically. In conjunction with the detection of specific fecal anti-Ent and anti-GlcEnt antibodies (Fig. 1 C–G), we detected specific fecal anti-CTB IgA in mice immunized with CTB-Ent; however, anti-CTB IgA was also detected in mice immunized with unmodified CTB, as expected (Fig. 1H). Moreover, IgA that recognized unmodified CTB was secreted by B cells isolated from the Peyer’s patches in both groups of mice (Fig. 1I). We found no evidence for the production of anti-Ent/GlcEnt IgG in the spleen or blood of mice immunized with CTB-Ent, even though we detected IgG and IgA recognizing unmodified CTB in the spleen (SI Appendix, Fig. S4C). Additionally, the mucosal response to the CTB-Ent antigen was independent of the route of immunization. Mice immunized i.p. with CTB-Ent also developed B cells secreting anti-Ent/GlcEnt IgA in the Peyer’s patches, but not antibodies recognizing these antigens in the spleen (SI Appendix, Fig. S5 A and B). We thus conclude that our immunization strategy generates mucosal IgA that recognize Ent and GlcEnt. These results are in agreement with the known effects of CTB in boosting mucosal immunity (25).
Immunization with Siderophore Conjugates Results in Lower Intestinal and Systemic Colonization of an Enteric Pathogen.
Once we established the antibody response, we investigated whether the production of anti-Ent/GlcEnt IgA would limit gut colonization and systemic dissemination of Salmonella. We used the mouse model of Salmonella colitis and infected mice (C57BL/6) that are highly susceptible to the pathogen (26). Groups of 10–20 C57BL/6 mice were purchased from Taconic Biosciences and immunized with either CTB or CTB-Ent. Subsequently, the mice were treated with a single oral dose of streptomycin, followed by infection with 109 cfu per mouse of S. enterica serovar Typhimurium. In the mouse model, pretreatment with streptomycin is necessary for development of cecal and colonic inflammation during Salmonella infection (27). The transient alteration of the microbiota (28), accompanied by the increase in inducible NOS (iNOS) production (29), renders mice more prone to colitis. In this model, the development of intestinal inflammation and the recruitment of neutrophils to the gut peak at day 3–4 postinfection. We previously demonstrated that iron is limited in the inflamed gut at these time points (18, 19) and that Salmonella uses GlcEnt to evade iron sequestration by LCN2 and thereby thrive in this environment and outcompete the microbiota (18).
At both days 1 and 2 postinfection, when levels of inflammation are low and iron is more abundant, we observed similar levels of Salmonella colonization in the feces of mice immunized with CTB or with CTB-Ent (Fig. 2A and SI Appendix, Fig. S6A). In contrast, we found a significant reduction of Salmonella in the feces of mice immunized with CTB-Ent at day 3, and especially in the cecum content at day 4, postinfection (Fig. 2A). These time points correspond to the peak of inflammatory response and of iron limitation during Salmonella infection (18, 19). Salmonella colonization was lower in mice with the highest anti-Ent antibody titer, with some mice showing a 20,000-fold reduction of Salmonella in the cecal content (Fig. 2B). These results demonstrate that our immunization strategy successfully hindered the growth advantage of Salmonella in the inflamed gut.
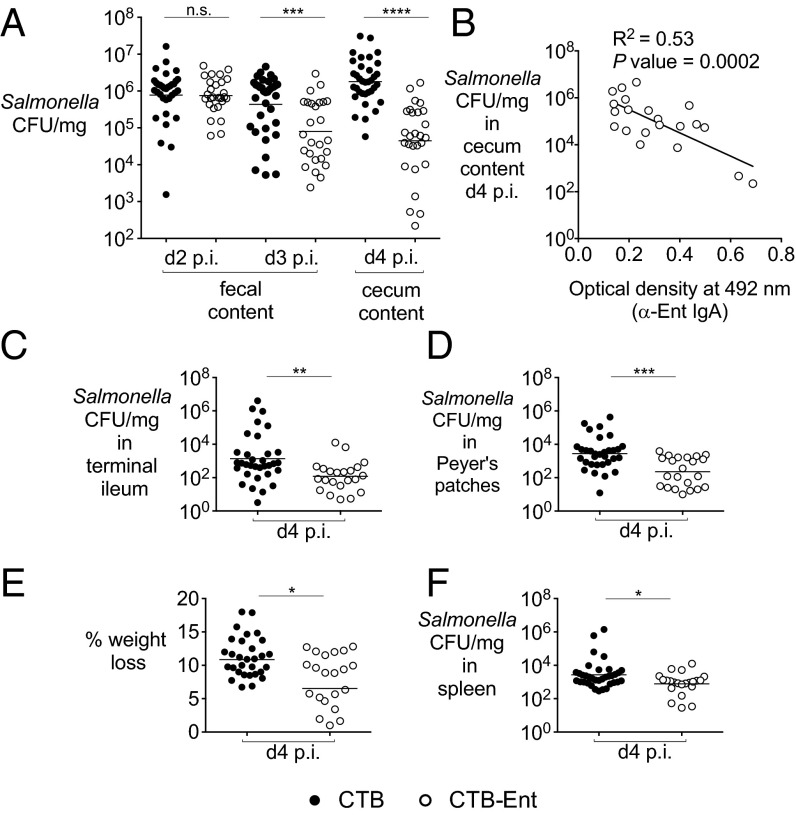
Fig. 2.
Immunization of mice with CTB-Ent reduces Salmonella gut colonization and systemic dissemination. (A) Salmonella colonization in the colon content of mice immunized intranasally with either CTB (n = 35) or CTB-Ent (n = 27) and then infected with 109 colony-forming units (cfu) of Salmonella by oral gavage. (B) Scatter plot showing an inverse correlation between Salmonella colony-forming units in the colon content at day 4 postinfection and anti-Ent IgA detected by ELISA in individual mice that were immunized with CTB-Ent before infection (n = 21). Salmonella cfu per milligram in the terminal ileum (C) and Peyer’s patches (D) of mice immunized with CTB (n = 33) or CTB-Ent (n = 22) at day 4 postinfection are shown. (E) Weight loss of mice immunized with either CTB or CTB-Ent at day 4 postinfection (n = 15 per group). (F) Salmonella cfu per milligram in the spleen of mice immunized with either CTB (n = 33) or CTB-Ent (n = 22) at day 4 postinfection. In A, C, D, and F, each circle represents an individual mouse. Bars represent the geometric mean. In E, bars represent the mean ± SE. Immunization with challenge experiments were repeated six times. A different batch of CTB/CTB-Ent was used each time. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. p.i., postinfection.
Salmonella colonization of the terminal ileum and Peyer’s patches (Fig. 2 C and D), but not of the mesenteric lymph nodes (SI Appendix, Fig. S6D), was also reduced in mice immunized with CTB-Ent. Moreover, mice immunized with CTB-Ent exhibited reduced weight loss (Fig. 2E) and reduced Salmonella burden in the spleen (Fig. 2F) compared with mock-immunized mice, indicating that CTB-Ent immunization also slows disease progression. Because C57BL/6 mice are very susceptible to lethal infection by Salmonella, all mock-immunized mice had to be euthanized at day 4 because of marked weight loss and high bacterial burden (SI Appendix, Fig. S6A). In contrast, mice immunized with CTB-Ent exhibited less severe weight loss (Fig. 2E) and could survive infection for two additional days (SI Appendix, Fig. S6A). Moreover, when mice were immunized i.p. with CTB-Ent, we observed similar reductions of Salmonella colonization of the gut and of the spleen (SI Appendix, Fig. S5 C and D).
To ensure that the success of our immunization strategy was independent of housing conditions and differences in the gut microbiota, we immunized and infected mice from two colonies bred in our vivarium. The first colony consisted of conventional C57BL/6 mice, and the second colony consisted of C57BL/6 mice engineered to express functional natural resistance-associated macrophage protein 1 (NRAMP1) (30). In our hands, these C57BL/6 NRAMP1+ mice survive up to 8–9 d postinfection. Analogous to the mice purchased from Taconic, the C57BL/6 mice from the first colony produced anti-Ent IgA in the Peyer’s patches and exhibited reduced Salmonella colonization in the feces at day 4 postinfection (SI Appendix, Fig. S6E). Moreover, in the C57BL/6 NRAMP1+ mice, we observed a significant reduction in Salmonella burden in the cecum at day 8 postinfection as a result of CTB-Ent immunization (SI Appendix, Fig. S6F). Taken together, these results indicate that our immunization strategy was reproducible in different mouse lines.
Based on our findings suggesting that anti-Ent/GlcEnt IgA produced in response to CTB-Ent immunization sequesters GlcEnt in the gut lumen, we evaluated whether the competitive advantage of wild-type Salmonella over a mutant in the GlcEnt receptor IroN is reduced in mice immunized with CTB-Ent. We have previously shown that the iroN mutant is defective in colonization of the inflamed gut, where LCN2 is highly induced (18). Moreover, the iroN mutant is outcompeted by Salmonella wild-type 10- to 100-fold during intestinal inflammation (18). We reasoned that anti-Ent/GlcEnt IgA produced in response to CTB-Ent immunization would bind to GlcEnt in the gut lumen, thereby inhibiting growth of wild-type Salmonella. Consistent with our hypothesis, the competitive advantage of wild-type Salmonella over the iroN mutant was significantly reduced by ∼10-fold in mice immunized with CTB-Ent (SI Appendix, Fig. S6C). These results further support the model that sequestration of GlcEnt by specific IgA contributes to the reduction of Salmonella colonization in mice immunized with CTB-Ent.
Pathology and Inflammation Are Similar in Mice Immunized with Either Carrier Proteins or Siderophore Conjugates.
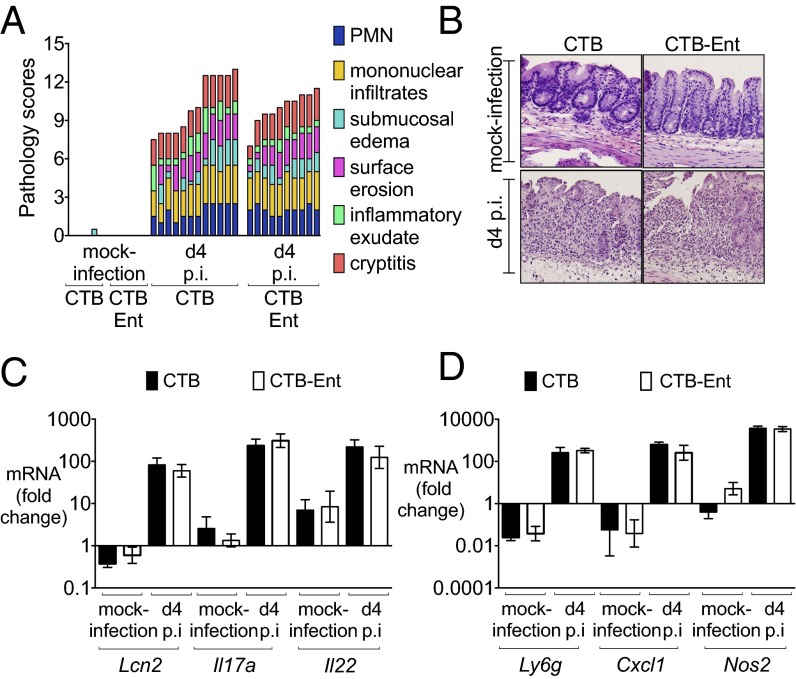
Because Salmonella thrives in the inflamed gut and benefits from inflammatory responses from the host during infection (31), we questioned whether the lower Salmonella levels in the colon content resulted from reduced inflammation in mice immunized with CTB-Ent (32, 33). Blinded histological analysis ruled out this possibility because similar levels of intestinal inflammation were observed in mock-immunized and CTB-Ent–immunized mice upon Salmonella infection (Fig. 3 A and B and SI Appendix, Fig. S6B). Moreover, several inflammatory markers, including genes encoding LCN2, interleukin (IL)-17A, IL-22, CXCL1, Ly-6G, and iNOS, were similarly up-regulated in both groups of mice after infection (Fig. 3 C and D). Taken together, these results demonstrate that lower levels of Salmonella gut colonization occurred in mice immunized with CTB-Ent despite similarly high levels of intestinal inflammation.
Fig. 3.
Mice immunized with CTB-Ent show similar pathology and inflammation as CTB-immunized mice. (A) Blinded histopathology scores of mice immunized with CTB or CTB-Ent at day 4 postinfection. (B) H&E-stained sections from representative animals for each group (40×). (C) Relative expression of Lcn2, Il17a, and Il22 in cecal samples of mice immunized with CTB (n = 7) or CTB-Ent (n = 7) and then mock-infected (n = 12) or infected (n = 12) with Salmonella for 4 d. (D) Relative expression of Ly6g, Cxcl1, and Nos2 in cecal samples of mice immunized with CTB (n = 7) or CTB-Ent (n = 7) and then mock-infected (n = 12) or infected (n = 12) with Salmonella for 4 d. In C and D, bars represent the mean ± SE.
Lactobacillus spp. Expand in the Context of Diminished Salmonella Intestinal Colonization.
One consequence of intestinal inflammation, whether induced by Salmonella infection or by other infectious and noninfectious etiologies, is a profound alteration of the gut microbiota, termed “dysbiosis.” The highly oxidative environment in the inflamed gut (34) reduces the growth of obligate anaerobes, such as Bacteroidetes and Clostridiales, which constitute ∼99% of microbes in the normal, noninflamed gut. In contrast, inflammation promotes the bloom of Enterobacteriaceae, which usually constitute less than 1% of the microbiota in the absence of intestinal inflammation (35). Commensal and pathogenic facultative anaerobes, including Salmonella, can respire novel electron acceptors (e.g., nitrate, tetrathionate) that only become available in the inflamed gut, thus outcompeting the resident microbiota (36, 37). Moreover, acquisition of metal ions, including iron in the inflamed gut, enhances the capability of Salmonella to thrive in this environment (38). Because we observed lower levels of Salmonella in mice immunized with CTB-Ent (Fig. 2A), we questioned the impact of our immunization strategy on the microbiota in the inflamed gut. In particular, we sought to identify which bacterial species thrive in the inflamed gut when Salmonella growth was hindered by CTB-Ent immunization.
The composition of the gut microbiota was not significantly different in mice that were immunized with either CTB or CTB-Ent at 36–51 d postimmunization (Fig. 4A and SI Appendix, Tables S1 and S2). The microbiota in these mice was largely composed of Bacteroidetes and Firmicutes, whereas Proteobacteria (Proteus and Desulfovibrio) constituted only a small fraction of the gut microbiota (SI Appendix, Table S1). Overall, the microbiota composition in these mice was comparable to prior studies in naive mice in the absence of intestinal inflammation (32).
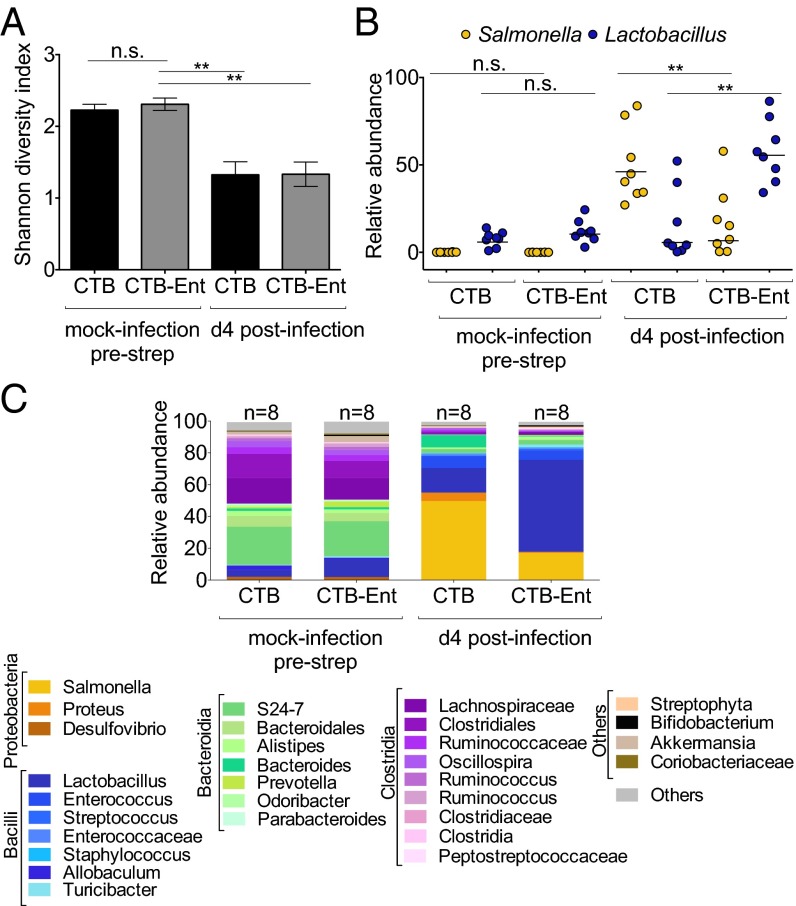
Fig. 4.
Analysis of the gut microbiota in mice immunized with CTB or CTB-Ent before and after infection with Salmonella. (A) Gut bacterial diversity, measured by the Shannon diversity index, in mice immunized with CTB or CTB-Ent, before infection and 4 d after infection with Salmonella (n = 8 per group). (B) Analysis of the relative abundance of Salmonella and Lactobacillus in individual mice (n = 8 per group) measured by Illumina MiSeq in mice immunized with CTB or CTB-Ent, before infection and 4 d after infection with Salmonella. (C) Relative abundance of order/family/genus 16S rDNA sequence assignments in fecal samples from mice immunized with CTB or CTB-Ent, collected before infection (mock, n = 8) and at day 4 postinfection (infected, n = 8). In A, bars represent the mean ± SE. In B, each circle represents an individual mouse. Bars represent the geometric mean. **P < 0.01.
In agreement with earlier studies in naive mice (32), we found that Salmonella infection induced a significant decrease in bacterial diversity at day 4 postinfection in mice immunized with either CTB or CTB-Ent (Fig. 4A and SI Appendix, Table S2). As observed previously in naive mice (32), Salmonella constituted an average of 50% of the gut microbes in mock-immunized mice (Fig. 4 B and C and SI Appendix, Fig. S7). Notably, this value dropped to an average of 15% in mice immunized with CTB-Ent (Fig. 4 B and C and SI Appendix, Fig. S7), which was in accord with the reduction of Salmonella cecal colony-forming units in these mice (Fig. 2A). Commensal Enterobacteriaceae (e.g., Proteus spp.) increased only slightly in CTB-immunized mice during Salmonella infection. This result agrees with prior studies (32), which showed that Salmonella outcompetes other Proteobacteria in the inflamed gut. Moreover, commensal Enterobacteriaceae did not expand in the inflamed gut of mice immunized with CTB-Ent 4 d after Salmonella infection. Because all Enterobacteriaceae that experience conditions of iron limitation deploy Ent to acquire this nutrient, we reason that the production of anti-Ent IgA limits the expansion of this family in the inflamed gut by sequestering this common siderophore.
Because of the similarly high levels of intestinal inflammation (Fig. 3), most other bacterial genera were similarly affected during Salmonella infection in both groups of mice (Fig. 4C and SI Appendix, Table S1). However, we detected a significant expansion of Lactobacillus spp. in the inflamed gut of mice immunized with CTB-Ent (50% of the microbiota), but not in CTB-immunized mice (15% of the microbiota) (Fig. 4 B and C and SI Appendix, Fig. S7). These results are in agreement with previous findings showing that Lactobacillus spp. also replicate in a similar inflamed environment as Enterobacteriaceae and also benefit from inflammation, albeit by unknown mechanisms (32). Consistent with this idea, Lactobacillus was recently shown to expand during noninfectious colitis and to up-regulate TonB-dependent receptor components in the inflamed gut, suggesting that this genus can also acquire essential metal nutrients in this environment (39). Importantly, Lactobacillus spp. are considered beneficial microbes, and a few species are even administered as therapeutic agents (40). By reducing Salmonella colonization without reducing intestinal inflammation, it appears that our immunization strategy indirectly promoted the expansion of beneficial microbes that also thrive in the inflamed gut, thereby providing additional benefits to the host.
Summary and Conclusions
For many years, siderophores, as well as the biosynthetic and transport machineries for these virulence factors, have garnered significant interest as targets for new antibiotics. Reported efforts include the design and application of siderophore–antibiotic conjugates for targeted drug delivery, the identification of small-molecule inhibitors of siderophore biosynthesis, and the inhibition of siderophore uptake by immunization against siderophore receptors or siderophores (9, 41–43). A prior study described the development of antibodies against vibriobactin; however, that study did not evaluate whether the vaccination provided protection to the host during infection with Vibrio cholerae (44). Our work establishes that immunization against bacterial siderophores results in the production of anti-siderophore antibodies and affords reduced intestinal colonization (up to 20,000-fold) during infection with the enteric pathogen Salmonella. These results are particularly significant because Salmonella is known to thrive in the inflamed gut and to outcompete the microbiota in this environment. Our immunization shifted the balance in favor of the microbiota, promoting the expansion of beneficial microbes (e.g., Lactobacillus) that thrive in a similar environment. Moreover, because only mice that shed the highest level of Salmonella (the so-called “supershedders”) are able to achieve successful transmission of the pathogen to naive hosts (45), we reason that our strategy, which reduces fecal shedding, could be used to hinder Salmonella transmission. Because Salmonella intestinal colonization was most highly reduced in mice that developed the highest antibody titers, future studies will address further optimization of the immunization strategy to increase antibody production.
In contrast to the mouse model, nontyphoidal Salmonella remains localized to the gut in most patients. We predict that anti-siderophore antibodies, either generated in the gut in response to immunization or therapeutically administered to patients infected with Salmonella, could reduce fecal shedding of the pathogen, thereby limiting its spread and reducing recovery time. Moreover, future immunization studies aimed at generating a systemic antibody response to siderophores in mice will determine whether this strategy could lead to greater protection at systemic sites, which would be informative for developing new preventive or therapeutic strategies for systemic salmonellosis.
This immunization strategy could be further developed as a therapy for infections caused by pathogens that use other siderophores to thrive in the mammalian host. We expect that development of resistance against antibody-based immunization would develop only if a new siderophore biosynthesis gene cluster is acquired by horizontal gene transfer, and future studies will address this notion. In principle, our strategy can be applied to every siderophore, which would be useful in the event that resistance develops, or to target pathogens that use more than one siderophore. Moreover, the development of anti-siderophore antibodies will likely provide narrow-spectrum antimicrobial activity by targeting only microbes that use that subset of siderophores while having a minimal impact on the microbiota (in contrast to broad- and even narrow-spectrum antibiotics). Broadly, our work indicates that new preventive and therapeutic strategies for microbial infections should target siderophore-mediated iron acquisition, a virulence trait shared by almost all bacterial and fungal pathogens (9). Moreover, because recent studies demonstrate additional, nonclassical roles for siderophores (46, 47), including protection of bacteria from oxidative stress (48) and modulation of host gene expression (49), it is possible that antibody-mediated sequestration of siderophores may inhibit bacterial growth by iron-independent mechanisms and may provide additional benefits to the host.
Methods
The preparation and characterization of CTB-Ent, Ent-PEG3-biotin, GlcEnt-PEG3-biotin, and native siderophores are described in SI Appendix. Six- to eight-week-old C57BL/6 female mice were purchased from Taconic. When indicated, 6- to 8-wk-old male or female C57BL/6 or C57BL/6 NRAMP1+ mice bred in our vivarium were also used for the experiments; 8 to 10 mice per experimental group were used for each experiment. The Institutional Animal Care and Use Committee at the University of California, Irvine approved all of the mouse experiments. Mucosal immunization with CTB, CTB-Ent conjugates, analysis of antibody production by ELISA and ELISPOT, infection models, histopathology, and analysis of gene expression and of the microbiota are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Matthew Rolston (University of California, Davis, Host-Microbe Systems Biology Core) for processing samples for Illumina MiSeq analysis. This work was supported by the Pacific Southwest Regional Center of Excellence for Biodefense and Emerging Infectious Disease [supported by Public Health Service (PHS) Grant U54AI065359] and by PHS Grants AI101784 and AI114625 (to M.R. and E.M.N.). Work in the laboratory of M.R. is also supported by PHS Grants AI105374, AI126277, AI121928, and DK058057. Work in the laboratory of E.M.N. is also supported by the Kinship Foundation Searle Scholar Award (to E.M.N.) and the Massachusetts Institute of Technology (MIT) Department of Chemistry. M.R. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. A.P.-L. was funded by a University of California Institute for Mexico and the United States (MEXUS) and El Consejo Nacional de Ciencia y Tecnología (CONACYT) (MEXUS-CONACYT) award. P.C. is a recipient of a Royal Thai Government Fellowship. NMR instrumentation in the MIT Department of Chemistry Instrumentation Facility is supported by National Science Foundation Grants CHE-9808061 and DBI-9729592.
Footnotes
Conflict of interest statement: M.S.-C., P.C., A.P.-L., E.M.N., and M.R. hold a patent related to this work. M.D.G. is an employee of Roche.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606290113/-/DCSupplemental.
References
- 1.World Health Organization (2014) Antimicrobial Resistance: Global Report on Surveillance 2014 (World Health Organization, Geneva)
- 2. Anonymous (2013) The antibiotic alarm. Nature 495(7440):141. [DOI] [PubMed]
- 3.Rohmer L, Hocquet D, Miller SI. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 2011;19(7):341–348. doi: 10.1016/j.tim.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66(2):223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol. 2006;2(3):132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 6.Leong J, Neilands JB. Mechanisms of siderophore iron transport in enteric bacteria. J Bacteriol. 1976;126(2):823–830. doi: 10.1128/jb.126.2.823-830.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71(3):413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratledge C. Iron metabolism and infection. Food Nutr Bull. 2007;28(4) Suppl:S515–S523. doi: 10.1177/15648265070284S405. [DOI] [PubMed] [Google Scholar]
- 9.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13(5):509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raymond KN, Dertz EA, Kim SS. Enterobactin: An archetype for microbial iron transport. Proc Natl Acad Sci USA. 2003;100(7):3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberg ED. Nutritional immunity. Host’s attempt to withold iron from microbial invaders. JAMA. 1975;231(1):39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- 12.Goetz DH, et al. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10(5):1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 13.Flo TH, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432(7019):917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 14.Fischbach MA, Lin H, Liu DR, Walsh CT. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc Natl Acad Sci USA. 2005;102(3):571–576. doi: 10.1073/pnas.0408463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci USA. 2003;100(7):3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bäumler AJ, et al. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J Bacteriol. 1998;180(6):1446–1453. doi: 10.1128/jb.180.6.1446-1453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischbach MA, et al. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc Natl Acad Sci USA. 2006;103(44):16502–16507. doi: 10.1073/pnas.0604636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raffatellu M, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5(5):476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deriu E, et al. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14(1):26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachman MA, et al. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun. 2011;79(8):3309–3316. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sánchez J, Holmgren J. Cholera toxin - a foe & a friend. Indian J Med Res. 2011;133(2):153–163. [PMC free article] [PubMed] [Google Scholar]
- 22.Landrø L, et al. Decreased serum lipocalin-2 levels in human immunodeficiency virus-infected patients: Increase during highly active anti-retroviral therapy. Clin Exp Immunol. 2008;152(1):57–63. doi: 10.1111/j.1365-2249.2008.03592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng T, Bullock JL, Nolan EM. Siderophore-mediated cargo delivery to the cytoplasm of Escherichia coli and Pseudomonas aeruginosa: Syntheses of monofunctionalized enterobactin scaffolds and evaluation of enterobactin-cargo conjugate uptake. J Am Chem Soc. 2012;134(44):18388–18400. doi: 10.1021/ja3077268. [DOI] [PubMed] [Google Scholar]
- 24.Zheng T, Nolan EM. Enterobactin-mediated delivery of β-lactam antibiotics enhances antibacterial activity against pathogenic Escherichia coli. J Am Chem Soc. 2014;136(27):9677–9691. doi: 10.1021/ja503911p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmgren J, Czerkinsky C. Mucosal immunity and vaccines. Nat Med. 2005;11(4) Suppl:S45–S53. doi: 10.1038/nm1213. [DOI] [PubMed] [Google Scholar]
- 26.Hapfelmeier S, Hardt WD. A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol. 2005;13(10):497–503. doi: 10.1016/j.tim.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Barthel M, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71(5):2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stecher B, et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5(10):2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spees AM, et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio. 2013;4(4):e00430-13. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barton CH, Biggs TE, Baker ST, Bowen H, Atkinson PGP. Nramp1: A link between intracellular iron transport and innate resistance to intracellular pathogens. J Leukoc Biol. 1999;66(5):757–762. doi: 10.1002/jlb.66.5.757. [DOI] [PubMed] [Google Scholar]
- 31.Santos RL, et al. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 2009;17(11):498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behnsen J, et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40(2):262–273. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winter SE, Lopez CA, Bäumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14(4):319–327. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rigottier-Gois L. Dysbiosis in inflammatory bowel diseases: The oxygen hypothesis. ISME J. 2013;7(7):1256–1261. doi: 10.1038/ismej.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter SE, Bäumler AJ. Why related bacterial species bloom simultaneously in the gut: Principles underlying the ‘Like will to like’ concept. Cell Microbiol. 2014;16(2):179–184. doi: 10.1111/cmi.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winter SE, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339(6120):708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiennimitr P, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA. 2011;108(42):17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raffatellu M, Bäumler AJ. Salmonella’s iron armor for battling the host and its microbiota. Gut Microbes. 2010;1(1):70–72. doi: 10.4161/gmic.1.1.10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ilott NE, et al. Defining the microbial transcriptional response to colitis through integrated host and microbiome profiling. ISME J. 2016;10(10):2389–2404. doi: 10.1038/ismej.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behnsen J, Deriu E, Sassone-Corsi M, Raffatellu M. Probiotics: Properties, examples, and specific applications. Cold Spring Harb Perspect Med. 2013;3(3):a010074. doi: 10.1101/cshperspect.a010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji C, Juárez-Hernández RE, Miller MJ. Exploiting bacterial iron acquisition: Siderophore conjugates. Future Med Chem. 2012;4(3):297–313. doi: 10.4155/fmc.11.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolin CA, Jensen AE. Passive immunization with antibodies against iron-regulated outer membrane proteins protects turkeys from Escherichia coli septicemia. Infect Immun. 1987;55(5):1239–1242. doi: 10.1128/iai.55.5.1239-1242.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clifton-Hadley FA, et al. A laboratory study of an inactivated bivalent iron restricted Salmonella enterica serovars Enteritidis and Typhimurium dual vaccine against Typhimurium challenge in chickens. Vet Microbiol. 2002;89(2-3):167–179. doi: 10.1016/s0378-1135(02)00169-4. [DOI] [PubMed] [Google Scholar]
- 44.Bergeron RJ, et al. Vibriobactin antibodies: A vaccine strategy. J Med Chem. 2009;52(12):3801–3813. doi: 10.1021/jm900119q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gopinath S, Carden S, Monack D. Shedding light on Salmonella carriers. Trends Microbiol. 2012;20(7):320–327. doi: 10.1016/j.tim.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Holden VI, Bachman MA. Diverging roles of bacterial siderophores during infection. Metallomics. 2015;7(6):986–995. doi: 10.1039/c4mt00333k. [DOI] [PubMed] [Google Scholar]
- 47.Johnstone TC, Nolan EM. Beyond iron: Non-classical biological functions of bacterial siderophores. Dalton Trans. 2015;44(14):6320–6339. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adler C, et al. The alternative role of enterobactin as an oxidative stress protector allows Escherichia coli colony development. PLoS One. 2014;9(1):e84734. doi: 10.1371/journal.pone.0084734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holden VI, et al. Bacterial siderophores that evade or overwhelm lipocalin 2 induce hypoxia inducible factor 1α and proinflammatory cytokine secretion in cultured respiratory epithelial cells. Infect Immun. 2014;82(9):3826–3836. doi: 10.1128/IAI.01849-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.