Significance
Chloroplasts arose from a cyanobacterial endosymbiont, which introduced photosynthesis into eukaryotes. It is widely believed that synchronization of division in the eukaryotic host cell and in the endosymbiont was critical for the host cell to maintain the endosymbiont/chloroplast permanently. However, it is unclear how the division of the endosymbiont (the chloroplast) and host cell became synchronized. Using the unicellular red alga Cyanidioschyzon merolae, we show that the host cell enters into the metaphase only when chloroplast division has commenced. A similar phenomenon also was observed in the glaucophyte alga Cyanophora paradoxa. It thus seems likely that the acquisition of the cell-cycle checkpoint of chloroplast division played an important role in the establishment of the chloroplast in ancient algae.
Keywords: chloroplast division, algal cell cycle, Cyanidioschyzon merolae, Cyanophora paradoxa
Abstract
Chloroplasts evolved from a cyanobacterial endosymbiont. It is believed that the synchronization of endosymbiotic and host cell division, as is commonly seen in existing algae, was a critical step in establishing the permanent organelle. Algal cells typically contain one or only a small number of chloroplasts that divide once per host cell cycle. This division is based partly on the S-phase–specific expression of nucleus-encoded proteins that constitute the chloroplast-division machinery. In this study, using the red alga Cyanidioschyzon merolae, we show that cell-cycle progression is arrested at the prophase when chloroplast division is blocked before the formation of the chloroplast-division machinery by the overexpression of Filamenting temperature-sensitive (Fts) Z2-1 (Fts72-1), but the cell cycle progresses when chloroplast division is blocked during division-site constriction by the overexpression of either FtsZ2-1 or a dominant-negative form of dynamin-related protein 5B (DRP5B). In the cells arrested in the prophase, the increase in the cyclin B level and the migration of cyclin-dependent kinase B (CDKB) were blocked. These results suggest that chloroplast division restricts host cell-cycle progression so that the cell cycle progresses to the metaphase only when chloroplast division has commenced. Thus, chloroplast division and host cell-cycle progression are synchronized by an interactive restriction that takes place between the nucleus and the chloroplast. In addition, we observed a similar pattern of cell-cycle arrest upon the blockage of chloroplast division in the glaucophyte alga Cyanophora paradoxa, raising the possibility that the chloroplast division checkpoint contributed to the establishment of the permanent organelle.
Chloroplasts trace their origin to a primary endosymbiotic event that took place more than a billion years ago, a process in which an ancestral cyanobacterium became integrated into a previously nonphotosynthetic eukaryote. The ancient alga that resulted from this primary endosymbiotic event evolved into the Glaucophyta (glaucophyte algae), Rhodophyta (red algae), and Viridiplantae (green algae, streptophyte algae, and land plants), which together are grouped as the Plantae (sensu stricto) or Archaeplastida. After these primitive green and red algae had become established, chloroplasts then spread into other eukaryote lineages through secondary endosymbiotic events in which a red or green alga became integrated into previously nonphotosynthetic eukaryotes (1).
The continuity of chloroplasts has been maintained for more than a billion years. The majority of algae (both unicellular and multicellular, with both possessing chloroplasts of primary and secondary endosymbiotic origin) have one or at most only a few chloroplasts per cell. Thus, chloroplast division is synchronized with the host cell cycle so that the chloroplast divides before cytokinesis and is thus transmitted into each daughter cell (2). In contrast, land plants and certain algal species contain dozens of chloroplasts per cell that divide asynchronously, even within the same cell (3). Because land plants evolved from algae, there should be a linkage between the cell cycle and chloroplast division in their algal ancestor that subsequently was modified or lost during the course of land plant evolution. Thus, it is probable that the continuity of chloroplasts in host cells was established originally by the synchronization of endosymbiotic cell division with host cell division in an ancient alga (4).
The requirement that division be synchronized to ensure permanent retention of either endosymbionts or endosymbiotic organelles is supported by the findings for several other endosymbiotic relationships. Hatena arenicola (Katablepharidophyta) has a transient green algal endosymbiont, and this photosynthetic endosymbiont is inherited by only one daughter cell during cell division. Daughter cells that have lost the endosymbiont engulf the green alga once again (5). Certain species of heterotrophic dinoflagellates engulf eukaryotic algae and use them as temporary chloroplasts (called “kleptoplasts”) for a period of days to weeks before digesting them. In certain cases, the kleptoplast divides in accord with dinoflagellate cell division and is inherited for a number of generations (6). A few dinoflagellate species maintain a eukaryotic algal unicell (i.e., containing a nucleus, mitochondria, Golgi apparatus, and so forth) as a permanent endosymbiont by synchronizing the endosymbiont cell division to the host cell cycle (7, 8). There also are eukaryotes that possess permanent cyanobacterial endosymbionts, such as Paulinella chromatophora (Cercozoa). This endosymbiont is persistently inherited by progeny cells as a consequence of the tight synchronization of the host and endosymbiotic cell cycles (9).
Currently, it is largely unknown how chloroplast division came to be coupled with cell-cycle progression in algae. However, studies over the last decade have provided information on the mechanisms underlying chloroplast division. In both algae and land plants, chloroplast division is performed by the constrictive action of a macromolecular ring-like division machinery that is comprised of a self-assembling GTPase Filamenting temperature-sensitive (Fts) Z (Fts7) of cyanobacterial endosymbiotic origin and another self-assembling GTPase dynamin, dynamin-related protein 5B (DRP5B), of eukaryotic host origin (10). Before chloroplast division, the FtsZ ring forms on the stromal side of the provisional chloroplast division site, followed by the formation of the inner PD ring of unknown molecular composition (but detectable by transmission electron microscopy) on the stromal side. Then the glucan-based outer PD ring, which is synthesized by the PDR1 protein, forms on the cytosolic side. Finally, DRP5B is recruited to the cytosolic side of the division site, and the competent chloroplast-division machinery begins to constrict (10).
We previously showed by means of various lineages of algae that possess chloroplasts of primary cyanobacterial endosymbiotic origin (glaucophyte, red, green, and streptophyte algae) that the onset of chloroplast division is restricted to the S phase by the S-phase–specific expression of some, but not all, nucleus-encoded components of the chloroplast-division machinery (11). When cell-cycle progression is arrested at the S phase, chloroplast-division genes and proteins continue to be expressed in the red alga Cyanidioschyzon merolae (11). In such S-phase–arrested cells, the chloroplast divides more than once, resulting in the emergence of abnormal cells that possess four to eight chloroplasts, in contrast to normal cells, which possess one or two chloroplasts (11, 12). Thus, it is likely that an as-yet-unknown mechanism restricts the number of chloroplast-division rounds. A plausible scenario is that the cell cycle progresses only upon the progression of chloroplast division and thereby terminates the expression of the chloroplast-division proteins.
To test this possibility, we examined the effect of blocking chloroplast division on host cell-cycle progression. We sought to determine whether cell-cycle progression is stalled until chloroplast division either progresses or is completed. The unicellular red alga C. merolae was chosen as the study organism because the molecular mechanism of chloroplast division has been well studied in this alga (2), and the nuclear and organelle genomes are completely sequenced (13–16). In addition, a procedure for nuclear gene targeting by homologous recombination has been developed (17, 18). Inducible gene-expression systems also were developed recently (19, 20).
By impairing chloroplast division in C. merolae with an inducible gene-expression system, we show that the cell cycle progresses only when chloroplast division commences. When chloroplast division was arrested before FtsZ ring formation, the host cell cycle was arrested at the prophase. In contrast, when chloroplast division was arrested during the constriction of the division site, the cell cycle progressed. These results suggest that the host cell cycle progresses to the metaphase by sensing some signal of the onset of chloroplast division to coordinate progression of the host cell cycle and chloroplast division. We have observed a similar phenomenon in the glaucophyte alga Cyanophora paradoxa. These results raise the possibility that the mechanism of the chloroplast-division checkpoint was established in ancient algae and contributed to the establishment of the permanent organelle.
Results
Experimental Design.
We planned to inhibit chloroplast division at certain stages to investigate whether host cell-cycle progression is stalled until chloroplast division progresses or is completed. However, unlike cell-cycle inhibitors, inhibitors specific to chloroplast division are not available. Thus, we applied an inducible gene-expression system using a heat-shock promoter in the unicellular red alga C. merolae (19). It is known that overexpression of FtsZ impairs FtsZ ring formation and subsequent chloroplast division in land plants (21, 22) and cell division in bacteria (23). In the case of dynamin, the expression of a dominant-negative form of human dynamin 1 (K44A) and of dynamin-related proteins with a relevant mutation that results in a defect in GTP binding and hydrolysis has been widely used to inhibit the function of the endogenous dynamin or of dynamin-related proteins, respectively (24). In addition, we previously reported that the expression of DRP5B/CmDnm2 K135A (which corresponds to K44A of human dynamin 1) inhibits chloroplast division in C. merolae cells (19), although its effect on the chloroplast-division machinery was not examined.
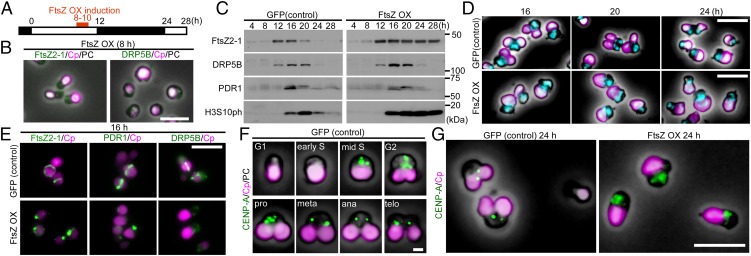
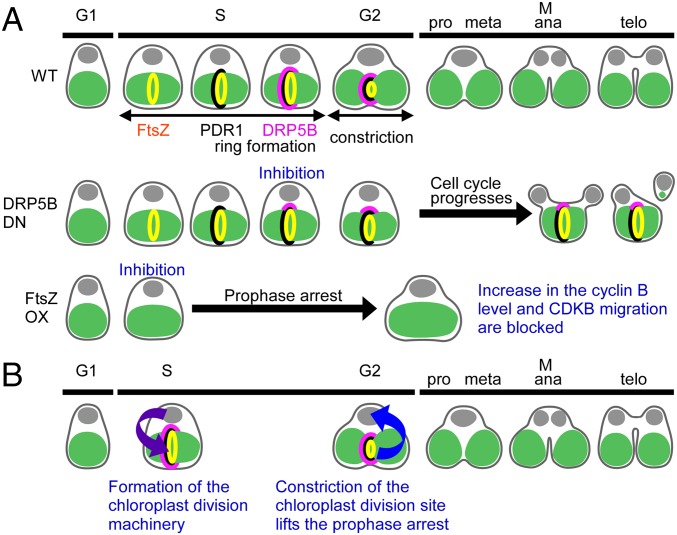
We integrated the heat-shock promoter (the promoter of HSP20, CMJ101C) and the FtsZ or DRP5B K135A ORF fusion into a C. merolae chromosomal locus and induced protein expression by shifting the temperature from 42 °C, which is optimal for growth, to 50 °C. Then we examined the effect of the overexpression of the respective proteins on the formation and constriction of the chloroplast-division machinery and on cell-cycle progression. The chloroplast-division stage was examined by the localization of FtsZ, PDR1, and DRP5B and by the degree of constriction of the chloroplast-division site. The cell-cycle stage was defined based on the cellular and chloroplast shape, the expression of cell-cycle marker genes and on the expression and localization of cell-cycle marker proteins based on previous studies (Fig. 1A) (25). In brief, previous studies showed that the chloroplast exhibits a spherical shape in the S phase. Nucleus-encoded components of the chloroplast-division machinery are expressed specifically in the S phase and are localized as a ring at the provisional chloroplast division site. Then chloroplast division commences. Chloroplast division progresses and is completed during the G2 phase (Fig. 1A) (11, 25).
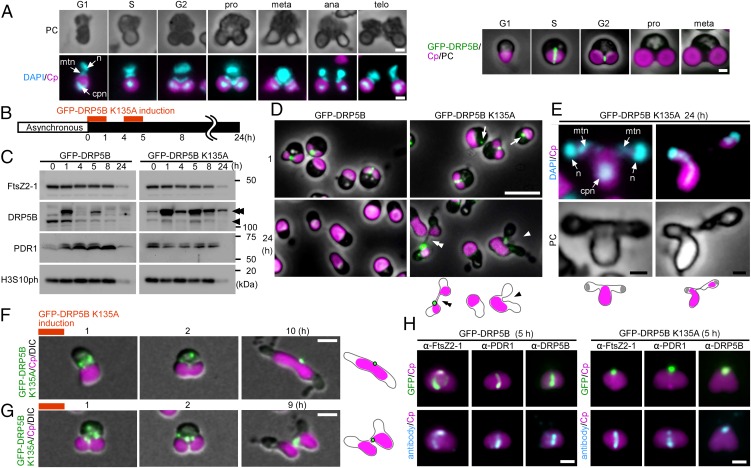
Fig. 1.
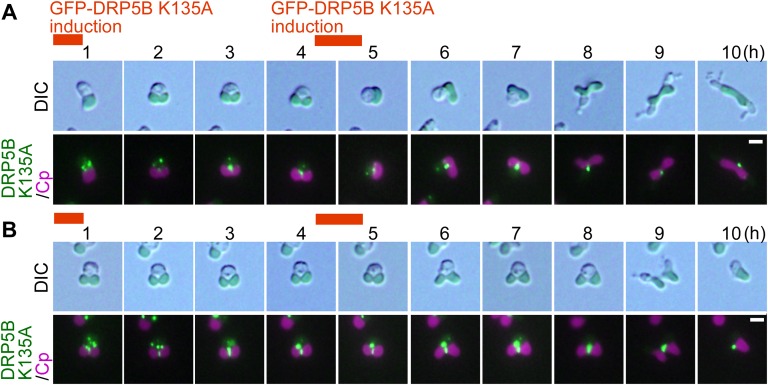
Effect of the dominant-negative DRP5B K135A on chloroplast division and cell-cycle progression in C. merolae. (A) Representative DAPI-stained images of C. merolae cells and DRP5B localization during cell-cycle progression. Magenta, autofluorescence of the chloroplast; cyan, DAPI-stained DNA; green, GFP-DRP5B; cpn, chloroplast nucleoid; mtn, mitochondrial nucleoid; n, nucleus. For the DAPI-stained images, the corresponding phase-contrast (PC) images are also shown. (Scale bars: 1 µm.) (B) Schematic diagram of the culture conditions. GFP-DRP5B or GFP-DRP5B K135A cells cultured at 42 °C under light were transferred to dark to stop cell growth and entrance into the S phase. Then GFP-DRP5B or GFP-DRP5B K135A was expressed by two rounds of heat shock at 50 °C for 1 h. (C) Immunoblot analyses showing the change in the levels of the chloroplast-division proteins FtsZ2-1, DRP5B, and PDR1 and the M-phase marker H3S10ph in GFP-DRP5B and GFP-DRP5B K135A cells. The double arrowhead indicates GFP-DRP5B or GFP-DRP5B K135A, and the single arrowhead indicates endogenous DRP5B. GFP-DRP5B and GFP-DRP5B K135A samples were blotted on the same membrane. (D) Microscopic images of GFP-DRP5B or GFP-DRP5B K135A cells 1 and 24 h after the onset of the heat-shock treatment. The double arrowhead indicates the two cells connected by a GFP-positive tube-like structure. The arrowhead indicates the cell with one chloroplast and two nuclei. The arrows indicate aggregated GFP-DRP5B K135A. Green, GFP-DRP5B or GFP-DRP5B K135A; magenta, autofluorescence of the chloroplast. The images obtained by fluorescence and phase-contrast microscopy are overlaid. (Scale bars: 5 µm.) (E) DAPI-stained images of a cell with one chloroplast and two nuclei (Left) and a cell during cytokinesis in which chloroplasts are unequally inherited by daughter cells (Right). Magenta, autofluorescence of the chloroplast; cyan, DAPI-stained DNA. (Scale bar: 1 µm.) (F and G) Time-lapse observation of GFP-DRP5B K135A cells after the induction of expression. (F) GFP-DRP5B K135A was expressed before the onset of chloroplast division-site constriction. (G) GFP-DRP5B K135A was expressed during the course of chloroplast division-site constriction. Green, GFP-DRP5B K135A; magenta, autofluorescence of the chloroplast. The images obtained by fluorescence and differential interference contrast (DIC) microscopy are overlaid. (Scale bars: 1 µm.) The results are the same as shown in Fig. S2 in more detail. (H) Images obtained by immunofluorescence microscopy showing FtsZ2-1, PDR1, and DRP5B localization in the GFP-DRP5B– or GFP-DRP5B K135A–expressing cells. Cyan, FtsZ2-1, PDR1, or DRP5B (the anti-DRP5B antibody detects both GFP-tagged and endogenous DRP5B) detected by the respective antibodies (originally detected by orange fluorescence and converted to cyan); magenta, autofluorescence of the chloroplast; green, GFP fluorescence of GFP-DRP5B or GFP-DRP5B K135A. (Scale bars: 1 µm.) Two independent experiments produced similar results. The results from one experiment are shown.
Expression of the Dominant-Negative DRP5B K135A Arrests Chloroplast Division at Either the Early or Final Stage of the Division-Site Constriction Event but Does Not Arrest Cell-Cycle Progression.
To examine the effect of the dominant-negative DRP5B K135A on chloroplast division and cell-cycle progression, transformed cells that were exponentially proliferating under light were transferred to dark to stop the cell growth and entrance into the S phase. Then the cells were heat shocked twice at 50 °C for 1 h to express GFP-DRP5B (as a control) or GFP-DRP5B K135A (Fig. 1B).
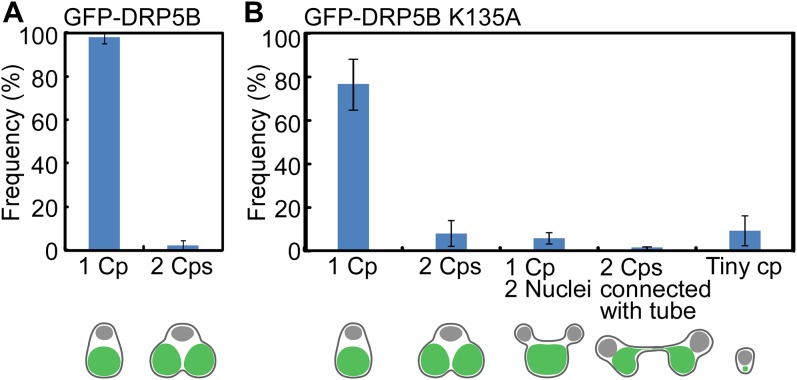
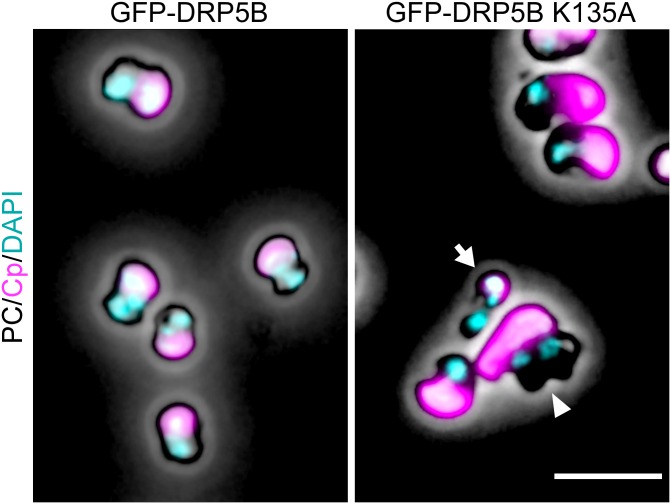
Immunoblot analysis showed that GFP-DRP5B and GFP-DRP5B K135A were expressed specifically after the heat-shock treatment (1 and 5 h after the transfer to dark) (Fig. 1C). GFP-DRP5B and GFP-DRP5B K135A were both detected at the chloroplast division site just after the heat-shock treatment (1 h posttransfer) using fluorescence microscopy (Fig. 1D, Upper). GFP-DRP5B K135A also was detected in the cytosol as an aggregate (Fig. 1D, Upper Right, arrows). At 24 h after the transfer to dark, 98 ± 2.4% of the GFP-DRP5B cells contained a single, nondividing chloroplast (Fig. 1D, Lower Left and Fig. S1A), and GFP-DRP5B was not detected by either fluorescence microscopy (Fig. 1D, Lower Left) or immunoblotting (Fig. 1C, 24-h lane). These results showed that the chloroplasts divided normally and that GFP-DRP5B was degraded, as was endogenous DRP5B. In contrast, GFP-DRP5B K135A was still detected in 15 ± 4.4% of the cells by fluorescence microscopy 24 h after the transfer to dark (Fig. 1D, Lower Right). Consistent with this observation, GFP-DRP5B K135A was still detected by immunoblotting even though FtsZ2-1 and PDR1 were not detected 24 h after the transfer to dark (Fig. 1C, 24-h lane).
Fig. S1.
Frequency of the abnormal GFP-DRP5B K135A cells after the induction of GFP-DRP5B K135A expression. (A) The frequency of the GFP-DRP5B cells with a single chloroplast or two divided chloroplasts 24 h after the onset of the heat-shock treatment. (B) The frequency of the GFP-DRP5B K135A cells with a single chloroplast, two divided chloroplast, a single chloroplast and two nuclei, two chloroplasts connected with a tubular bridge and two nuclei, or a single tiny chloroplast remnant 24 h after the onset of the heat-shock treatment. The error bars represent the SD (n = 5).
As reported previously (19), chloroplast division was apparently blocked at the final fission stage in 1.0 ± 0.7% of the GFP-DRP5B K135A cells, in which two daughter chloroplasts were still connected by a GFP-DRP5B K135A–positive tubular bridge (Fig. 1D, Lower Right, double arrowhead and Fig. S1B). In addition, 6.0 ± 3.1% of the GFP-DRP5B K135A cells contained a chloroplast that exhibited only a slight constriction at the division site, and cytokinesis in these cells was blocked by the undivided chloroplast (Fig. 1D, Lower Right, arrowhead and Fig. S1B). Cytokinesis was evident based on the separation of cytosolic compartments other than the chloroplast. DAPI staining showed that the culture contained cells with two divided nuclei and an undivided chloroplast (Fig. 1E, Left) and cells at the final phase of cytokinesis in which chloroplasts had been unequally inherited by daughter cells (Fig. 1E, Right).
Time-lapse observation of the cells after GFP-DRP5B K135A expression was performed to investigate how the expression of GFP-DRP5B K135A results in these two types of chloroplast-division arrest (i.e., blockage at an early stage or at the final stage of constriction). When GFP-DRP5B K135A was expressed in cells before the onset of chloroplast division, the protein appeared as dots in the cytoplasm and then migrated to the nuclear side of the chloroplast division site at an early stage of constriction (Fig. 1F and Fig. S2A). The chloroplast constricted slightly on the nuclear side where GFP-DRP5B K135A localized, but the constriction did not progress, even up to and during cytokinesis, which was evident based on the separation of cytosol other than the chloroplast (Fig. 1F and Fig. S2A). When GFP-DRP5B K135A was expressed in cells during the constriction of the chloroplast division site, the protein appeared both as dots in the cytoplasm and as a ring at the chloroplast division site (Fig. 1G and Fig. S2B). In this case, constriction at the chloroplast division site progressed, but the final fission of the daughter chloroplasts, which occurs before nuclear division and cytokinesis in the wild-type cell (Fig. 1A), was blocked until cytokinesis (Fig. 1G and Fig. S2B). However, the two daughter chloroplasts were separated in accord with cytokinesis (Fig. S2B), probably by being physically torn by the constriction of the cell-division plane.
Fig. S2.
Time-lapse observation of GFP-DRP5B K135A cells after the induction of expression. (A) GFP-DRP5B K135A was expressed before the onset of chloroplast division-site constriction. (B) GFP-DRP5B K135A was expressed during the constriction of the chloroplast division site. Images obtained by DIC and fluorescence microscopy are shown. Green, GFP-DRP5B K135A; magenta, autofluorescence of the chloroplast. (Scale bars: 1 µm.) Two independent experiments produced similar results. The results from one experiment are shown.
To examine the effect of GFP-DRP5B K135A expression on the chloroplast-division machinery, the localization of FtsZ, PDR1, and endogenous DRP5B was examined in GFP-DRP5B K135A–expressing cells. In both the GFP-DRP5B–expressing control cells and GFP-DRP5B K135A–expressing cells, FtsZ and PDR1 rings were clearly evident by immunofluorescence microscopy 5 h after the transfer to dark (Fig. 1H). In contrast, in GFP-DRP5B K135A–expressing cells, the endogenous DRP5B ring (detected by the anti-DRP5B antibody) was not found, and only a DRP5B dot was observed on the nuclear side of the chloroplast division site (corresponding to GFP-DRP5B K135A localization) (Fig. 1H). Thus, the expression of GFP-DRP5B K135A impairs the formation of the DRP5B ring but not the of the FtsZ and PDR1 rings, blocking either the constriction of the division site or the final fission of the daughter chloroplast, depending on the timing of GFP-DRP5B K135A expression.
Although GFP-DRP5B K135A blocked chloroplast division, the nucleus divided (Fig. 1E), and cells performed cytokinesis (Fig. 1 D, Lower, Fig. 1 F and G, and Fig. S2), resulting in the emergence of cells that contained a single undivided chloroplast and two divided cytosol and nuclei (5.3 ± 2.7%) (Fig. 1E, Left and Fig. S1B) or of daughter cells that contained only a tiny remnant of the chloroplast (8.7 ± 5.2%) (Figs. S1B and S3), a result produced by the asymmetric inheritance of chloroplasts during cytokinesis (Fig. 1E, Right). An M-phase marker, histone H3 phosphorylated at serine 10 (H3S10ph) (25), was undetectable by immunoblotting at 24 h after the transfer to dark in both the GFP-DRP5B–expressing control cells and GFP-DRP5B K135A–expressing cells (Fig. 1C), indicating that both had completed the cell cycle. These results show that the blockage of chloroplast division by dominant negative GFP-DRP5B K135A does not arrest cell-cycle progression, although the progression of cytokinesis is hampered by the undivided chloroplast in certain cells.
Fig. S3.
DAPI-stained images of GFP-DRP5B and GFP-DRP5B K135A cells 24 h after the onset of the heat-shock treatment. The arrowhead indicates a cell with one chloroplast and two nuclei. The arrow indicates a cell with the tiny chloroplast remnant. Magenta, autofluorescence of the chloroplast; cyan, DNA stained with DAPI. Images obtained by fluorescence and phase-contrast microscopy are overlaid. (Scale bar: 5 µm.)
Overexpression of FtsZ Results in Two Phenotypes Depending on the Timing of Overexpression.
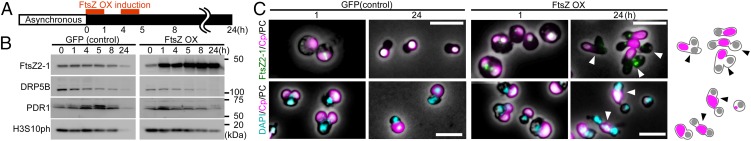
To examine the effect of FtsZ overexpression on chloroplast division and cell-cycle progression, we transferred the transformed cells were exponentially proliferating under light to the dark to stop cell growth and entry into the S phase. Then the cells were heat shocked twice at 50 °C for 1 h to overexpress GFP (19) as a control or FtsZ2-1 (Fig. 2A) [the C. merolae genome encodes FtsZ2-1 and FtsZ2-2, which are involved in chloroplast division, and FtsZ1-1 and FtsZ1-2, which are involved in mitochondrial division (26)].
Fig. 2.
Effect of FtsZ2-1 overexpression on chloroplast division and cell-cycle progression in asynchronously cultured C. merolae. (A) Schematic diagram of the culture conditions. The control GFP heat-inducible (GFP) or FtsZ heat-inducible (FtsZ OX) cells cultured at 42 °C under light were transferred to dark to stop cell growth and entrance into the S phase. Then GFP or FtsZ from the transgene was expressed by two rounds of heat shock at 50 °C for 1 h. (B) Immunoblot analyses showing the change in the levels of the chloroplast-division proteins FtsZ2-1, DRP5B, and PDR1 and the M-phase marker H3S10ph in GFP and FtsZ OX cells. The GFP and FtsZ OX samples were blotted on the same membrane. (C) Microscopic images of GFP and FtsZ OX cells 1 and 24 h after the onset of heat-shock treatment. (Upper) Cells immunostained with the anti–FtsZ2-1 antibody. (Lower) DAPI-stained images of DNA. Green, immunostained FtsZ2-1 (the GFP expressed in the GFP cell cytosol had been extracted before the antibody reaction; thus the green fluorescence specifically indicates FtsZ2-1); magenta, autofluorescence of the chloroplast; cyan, DNA stained with DAPI. The images obtained by fluorescence and phase-contrast microscopy are overlaid. (Scale bars: 5 µm.) The arrowheads indicate the cells that possess a single chloroplast and two nuclei. Two independent experiments produced similar results. The results from one experiment are shown.
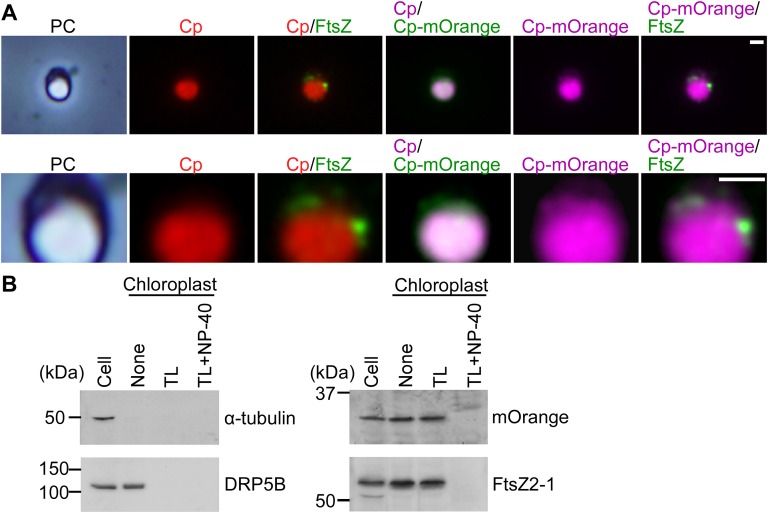
Immunoblot analysis showed that FtsZ2-1 was specifically overexpressed after the heat-shock treatment (Fig. 2B). The size of the overexpressed FtsZ2-1 was the same as that of endogenous FtsZ2-1 (i.e., the FtsZ2-1 band in the GFP-expressing control cells and the band in the FtsZ2-1–overexpressing cells before the heat-shock treatment) (Fig. 2B), suggesting that the chloroplast-targeted transit peptide of the overexpressed FtsZ was processed and that the protein translocated into the chloroplast. Immunofluorescence microscopy showed that the overexpressed FtsZ2-1 localized in the space between the thylakoid and envelope membranes (Fig. S4A; the thylakoid membrane was observed as red autofluorescence and the stroma was visualized by chloroplast-targeted mOrange). In addition, immunoblot analysis showed that the overexpressed FtsZ2-1 was enriched in the isolated intact chloroplast fraction, as were DRP5B and the stroma-targeted mOrange (Fig. S4B). When the isolated chloroplasts were treated with thermolysin, which cannot penetrate the outer envelope, DRP5B (on the cytosolic side of the outer envelope membrane) was digested, and the overexpressed FtsZ2-1 and the stroma-targeted mOrange were protected from thermolysin (Fig. S4B). However, when the envelope membranes were solubilized with the nonionic detergent Nonidet P-40, overexpressed FtsZ2-1 and the stroma-targeted mOrange were digested by thermolysin (Fig. S4B). These results indicate that the overexpressed FtsZ2-1 was translocated into the chloroplast stroma. In the GFP-expressing control cells, the chloroplast-division proteins FtsZ2-1, DRP5B, and PDR1 and the M-phase marker H3S10ph became undetectable by immunoblotting 24 h after the transfer to dark (Fig. 2B). Immunofluorescence microscopy detected the FtsZ ring 1 h after the transfer to dark in some of the control cells (Fig. 2C; GFP was extracted during the process of immunostaining, and thus the green fluorescence specifically indicates the FtsZ2-1 protein). No FtsZ ring was detected in the control cells at 24 h after the transfer to dark (Fig. 2C). These results indicate that the chloroplast division and cell-cycle progression in the control GFP cells had been completed by 24 h after the transfer to dark.
Fig. S4.
FtsZ2-1 localization in the FtsZ2-1–overexpressing cells. (A) Immunofluorescence microscopy showing the localization of the chloroplast-targeted mOrange and FtsZ2-1 in the FtsZ2-1–overexpressing cells 24 h after the onset of the induction of FtsZ2-1 overexpression by heat shock. mOrange was targeted to the chloroplast by fusing the protein with the N-terminal chloroplast-transit peptide CmSecA (50). FtsZ2-1 was detected with the anti–FtsZ2-1 antibody. Enlarged images are shown in the lower panels. (Scale bars: 1 µm.) (B) Intact chloroplasts were isolated from FtsZ2-1–overexpressing cells. Then the intact chloroplasts were incubated with thermolysin (TL) in the absence or presence of the detergent Nonidet P-40. Proteins were extracted from the cells (20 µg protein), and the chloroplasts (20 µg protein) treated with thermolysin were loaded. The cytosolic protein α-tubulin, the outer envelope peripheral protein DRP5B, chloroplast-targeted mOrange, and FtsZ2-1 were detected with respective antibodies.
In contrast, no normal FtsZ ring was detected in the FtsZ2-1–overexpressing cells, although an aggregation of FtsZ2-1 was observed between the envelope and thylakoid membranes both 1 h and 24 h after the transfer to dark (Fig. 2C and Fig. S4A). At 24 h after the transfer to dark, DRP5B and PDR1 became undetectable, but FtsZ2-1 kept accumulating, and the M-phase marker H3S10ph was still detected in the FtsZ2-1–overexpressing cells by immunoblotting (Fig. 2B). These results suggest that at least some of the FtsZ2-1–overexpressing cells are arrested in the M-phase.
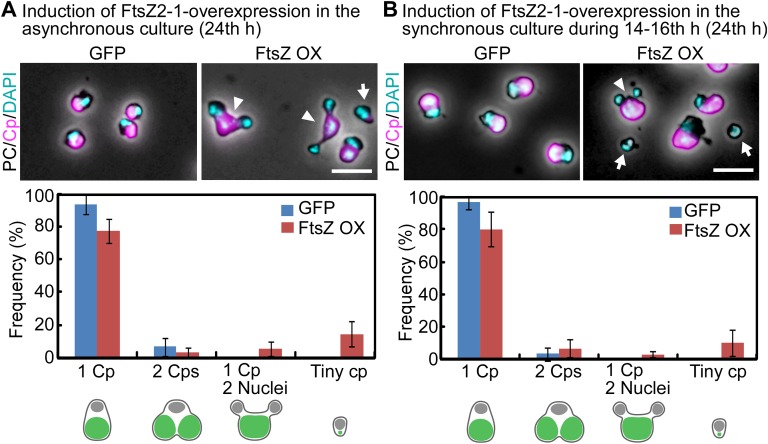
However, we found that the FtsZ2-1–overexpressing culture contained cells with a single chloroplast but two nuclei 24 h after the transfer to dark (Fig. 2C, 24 h, and Fig. S5A). In addition, 8.7 ± 5.2% of the cells contained a tiny chloroplast remnant (Fig. S5A). These observations suggest that some of the cells had completed the cell cycle and that cytokinesis was physically blocked by the obstruction of the undivided chloroplast in certain cells. Thus, there probably were two distinct populations of FtsZ2-1–overexpressing cells: one population (H3S10ph-positive cells) was arrested at a certain point during the M phase, whereas the other population (H3S10ph-negative cells that had entered into cytokinesis) completed the cell cycle, even though chloroplast division was impaired in both populations.
Fig. S5.
DAPI-stained images of the GFP-expressing control cells and FtsZ2-1–overexpressing cells. (A) The control GFP heat-inducible (GFP) or FtsZ2-1 heat-inducible (FtsZ OX) cells cultured asynchronously at 42 °C under light were transferred to dark. Then GFP or FtsZ2-1 from the transgene was expressed by two rounds of heat shock at 50 °C for 1 h, as shown in Fig. 2A. (Upper) DAPI-stained images of the cells at 24 h after the onset of FtsZ2-1 overexpression by heat shock. Magenta, autofluorescence of the chloroplast; cyan, DNA stained with DAPI. The images obtained by fluorescence and phase-contrast microscopy are overlaid. The arrowheads indicate cells with one chloroplast and two nuclei. The arrow indicates a cell with a tiny chloroplast remnant. (Scale bar: 5 µm.) (Lower) The frequency of each cell type in control GFP-expressing cells or FtsZ2-1–overexpressing cells. The error bars represent the SD (n = 3). (B) FtsZ heat-inducible (FtsZ OX) cells were synchronized by a 12-h/12-h light/dark cycle at 42 °C and were heat shocked at 50 °C at hour 14 (during constriction of the chloroplast division site) for 2 h, as shown in Fig. 4A. (Upper) DAPI-stained images of the cells at hour 24 in the synchronous culture. The arrowhead indicates a cell with one chloroplast and two nuclei. The arrows indicate cells with a tiny remnant of the chloroplast. (Scale bar: 5 µm.) (Lower) The frequency of each cell type in control GFP-expressing cells or FtsZ2-1–overexpressing cells. The error bars represent the SD (n = 3).
Overexpression of FtsZ During Chloroplast Division Impairs the Completion of Chloroplast Division but Does Not Arrest Cell-Cycle Progression.
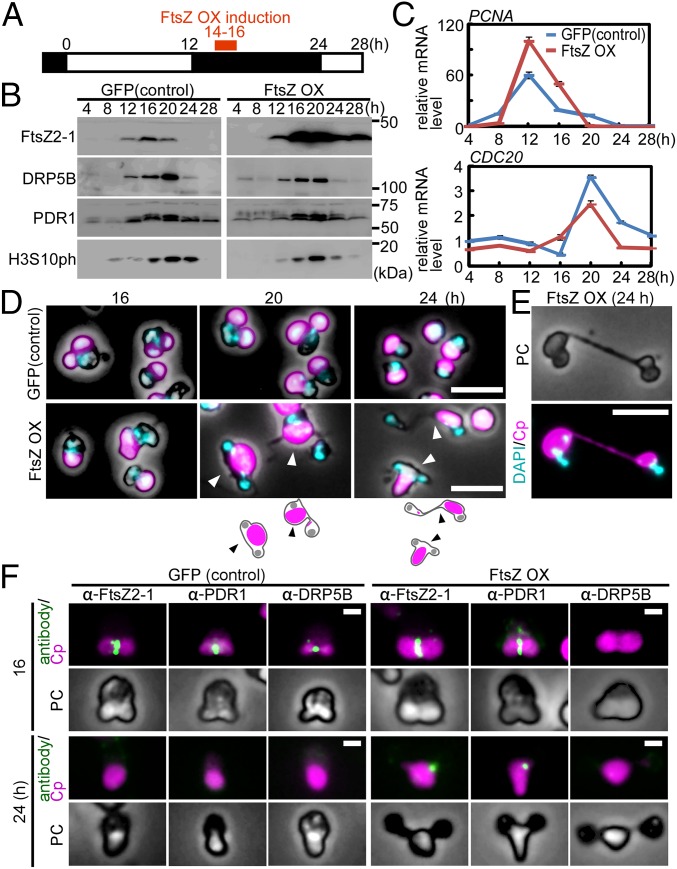
To examine the two populations of FtsZ2-1–overexpressing cells separately, we applied synchronous culture to overexpress FtsZ2-1 at a specific phase of the chloroplast-division cycle. At first, FtsZ2-1 was overexpressed during constriction of the chloroplast division site. Stable transformants were synchronized with a 12-h/12-h light/dark cycle and GFP as a control, or FtsZ2-1 was overexpressed with heat shock for 2 h at hour 14 in a synchronous culture in which chloroplasts were undergoing division-site constriction (Fig. 3A).
Fig. 3.
Effect of FtsZ2-1 overexpression during the constriction of the chloroplast division site on chloroplast division and cell-cycle progression in synchronously cultured C. merolae. (A) Schematic diagram of the culture conditions. The control GFP heat-inducible (GFP) or FtsZ heat-inducible (FtsZ OX) cells were synchronized by a 12-h/12-h light/dark cycle at 42 °C and were heat shocked at 50 °C at hour 14 (during the constriction of the chloroplast division site) for 2 h. (B) Immunoblot analyses showing the change in the levels of the chloroplast-division proteins FtsZ2-1, DRP5B, and PDR1 and the M-phase marker H3S10ph in GFP and FtsZ OX cells. The GFP and FtsZ OX samples were blotted on the same membrane. (C) Quantitative RT-PCR analyses showing the change in the mRNA levels of an S-phase marker (PCNA) and an M-phase marker (CDC20) in GFP (blue trace) and FtsZ OX (red trace) cells. DRP3 was used as the internal control. The error bars indicate the SD (n = 3). The expression levels in the control cells at hour 4 were defined as 1.0. (D) Microscopic images of DAPI-stained GFP and FtsZ OX cells at 16, 20, and 24 h in the synchronous culture. The arrowheads indicate cells that possess a single chloroplast and two nuclei. Magenta, autofluorescence of the chloroplast; cyan, DNA stained with DAPI. The fluorescence and phase-contrast images are overlaid. (Scale bars: 5 μm.) (E) DAPI-stained image of two FtsZ OX cells connected by a long narrow bridge that contains a tube-like structure of the undivided chloroplast. PC, phase contrast; magenta, autofluorescence of the chloroplast; cyan, DNA stained with DAPI. (Scale bar: 5 μm.) (F) Immunofluorescent images of GFP and FtsZ OX cells showing the localization of FtsZ2-1, PDR1, and DRP5B just after the heat shock (hour 16 in synchronous culture) and 8 h later (hour 24 in synchronous culture). (Upper) Cells with one dividing chloroplast and one nucleus at hour 16. (Lower) FtsZ OX cells with one undivided chloroplast and two nuclei at hour 24. Green, immunostained FtsZ2-1, PDR1, or DRP5B; magenta, autofluorescence of the chloroplast; PC, phase-contrast. (Scale bars: 1 μm.) Two independent experiments produced similar results. The results from one experiment are shown.
When FtsZ2-1 was overexpressed, DRP5B and PDR1 exhibited S-phase–specific expression, as in the control GFP-expressing cells (Fig. 3B). However, FtsZ2-1 still accumulated at hour 28 in the FtsZ2-1–overexpressing cells, unlike the GFP-expressing cells (Fig. 3B). The M-phase marker H3S10ph accumulated and then decreased during the dark period in both the control and FtsZ2-1–overexpressing cells (Fig. 3B). Quantitative RT-PCR analysis showed that the mRNA levels of the S-phase marker PCNA and the M-phase marker CDC20 (11, 25, 27) exhibited similar patterns of expression, peaking at 12 and 20 h, respectively, in the FtsZ2-1–overexpressing and GFP-expressing cells (Fig. 3C). These results suggest that FtsZ2-1–overexpressing cells had completed the cell cycle in a manner similar to the control cells, except for the prolonged retention of the overexpressed FtsZ2-1.
Abnormal cells were observed at 20 and 24 h in the FtsZ2-1–overexpressing cells that possessed a single undivided chloroplast and two nuclei (3.4 ± 4.4% of the cells at hour 24) (Fig. 3D and Fig. S5B). The culture also included two cells connected by a long, autofluorescent (chlorophyll)-positive tube-like structure (Fig. 3E). In addition, we also observed that 10 ± 5.5% of the cells had received a very small remnant of the chloroplast (Fig. S5B). These results suggest that chloroplast division is arrested by FtsZ2-1 overexpression, but the cell cycle progresses, and in some cases the progression of cytokinesis is hampered by the obstruction of the undivided chloroplast.
Immunofluorescence microscopy showed that a portion of the overexpressed FtsZ2-1 at 16 h localized to the chloroplast division site (Fig. 3F, 16 h). In the FtsZ2-1–overexpressing cells at 16 h, PDR1 also was localized to the division site but DRP5B was not detected (Fig. 3F, 16 h). This localization occurred even though the DRP5B level, which was detected by immunoblotting of the whole-cell lysate, was comparable to the level in control cells (Fig. 3B). At hour 24, FtsZ2-1 and PDR1 were still detected as dots on the periphery of the chloroplast in the FtsZ2-1–overexpressing cells (Fig. 3F, 24 h). These results suggest that the overexpression of FtsZ2-1 during the constriction of the chloroplast division site impairs DRP5B localization to the division site even when the division site is undergoing constriction; this impairment inhibits any further constriction of the chloroplast division site.
In summary, overexpression of FtsZ2-1 during the constriction of the chloroplast division site impairs further progression of the constriction process but does not block cell-cycle progression.
Overexpression of FtsZ Before the Onset of Chloroplast Division Impairs the Formation of the Chloroplast-Division Machinery and Arrests the Cell Cycle at the Prophase.
To examine whether a blockage of chloroplast division before the onset of division-site constriction affects cell-cycle progression, stable transformants were synchronized, and FtsZ2-1 or GFP as a control was overexpressed at hour 8 of synchronous culture (i.e., before the assembly of the chloroplast-division machinery) by heat shock for 2 h (Fig. 4A). At this point (hour 8 in Fig. 4A), FtsZ2-1, DRP5B, and PDR1 were not detected by immunoblotting (Fig. 4C), and the FtsZ and DRP5B rings were not detected by immunofluorescence microscopy (Fig. 4B).
Fig. 4.
Effect of FtsZ2-1 overexpression before the onset of chloroplast division on chloroplast division and cell-cycle progression in synchronously cultured C. merolae. (A) Schematic diagram of the culture conditions. The control GFP heat-inducible (GFP) or FtsZ heat-inducible (FtsZ OX) cells were synchronized by a 12-h/12-h light/dark cycle at 42 °C and were heat shocked at 50 °C at hour 8 (just before the formation of the FtsZ ring and the onset of chloroplast division) for 2 h. (B) Immunofluorescent images of FtsZ OX cells at hour 8 (just before the induction of FtsZ by heat shock) in the synchronous culture treated with the anti–FtsZ2-1 or DRP5B antibodies. Magenta, autofluorescence of the chloroplast; green, FtsZ2-1 or DRP5B. The images obtained by fluorescence and phase-contrast microscopy are overlaid. (Scale bar: 5 μm.) (C) Immunoblot analyses showing the change in the levels of the chloroplast-division proteins FtsZ2-1, DRP5B, and PDR1 and the M-phase marker H3S10ph in GFP and FtsZ OX cells. GFP and FtsZ OX samples were blotted on the same membrane. (D) Microscopic images of DAPI-stained GFP and FtsZ OX cells at 16, 20, and 24 h in synchronous culture. Magenta, autofluorescence of the chloroplast; cyan, DNA stained with DAPI. The images obtained by fluorescence and phase-contrast microscopy are overlaid. (Scale bars: 5 μm.) (E) Immunofluorescent images of the control GFP and FtsZ OX cells at hour 16 in the synchronous culture showing FtsZ2-1, PDR1, and DRP5B localization. Magenta, autofluorescence of the chloroplast; green, immunostained FtsZ2-1, PDR1, or DRP5B. (Scale bar: 5 μm.) (F) Immunofluorescent images of the control GFP cells showing changes in the level and localization of CENP-A during cell-cycle progression. The cell-cycle stage was defined based on the cell shape according to ref. 25. The fluorescence and phase-contrast images are overlaid. (Scale bar: 1 μm.) (G) Immunofluorescent images of GFP and FtsZ OX cells showing the localization of CENP-A at hour 24 in synchronous culture. Magenta, autofluorescence of the chloroplast; green, immunostained CENP-A. The fluorescence and phase-contrast images are overlaid. (Scale bar: 5 μm.) Two independent experiments produced similar results. The results from one experiment are shown.
When FtsZ2-1 was overexpressed at hour 8 of synchronous culture, the FtsZ, PDR1, and DRP5B rings did not form, whereas in the GFP-expressing control cells the FtsZ, PDR1, and DRP5B rings were observed at hour 16 by immunofluorescence microscopy (Fig. 4E). However, as in the control cells, PDR1 and DRP5B were specifically detected in the FtsZ2-1–overexpressing cells from hour 16 to hour 20 and from hour 12 to hour 20, respectively, by immunoblotting (Fig. 4C). We did not find any FtsZ2-1–overexpressing cells with dividing chloroplasts by microscopy throughout the course of one round (24 h) of synchronous culture (Fig. 4D). These results indicate that FtsZ2-1 overexpression impaired the formation of the chloroplast-division machinery, and thus the onset of chloroplast division, but not the expression of the individual components.
In the FtsZ2-1–overexpressing cells, DRP5B and PDR1 were degraded at hour 24, as they were in the control cells (Fig. 4C). The M-phase marker H3S10ph also was detected from hour 16, but it continued to accumulate, even at hour 28, unlike the control cells (Fig. 4C). DAPI staining showed that the FtsZ2-1–overexpressing cells possessed a single nucleus throughout the single round of synchronous culture (Fig. 4D). These results suggest that FtsZ2-1–overexpressing cells are arrested at a certain point in the M phase before nuclear division (the anaphase). To identify the point at which the cell cycle is arrested by FtsZ2-1 overexpression, the localization of centromere protein A (CENP-A) was examined in the FtsZ2-1–overexpressing cells at hour 24. In the control cells, CENP-A was specifically expressed in the nucleus in the middle of the S phase and then was converted into two discrete clusters in the metaphase (Fig. 4F) (25). At hour 24, in contrast to the control cells, in some of which CENP-A was localized in two discrete clusters, CENP-A was still dispersed throughout the nucleus in all the FtsZ2-1–overexpressing cells (Fig. 4G), suggesting that the cells remained stuck at a certain point before the metaphase. Combined with H3S10ph (Fig. 4C), which was detected from the prophase to the telophase in control cells (25), the results suggest that blockage of the onset of chloroplast division by FtsZ2-1 overexpression leads to cell-cycle arrest at the prophase.
Blockage of the Onset of Chloroplast Division Inhibits Cyclin B Expression and Migration of Cyclin-Dependent Kinase B.
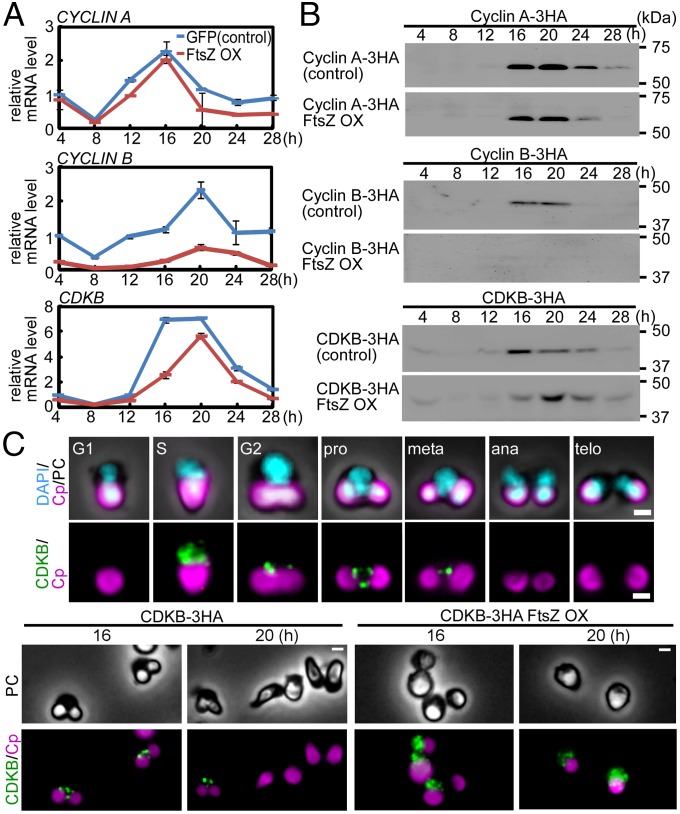
We next addressed how cell-cycle progression is arrested at the prophase by the blockage of the onset of chloroplast division. To this end, we examined the effect of FtsZ2-1 overexpression before the onset of chloroplast division on the level and localization of the regulators of G2/M transition, i.e., cyclin A, cyclin B, and cyclin-dependent kinase B (CDKB) (28).
Quantitative RT-PCR analyses showed that CYCLIN_A mRNA exhibited a similar pattern of expression in the control GFP-expressing and FtsZ2-1–overexpressing cells, peaking at hour 16 in synchronous culture (Fig. 5A). The CDKB mRNA level peaked at hour 16 in the control GFP-expressing cells and at hour 20 in the FtsZ2-1–overexpressing cells (Fig. 5A). In contrast, although a slight increase CYCLIN B mRNA level was observed at hour 20 in the FtsZ2-1–overexpressing cells, it was much lower than in control cells, where it peaked at hour 20 (Fig. 5A).
Fig. 5.
Effect of FtsZ2-1 overexpression before the onset of chloroplast division on the levels of cyclin A, cyclin B, and CDKB and on the localization of CDKB. Cells were synchronously cultured and heat shocked at hour 8 for 2 h, as shown in Fig. 4. (A) Quantitative RT-PCR analyses showing the change in the mRNA levels of CYCLIN A, CYCLIN B, and CDKB in the control GFP heat-inducible (GFP) or FtsZ heat-inducible (FtsZ OX) cells. DRP3 was used as the internal control. The error bars indicate the SD (n = 3). The expression levels in the control cells at 4 h were defined as 1.0. (B) Immunoblot analyses showing the change in the levels of cyclin A, cyclin B, and CDKB in control GFP or FtsZ OX cells. To detect the respective proteins, 3×HA-epitope–coding DNA was inserted into the respective chromosomal loci. C-terminal 3×HA-tagged proteins expressed by the respective endogenous promoters were detected with an anti-HA antibody. Control and FtsZ OX samples were blotted on the same membrane. (C) Immunofluorescent images of the control and FtsZ OX cells (in both strains CDKB-3HA was knocked into the CDKB locus) showing the change in the level and localization of CDKB. CDKB was detected with an anti-HA antibody. (Upper) The change in the level and localization of CDKB in the control cells during cell-cycle progression. The cell-cycle stage was defined based on the cell shape according to ref. 25. (Lower) The localization of CDKB in the control and FtsZ OX cells at hours 16 and 20 in the synchronous culture. Magenta, autofluorescence of the chloroplast; green, immunostained CDKB-3HA; cyan, DNA stained with DAPI; PC, phase-contrast. (Scale bars: 1 μm.) Two independent experiments produced similar results. The results from one experiment are shown.
To examine the cyclin A, cyclin B, and CDKB protein level and localization with an anti- HA antibody, a 3×HA-tag coding sequence was inserted into the chromosomal CYCLIN A, CYCLIN B, or CDKB locus just before the stop codon to express the respective C-terminal 3×HA fusion proteins. The C. merolae genome encodes single copies of these genes, and the transformants proliferated normally, suggesting that the cyclin A-3HA, cyclin B-3HA, and CDKB-3HA are fully functional in C. merolae.
Consistent with the change in the mRNA levels, immunoblot analyses showed a similar pattern of time-dependent expression of cyclin A in the control cells (expressing cyclin A-3HA, cyclin B-3HA, or CDKB-3HA but not overexpressing FtsZ2-1) and the FtsZ2-1–overexpressing cells expressing cyclin A-3HA, cyclin B-3HA, or CDKB-3HA (Fig. 5B). In the FtsZ2-1–overexpressing cells, the expression of CDKB (peaking at hour 20) was delayed compared with the control cells (peaking at hour 16) (Fig. 5B). No cyclin B expression was observed in the FtsZ2-1–overexpressing cells, whereas cyclin B was specifically expressed at hours 16 and 20 in the control cells (Fig. 5B).
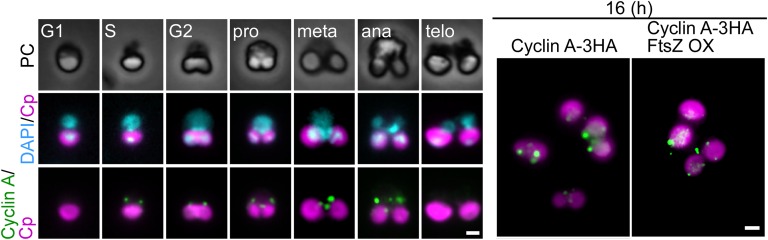
In the control cells, cyclin A was detected by immunofluorescence microscopy between the nucleus and the chloroplast specifically from the S phase to anaphase (Fig. S6). At hour 16, some chloroplasts in the control cells, but not in FtsZ2-1–overexpressing cells, had completed chloroplast division (the cells contained two chloroplasts). Despite this difference in chloroplast division, there was no difference in the localization of cyclin A, which appeared as a few dots in both the control and FtsZ2-1–overexpressing cells (Fig. S6).
Fig. S6.
Immunofluorescent images of the control and FtsZ2-1–overexpressing cells showing the localization of cyclin A. In both strains, CYCLIN A-3HA was integrated into the CYCLIN A locus. Cells were synchronously cultured and heat shocked at hour 8 for 2 h, as shown in Fig. 4A. Cyclin A was detected with an anti-HA antibody. (Left) The change in the level and localization of cyclin A in the control cells during cell-cycle progression. The cell-cycle stage was defined based on the cell shape according to ref. 25. (Right) The localization of cyclin A in the control cells and FtsZ2-1–overexpressing (FtsZ OX) cells at hour 16 in the synchronous culture. Magenta, autofluorescence of the chloroplast; green, immunostained cyclin A-3HA; cyan, DNA stained with DAPI; PC, phase-contrast image. (Scale bars: 1 µm.) Two independent experiments produced similar results. The results from one experiment are shown.
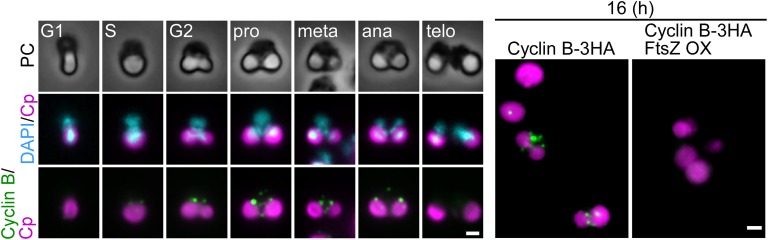
Cyclin B also localized as a few dots between the nucleus and the chloroplast from the G2 phase to anaphase in the control cells (Fig. S7). In contrast, cyclin B was not detected by immunofluorescence microscopy in FtsZ2-1–overexpressing cells (Fig. S7), as is consistent with the result of immunoblotting (Fig. 5B).
Fig. S7.
Immunofluorescent images of the control and FtsZ2-1–overexpressing cells showing the localization of cyclin B. In both strains, CYCLIN B-3HA was integrated into the CYCLIN B locus. Cells were synchronously cultured and were heat shocked at hour 8 for 2 h, as shown in Fig. 4. Cyclin B was detected with the anti-HA antibody. (Left) The change in the level and localization of cyclin B in the control cells during cell-cycle progression. The cell-cycle stage was defined based on the cell shape according to ref. 25. (Right) The localization of cyclin B in the control and FtsZ2-1–overexpressing (FtsZ OX) cells at hour 16 in the synchronous culture. Magenta, autofluorescence of the chloroplast; green, immunostained cyclin B-3HA; cyan, DNA stained with DAPI; PC, phase-contrast image. (Scale bars: 1 µm.) Two independent experiments produced similar results. The results from one experiment are shown.
The cyclin B partner CDKB localized in the vicinity of the nucleus during the S phase in the control cells (Fig. 5C). Then CDKB migrated into the space between the nucleus and the chloroplast and localized as a few dots from the G2 to metaphase (Fig. 5C). At hours 16 and 20, CDKB localized as a few dots in the control cells (Fig. 5C). In contrast, CDKB was still present in the vicinity of the nucleus in the FtsZ2-1–overexpressing cells (Fig. 5C). These results suggest that blocking the onset of chloroplast division inhibits the migration of CDKB from the vicinity of the nucleus into the space between the nucleus and the chloroplast that occurs during the G2 phase in control cells and the subsequent expression of cyclin B, which in control cells is observed from the G2 to M phase.
Blockage of Chloroplast Division Before the Onset of Division-Site Constriction Also Arrests Cell-Cycle Progression in the Glaucophyte Alga C. paradoxa.
We then examined whether cell-cycle progression is arrested when chloroplast division is blocked in other lineages of eukaryotic algae. The glaucophyte algae belong to the Archaeplastida (Plantae sensu stricto; i.e., eukaryotes that possess the chloroplast of cyanobacterial primary endosymbiotic origin). Evolutionary studies have suggested that the glaucophyte algae were the earliest to branch off from the common ancestor of the Archaeplastida, before the divergence of the red algae and Viridiplantae (green algae and land plants) (29). The chloroplasts in the glaucophyte algae (called “cyanelles”) have retained a peptidoglycan (PG) layer between the two envelope membranes that is descended from the cyanobacterial endosymbiont. In bacterial cell division, the proteins involved in PG synthesis interact with the cell-division machinery, and PG ingrowth at the division site is required for the progression of cell division (30). Thus, PG-targeting antibiotics such as ampicillin (31) effectively inhibit chloroplast division in glaucophyte algae, although these algae are not genetically tractable at present.
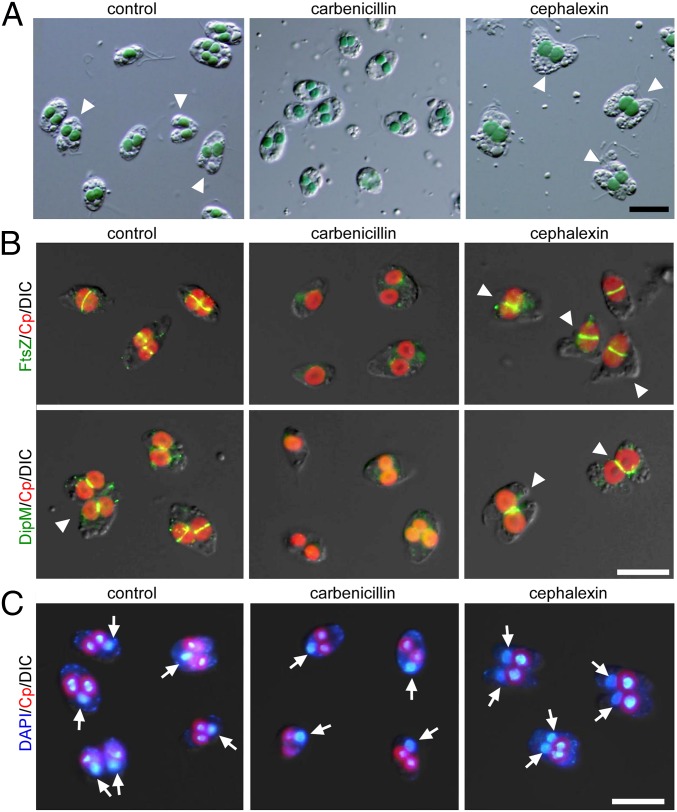
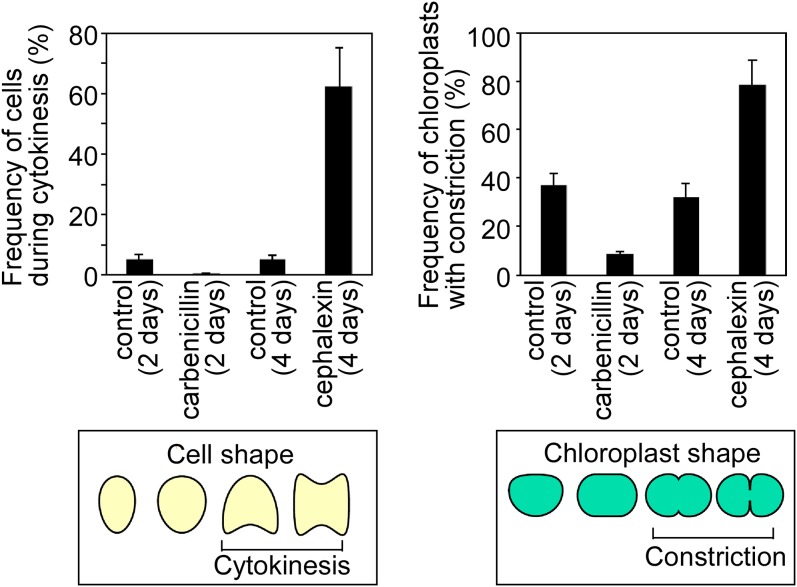
When the exponentially proliferating glaucophyte alga C. paradoxa was treated with carbenicillin (an inhibitor of DD-transpeptidase) for 48 h (under our culture conditions the doubling time was ∼48 h), 90 ± 1.0% of the chloroplasts became round in shape, in contrast to cells without antibiotics, in which dumbbell-shaped chloroplasts were often observed (Fig. 6 A and B and Fig. S8). Immunofluorescence microscopy showed that FtsZ and DipM (which forms a ring at the division site after the formation of the FtsZ ring in C. paradoxa) (27) rings were not formed in the chloroplast when the cells were treated with carbenicillin (Fig. 6B). Thus, carbenicillin treatment arrested chloroplast division before the formation of the FtsZ ring and constriction of the division site. In contrast to the cells without any antibiotics, 5.3 ± 1.7% of which exhibited cytokinesis, only 0.3 ± 0.5% of cells treated with carbenicillin exhibited cytokinesis in culture (Fig. 6 A and B and Fig. S8). DAPI staining showed that the carbenicillin-treated cells possessed a single nucleus (Fig. 6C), suggesting that the cells were arrested at a certain point before the anaphase.
Fig. 6.
Effect of PG-targeted antibiotics on chloroplast division and cell division in the glaucophyte alga C. paradoxa. Exponentially proliferating cells were treated with 1 μg/mL of the DD-transpeptidase inhibitor carbenicillin or the FtsI inhibitor cephalexin and were cultured for 2 (carbenicillin) or 4 (cephalexin) days after the addition of antibiotics. (A) Images obtained by DIC microscopy. (Scale bar: 10 μm.) The arrowheads indicate cells undergoing cytokinesis. (B) Immunofluorescent images of the cells displaying the localization of FtsZ and DipM. Red, autofluorescence of the chloroplast; green, immunostained FtsZ or DipM. The arrowheads indicate cells undergoing cytokinesis. The images obtained by fluorescence and DIC are overlaid. (Scale bar: 5 μm.) (C) DAPI-stained images of the cells. The arrows indicate the nuclei. Red, autofluorescence of the chloroplast; blue, DNA stained with DAPI. (Scale bar: 10 μm.) Two independent experiments produced similar results. The results from one experiment are shown.
Fig. S8.
Effect of PG-targeted antibiotics on the frequency of cells during cytokinesis (Left) and chloroplasts with the division-site constriction (Right). The error bars represent the SD (n = 3).
When the cells were treated with the FtsI inhibitor cephalexin for 4 d, 78 ± 10% of the chloroplasts became dumbbell-shaped and were larger than those in cells not treated with antibiotics (Fig. 6 A and B and Fig. S8). In addition, 63 ± 13% of the cells exhibited cytokinesis and possessed two nuclei, but this cytokinesis apparently was blocked by a single dividing chloroplast (Fig. 6 A and C and Fig. S8). Immunofluorescence microscopy showed that both the FtsZ and DipM rings were formed at the chloroplast-division site in the cells treated with cephalexin (Fig. 6B). These observations suggest that cephalexin treatment arrested chloroplast division during the process of division-site constriction. However, the cell cycle progressed, leading to a blockage of cytokinesis by the obstruction of the undivided chloroplast. These results suggest that a checkpoint exists in the cell cycle of C. paradoxa that arrests cell-cycle progression at a certain point until chloroplast division commences.
Discussion
The Mechanism That Coordinates Cell and Chloroplast Division.
It has long been believed that the synchronization of division in the endosymbiotic and host cell was a critical step in establishing permanent endosymbiotic organelles, such as mitochondria and chloroplasts. The majority of both unicellular and multicellular algae have either one or only a few chloroplasts, and the division of the chloroplasts is tightly correlated with host cell-cycle progression. We previously showed that some, but not all, nucleus-encoded chloroplast-division genes/proteins are specifically expressed in the S phase in a variety of algal lineages that possess chloroplasts of primary cyanobacterial endosymbiotic origin (11). In addition, cell-cycle stage-specific expression of FtsZ mRNA was also reported in a diatom, which possesses chloroplasts of a secondary red algal endosymbiotic origin (32), and in a chlorarachniophyte alga, which possesses chloroplasts of a green algal secondary endosymbiotic origin (33). These studies suggest that the host cell restricts the onset of chloroplast division at a specific stage of the host cell cycle by the regulation of gene/protein expression.
In this study, using the red alga C. merolae, we have shown that (i) cell-cycle progression is arrested at the prophase when chloroplast division is blocked before the formation of the chloroplast-division machinery by the overexpression of FtsZ2-1, but (ii) the cell cycle progresses when chloroplast division is blocked during division-site constriction by the overexpression of either FtsZ2-1 or dominant-negative DRP5B K135A (Fig. 7A). These results suggest that chloroplast division restricts host cell-cycle progression so that the cell cycle progresses to the metaphase only when the chloroplast division has started. Thus, chloroplast division and host cell-cycle progression are synchronized by an interactive restriction between the nucleus, which restricts the formation of the chloroplast-division machinery to the S phase, and the chloroplast, which lifts the prophase arrest only upon the onset of chloroplast division (Fig. 7B).
Fig. 7.
Schematic diagram of the mechanism for the synchronization of cell and chloroplast division in C. merolae. (A) Summary of the results obtained in this study. Previous studies showed that FtsZ (yellow), PDR1 (black), and DRP5B (pink) localize to the chloroplast division site in that order and thereby form the competent division machinery during the S phase (34, 35). The chloroplast division is completed by the prophase (25). Although the inhibition of DRP5B ring formation impairs the constriction of the chloroplast division site, the cell cycle progresses. As a result, either the progression of cytokinesis is hampered by the undivided chloroplast or cytokinesis produces a daughter cell that inherits only a tiny remnant of the chloroplast. In contrast, when the FtsZ ring formation is inhibited (in this case, PDR1 and DRP5B are not recruited to the division site), cyclin B accumulation and CDKB migration do not occur, resulting in the arrest of cell-cycle progression at the prophase. (B) The interactive synchronization of division in the eukaryotic host cell and the chloroplast in algae. The host cell restricts the onset of chloroplast division to the S phase by S-phase–specific expression of nucleus-encoded chloroplast-division proteins. The formation of the competent chloroplast-division machinery and constriction of the chloroplast division site lift the prophase arrest so that the host cell enters into the metaphase only when chloroplast division progresses.
The Mechanism by Which the Onset of Chloroplast Division Lifts the Prophase Arrest.
The results of this study suggest the existence of a cell-cycle checkpoint that confirms the progression of chloroplast division. One unresolved question is which chloroplast-division event is exactly sensed by the host cell cycle. In C. merolae chloroplast division, FtsZ, PDR1, and DRP5B are recruited to the chloroplast division site in that order in the S phase, and once the competent division machinery is formed, the division site constricts (34, 35) from the S to G2 phase (Fig. 7A) (25).
When DRP5B ring formation was blocked before the initiation of the division-site constriction by the expression of DRP5B K135A, DRP5B dots formed on the nuclear side of the chloroplast division site, and the division site became slightly constricted (Fig. 1F and Fig. S2A). In this case, the cell cycle progressed even though cytokinesis was physically blocked in some cases by the undivided chloroplast (Figs. 1F and 7A and Fig. S2A). Likewise, blockage of chloroplast division during division-site constriction by the overexpression of FtsZ2-1 (Fig. 3) or blockage at the final fission stage by the overexpression of either FtsZ2-1 (Fig. 3) or DRP5B K135A (Fig. 1G) did not arrest cell-cycle progression. In contrast, cell-cycle progression was arrested at the prophase when the formation of the FtsZ ring (along with the recruitment of PDR1 and DRP5B to the division site) was blocked by the overexpression of FtsZ2-1 (Figs. 4 and 7A). These results suggest that the host cell cycle senses a certain event that occurs between FtsZ ring formation and the recruitment of DRP5B to the nuclear side of the chloroplast division site.
Another important question concerns the event or signal in the course of cell-cycle progression that induces the need to wait for the activity of chloroplast division. In prophase cells arrested by the overexpression of FtsZ2-1, CDKB (expressed from the S phase to metaphase in the control cells) was dispersed in the vicinity of the nucleus, whereas in the control cells it had migrated into the space between the nucleus and the chloroplast by the G2 phase (Fig. 5). The cyclin B protein, which was expressed from the G2 to metaphase in control cells, was not expressed in the prophase-arrested cells (Fig. 5). Because both CDKB and its partner cyclin B localize as a few dots between the nucleus and the dividing dumbbell-shaped chloroplast in the control cells during the G2 phase (Fig. 5), the blocking of the migration of CDKB from the nucleus is likely related to the blockage of cyclin B expression. The localization patterns of cyclin A (Fig. S6), cyclin B (Fig. S7), and CDKB (Fig. 5) in the G2 to metaphase are similar to that of the kinesin-like protein TOP, which localizes at the spindle poles and spindle microtubules and is involved in nuclear division in C. merolae (36). In fission yeast, CDK (Cdc2) and cyclin B colocalize to the spindle pole bodies (37). Thus, the blockage of CDKB migration from the nucleus and the blockage of cyclin B expression in the FtsZ2-1–overexpressing cells likely exert an effect on the function of the spindle poles, leading to prophase arrest.
In land plants, the MYB3R protein, which possesses three Myb motif repeats, activates CYCLIN B transcription at the G2/M transition by binding to the cis-acting MSA element (38, 39). It is proposed that a positive feedback mechanism in which the cyclin B induced by MYB3R forms a complex with CDKB and hyperactivates MYB3R activity and thus cyclin B expression (21). In the prophase-arrested C. merolae induced by FtsZ2-1 overexpression, the cyclin B protein was not detected, but a slight increase in CYCLIN B mRNA level was observed (Fig. 5). The C. merolae genome encodes a putative MYB3R protein (CMT134C). Taken together, these results indicate that the prophase arrest resulted from the blockage of MYB3R hyperactivation.
In cultured mammalian (HeLa and hTert-RPE1) cells, depletion of cyclin B arrests the cells at the G2 phase (40). Although cyclin B was not detected in the C. merolae cells when the formation of the chloroplast-division machinery was blocked, we concluded that the cell cycle is arrested at the prophase, not the G2 phase. It should be noted that this conclusion is based on the phosphorylation of histone H3 at serine 10 (which is detected from the prophase to anaphase in other eukaryotes) in the cell-cycle–arrested cells (Fig. 4). There likely are differences in the mechanisms underlying cell-cycle progression in algal and mammalian cells. Therefore, the definition of the arrest point is only tentative at this point.
How the state of chloroplast division is relayed as a retrograde signal to the cell-cycle regulators is also an unresolved question. In C. merolae, it was suggested that the kinesin-like protein TOP activates chloroplast-division machinery to start constriction by inducing phosphorylation of the chloroplast-division machinery by Aurora kinase after DRP5B ring formation (36). However, the host cell cycle most likely senses a certain event before DRP5B ring formation, as discussed above, so the phosphorylation of the chloroplast-division machinery by Aurora kinase probably is not the event that is sensed by the host cell cycle. In this study, we observed the migration of CDKB from the vicinity of the nucleus into the space between the nucleus and the chloroplast during the G2 phase, when the chloroplast division site was undergoing constriction (Fig. 5). Thus, a certain physical interaction between a cell-cycle regulator and a cytosolic component of the chloroplast-division machinery is likely to be involved in the retrograde signaling from the chloroplast-division site to the host cell cycle.
The composition of the chloroplast-division machinery in the glaucophyte alga C. paradoxa is considerably different from that in other algae, especially on the cytosolic side. The FtsZ ring and the inner PD ring were detected at the chloroplast division site (41, 42). However, the outer PD ring was not evident (41, 42), and DRP5B is not encoded in the C. paradoxa genome (43). It has been suggested that the constriction of the division site is accompanied by an ingrowth and subsequent degradation of the PG layer at the division site in glaucophyte chloroplast division, as in bacterial cell division (41, 43).
Despite these differences in the chloroplast-division machinery of glaucophytes and other algae, we previously showed that the nucleus-encoded MinD and MinE proteins, which regulate FtsZ ring formation (2), are expressed predominantly during the S phase (11). In addition, FtsZ ring formation is restricted to the S phase, even though the level of FtsZ, which is encoded in the nucleus, is constant throughout the cell cycle (11). In addition to these observations, this study suggests that in C. paradoxa cell-cycle progression is stalled at a certain point until chloroplast division progresses, as in the red alga C. merolae. When chloroplast division was arrested by carbenicillin before the division-site constriction, FtsZ and DipM ring formation was blocked, and host cell-cycle progression was arrested before nuclear division (Fig. 6). In contrast, when chloroplast division was arrested by cephalexin during the division-site constriction, nuclear division and cytokinesis occurred, although the cytokinesis was blocked during constriction because of the obstruction by the undivided chloroplast (Fig. 6). These results are similar to those obtained in the red alga C. merolae, in which cell-cycle progression was arrested when the formation of the FtsZ ring was blocked during the constriction of the division site, but chloroplast division continued. Thus, the mechanism underlying the chloroplast division checkpoint likely arose in ancient algae during the course of evolution.
Materials and Methods
The primer sequences used in this study are listed in Table S1.
Table S1.
Sequences of primers used in this study
| Primer name | Sequence |
| Primers used for plasmid construction | |
| Z_Bgl_Bam_Inf_F | CAGATCTGGAGGATCC ATGAACCACCGTACGCCTC |
| Z_Not_Inf_R | TGCAGCCGGGCGGCCGCTTACTTTTGGAATCGACG |
| 184_HSpro200_Inf_F | GTCCCGAGGGTGACG CTTATAGCTTACGTGGCGG |
| Nos_Inf_R | TCATGTTTGACAGCTTATCATCG |
| CMD184C_+25∼_R | CGTCACCCTCGGGACTTGATGTTTACGTTC |
| Nos_CmUra_Inf_F | AGCTGTCAAACATGA GAACTGAGGGGCGAACGC |
| D184(1270)F | ACACGAATCACACGGTGCTG |
| D184(+1448)R | TTGCCGATAACGCAGAAGAGA |
| CycA_−1620_F | CCGGTTATGGCCTATGGAGC |
| CycA_+2018_R | TTATCGCCTTCTGGACCACTG |
| CycA_1482_R_HA | AACATCGTATGGGTA GCGCCCGAAAGGGTTTTCGCTC |
| CycA_+1_F_Ura | AGTCAGCTGCTAGGG TGAACGATAACTTGAACGTGTGTGGGATTC |
| HA_4_F | TACCCATACGATGTTCCTGACTATG |
| Ura_+471_R | CCCTAGCAGCTGACTGTATC |
| CycA_−1506∼_F | GACGTGCGGGTAGGGAACG |
| CycA_∼+1991_R | CTTCTGGACCACTGGCAGCG |
| HSPro_F | CTTATAGCTTACGTGGCGGATTCGCAGCG |
| Z_∼1512_R | TTACTTTTGGAATCGACGCAGAAAATCC |
| Z_CycA3′_Inf_F | CGATTCCAAAAGTAA TGAACGATAACTTGAACG |
| HSpro_Ura_Inf_R | CACGTAAGCTATAAG CCCTAGCAGCTGACTGTA |
| CycB_−536∼_F | GATCCCATCCATCGGGCCAGTTC |
| CycB_∼+1994_R | TCAGTAACCAGAAGCTCCTCGA |
| Ura_CycB3′_Inf_F | AGTCAGCTGCTAGGG GTCGAGACCGCTGCGCTCCG |
| HA_F | TACCCATACGATGTTCCTGACTATGCG |
| CmUra3′+471_Inf_R | CCCTAGCAGCTGACTGTATC |
| Z_CycB3′_Inf_F | CGATTCCAAAAGTAA GTCGAGACCGCTGCGCTCCG |
| HA_CycBORF_Inf_R | AACATCGTATGGGTA GAACAGGTTCGCGACACCGA |
| CDKB_−951∼_F | CGGGATCGCGTGGAAGGAGC |
| CDKB_∼+1966_R | TCCTGCATCTGGAAGCGAGCTC |
| Ura_CDKB3′_Inf_F | AGTCAGCTGCTAGGG CGGTCGTCACTGCATCCGAT |
| HA_CDKBORF_Inf_R | AACATCGTATGGGTA CGGCAGCATGACCTCGTTCT |
| Z_CDKB3′_Inf_F | CGATTCCAAAAGTAA CGGTCGTCACTGCATCCGAT |
| BamHI-mORANGE-F | TGAACTCAGAGGATCCATGGTGAGCAAGGGCGAG |
| BamHI-mORANGE-R | CGTATGGGTAGGATCCCTTGTACAGCTCGTCCATG |
| CatP_Inf_F | AGCAGATAGTTCATAGAGTTAGGAGGGC |
| D184_Nos_R | TCGCTGAGCGGTTTC TCATGTTTGACAGCTTAT |
| CatP_Ura_inf_R | TATGAACTATCTGCT CCCTAGCAGCTGACTGTA |
| D184C_+28∼_F | GAAACCGCTCAGCGACCA |
| pET100_FtsZ2-1_F | CACCAACCACCGTACGCCTCTTTCG |
| pET100_FtsZ2-1_R | TTACTTTTGGAATCGACGCAGAAAATC |
| Primers used for quantitative RT-PCR | |
| PCNA_CMS101C_qRT_F | CTCCGTGCGCATTTCTGTAA |
| PCNA_CMS101C_qRT_R | TCGTCACCACGAACCGTAAA |
| CycA_CMI203C_qRT_F | TCACGCGAGGTTCTGCAA |
| CycA_CMI203C_qRT_R | CTTTGGCTGCAACGAGCAT |
| Cdc20_CMA138C_qRT_F | CAGTAGGGACCGCCTTTGG |
| Cdc20_CMA138C_qRT_R | CGCTTGTTCGCTTCAACATC |
| CycB_CMI227C_qRT_F | TGCAACTCGTCGGCGTTA |
| CycB_CMI227C_qRT_R | TCGGGCGCGTATATTTCC |
| CDKB_CMH128C_qRT_F | ACGCATGAGGTTGTGACTTTGT |
| CDKB_CMH128C_qRT_R | ACGCGTCCCGAGAAGGA |
| DRP3(1981)F | GCAGGCGAAGTATCGAGC |
| DRP3(2050)R | AGTAACTCGCAAGAAGCGTCT |
Algal Culture.
C. merolae 10D and its stable transformants were grown in 2× Allen's medium in a 500-mL flat bottle (60 mm thick, containing 300 mL of culture) with 5 L/min aeration by ambient air at 100 μE⋅m−2⋅s−1 (44).
C. paradoxa UTEX555 (NIES-547) was grown in C medium (mcc.nies.go.jp/02medium-e.html) in 100-mL Erlenmeyer flasks (containing 20 mL of culture) on a rotary shaker at 21 °C under continuous light (30 μmol photons⋅m−2⋅s−1). For treatment with antibiotics, a 1/1,000 volume of 1 mg/mL carbenicillin or 1 mg/mL cephalexin stock solution in distilled water was added to the culture (∼2 × 106 cells/mL). Cells were harvested by centrifugation at 1,000 ×g for 10 min 48 h after the addition of carbenicillin or 4 d after the addition of cephalexin and then were observed by microscopy.
Preparation of the Stable C. merolae Transformants.
DNA construction and the procedure for obtaining the stable transformants are provided in SI Materials and Methods. The PCR products prepared for the transformation as described in SI Materials and Methods were introduced by PEG-mediated protocol into C. merolae M4, which has a mutation in the URA gene, and the transformants were selected as described previously (45).
Quantitative RT-PCR Analysis.
Total RNA was extracted and subjected to quantitative RT-PCR analyses as described in ref. 19. See SI Materials and Methods for details.
Antibodies and Immunoblot Analyses.
The antibody against C. merolae FtsZ2-1 was raised in rabbits. Immunoblot analyses were performed as described in ref. 19. See SI Materials and Methods for details.
Immunofluorescence Microscopy.
Immunofluorescence staining of C. merolae and C. paradoxa was performed as described (11). See SI Materials and Methods for details.
Time-Lapse Imaging.
To follow the progression of chloroplast division in GFP-DRP5B K135A–expressing cells, the cells were synchronized and collected 12 h after the beginning of the light period. The cells then were sandwiched between two coverslips with a piece of surgical tape as a spacer. To avoid drying, the coverslips were surrounded with liquid paraffin (26137-85; Nacalai Tesque). The coverslips were placed on a Thermo Plate (TP-S-100; Tokai Hit), and the temperature was maintained at 42 °C for normal growth or was increased to 50 °C for heat shock. The images were captured with an epifluorescence microscope (BX51) and a CCD camera (DP70) every hour.
SI Materials and Methods
Construction of the Plasmids and Preparation of the PCR Products for the Stable Transformants.
To prepare a C. merolae stable transformant expressing FtsZ by the heat-shock inducible promoter, FtsZ2-1 ORF (CMS004C), which was amplified with the primers Z_Bgl_Bam_Inf_F and Z_Not_Inf_R, was digested with BamHI and NotI and cloned into the plasmid P-200 (19) with the In-fusion cloning kit (TaKaRa). The resultant plasmid was used as the template, and the fusion of the heat-shock promoter FtsZ2-1 ORF and the Nos terminator was amplified by 184_HSpro200_Inf_F and Nos_Inf_R. To integrate the construct into a neutral C. merolae chromosomal locus, the PCR product was inserted into the vector pD184-O250-EGFP-UraCm-Cm (46), which was amplified and linearized using the primers CMD184C_+25∼_R and Nos_CmUra_Inf_F, with the In-fusion cloning kit to produce the HSp_FtsZ_Ura_D184 plasmid. Five micrograms of the PCR product amplified from this plasmid by the primers D184(1270)F and D184(+1448)R were used for the transformation of C. merolae.
CYCLIN A-3HA was integrated into the C. merolae CYCLIN A locus to produce a C-terminal 3×HA-tagged cyclin A-expressing stable transformant by its own promoter. The CYCLIN A (CMI203C) ORF flanked with its 1,620-kb 5′ and 2,018 kb 3′ genomic sequence was amplified from genomic DNA with the primers CycA_−1620_F and CycA_+2018_R and was cloned into the vector pGEM-T easy (Promega). The resultant plasmid was linearized by PCR amplification with the primers CycA_1482_R_HA and CycA_+1_F_Ura. The 3×HA-coding sequence linked to the Nos terminator and CmURA transformation marker, which was amplified with the primers HA_4_F and Ura_+471_R with the pQED3HAnosUra vector (25) used as the template, was cloned into the linearized vector with the In-fusion cloning kit to produce the CycA_HA_Ura plasmid. Five micrograms of the PCR product amplified from this plasmid by the primers CycA_−1506∼_F and CycA_∼+1991_R were used for the transformation of C. merolae.
CYCLIN A-3HA and the heat-shock promoter-FtsZ2-1 were integrated into the C. merolae CYCLIN A locus to produce a C. merolae stable transformant that expresses cyclin A-3HA by its own promoter and FtsZ2-1 by the heat shock-inducible promoter. The heat-shock promoter and FtsZ2-1 ORF fusion was amplified with the primers HSPro_F and Z_∼1512_R and was inserted into the CycA_HA_Ura plasmid, which was amplified and linearized using the primers Z_CycA3′_Inf_F and HSpro_Ura_Inf_R from the In-fusion cloning kit to produce the CycA_HA_Ura_HSp_FtsZ plasmid. Five micrograms of the PCR product amplified from this plasmid by the primers CycA_−1506∼_F and CycA_∼+1991_R were used for the transformation of C. merolae.
CYCLIN B-3HA was integrated into the C. merolae CYCLIN B locus to produce a C-terminal 3×HA-tagged cyclin B-expressing stable transformant by its own promoter. The CYCLIN B (CMI227C) ORF flanked with its 536-bp 5′ and 1,994-bp 3′ genomic sequence was amplified from genomic DNA with the primers CycB_−536∼_F and CycB_∼+1994_R and was cloned into the vector pGEM-T easy. The resultant plasmid was linearized by PCR amplification with the primers Ura_CycB3′_Inf_F and HA_CycBORF_Inf_R. The set of the 3×HA-coding sequence and the CmURA transformation marker, which was amplified with the primers HA_F and CmUra3′+471_Inf_R with the CycA_HA_Ura plasmid used as the template, was cloned into the linearized vector with the In-fusion cloning kit to produce the CycB_HA_Ura plasmid. Five micrograms of the PCR product from this plasmid by the primers CycB_−536∼_F and CycB_∼+1994_R were used for the transformation of C. merolae.
CYCLIN B-3HA and the heat-shock promoter FtsZ2-1 were integrated into the C. merolae CYCLIN B locus to produce a stable transformant which expresses cyclin B-3HA by its own promoter and FtsZ by the heat shock-inducible promoter. The heat-shock promoter and FtsZ2-1 ORF fusion was amplified with the primers HSPro_F and Z_∼1512_R and was inserted into the CycB_HA_Ura plasmid, which was amplified and linearized using the primers Z_CycB3′_Inf_F and HA_CycBORF_Inf_R, with the In-fusion cloning kit to produce the CycB_HA_Ura_HSp_FtsZ plasmid. Five micrograms of the PCR product amplified from this plasmid by the primers CycB_−536∼_F and CycB_∼+1994_R were used for the transformation of C. merolae.
CDKB-3HA was integrated into the C. merolae CDKB locus to produce a C-terminal 3×HA-tagged CDKB-expressing stable transformant by its own promoter. The CDKB (CMH128C) ORF flanked with its 951-bp 5′ and 1,966-bp 3′ genomic sequence was amplified from genomic DNA with the primers CDKB_−951∼_F and CDKB_∼+1966_R and was cloned into the vector pGEM-T easy. The resultant plasmid was linearized by PCR amplification with the primers Ura_CDKB3′_Inf_F and HA_CDKBORF_Inf_R. The set of the 3×HA-coding sequence and the CmURA transformation marker, which was amplified with the primers HA_F and CmUra3′+471_Inf_R using the CycA-HA_Ura plasmid as the template, was cloned into a linearized vector with the In-fusion cloning kit to produce the CDKB_HA_Ura plasmid. Five micrograms of the PCR product amplified from this plasmid with the primers CDKB_−951∼_F and CDKB_∼+1966_R were used for the transformation of C. merolae.
CDKB-3HA and the heat-shock promoter FtsZ2-1 were integrated into the C. merolae CDKB locus to produce a stable transformant that expresses CDKB-3HA by its own promoter and FtsZ by the heat shock-inducible promoter. The heat-shock promoter and FtsZ2-1 ORF fusion was amplified with the primers HSPro_1 and Z_∼1512_R and was inserted into the plasmid CDKB_HA, which was amplified and linearized using the primers Z_CDKB3′_Inf_F and HA_CDKBORF_Inf_R, with the In-fusion cloning kit to produce the CDKB_HA_Ura_HSp_FtsZ plasmid. Five micrograms of the PCR product amplified from this plasmid by the primers CDKB_−951∼_F and CDKB_∼+1966_R were used for the transformation of C. merolae.
To prepare a C. merolae stable transformant that expresses chloroplast-targeted mOrange by the CATALASE promoter and FtsZ by the heat shock-inducible promoter, we first constructed a plasmid containing the CATALASE (CMI050C) promoter, sequences encoding the N-terminal 130 amino acids of CmSecA (CMQ393C), mOrange, the 3×HA tag, and the Nos terminator in that order. The mOrange ORF was amplified from the pmOrange Vector (Clontech) with the primers BamHI-mORANGE-F and BamHI-mORANGE-R and was cloned into the pUC18_CatP_SecA_AAR-3HA_Nos plasmid (44), which was digested with BamHI with the In-fusion cloning kit. The resultant plasmid was used as the template, and the fusion of the CATALASE promoter, sequences encoding the N-terminal 130 amino acids of CmSecA, mOrange, and the 3×HA tag along with the Nos terminator were amplified by CatP_Inf_F and D184_Nos_R. The PCR product was inserted into the HSp_FtsZ_Ura_D184 plasmid, which was amplified and linearized using the primers CatP_Ura_inf_R and D184C_+28∼_F with the In-fusion cloning kit to produce the HSp_FtsZ_Ura_CatP_SecA_mORANGE_Nos_D184 plasmid. Five micrograms of the PCR product amplified from this plasmid by the primers D184(1270)F and D184(+1448)R were used for the transformation of C. merolae.
All the constructs were verified by DNA sequencing before transformation. The primer sequences are listed in Table S1.
Quantitative RT-PCR Analysis.
Quantitative RT-PCR analyses were performed using a StepOnePlus Real-Time PCR system (Life Technologies) and a 20-µL reaction mixture containing 2 µL of template cDNA, 0.25 nM primers, and 10 µL of Power SYBR Green Master Mix (Life Technologies). Standard curves were constructed by using serially diluted solutions of cDNA prepared with the relevant set of primers from the cells at 4 h after the onset of the light period in synchronous culture. The values of PCNA (CMS101C), CYCLIN A (CMI203C), CDC20 (CMA138C), CYCLIN B (CMI227C), and CDKB (CMH128C) were normalized with the data of DRP3 (CME019C). The primer sequences for the quantitative-PCR analyses are shown in Table S1.
Preparation of Antibodies.
The antibody against C. merolae FtsZ2-1 was raised in rabbits using the recombinant protein. The cDNA sequence encoding the full-length ORF (CMS004C) was amplified by PCR using the primers pET100_FtsZ2-1_F and pET100_FtsZ2-1_R (Table S1). The PCR product was cloned into the vector pET100 (Life Technologies), and the 6×His-tagged protein was expressed in Escherichia coli Rosetta (DE3) cells by the addition of isopropyl-B-d-thiogalactopyranoside (IPTG) to the medium. The protein then was purified with a HisTrap HP column (GE Healthcare) and was used as the antigen. The antibody was affinity purified using the recombinant polypeptide coupled to a HiTrap NHS-activated HP column (GE Healthcare).
Immunoblot Analyses.
Cells (5 × 108) were harvested by centrifugation at 3,000 × g for 10 min and were stored at −80 °C. Cells were resuspended in SDS/PAGE sample buffer [50 mM Tris (pH 6.8), 6% 2-mercaptoethanol, 2% SDS, 10% glycerol] and were sonicated. After centrifugation at 15,000 × g for 10 min at 4 °C, the supernatant fraction was separated by SDS/PAGE and transferred onto PVDF membranes. For immunoblotting, the following primary antibodies were used: rabbit polyclonal anti–FtsZ2-1 (1:5,000; prepared in this study), rabbit polyclonal anti-DRP5B [1:2,000 (34)], rat polyclonal anti-PDR1 [1:2,000 (35)], rabbit polyclonal anti-H3S10ph [1:1,000 (47)], mouse monoclonal anti-HA (1:1,000; HA-7; Sigma-Aldrich), mouse monoclonal anti–α-tubulin (1:2,000; DM1A; Sigma), and rabbit polyclonal anti-mOrange (1:2,000; PM005; MBL). Goat anti-mouse, -rabbit, or -rat IgG-HRP antibody (Thermo Fisher Scientific) was used as the secondary antibody at a dilution of 1:2,000.
Immunofluorescence Microscopy.
Immunofluorescence staining for C. merolae and C. paradoxa was performed as described (11). For C. merolae, the following primary antibodies were used at the indicated concentrations: rabbit polyclonal anti–FtsZ2-1 (1:1,000; prepared in this study), rat polyclonal anti-PDR1 [1:500 (35)], rabbit polyclonal anti-DRP5B [1:500 (34)], rabbit polyclonal anti–CENP-A [1:500 (48)], and mouse monoclonal anti-HA (1:100; HA-7; Sigma-Aldrich). For C. paradoxa, the following primary antibodies were used at the indicated concentrations: rabbit polyclonal anti-FtsZ [1:500 (43)] and rabbit polyclonal anti-DipM [1:500 (43)]. As secondary antibody, Alexa Fluor 488 goat anti-rabbit, -mouse, or -rat IgG antibody or Alexa Fluor 555 goat anti-rabbit IgG antibody (Life Technologies) was used at a dilution of 1:1,000. The cells were examined using an epifluorescence microscope (BX51; Olympus) equipped with a CCD camera (DP70; Olympus).
Isolation of C. merolae Intact Chloroplasts and the Thermolysin Protection Assay.
Intact C. merolae chloroplasts were isolated as described previously (49) with some modifications. The FtsZ2-1–overexpressing cells were collected from the 500-mL culture by centrifugation and were suspended in the isolation medium [20 mM Tris⋅HCl (pH 7.6), 5 mM MgCl2, 5 mM KCl] containing 180 mM sucrose (hypotonic isolation medium). Then the cells were lysed in a French Pressure Cell (SLM Aminco) at 1,500 psi. After the addition of DNase I (100 µg/mL), the lysate was incubated for 2 h at 4 °C and then was layered on three step gradients of Percoll prepared in 50-mL tubes: 8 mL of 80%, 8 mL of 60%, and 8 mL of 40% Percoll dissolved in the isolation medium containing 300 mM sucrose (isotonic isolation medium). After 3-h centrifugation at 3,000 × g in a swinging-bucket rotor, intact chloroplasts were found to band at the 60/80% Percoll interface. This fraction was washed with the isotonic isolation medium by centrifugation at 700 × g, and the intact chloroplasts were suspended in the isotonic isolation medium to give a protein concentration of 2 mg/mL.
The isolated chloroplasts were incubated for 60 min in the isotonic isolation medium with 2 mM CaCl2 and 0.2 mg/mL thermolysin (Wako) in the absence or presence of 0.2% Nonidet P-40. Thermolysin reactions were quenched by adding EDTA to a final concentration of 10 mM and a protease inhibitor mixture.
Acknowledgments
We thank Dr. Kuroiwa (Japan Women's University) for the PDR1 antibody, Dr. Tanaka (Tokyo Institute of Technology) for the CENP-A antibody, Dr. Hirano (RIKEN) for the H3S10ph antibody, and Ms. Yamashita and Ms. Hashimoto (National Institute of Genetics) for technical assistance. This study was supported by Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research 25251039 (to S.-y.M.) and by the Core Research for Evolutional Science and Technology Program of the Japan Science and Technology Agency (S.-y.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612872113/-/DCSupplemental.
References
- 1.Reyes-Prieto A, Weber AP, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41(1):147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- 2.Miyagishima SY. Mechanism of plastid division: From a bacterium to an organelle. Plant Physiol. 2011;155(4):1533–1544. doi: 10.1104/pp.110.170688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Possingham JV, Lawrence ME. Controls to plastid division. Int Rev Cytol. 1983;84:1–56. [Google Scholar]
- 4.Rodríguez-Ezpeleta N, Philippe H. Plastid origin: Replaying the tape. Curr Biol. 2006;16(2):R53–R56. doi: 10.1016/j.cub.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Okamoto N, Inouye I. A secondary symbiosis in progress? Science. 2005;310(5746):287–287. doi: 10.1126/science.1116125. [DOI] [PubMed] [Google Scholar]
- 6.Onuma R, Horiguchi T. Kleptochloroplast enlargement, karyoklepty and the distribution of the cryptomonad nucleus in Nusuttodinium (= Gymnodinium) aeruginosum (Dinophyceae) Protist. 2015;166(2):177–195. doi: 10.1016/j.protis.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Wouters J, Raven JA, Minnhagen S, Janson S. The luggage hypothesis: Comparisons of two phototrophic hosts with nitrogen-fixing cyanobacteria and implications for analogous life strategies for kleptoplastids/secondary symbiosis in dinoflagellates. Symbiosis. 2009;49(2):61–70. [Google Scholar]
- 8.Imanian B, Pombert JF, Keeling PJ. The complete plastid genomes of the two ‘dinotoms’ Durinskia baltica and Kryptoperidinium foliaceum. PLoS One. 2010;5(5):e10711. doi: 10.1371/journal.pone.0010711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowack EC. Paulinella chromatophora-rethinking the transition from endosymbiont to organelle. Acta Soc Bot Pol. 2014;83(4):387–397. [Google Scholar]
- 10.Yoshida Y, Miyagishima SY, Kuroiwa H, Kuroiwa T. The plastid-dividing machinery: Formation, constriction and fission. Curr Opin Plant Biol. 2012;15(6):714–721. doi: 10.1016/j.pbi.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Miyagishima SY, Suzuki K, Okazaki K, Kabeya Y. Expression of the nucleus-encoded chloroplast division genes and proteins regulated by the algal cell cycle. Mol Biol Evol. 2012;29(10):2957–2970. doi: 10.1093/molbev/mss102. [DOI] [PubMed] [Google Scholar]
- 12.Itoh R, Takahashi H, Toda K, Kuroiwa H, Kuroiwa T. Aphidicolin uncouples the chloroplast division cycle from the mitotic cycle in the unicellular red alga Cyanidioschyzon merolae. Eur J Cell Biol. 1996;71(3):303–310. [PubMed] [Google Scholar]
- 13.Ohta N, Sato N, Kuroiwa T. Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res. 1998;26(22):5190–5198. doi: 10.1093/nar/26.22.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohta N, et al. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2003;10(2):67–77. doi: 10.1093/dnares/10.2.67. [DOI] [PubMed] [Google Scholar]
- 15.Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428(6983):653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 16.Nozaki H, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5(1):28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minoda A, Sakagami R, Yagisawa F, Kuroiwa T, Tanaka K. Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2004;45(6):667–671. doi: 10.1093/pcp/pch087. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara T, Ohnuma M, Yoshida M, Kuroiwa T, Hirano T. Gene targeting in the red alga Cyanidioschyzon merolae: Single- and multi-copy insertion using authentic and chimeric selection markers. PLoS One. 2013;8(9):e73608. doi: 10.1371/journal.pone.0073608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sumiya N, Fujiwara T, Kobayashi Y, Misumi O, Miyagishima SY. Development of a heat-shock inducible gene expression system in the red alga Cyanidioschyzon merolae. PLoS One. 2014;9(10):e111261. doi: 10.1371/journal.pone.0111261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujiwara T, et al. A nitrogen source-dependent inducible and repressible gene expression system in the red alga Cyanidioschyzon merolae. Front Plant Sci. 2015;6:657. doi: 10.3389/fpls.2015.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Araki Y, Takio S, Ono K, Takano H. Two types of plastid ftsZ genes in the liverwort Marchantia polymorpha. Protoplasma. 2003;221(3-4):163–173. doi: 10.1007/s00709-002-0080-1. [DOI] [PubMed] [Google Scholar]
- 22.Stokes KD, McAndrew RS, Figueroa R, Vitha S, Osteryoung KW. Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 2000;124(4):1668–1677. doi: 10.1104/pp.124.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174(19):6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Bliek AM, et al. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122(3):553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujiwara T, Tanaka K, Kuroiwa T, Hirano T. Spatiotemporal dynamics of condensins I and II: Evolutionary insights from the primitive red alga Cyanidioschyzon merolae. Mol Biol Cell. 2013;24(16):2515–2527. doi: 10.1091/mbc.E13-04-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyagishima SY, et al. Two types of FtsZ proteins in mitochondria and red-lineage chloroplasts: The duplication of FtsZ is implicated in endosymbiosis. J Mol Evol. 2004;58(3):291–303. doi: 10.1007/s00239-003-2551-1. [DOI] [PubMed] [Google Scholar]
- 27.Miyagishima SY, et al. Translation-independent circadian control of the cell cycle in a unicellular photosynthetic eukaryote. Nat Commun. 2014;5:3807. doi: 10.1038/ncomms4807. [DOI] [PubMed] [Google Scholar]
- 28.Scofield S, Jones A, Murray JAH. The plant cell cycle in context. J Exp Bot. 2014;65(10):2557–2562. doi: 10.1093/jxb/eru188. [DOI] [PubMed] [Google Scholar]
- 29.Keeling PJ. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol. 2013;64(1):583–607. doi: 10.1146/annurev-arplant-050312-120144. [DOI] [PubMed] [Google Scholar]
- 30.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10(2):123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato M, Nishikawa T, Kajitani H, Kawano S. Conserved relationship between FtsZ and peptidoglycan in the cyanelles of Cyanophora paradoxa similar to that in bacterial cell division. Planta. 2007;227(1):177–187. doi: 10.1007/s00425-007-0605-0. [DOI] [PubMed] [Google Scholar]
- 32.Gillard J, et al. Physiological and transcriptomic evidence for a close coupling between chloroplast ontogeny and cell cycle progression in the pennate diatom Seminavis robusta. Plant Physiol. 2008;148(3):1394–1411. doi: 10.1104/pp.108.122176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirakawa Y, Ishida K. Prospective function of FtsZ proteins in the secondary plastid of chlorarachniophyte algae. BMC Plant Biol. 2015;15:276. doi: 10.1186/s12870-015-0662-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyagishima SY, et al. A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell. 2003;15(3):655–665. doi: 10.1105/tpc.009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida Y, et al. Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science. 2010;329(5994):949–953. doi: 10.1126/science.1190791. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida Y, et al. The kinesin-like protein TOP promotes Aurora localisation and induces mitochondrial, chloroplast and nuclear division. J Cell Sci. 2013;126(Pt 11):2392–2400. doi: 10.1242/jcs.116798. [DOI] [PubMed] [Google Scholar]
- 37.Alfa CE, Ducommun B, Beach D, Hyams JS. Distinct nuclear and spindle pole body population of cyclin-cdc2 in fission yeast. Nature. 1990;347(6294):680–682. doi: 10.1038/347680a0. [DOI] [PubMed] [Google Scholar]
- 38.Ito M, et al. A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell. 1998;10(3):331–341. doi: 10.1105/tpc.10.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito M, et al. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell. 2001;13(8):1891–1905. doi: 10.1105/TPC.010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soni DV, Sramkoski RM, Lam M, Stefan T, Jacobberger JW. Cyclin B1 is rate limiting but not essential for mitotic entry and progression in mammalian somatic cells. Cell Cycle. 2008;7(9):1285–1300. doi: 10.4161/cc.7.9.5711. [DOI] [PubMed] [Google Scholar]
- 41.Iino M, Hashimoto H. Intermediate features of cyanelle division of Cyanophora paradoxa (Glaucocystophyta) between cyanobacterial and plastid division. J Phycol. 2003;39(3):561–569. [Google Scholar]
- 42.Sato M, et al. The dynamic surface of dividing cyanelles and ultrastructure of the region directly below the surface in Cyanophora paradoxa. Planta. 2009;229(4):781–791. doi: 10.1007/s00425-008-0872-4. [DOI] [PubMed] [Google Scholar]
- 43.Miyagishima SY, Kabeya Y, Sugita C, Sugita M, Fujiwara T. DipM is required for peptidoglycan hydrolysis during chloroplast division. BMC Plant Biol. 2014;14(1):57. doi: 10.1186/1471-2229-14-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sumiya N, et al. Expression of cyanobacterial acyl-ACP reductase elevates the triacylglycerol level in the red alga Cyanidioschyzon merolae. Plant Cell Physiol. 2015;56(10):1962–1980. doi: 10.1093/pcp/pcv120. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi Y, Ohnuma M, Kuroiwa T, Tanaka K, Hanaoka M. The basics of cultivation and molecular genetic analysis of the unicellular red alga Cyanidioschyzon merolae. Endocytobiosis Cell Res. 2010;20:53–61. [Google Scholar]
- 46.Fujiwara T, Ohnuma M, Yoshida M, Kuroiwa T, Hirano T. Gene targeting in the red alga Cyanidioschyzon merolae: Single- and multi-copy insertion using authentic and chimeric selection markers. PLoS One. 2013;8(9):e73608. doi: 10.1371/journal.pone.0073608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura K, Hirano T. Dual roles of the 11S regulatory subcomplex in condensin functions. Proc Natl Acad Sci USA. 2000;97(22):11972–11977. doi: 10.1073/pnas.220326097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maruyama S, Kuroiwa H, Miyagishima SY, Tanaka K, Kuroiwa T. Centromere dynamics in the primitive red alga Cyanidioschyzon merolae. Plant J. 2007;49(6):1122–1129. doi: 10.1111/j.1365-313X.2006.03024.x. [DOI] [PubMed] [Google Scholar]
- 49.Miyagishima SY, Itoh R, Aita S, Kuroiwa H, Kuroiwa T. Isolation of dividing chloroplasts with intact plastid-dividing rings from a synchronous culture of the unicellular red alga cyanidioschyzon merolae. Planta. 1999;209(3):371–375. doi: 10.1007/s004250050645. [DOI] [PubMed] [Google Scholar]
- 50.Koyama Y, et al. Characterization of the nuclear- and plastid-encoded secA-homologous genes in the unicellular red alga Cyanidioschyzon merolae. Biosci Biotechnol Biochem. 2011;75(10):2073–2078. doi: 10.1271/bbb.110338. [DOI] [PubMed] [Google Scholar]