Significance
Current dogma states that the integrated HIV type 1 provirus encodes a single RNA transcript that serves as both mRNA for generating viral proteins and as genomic RNA that is packaged and used for reverse transcription. We now show that multiple transcripts with different functions are generated in infected cells, a consequence of heterogeneous transcriptional start site usage. Transcripts that begin with a single capped guanosine are specifically selected for packaging, whereas those that begin with two or three capped guanosines are enriched on polysomes and used for translation. In vitro studies with recombinant 5′-leader transcripts reveal a mechanism by which the incorporation of a single 5′ guanosine dramatically alters the structure, function, and fate of the viral RNA.
Keywords: HIV-1, 5′-leader, transcription, RNA, structure
Abstract
The promoter in HIV type 1 (HIV-1) proviral DNA contains three sequential guanosines at the U3–R boundary that have been proposed to function as sites for transcription initiation. Here we show that all three sites are used in cells infected with HIV-1 and that viral RNAs containing a single 5′ capped guanosine (Cap1G) are specifically selected for packaging in virions, consistent with a recent report [Masuda et al. (2015) Sci Rep 5:17680]. In addition, we now show that transcripts that begin with two or three capped guanosines (Cap2G or Cap3G) are enriched on polysomes, indicating that RNAs synthesized from different transcription start sites have different functions in viral replication. Because genomes are selected for packaging as dimers, we examined the in vitro monomer–dimer equilibrium properties of Cap1G, Cap2G, and Cap3G 5′-leader RNAs in the NL4-3 strain of HIV-1. Strikingly, under physiological-like ionic conditions in which the Cap1G 5′-leader RNA adopts a dimeric structure, the Cap2G and Cap3G 5′-leader RNAs exist predominantly as monomers. Mutagenesis studies designed to probe for base-pairing interactions suggest that the additional guanosines of the 2G and 3G RNAs remodel the base of the PolyA hairpin, resulting in enhanced sequestration of dimer-promoting residues and stabilization of the monomer. Our studies suggest a mechanism through which the structure, function, and fate of the viral genome can be modulated by the transcriptionally controlled presence or absence of a single 5′ guanosine.
The assembly of HIV type 1 (HIV-1) particles in infected cells is initiated by the transcription of viral RNA molecules encoded within integrated proviral DNA (1). Like all eukaryotic cellular mRNAs, HIV-1 transcripts are cotranscriptionally capped by a 5′-5′ triphosphate-linked 7-methylguanosine (7MeG) moiety shortly after initiation of RNA polymerase II-dependent synthesis (2–5). Some transcripts undergo splicing to produce mRNAs that direct ribosomal synthesis of the viral envelope (Env) protein and accessory proteins, whereas others are not spliced and instead function as mRNAs for the viral Gag and, via frameshifting, the Gag-Pol polyproteins (1). A subset of unspliced viral RNAs does not appear to function as mRNAs, instead being recognized by Gag proteins and incorporated into assembling virions. These RNAs serve as the viral genome (gRNA) and are used as templates for reverse transcription during an early stage of the replication cycle. gRNAs are selected for packaging as dimers (6–22), enabling strand-transfer–mediated recombination during reverse transcription and facilitating genetic evolution under environmental and chemotherapeutic pressures (23).
The current understanding of the factors that control the diverse functions of the viral genome is limited. However, the structure of the RNA itself, particularly the highly conserved 5′-leader (5′-L) (1, 24), appears to play an important role. The HIV-1 5′-L contains discrete nucleotide sequences that are important for transcriptional activation (TAR), tRNA primer binding for initiation of reverse transcription (PBS), initiation of RNA dimerization (DIS), splicing (SD), and packaging (Ψ) (Fig. 1). Residues at the 3′ end of the 5′-L that overlap the gag gene start site (AUG) function in both translation and dimerization. Studies have shown that, in vitro, HIV-1 5′-L RNAs can exist as an equilibrium mixture of monomeric and dimeric species and that the equilibrium can be shifted by rational mutagenesis (25, 26). Mutations that favor the monomeric fold in vitro also inhibit nucleocapsid (NC) protein binding, and vector RNAs with these mutations are unable to compete with wild-type 5′-L sequences for encapsidation into viruses (26). Conversely, mutations that favor dimerization and NC binding in vitro also promote competitive RNA packaging (26). These and other data collectively support models in which a single viral transcript directs multiple functions via a monomer–dimer RNA structural switch (8–18, 25–33).
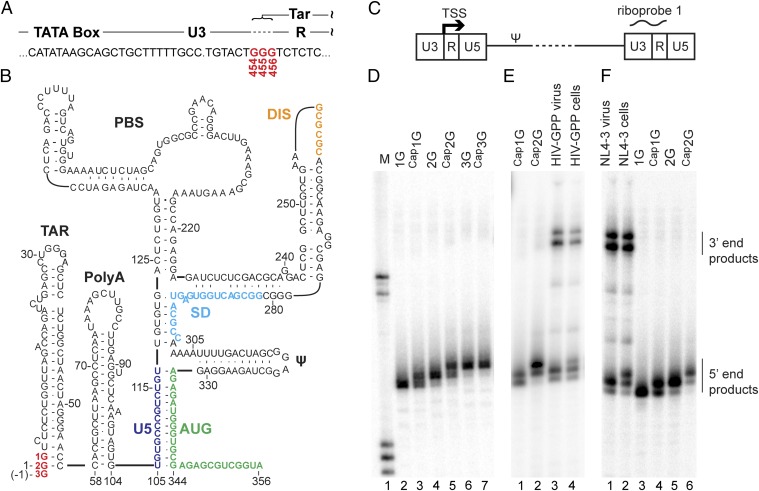
Fig. 1.
HIV-1 RNA 5′-end heterogeneity in virions and cells. (A) The HIV-1NL4-3 provirus U3–R boundary showing the three guanosines (G454, G455, and G456) that can serve as the TSS (and encode the 5′ end of the TAR element of the viral transcript). (B) Secondary structure of the dimer-promoting form of HIV-1 RNA (adapted from ref. 42). 1G, 2G, and 3G guanosines (red) correspond to G456, G455, and G454 TSS residues. (C) Schematic representation of an HIV-1 provirus (not to scale). Ψ indicates the packaging signal. Riboprobe 1 indicates the location of the probe used in D–F and Fig. 3. (D) RNase protection products of in vitro-generated HIV-1 5′-L RNA size standards corresponding to the use of alternate TSS, with or without capping. Lane 1: molecular size standards. Lanes 2–7: protected products for 1G, Cap1G, 2G, Cap2G, 3G, and Cap3G RNAs, respectively. (E) RNase protection analysis of cell and virus samples from transiently transfected cells. Lanes 1 and 2: protected fragments for Cap1G and Cap2G RNA standards. Lanes 3 and 4: fragments protected by virion and cell RNA, respectively. (F) RNase protection analysis of RNA samples harvested from CEM-SS cells chronically infected with HIV-1 strain NL4-3. Lanes 1 and 2: products protected by RNA samples from virus or cells, respectively. Lanes 3–6: protected fragments generated from the indicated RNA size standards. Mobilities of products protected from the 5′ and 3′ ends of viral RNA are indicated on the right.
To identify molecular determinants of retroviral RNA structure, we recently examined the dimerization properties of native and mutant 5′-L RNAs of several lentiviruses (31). Unexpectedly, RNAs reported to start with three 5′ guanosines exhibited dimerization properties substantially different from those that begin with two guanosines. 5′-Capped HIV-1 transcripts that begin with one, two, or three 5′ guanosines (herein called “Cap1G,” “Cap2G,” and “Cap3G” genomes) (Fig. 1) have been predicted based on published HIV-1 proviral DNA sequences [Los Alamos National Laboratory (LANL) HIV sequence database]. These variations are caused by inconsistencies in defining the position of the HIV-1 transcription start site (TSS) (4, 34–36), which is defined genetically as the first residue in the repeat (R) element of the HIV-1 LTR (Fig. 1A). HIV-1 proviruses typically contain a conserved stretch of three guanosines at the U3/R junction (LANL HIV sequence database), any of which potentially could function as the TSS. For the widely studied NL4-3 strain of HIV-1 (HIV-1NL4-3), these residues correspond to G454, G455, and G456 of the proviral DNA and to residues −1, 1, and 2 of the transcribed RNA according to the traditional numbering for the HIV-1 5′-L RNA (Fig. 1A). Early nuclease digestion experiments indicated that G454 serves as the TSS in HIV-1NL4-3 (35). However, subsequent primer extension experiments indicated that G455 serves as the TSS (34), and a more recent study that used RNA ligase-mediated 5′-RACE identified G456 as the TSS (4). However, a very recent study that also probed transcript 5′ ends by 5′-RACE indicated that all three start sites are used, but that only one transcript (1G) is enriched in virions and functions efficiently in reverse transcription (36).
Here we used an RNase protection assay to probe for TSS use in cells and viruses and tested the selectivity of viral replication steps for specific RNA species among experimentally altered intracellular RNA populations. This approach has the advantage of avoiding the 5′-end deletions or additions that can be generated during 5′-RACE procedures and that were reported to occur in earlier studies (36). Our findings confirm that all three transcripts are synthesized in cells and that the Cap1G RNA is specifically selected for packaging. In addition, we now show that the Cap2G and Cap3G transcripts are enriched on polysomes, demonstrating that transcripts with different TSS have distinct functions. We also examined the effects of TSS selection and 5′ capping on the monomer–dimer equilibrium behavior of recombinant HIV-1NL4-3 5′-L RNAs. In vitro, the 5′-L RNAs exhibit strikingly different behavior, with both the 1G and 2G RNAs favoring dimerization and the 3G RNA favoring the monomer. Thus the inclusion of a single 5′ guanosine is sufficient to alter RNA structure dramatically. Surprisingly, the influence of the 5′-Cap moiety on dimerization was similar to that of adding a single phosphodiester-linked 5′ guanosine. Our findings support proposals that genome dimerization is a major determinant of RNA function (8–18, 25–33) and suggest a paradigm in which the structure, function, and fate of the viral transcripts are modulated by heterogeneous TSS selection.
Results
Heterogeneous HIV-1 TSS Use in Infected and Transfected Cells.
The unexpected discovery of differing dimerization properties for 5′-L RNAs with different 5′ ends (see above) made identifying the authentic TSS used by replicating HIV-1 a priority. Because RNase protection assays yielded the cleanest results of several approaches tested here and were readily applicable to both in vitro-transcribed RNAs and samples extracted from viruses and cells, this approach was the primary method used for 5′-end determination. All approaches for determining 5′ ends are affected by the substrate preferences of analytic enzymes, and these and other experimental variables likely have contributed to the literature’s conflicting reports about the 5′ ends of HIV-1 RNA. Therefore, to ensure the accuracy of the RNA-end assignments here, six in vitro-synthesized RNAs with known 5′ ends served as RNase protection assay controls to provide signatures for protected products of specific RNA 5′ ends (Fig. 1D). These RNA standards were prepared by in vitro transcription and were capped using a recombinant Vaccinia virus capping enzyme (37, 38).
RNase protection of the six in vitro-generated 5′-L RNAs yielded distinct protected products for the Cap1G, Cap2G, and Cap3G RNAs (Fig. 1D, lanes 2, 4, and 6). Cap1G and Cap2G RNAs each generated a mixture of two protected products, likely reflecting partial base pairing or breathing between the Cap moiety and the riboprobe’s corresponding cytosine residue (Fig. 1D, lanes 3 and 5). With this demonstration that this riboprobe allowed accurate assignment of RNA 5′ ends that differed by a single base, studies with viral RNAs were begun. Note that, because the riboprobe used here (probe 1 in Fig. 1C) was complementary to the native U3/R junction to avoid confounding signal from U3/R sequences near the 3′ end of viral RNAs, this probe could not distinguish uncapped 3G RNA from Cap3G RNA (Fig. 1A), and thus the 3G RNA and its capped derivative protected the same-length product (Fig. 1D, lanes 6 and 7).
To define the 5′ ends of virus-generated RNAs, the mobilities of RNase protection products of cell and virus samples were compared with those of the 5′-end standards (Fig. 1 D and E). RNA samples were harvested from 293T cells transfected with the Env-deletion HIV-1 derivative NL4-3-GPP (39) and from virions produced by these cells. Analysis of these samples revealed that cell RNA appeared to contain a mixture of the Cap1G and Cap2G/ Cap3G classes of HIV RNA (Fig. 1E, lane 4). In contrast, protected products generated from NL4-3-GPP virion RNA resembled only the products of the Cap1G standard (Fig. 1E, lanes 1 and 3).
Overexpression of HIV-1–derived plasmids in 293T cells, as used in these initial experiments, can generate viral phenotypes that differ from those observed during natural infection. Thus, to test whether findings with NL4-3-GPP expressed in 293T cells resembled those of infectious virus, we next assayed cell and viral samples from CEM-SS T cells supporting a spreading infection initiated by the HIV-1 molecular clone NL4-3 (Fig. 1F). The results (lanes 1 and 2) confirmed a marked difference in HIV-1 cell and virion RNA populations, with an even more pronounced difference in RNA populations than observed for transiently transfected 293T cells.
Although our experiments do not rule out the possibility that 5′-end heterogeneity is caused by the presence of some noncapped RNAs, previous 5′-end–mapping studies failed to detect evidence for a decapped form of HIV-1 RNA in either cells or virions (4). The possibility that HIV-1 transcription partially bypasses mechanisms evolved to promote capping (2) or that HIV-1 RNAs become decapped in cells or virions appears remote (4).
HIV-1 RNA Packaging Is Selective for Cap1G RNAs.
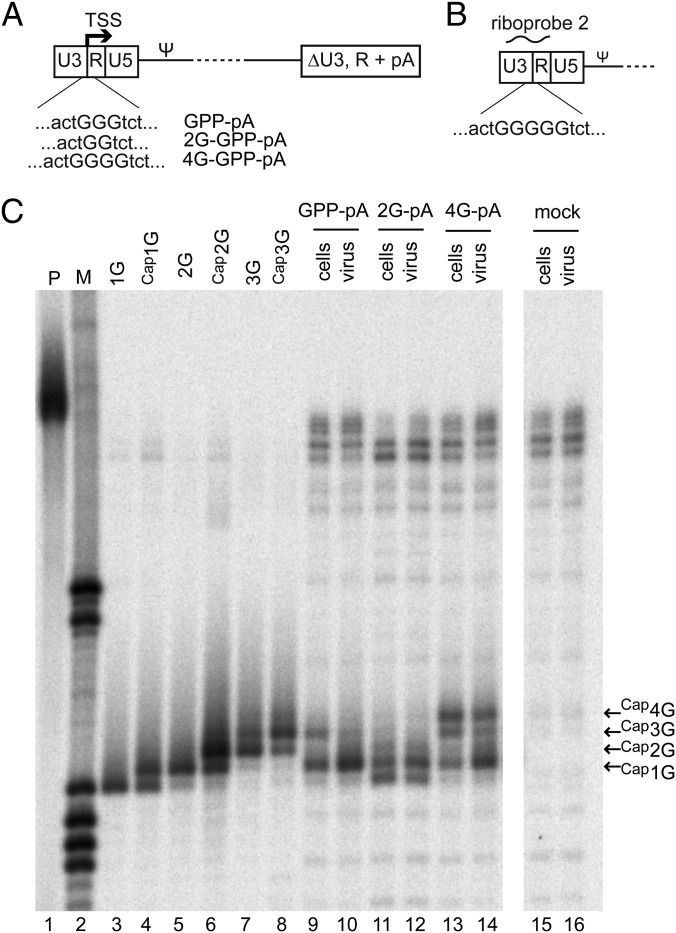
The data presented above showed that Cap1G RNAs predominated in virions but also comprised a significant proportion of the intracellular species. Thus, it seemed possible that the prominence of these RNAs within virions might reflect their high intracellular prevalence rather than selectivity for a particular RNA species per se. To address this possibility, the intracellular distribution of HIV-1 RNA 5′-end classes was skewed by expressing NL4-3 GPP derivatives with altered numbers of guanosine residues (denoted “2G-GPP” and “4G-GPP” to indicate the subtraction or addition of a guanosine residue) (Fig. 2A) at their U3/R junctions. To distinguish RNA 5′ ends from 3′ repeats in RNase protection assays, portions of the downstream LTR were replaced with a simian virus 40 polyadenylation (pA) signal. These constructs were assayed using a modified riboprobe capable of differentiating RNAs with up to five -terminal guanosines (riboprobe 2 in Fig. 2B). Virion and cell RNA populations for these derivatives were compared with native 3G GPP-pA (Fig. 2C).
Fig. 2.
Selectivity for packaging of Cap1G RNAs from cells with altered RNA proportions. (A) Schematic representation of constructs with altered numbers of guanosines at the U3/R junction used to skew intracellular RNA populations. ΔU3, R + pA indicates replacement of downstream LTR sequences with an SV40 polyadenylation signal. (B) Schematic representation of the portion of the HIV-1 genome to which riboprobe 2 is complementary. Note the five guanosines at the U3/R border, which allowed discrimination among more products than the 3G riboprobe. (C) RNase protection assay of constructs with altered numbers of guanosines. Lane 1: undigested probe. Lane 2: size standards. Lanes 3–8: fragments protected by RNA standards. Lanes 9–16: products protected by the indicated cell and virus RNA samples from cells transfected with the indicated HIV-1 GPP derivatives. Mobilities of products protected by Cap1G, Cap2G, Cap3G, and Cap4G RNAs are indicated at the right.
Consistent with a mechanism for TSS selection based in part on distance counting from upstream promoter elements, the RNAs generated in cells expressing 2G-GPP-pA included an RNA one base shorter than Cap1G (Fig. 2C, lane 11). Similarly, most intracellular RNA species produced by 4G-GPP-pA appeared to be one base longer than the corresponding products in cells expressing a native 3G genome (compare lanes 9 and 13). However, although the Cap1G RNA was a relatively minor intracellular species when 2G or 4G vectors were expressed (lanes 11 and 13), the Cap1G RNA nonetheless was the predominant species in extracellular virions (lanes 12 and 14).
Interestingly, although virion production levels from 293T cells transiently transfected with 2G-GPP-pA, 4G-GPP-pA, and parental NL4-3-GPP-pA were indistinguishable, when these mutations were built into both LTRs of NL4-3 and equal amounts of virus were used to infect CEM-SS cells, both the 2G and 4G forms showed significant delay in a spreading assay.
Capped 2G/3G HIV-1 RNAs Are Enriched on Polysomes.
The correlation between RNAs that preferentially adopted a packaging-competent fold and the RNA subpopulation that was enriched in virions (Figs. 1 and 2) suggested that the nature of an HIV-1 RNA’s 5′ end may dictate its functional fate. Because infected cells contain both Cap1G and longer (Cap2G/ Cap3G) RNAs, a further prediction of this hypothesis was that the longer unspliced HIV-1 RNAs that are not packaged instead adopt an mRNA fate.
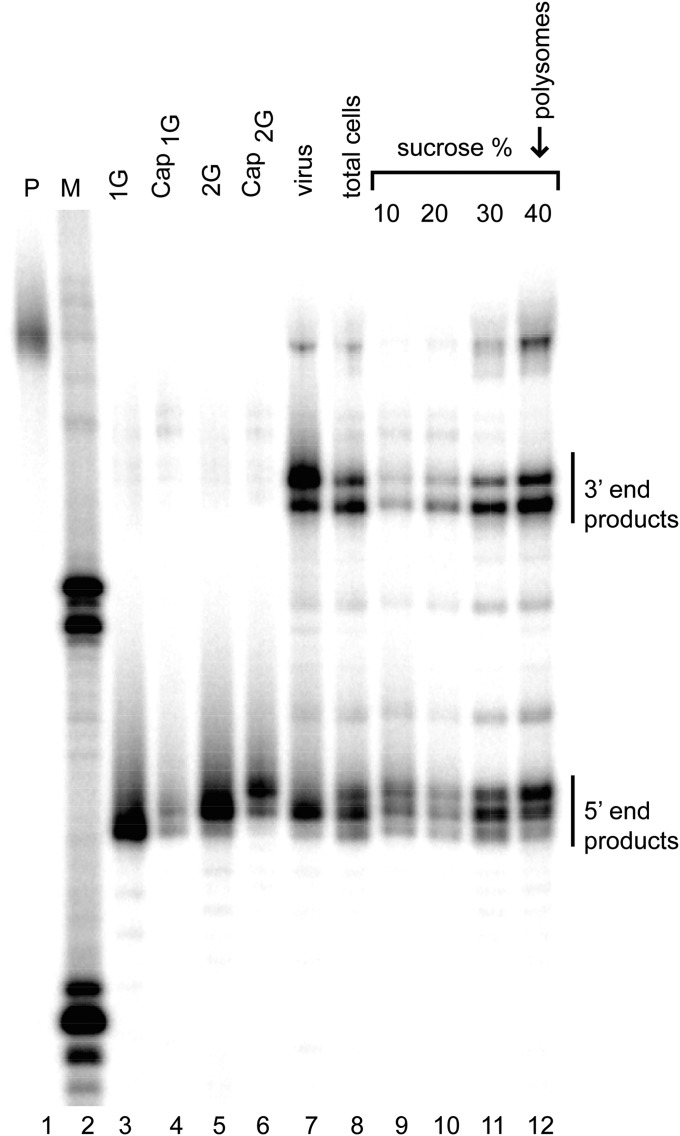
To test this hypothesis, a spreading infection using the HIV-1 molecular clone NL4-3 was established in CEM-SS T cells, and both infected cells and cell-free virions were harvested (Fig. 3). Cells were lysed under conditions that stabilize polyribosomes and were fractionated on sucrose step gradients (Fig. 3). This process yielded fractions containing material more buoyant than ribosomes (10 and 20% sucrose fractions), a fraction that likely includes ribosome subunits and some nonribosomal ribonucleoprotein complexes (30% sucrose fraction), and the major polysome-containing fraction (40% sucrose fraction; lane 12). Consistent with earlier results, virion RNA was dominated by Cap1G RNA, whereas RNA harvested from unfractionated cells contained both Cap1G and longer (Cap2G/ Cap3G) HIV RNAs (lanes 7 and 8). Both RNA classes were also observed in each sucrose gradient fraction of the infected cells (lanes 9–12). However, in contrast to the nonpolysomal fractions, which display Cap1G /total RNA ratios reminiscent of the total cell extract, RNAs in the polysomal fraction displayed a marked bias toward the nonpackaged longer RNAs.
Fig. 3.
Analysis of HIV-1 RNA forms associated with polyribosomes. RNase protection assay on cell lysate fractions from chronically infected CEM-SS cells, analyzed with riboprobe 1 (Fig. 1). Lane 1: undigested probe. Lane 2: size standards. Lanes 3–6: fragments protected by the indicated RNA standards. Lanes 7–8: fragments protected by chronically infected cell medium (virus) and total cell lysate. Lanes 9–12: probe fragments protected by RNA from 10, 20, 30, and 40% sucrose step gradient fractions. Mobilities of protected products from the 5′ and 3′ ends of viral RNA are indicated on the right.
Influence of TSS Selection and Capping on Dimerization.
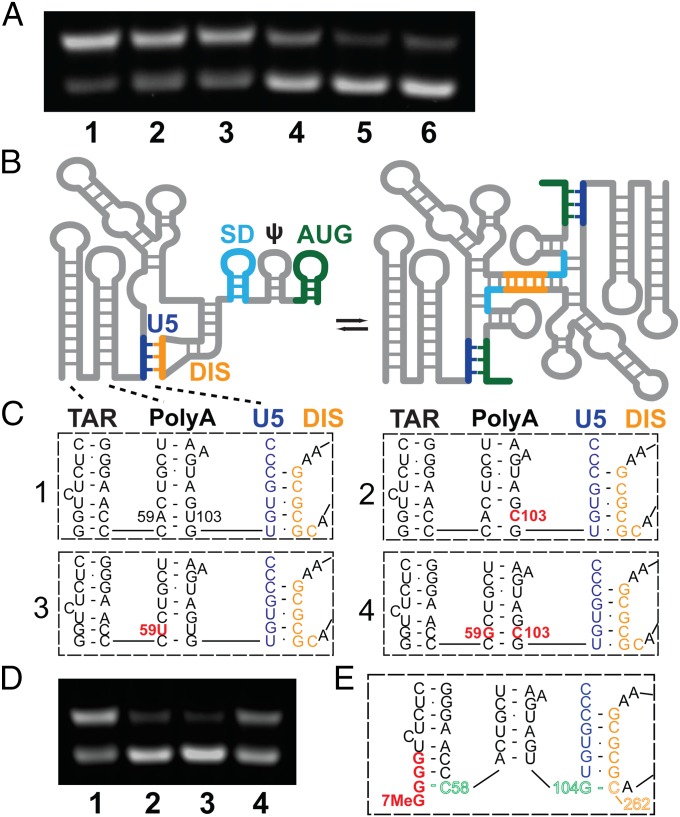
Because prior studies have established that genome dimerization likely controls packaging and other RNA-dependent functions (8–18, 25–33), we examined the influence of TSS use on the in vitro dimerization properties of the HIV-1NL4-3 5′-L (Fig. 4A). 1G, 2G, and 3G forms of the RNA were prepared by in vitro transcription, and their relative propensities to form dimers were determined by native gel electrophoresis after incubation under conditions of physiological-like ionic strength (Materials and Methods). As shown in Fig. 4, all existed as equilibrium mixtures of monomeric and dimeric species. However, although the 1G and 2G 5′-L RNAs adopted predominantly dimeric structures, the 3G 5′-L RNA existed predominantly as a monomer.
Fig. 4.
(A) Influence of 5′-guanosine number and capping on dimerization, as follows: lane 1: 1G 5′-L RNA; lane 2: Cap1G 5′-L; lane 3: 2G 5′-L; lane4: Cap2G 5′-L; lane 5: 3G 5′-L; and lane 6: Cap3G 5′-L. Under the conditions used, RNAs in lanes 1–3 favor the dimer, whereas those in lanes 4–6 favor the monomer. (B–D) Mutations engineered to destabilize the base of PolyA favor the monomer. (B) Working model for secondary structural changes associated with the monomer–dimer equilibrium (adapted from ref. 26). (C) Proposed base pairing of residues in the lower stems of the TAR and PolyA hairpins and the U5:DIS pseudoknot for wild-type (1), U103C (2), A59U (3), and A59G/U103C (4) constructs. (D) Influence of point mutations on dimerization; lane numbers correspond to the panel labels in C. (E) Model for TSS-dependent dimerization control. In Cap3G transcripts, G(−1)–C58 base pairing disrupts and remodels the PolyA hairpin, allowing G104 to base pair with C262 of the DIS palindrome and thereby stabilizing the monomeric conformer.
The dimerization propensities of 5′-capped forms of the above RNAs (Cap1G, Cap2G, and Cap3G 5′-L) were also determined. Interestingly, the influence of the Cap moiety was similar to that observed with the transcriptional inclusion of an additional 5′ guanosine. Thus, 5′-L RNAs containing two 5′ guanosines and no Cap (2G 5′-L) or one 5′ guanosine plus a Cap (Cap1G 5′-L) behaved similarly, both exhibiting strong propensities to form dimers. In contrast, RNAs that contain three 5′ guanosines or two 5′ guanosines plus a Cap exhibited similar propensities to form monomers (Fig. 4A). These findings demonstrate that the presence of a single additional 5′ guanosine can strongly perturb the monomer–dimer equilibrium of the HIV-1 5′-L and that a Cap moiety influences dimerization in a manner similar to that of a phosphodiester-linked 5′ guanosine.
Probing the Mechanism of 5′ Guanosine- and Cap-Dependent Dimerization Control.
To understand how the 5′ guanosine and 5′-Cap residues influence dimerization, we prepared a series of 5′-L RNAs with point mutations designed to stabilize the monomeric or dimeric conformers differentially. Experiments were guided by a recently proposed dimerization mechanism, in which the monomeric conformer is stabilized by U5:DIS base pairing that sequesters the DIS and dimerization is promoted by the formation of U5:AUG base pairs that displace and expose the DIS (Fig. 4B) (26). Comparative studies were conducted with native and mutant forms of the 2G form of the 5′-L, which exists predominantly as a dimer under the ionic conditions used (Fig. 4D, lane 1). The replacement of U103 by cysteine, designed to destabilize the lower PolyA stem (Fig. 4C, lane 2), led to a significant equilibrium shift in favor of the monomer (Fig. 4D, lane 2). The replacement of A59 by uracil (Fig. 4C, subpanel 3) also shifted the equilibrium toward the monomer (Fig. 4D, lane 3). Moreover, the compensatory replacement of A59 by guanosine in the previously mutated U103C construct, designed to re-establish base pairing in the lower PolyA stem (Fig. 4C, subpanel 4), resulted in an equilibrium shift back toward the dimer (Fig. 4D, lane 4). These findings reveal that the propensity of the 5′-L RNA to adopt the monomeric species is inversely correlated with the stability of the lower stem of the PolyA hairpin. Note that, although the mutagenesis results are compatible with the monomeric 5′-L model shown in Fig. 4B (26), they do not appear to be compatible with an alternative long-distance interactive model (28) in which all mutations but one (U103C) are predicted to reside in unstructured loops and have little influence on the equilibrium.
Discussion
Viruses often evolve genetic simplicity by diversifying the functions of individual molecules. A classic example has been the use by retroviruses of a single unspliced transcript not only as the genome in progeny virions but also as the mRNA for the viral structural proteins. Although much remains to be learned about the factors that modulate these functions, there is considerable biological and biophysical evidence that the bifurcation of retroviral RNA into genome and mRNA roles is dictated by dimerization-dependent structural changes (9, 12, 16, 26, 29). When RNA mutations that destabilize the dimer-competent fold of the HIV-1 5′-L are reconstituted into viral genomes, and these are coexpressed with wild-type genomes, the mutant RNAs fail to compete with the wild-type RNA for packaging (26). Similarly, mutations that prevent dimerization of the recombinant 5′-L RNA of the Moloney murine leukemia virus (MLV) block packaging (40), suggesting that dimerization-dependent control mechanisms are conserved among evolutionarily distant retroviruses (40, 41). MLV-derived vectors harboring monomer-stabilizing mutations remain unpackaged in the absence of competing wild-type leader RNAs (40), whereas mutations that shift the equilibrium of the HIV-1 5′-L in favor of the monomer but do not prevent dimerization do not prevent the HIV-1 RNA from functioning as both genome and mRNA in the absence of wild-type competition (26, 39, 42). These and other observations support a mechanism in which a single viral transcript equilibrates between structures associated with alternate replication functions (9, 12, 16, 26, 29). In these models, the equilibration is not rate limiting, and HIV-1 RNA fates are sealed by the binding of alternate sets of proteins associated with differing replication roles (8–18, 33).
The present findings suggest a modified paradigm in which the functions and fates of HIV-1 transcripts are encoded not by a preexisting monomer–dimer equilibrium but instead by the intrinsic propensities of RNAs with different 5′ ends to adopt a monomeric or a dimeric conformation preferentially. In this model, the differential propensity of the capped 1G transcript to dimerize and of the 2G/3G RNAs to remain monomeric establishes the function and fate of the transcripts, with the capped 1G RNAs being selected for packaging and functioning as gRNA and the capped 2G/3G RNAs preferentially accumulating on polysomes and functioning as mRNAs.
Our studies also show that destabilizing the lower stem of PolyA by mutagenesis can cause a dramatic equilibrium shift in favor of the monomer. Based on secondary structure analyses, we propose a model in which the overhanging 5′ guanosine in the 3G and Cap3G transcripts [position (−1)] (Fig. 1B) remodels the lower stem of PolyA by base pairing with C58 (Fig. 4E). Disruption of the C58–G104 base pair would enable G104 to form a new base pair with C262 of the DIS, thereby favoring the monomeric conformer by stabilizing the U5:DIS helix by ∼3–10 kcal/mol (Fig. 4E) (43). Because the 5′-Cap and 5′-G moieties have similar influences on the monomer–dimer equilibrium, it seems plausible that the overhanging 5′ Cap of the Cap2G transcript also stabilizes the monomer by interacting with C58 and disrupting the PolyA helix. This model is compatible with studies showing that deletion of the TAR or PolyA hairpins does not significantly affect 5′-L dimerization or vector packaging (39), whereas mutations that open the lower helix of TAR inhibit both dimerization and packaging (44, 45).
Our studies reveal that transcriptional addition of only a single guanosine to Cap1G RNA can dramatically perturb the monomer–dimer equilibrium in vitro, but the predominant species generated in cells and used in replication are the Cap1G and Cap3G transcripts. Greater production of Cap3G (versus Cap2G) transcripts may reflect an evolutionary preference for promoters that contain three sequential guanosines, which can afford optimal transcription in eukaryotic cells (36). In addition, because our models predict that the Cap is sequestered by base pairing in Cap2G transcripts but is unpaired and exposed in Cap3G RNAs, preferential Cap3G use could result from the enhanced ability of Cap3G transcripts to bind the eukaryotic Cap-binding protein eIF4E and thereby promote translation (46).
It is not unusual for polymerase II promoters to use multiple TSS (47). A recent genome-wide study of mammalian promoter architecture by cap analysis of gene expression identified subsets of TATA-driven promoters, termed “twinned TSS promoters,” that have two dominant start sites separated by 0–3 nt (48, 49). Interestingly, those with adjacent dominant start sites display a GGG consensus sequence and weak TATA conservation, and those with TSS sites separated by a single nucleotide also exhibit a relatively strong GGG preference and strong TATA use (49). Importantly, TSS use varied among cell types and between normal and tumoral cell lines, indicating that factors other than sequence are important for TSS selection (49, 50). Thus it is conceivable that HIV-1 TSS use may be affected by intracellular conditions or change temporally during replication, enabling early production of the longer Cap2G/Cap3G mRNAs used for viral protein synthesis followed by an increase in the production of Cap1G gRNA transcripts relegated for packaging. Efforts to test this hypothesis are underway.
Materials and Methods
RNAs for in vitro dimerization studies were prepared by mutating a previously described plasmid containing the 2G 5′-L sequence (51). In vitro transcription was performed using a method that inhibits self-templated run-on (51, 52). The 5′-capped RNAs were prepared using a Vaccinia virus capping enzyme prepared in house with plasmid kindly provided by Stephen Cusack, European Molecular Biology Laboratory (EMBL), Grenoble, France (38). Cells were propagated and treated and plasmids and RNAs for viral and cellular assays were prepared using modifications of previously described approaches (39). Further details regarding the methods used can be found in SI Materials and Methods.
SI Materials and Methods
RNA Preparation.
A pUC19 plasmid containing the T7 promoter followed by the 5′-L construct that started with two guanosines (G455 start site) (46) was mutated to contain either a single guanosine (G456 start site) or three guanosines (G454 start site) using the QuikChange site-directed mutagenesis kit (Agilent Technologies) and the following primers (IDT): 5′-ATA CGA CTC ACT ATA GGG TCT CTC TGG TTA G-3′ and 5′-CTA ACC AGA GAG ACC CTA TAG TGA GTC GTA T-3′ for the G454 start site and 5′-ATA CGA CTC ACT ATA GTC TCT CTG GTT AG-3′ and 5′-CTA ACC AGA GAG ACT ATA GTG AGT CGT AT-3′ for the G456 start site. The plasmids were sequenced (Genewiz) to confirm the mutations. DNA templates for transcription were obtained by PCR amplification using Lucigen EconoTaq PLUS 2X Master Mix and the primers 5′-GGGATGTGCTGCAAGGCGATTAAGTTGGG-3′ and 5′-mUmACCGACGCTCTCGC-ACCCATCTCTC-3′ (“m” preceding a base indicates that the base is 2′-O-methylated) (IDT).
In Vitro Transcription.
In-house purified T7 RNA polymerase was used to synthesize RNA in reactions containing 40–53.3 ng/μL purified DNA template, 2 mM spermidine, 80 mM Tris⋅HCl (pH 8), 2 mM DTT, 10–20 mM MgCl2, 3–6 mM nucleoside triphosphates (NTPs), and 20% (vol/vol) DMSO. Trial reactions were used to optimize MgCl2 and NTP ratios for each construct. Transcription reactions were incubated for 2.5 h at 37 °C and were halted by the addition of 1 mmol of EDTA followed by boiling for 3 min and snap cooling on ice. RNA was purified on a 6% denaturing acrylamide gel (SequaGel; National Diagnostics), visualized by UV shadowing, and eluted using the Elutrap electroelution system (Whatman). The eluted RNA was washed twice with 2-M high-purity NaCl (99.999%; Acros) followed by washing eight times with ultrapure water on a 30K Amicon Ultra centrifugal filter device.
Enzymatic Capping.
An in-house Vaccinia virus capping system was developed based on the New England Biolabs system, using a plasmid encoding His-tagged Vaccinia virus capping enzyme from Stephen Cusack’s laboratory at the EMBL, Grenoble, France (31). The plasmid was transformed into BL21(DE3)pLysS cells (Life Technologies), which were incubated in Terrific broth at 37 °C, 250 rpm shaking overnight. Four 1-L Terrific broth cultures with ampicillin and chlorampicillin were inoculated from this starter culture and were grown at 37 °C with 150 rpm shaking to an OD of ∼0.88. Cultures were placed on ice for 15–45 min and then were induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (500 µL of 1 M) and incubated at 20 °C overnight with 150 rpm shaking. Frozen cell pellets were resuspended in 100 mL lysis buffer (40 mM Tris⋅HCl, 200 mM NaCl, 10 mM imidazole, 5 mM TCEP, pH 8.0), 300 µL of protease inhibitor was added, and cells were lysed. Cobalt resin slurry (10 mL) was equilibrated in lysis buffer by washing twice with a 10-mL volume. The lysate was cleared by centrifugation for 30 min at 35,000 × g at 4 °C and was bound to the resin for 1 h at 4 °C. The lysate was allowed to flow through, and then the resin was washed two times with 50 mL of lysis buffer and four times with 10-mL aliquots of lysis buffer. The enzyme was eluted with five 10-mL aliquots of elution buffer (40 mM Tris⋅HCl, 200 mM NaCl, 250 mM imidazole, 5 mM TCEP, pH 8.0). The protein concentrations of all fractions were determined, and those with concentrations >0.05 mg/mL were combined and dialyzed overnight at 4 °C in 20 mM Tris⋅HCl, 100 mM NaCl, 0.100 mM EDTA, 10% glycerol, 1 mM DTT (pH 8.0) and then were tested for activity by comparison with the New England Biolabs Vaccinia capping system enzyme using a 19-nt hairpin. Products were analyzed on a 20% denaturing acrylamide gel (SequaGel; National Diagnostics) and were visualized, eluted, and washed as described above.
Native Gel Electrophoresis Studies.
RNAs were prepared for native gel electrophoresis from purified stocks by dilution to the desired concentration (0.15 µM) in RNase-free water (autoclaved Millipore purified water), followed by the addition of 10% of the final volume of a 100 mM Hepes buffer, pH 7.0. Ten percent of the sample was used to verify the concentration. The remainder was transferred to a fresh RNase-free 1.5-mL centrifuge tube, physiological ionic salts were added to a final concentration of 140 mM KCl, 10 mM NaCl, and 1 mM MgCl2, and the sample was incubated at 37 °C for 24 h. Samples were analyzed on native Tris base-boric acid agarose gels and visualized a Kodak Gel Logic 200 Imaging System.
Plasmids for Virology.
pNL4-3 contains an infectious HIV-1 molecular clone (48). Construction of the HIV-GPP retroviral vector, which encodes all HIV proteins except Env, has been described previously (23). The 2G and 4G TSS mutations were introduced into HIV vectors using sequence-verified PCR fragments containing the desired HIV-1 mutations and were cloned into intermediate plasmids and then into pNL4-3 using AatII and SphI to generate pNL4-3 TS-2G and pNL4-3 TS-4G. GPP-pA, 2G-GPP-pA, and 4G-GPP-pA plasmids were constructed by replacing the XhoI-NgoMIV fragment of pCMV-ΔR8.2 with the SV40 virus pA signal (49).
Viruses and Cells.
293T and CEM-SS cells were grown at 37 °C and 5% CO2 in DMEM and RPMI1640, respectively, containing 10% FBS and 50 µg/mL gentamicin. For production of virus stocks, 293T cells were cultured to 70% confluence in 10-cm2 dishes and were transfected with 3 μg pNL4-3 DNA and 12 μg polyethylenimine (PEI) (50). Virus-containing supernatant was harvested after 48 h. The virus level in the culture medium was determined using a real-time PCR-based reverse transcriptase (RT) assay modified from ref. 51, in which HIV-1–containing medium with known concentrations of CA-p24 was used as the standard. Amounts of virus equivalent to 1 ng CA-p24 were used to infect CEM-SS cells (4 × 105 per mL), which were fed with fresh culture medium twice a week. Establishment of a chronically infected culture was monitored by the real-time PCR-based RT assay. Once virus concentrations plateaued, virus and cells were harvested.
For transient transfection with HIV-1 vectors, 293T cells were cultured to 70% confluence in 10-cm2 dishes and were transfected using 6 μg of plasmid DNA and 4 μg PEI per microgram of DNA (50). Virus medium and cells were harvested 48 h after transfection.
Polysome Fractionation.
Chronically infected CEM-SS cells were incubated in medium containing 50 μg/mL cycloheximide 30 min before harvesting. Cells were harvested by centrifugation at 3,000 × g for 5 min. Cell pellets were washed twice with ice-cold PBS containing 50 μg/mL cycloheximide. Clarified cell lysates for polysome fractionation were obtained as described (52), with modifications. Specifically, cell pellets were resuspended in 0.5 mL of cold resuspension buffer (RSB) [10 mM Tris⋅HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2) containing 250 units of RNasin (Promega) and were incubated for 2 min on ice. After the addition of 0.5 mL RSB containing 1% Triton X-100, 1% deoxycholate, and 2% Tween 20, the cell suspension was incubated for an additional 7 min on ice and then was centrifuged for 5 min at 3,000 × g. The supernatant was clarified by additional centrifugation for 10 min at 10,000 × g, and sucrose step gradient fractionation was performed as described (53). RNA was extracted from each sucrose fraction using TRIzol LS Reagent (Ambion) following the manufacturer’s protocol.
RNA Extraction.
Virus-containing medium was filtered through 0.22-μm filters and was ultracentrifuged through a 20% sucrose cushion. Pellets were lysed in TRIzol Reagent (Ambion). To obtain cellular RNA from chronically infected CEM-SS cells, the cells were collected from 8 mL of culture by centrifugation, washed once with PBS, and lysed in 1 mL TRIzol Reagent (Ambion). To obtain cellular RNA from transfected 293T cells, the cells were washed with PBS and lysed by the addition of 2.5 mL of TRIzol Reagent (Ambion) per 10-cm2 plate. After precipitation, RNA extracted with TRIzol was dissolved in Tris-EDTA buffer and treated with RQ1 DNase (Promega) to remove possible DNA traces. RNA then was re-extracted with phenol-chloroform, precipitated with EtOH, dissolved in TENS buffer (10 mM Tris, 1 mM EDTA, 1% SDS, 100 mM NaCl), and stored at −80 °C.
RNase Protection Assay.
RNase protection assays were performed as previously described (54) with minor modifications. Riboprobes were transcribed with SP6 RNA polymerase using PCR product templates. RNase A was used for digestion of nonhybridized RNA. RNase protection assay products were resolved by denaturing 15% PAGE (29:1 acrylamide/bis-acrylamide) at 1,700 V for ∼5 h.
Acknowledgments
We thank the Howard Hughes Medical Institute (HHMI) staff at the University of Maryland Baltimore County (UMBC) for technical support; Cleo Burnett (University of Michigan) for help with illustrations; and Stephen Cusack and Delphine Guilligay (EMBL) for providing Vaccinia virus capping enzyme plasmid and technical advice. This research was supported by National Institute of General Medical Sciences Grants R01 GM42561 (to M.F.S.) and P50 GM103297 (to A.T.). J.D.B. was supported by NIH Grant for Promoting Graduate Diversity 2R25 GM055036, and N.C.B. was supported by an HHMI Education Grant to UMBC and a National Institute of Biomedical Imaging and Bioengineering/National Institute on Drug Abuse contract to support science, technology, engineering, and mathematics diversity.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616627113/-/DCSupplemental.
References
- 1.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Lab Press; Plainview, NY: 1997. [PubMed] [Google Scholar]
- 2.Chiu YL, Coronel E, Ho CK, Shuman S, Rana TM. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J Biol Chem. 2001;276(16):12959–12966. doi: 10.1074/jbc.M007901200. [DOI] [PubMed] [Google Scholar]
- 3.Zhou M, et al. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc Natl Acad Sci USA. 2003;100(22):12666–12671. doi: 10.1073/pnas.1835726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menees TM, Müller B, Kräusslich HG. The major 5′ end of HIV type 1 RNA corresponds to G456. AIDS Res Hum Retroviruses. 2007;23(8):1042–1048. doi: 10.1089/aid.2006.0275. [DOI] [PubMed] [Google Scholar]
- 5.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. Thriving under stress: Selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012;8(3):e1002612. doi: 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz S, Felber BK, Benko DM, Fenyö EM, Pavlakis GN. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64(6):2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikolaitchik OA, et al. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 2013;9(3):e1003249. doi: 10.1371/journal.ppat.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell RS, Liang C, Wainberg MA. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology. 2004;1(23):23. doi: 10.1186/1742-4690-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paillart J-C, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: An inseparable pair. Nat Rev Microbiol. 2004;2(6):461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 10.Paillart J-C, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie. 1996;78(7):639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 11.Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol. 2011;410(4):609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuzembayeva M, Dilley K, Sardo L, Hu W-S. Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology. 2014;454-455:362–370. doi: 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jewell NA, Mansky LM. In the beginning: Genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J Gen Virol. 2000;81(Pt 8):1889–1899. doi: 10.1099/0022-1317-81-8-1889. [DOI] [PubMed] [Google Scholar]
- 14.Hellmund C, Lever AM. Coordination of genomic RNA packaging with viral assembly in HIV-1. Viruses. 2016;8(7):E192. doi: 10.3390/v8070192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greatorex J, Lever A. Retroviral RNA dimer linkage. J Gen Virol. 1998;79(Pt 12):2877–2882. doi: 10.1099/0022-1317-79-12-2877. [DOI] [PubMed] [Google Scholar]
- 16.Greatorex J. 2004. The retroviral RNA dimer linkage: Different structures may reflect different roles. Retrovirology 1(22):(18 August edition)
- 17.D’Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3(8):643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 18.Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 19.Sakuragi J, Sakuragi S, Shioda T. Minimal region sufficient for genome dimerization in the human immunodeficiency virus type 1 virion and its potential roles in the early stages of viral replication. J Virol. 2007;81(15):7985–7992. doi: 10.1128/JVI.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakuragi J, Iwamoto A, Shioda T. Dissociation of genome dimerization from packaging functions and virion maturation of human immunodeficiency virus type 1. J Virol. 2002;76(3):959–967. doi: 10.1128/JVI.76.3.959-967.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakuragi J, Shioda T, Panganiban AT. Duplication of the primary encapsidation and dimer linkage region of human immunodeficiency virus type 1 RNA results in the appearance of monomeric RNA in virions. J Virol. 2001;75(6):2557–2565. doi: 10.1128/JVI.75.6.2557-2565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakuragi J, Ueda S, Iwamoto A, Shioda T. Possible role of dimerization in human immunodeficiency virus type 1 genome RNA packaging. J Virol. 2003;77(7):4060–4069. doi: 10.1128/JVI.77.7.4060-4069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev. 2009;73(3):451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lever AM. HIV-1 RNA packaging. Adv Pharmacol. 2007;55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- 25.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44(25):9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 26.Lu K, et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334(6053):242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miele G, Mouland A, Harrison GP, Cohen E, Lever AM. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J Virol. 1996;70(2):944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbink TEM, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J Biol Chem. 2003;278(13):11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 29.Abbink TEM, Berkhout B. RNA structure modulates splicing efficiency at the human immunodeficiency virus type 1 major splice donor. J Virol. 2008;82(6):3090–3098. doi: 10.1128/JVI.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jablonski JA, Buratti E, Stuani C, Caputi M. The secondary structure of the human immunodeficiency virus type 1 transcript modulates viral splicing and infectivity. J Virol. 2008;82(16):8038–8050. doi: 10.1128/JVI.00721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran T, et al. Conserved determinants of lentiviral genome dimerization. Retrovirology. 2015;12:83. doi: 10.1186/s12977-015-0209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mailler E, et al. The life-cycle of the HIV-1 Gag-RNA complex. Viruses. 2016;8(9):E248. doi: 10.3390/v8090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rein A. Retroviral RNA packaging: A review. Arch Virol Suppl. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 34.Muesing MA, et al. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature. 1985;313(6002):450–458. doi: 10.1038/313450a0. [DOI] [PubMed] [Google Scholar]
- 35.Starcich B, et al. Characterization of long terminal repeat sequences of HTLV-III. Science. 1985;227(4686):538–540. doi: 10.1126/science.2981438. [DOI] [PubMed] [Google Scholar]
- 36.Masuda T, et al. Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci Rep. 2015;5:17680. doi: 10.1038/srep17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuchs AL, Neu A, Sprangers R. A general method for rapid and cost-efficient large-scale production of 5′ capped RNA. RNA. 2016;22(9):1454–1466. doi: 10.1261/rna.056614.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De la Peña M, Kyrieleis OJ, Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 2007;26(23):4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heng X, et al. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol. 2012;417(3):224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyazaki Y, et al. An RNA structural switch regulates diploid genome packaging by Moloney murine leukemia virus. J Mol Biol. 2010;396(1):141–152. doi: 10.1016/j.jmb.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J Mol Biol. 2004;337(2):427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 42.Keane SC, et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science. 2015;348(6237):917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C, Jiang L, Michalczyk R, Russu IM. Structural energetics and base-pair opening dynamics in sarcin-ricin domain RNA. Biochemistry. 2006;45(45):13606–13613. doi: 10.1021/bi060908n. [DOI] [PubMed] [Google Scholar]
- 44.Helga-Maria C, Hammarskjöld M-L, Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J Virol. 1999;73(5):4127–4135. doi: 10.1128/jvi.73.5.4127-4135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das AT, Vrolijk MM, Harwig A, Berkhout B. Opening of the TAR hairpin in the HIV-1 genome causes aberrant RNA dimerization and packaging. Retrovirology. 2012;9:59. doi: 10.1186/1742-4690-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin Cell Dev Biol. 2005;16(1):39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol. 2012;1(1):40–51. doi: 10.1002/wdev.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38(6):626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 49.Ponjavic J, et al. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7(8):R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frith MC, et al. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18(1):1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keane SC, et al. NMR detection of intermolecular interaction sites in the dimeric 5′-leader of the HIV-1 genome. Proc Natl Acad Sci USA. 2016;113(46):13033–13038. doi: 10.1073/pnas.1614785113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helmling C, et al. Rapid NMR screening of RNA secondary structure and binding. J Biomol NMR. 2015;63(1):67–76. doi: 10.1007/s10858-015-9967-y. [DOI] [PubMed] [Google Scholar]
- 53.Pioli PA, Hamilton BJ, Connolly JE, Brewer G, Rigby WF. Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. J Biol Chem. 2002;277(38):35738–35745. doi: 10.1074/jbc.M204002200. [DOI] [PubMed] [Google Scholar]
- 54.Onafuwa-Nuga AA, King SR, Telesnitsky A. Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J Virol. 2005;79:13528–13537. doi: 10.1128/JVI.79.21.13528-13537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]