The genomes of retroviruses, such as HIV, consist of two single-stranded RNAs held together as a dimer through noncovalent interactions near the 5′-end. Why retroviruses carry two copies of their genetic material when most other viruses carry a single copy is something of a mystery. The single-stranded genomic RNA (gRNA) is susceptible to damage; having two copies of the RNA may help preserve the genetic information during reverse transcription (1, 2). The dimer also promotes genetic diversity through frequent recombination events, allowing efficient production of variants that can evade the host immune response or resist antiviral treatments (1, 3). In addition, gRNA dimerization has long been suspected to play structural roles, particularly during selective packaging of the viral gRNA (4, 5). A recent study by Keane et al. provides new insight into how the two ∼9-kb HIV gRNA copies are held together in the dimer and how dimerization involves a structural switch that regulates packaging and other steps during viral replication (6).
The structure of the retroviral gRNA dimer has been pursued ever since it was initially proposed, ∼50 y ago, to explain the observation that retroviral particles contain RNA with twice the buoyant density as expected for the single gRNA (7). Early electron microscopy studies indicated that the two RNAs are held together near their 5′-ends at a region referred to as the dimer linkage structure (8). The dimer linkage structure is located within the ∼350-nt 5′-leader of the HIV gRNA, which contains several highly structured and conserved stem-loops that play essential roles at distinct steps during viral replication (9). Among these stem-loops, the dimerization initiation site (DIS) was identified early on as important for dimerization (10–12). Isolated DIS stem-loops spontaneously form “kissing” dimers through self-pairing of the palindromic apical loop. This labile kissing dimer converts into a more stable duplex dimer (Fig. 1B) through heat treatment or with the aid of the nucleocapsid (NC) protein, a small RNA chaperone necessary for gRNA maturation and packaging, which is generated by cleavage of the viral Gag polyprotein (13, 14). In addition to the DIS, early in vitro reactivity experiments suggested that the dimer is stabilized by other elements in the leader, including the transactivation response element (TAR), the major splice-site donor stem (SD), and long-range base pairing between residues overlapping the gag start codon (AUG) and the unique-5′ element (U5) (15–17). However, in the absence of high-resolution structural information, conflicting structural models for the monomeric and dimeric leader could not be reconciled, and the role of dimerization in packaging and other viral replication steps remained elusive.
Fig. 1.
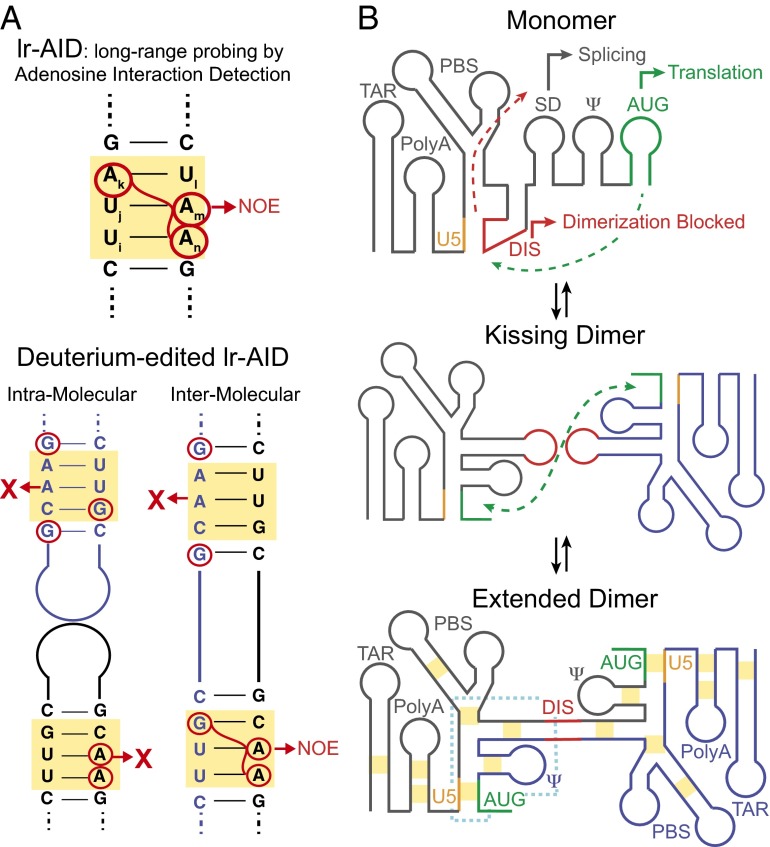
New NMR strategy allows characterization of the dimeric interface in the HIV 5′-leader. (A) lr-AID relies on the unique NMR chemical shifts produced by specific base-pair sequences to resolve signals from congested NMR spectra of large RNAs. By differentially silencing all residues other than adenosine or guanosine with deuterium, the deuterium-edited lr-AID approach can distinguish between inter- or intramolecular base-pairing interactions. (B) Proposed mechanism of HIV leader dimerization. In the monomer, the SD and AUG are exposed to promote splicing and translation, respectively. Conversely, the DIS is sequestered by U5, preventing dimerization. In the dimer, DIS dimerization is extended and intermolecular U5:AUG interactions are formed. Yellow boxes indicate the regions probed by deuterium-edited lr-AID and the dotted blue line indicates the core encapsidation signal. The proposed intermediate features a kissing DIS dimer, which was not observed in the present study possibly because it is too low populated and short-lived.
Obtaining high-resolution structures for large RNAs, such as the ∼350-nt HIV leader, presents a significant challenge to techniques, such as X-ray crystallography and NMR spectroscopy. These RNAs are typically too flexible and heterogeneous to be crystallized for X-ray crystallography and too large for solution-state NMR spectroscopy. To circumvent these problems, structural biologists have pursued a divide-and-conquer strategy in which high-resolution structures are obtained for individual stem-loops excised from their native context. Although this strategy was justified by studies showing that individual stem-loops fold autonomously to carry out distinct functions, there was also evidence for overlapping functions that could be encoded at the level of the entire leader structure.
A recent series of studies by Summers and coworkers sought to overcome the size limitations of NMR, culminating in the recent structural characterization of the 688-nt full-length HIV-1 RNA 5′-leader dimer by Keane et al. (6). Any NMR spectroscopist in their right mind would not think of tackling an RNA this large: more than an order-of-magnitude larger than the largest RNAs typically studied by NMR. Even if appropriate samples could be prepared, severe spectral overcrowding and decreased sensitivity because of slower overall molecular tumbling would surely guarantee a featureless, undecipherable blob of many unresolved signals. So how can any structural information be obtained on RNAs this large using NMR?
Many years ago, Summers and coworkers recognized that helical stretches of Watson–Crick base pairs with particular sequence (e.g., [UiUjAk]:[UlAmAn]) give rise to unique upfield shifted NMR 1H signals that self-separate from the otherwise congested spectrum (Fig. 1A) (18). This result allows for resolution of cross-strand distance-dependent 1H-1H nuclear Overhauser effects (NOEs) that can be used to verify base pairing. By mutating residues in suspected helices into these NMR-friendly motifs, the authors could assess long-range interactions at specific regions in the monomeric and dimeric leader (18). Control experiments were used to ensure that the substitution mutations do not affect the structural and functional integrity of the leader. This strategy, termed “long-range probing by adenosine interaction detection” (lr-AID), revealed that in the monomeric leader, the DIS hairpin is sequestered through pairing with U5, whereas the AUG hairpin is accessible for translation (Fig. 1B). Upon dimerization, AUG pairs with U5, thus blocking translation while also displacing DIS, such as to make it available for dimerization and packaging (Fig. 1B). This conformational switch was further supported by NMR data showing that NMR signals of AUG in the monomeric leader match those of an isolated AUG hairpin, and AUG signals in the dimeric leader match those of an isolated AUG–U5 duplex, and by experiments showing that mutations that inhibit or promote these interactions have the expected effects on dimerization and packaging (18). Recent studies on HIV-2 and simian immunodeficiency virus suggest that this molecular switch is highly conserved among retroviruses (19).
Additional mutational studies helped define a minimal core encapsidation signal (ΨCES) within the leader that retains the key structural features of the dimer, as well as elements needed to direct packaging (Fig. 1B) (20). Using a segmental labeling strategy, the authors went on to solve a high-resolution structure of the 155-nt ΨCES, which is the largest RNA structure determined by NMR to date (21). To minimize the size of the RNA, TAR, polyadenylation (PolyA) signal, and the upper region of the primer binding site (PBS) were removed and the DIS apical loop was replaced with a GAGA loop, thus abrogating dimerization. The structure adopted an unexpected tandem three-way junction architecture, exposing guanosines essential for packaging and high-affinity binding to the NC. The AUG start codon (in AUG) and the SD are both sequestered through long-range pairing, providing a mechanism by which dimerization can suppress both translation and splicing (Fig. 1B). The structure also helps explain the exquisite selectivity of packaging toward a full-length, unspliced dimer; neither spliced mRNA nor monomeric gRNA are capable of adopting this tandem three-way junction architecture.
Despite these advances, important details were missing regarding the dimeric interface. It was yet to be conclusively demonstrated that the DIS hairpin does indeed mediate pairing between the two leaders. Moreover, it was unclear whether the U5:AUG interaction observed in the dimer was intramolecular, or possibly intermolecular, thereby extending the dimeric interface beyond the DIS. Finally, it remained unclear whether or not other elements in the leader, such as the TAR and SD, are involved in dimerization.
To address the above questions, Keane et al. adapted the lr-AID strategy to make it possible to distinguish intra- from intermolecular base paring (Fig. 1A) (6). In this approach, a 1:1 mixture is prepared, consisting of two leader RNA constructs containing distinct NMR labeling patterns. In one construct, all residues except adenosine are “NMR-silenced” via deuteration, whereas in the second construct, all residues except guanosine are silenced. In this manner, 1H-1H NOEs between guanosine and adenosine residues in the dimeric leader are only observed if the two molecules are brought into proximity via intermolecular pairing (Fig. 1A). Conversely, the corresponding intramolecular NOEs are abolished because one of the NOE partners is silenced through deuteration (Fig. 1A).
Using this approach, Keane et al. probed for intermolecular pairing in the dimeric leader at a number of positions previously proposed to be involved in dimerization (Fig. 1B) (6). This approach confirmed formation of an extended duplex DIS dimer within the context of the dimeric leader. Interestingly, unlike the isolated DIS, the extended duplex dimer formed spontaneously within the leader context at physiological temperatures without the need for the NC. The NMR data did not provide evidence for the kissing DIS dimer species (Fig. 1B), suggesting that if it is present, it exists in low abundance or transiently before duplex formation.
Strikingly, this NMR approach revealed that the U5:AUG pairing observed previously in the leader dimer is intermolecular, with one molecule contributing U5 and another AUG (Fig. 1B). This finding helps explain why mutations in U5 and AUG that disrupt U5:AUG pairing inhibit dimerization (16, 18). These effects were previously attributed to an intramolecular switch that promotes dimerization by exposing the DIS (18). The present study indicates that in addition to liberating the DIS, the intermolecular U5:AUG can directly stabilize the dimer, which forms on a timescale comparable to overall leader dimerization based on time-resolved NMR. No evidence was obtained for intermolecular pairing involving PolyA, PBS, TAR, or the Ψ packaging element. However, Keane et al. (6) point out that intermolecular pairing outside the probed region cannot be ruled out. For example, although no evidence was obtained for intermolecular pairing in the TAR hairpin below the bulge (Fig. 1B), this does not rule out intermolecular pairing involving the TAR upper stem or apical loop. Further studies are needed to fully visualize the dimeric interface and how it might change upon binding to the NC during encapsidation.
By revealing the structure of the dimeric interface of the HIV leader, the study by Keane et al. (6) caps a series of NMR investigations that have illuminated a structural role for HIV gRNA dimerization as a molecular switch that converts a monomeric species competent for translation into a dimeric species stabilized by an extended interface that suppresses translation and splicing and that promotes selective packaging (Fig. 1B). The NMR strategy introduced by Keane et al. provides a new approach to gain high-resolution structural insights into RNA–RNA interactions in systems that are inaccessible to conventional high-resolution structure-determination methods.
Footnotes
The authors declare no conflict of interest.
See companion article on page 13033 in issue 46 of volume 113.
References
- 1.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev. 2009;73(3):451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King SR, Duggal NK, Ndongmo CB, Pacut C, Telesnitsky A. Pseudodiploid genome organization AIDS full-length human immunodeficiency virus type 1 DNA synthesis. J Virol. 2008;82(5):2376–2384. doi: 10.1128/JVI.02100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nora T, et al. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J Virol. 2007;81(14):7620–7628. doi: 10.1128/JVI.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol. 2011;410(4):609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudin F, et al. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229(2):382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 6.Keane SC, et al. NMR detection of intermolecular interaction sites in the dimeric 5′-leader of the HIV-1 genome. Proc Natl Acad Sci USA. 2016;113(46):13033–13038. doi: 10.1073/pnas.1614785113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson WS, Robinson HL, Duesberg PH. Tumor virus RNA’s. Proc Natl Acad Sci USA. 1967;58(3):825–834. doi: 10.1073/pnas.58.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender W, Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976;7(4):595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- 9.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 10.Laughrea M, Jetté L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33(45):13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 11.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91(11):4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paillart JC, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269(44):27486–27493. [PubMed] [Google Scholar]
- 13.Muriaux D, De Rocquigny H, Roques BP, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J Biol Chem. 1996;271(52):33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 14.Muriaux D, Fossé P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35(15):5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 15.Deforges J, Chamond N, Sargueil B. Structural investigation of HIV-1 genomic RNA dimerization process reveals a role for the Major Splice-site Donor stem loop. Biochimie. 2012;94(7):1481–1489. doi: 10.1016/j.biochi.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Song R, Kafaie J, Laughrea M. Role of the 5′ TAR stem-loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry. 2008;47(10):3283–3293. doi: 10.1021/bi7023173. [DOI] [PubMed] [Google Scholar]
- 17.Nikolaitchik O, Rhodes TD, Ott D, Hu WS. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J Virol. 2006;80(10):4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu K, et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334(6053):242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran T, et al. Conserved determinants of lentiviral genome dimerization. Retrovirology. 2015;12(83):83. doi: 10.1186/s12977-015-0209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heng X, et al. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol. 2012;417(3):224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keane SC, et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science. 2015;348(6237):917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]