Significance
This paper identifies a significant relationship between cortical myeloarchitecture and functional connectivity in the human brain. This is a significant step toward understanding the role of myelin in shaping large-scale neural networks. Our results extend recent work showing that electrical activity promotes myelination and add significant weight to the argument that neural oscillations are a core mediator of brain connectivity.
Keywords: network, functional connectivity, myelination, magnetoencephalography, MRI
Abstract
The human brain relies upon the dynamic formation and dissolution of a hierarchy of functional networks to support ongoing cognition. However, how functional connectivities underlying such networks are supported by cortical microstructure remains poorly understood. Recent animal work has demonstrated that electrical activity promotes myelination. Inspired by this, we test a hypothesis that gray-matter myelin is related to electrophysiological connectivity. Using ultra-high field MRI and the principle of structural covariance, we derive a structural network showing how myelin density differs across cortical regions and how separate regions can exhibit similar myeloarchitecture. Building upon recent evidence that neural oscillations mediate connectivity, we use magnetoencephalography to elucidate networks that represent the major electrophysiological pathways of communication in the brain. Finally, we show that a significant relationship exists between our functional and structural networks; this relationship differs as a function of neural oscillatory frequency and becomes stronger when integrating oscillations over frequency bands. Our study sheds light on the way in which cortical microstructure supports functional networks. Further, it paves the way for future investigations of the gray-matter structure/function relationship and its breakdown in pathology.
The way in which integration of functionally specific brain regions supports ongoing cognition is one of the most important questions in neuroscience, and noninvasive in vivo imaging provides a tool to investigate this interregional connectivity in terms of both brain function and structure. Functional connectivity refers to statistical interdependencies between patterns of brain “activity” measured at separate cortical locations (1) and, even in the “resting state,” measured spontaneous brain activity defines nonrandom networks that are related to cognitive processes (2). The way in which these functional networks are supported by structural white-matter pathways is reasonably well understood (3). However, it is likely that the structure–function association extends to gray-matter morphology, for which fundamental understanding is lacking. Structural morphology of the cortex is known to vary significantly between individuals, and does so in an organized fashion. For example, individuals with a high cortical volume in Broca’s area typically exhibit high cortical volume in Wernicke’s area, reflecting a language network (4). Similar observations can be made between other associated cortical regions (5, 6). This is known as structural covariance (7–9) and it allows the formation of matrices showing how structural properties of individual brain regions covary over subjects. In this paper, we assess structural covariance based upon cortical myeloarchitecture and probe its relationship to functional networks that are assessed based upon measured spontaneous brain activity.
Noninvasive mapping of cortical myeloarchitecture (10, 11) has grown in popularity in recent years, fueled by an increased interest in myelination-based parcellation (12). The general finding is that primary sensory cortices tend to be heavily myelinated whereas regions associated with multisensory integration are less myelinated (10, 13). More localized spatial changes in myelination also exist; for example, subtle subdivisions between regions are apparent in visual (14, 15), somatosensory (16), and auditory (17) cortices. There is evidence of cross-species changes in myelination (18), with the brains of nonhuman primates more heavily myelinated than those of humans. Although still unproven, findings suggest that cortical myelin may inhibit plasticity; for example, early sensory areas may require less plasticity, and therefore more myelin, whereas higher-order areas have less myelination, which might enable greater plasticity (10). Cortical myelination has been shown to change throughout development, and is not established fully until the third decade of life (19, 20). However, no studies have yet used myelin mapping to examine structural covariance or the link between a cortical myelin network and functional connectivity. Previous work does show a direct link between myeloarchitecture and function (21–23). For example, a recent study (24) showed that optogenetic stimulation directed to increase neuronal firing in premotor cortex of mice promotes oligodendrogenesis and therefore myelination, thus providing a link between neuroelectrical activity and myeloarchitecture. Further work (25) suggests that the amount of cortical myelin in a region predicts the magnitude of electrophysiological responses. Taken together, the evidence converges to a hypothesis that, if neuronal firing acts to shape the spatial signature of myeloarchitecture, then networks reflecting the brain’s primary pathways of functional connectivity should be predictive of structural networks of intracortical myelin.
Magnetoencephalography (MEG) characterizes electrical activity in the brain via measurement of extracranial magnetic fields (26). The MEG signal from any one brain region is dominated by neural oscillations (rhythmic changes in electrical activity) that are observable in the 1- to 200-Hz frequency range. Evidence suggests that these oscillations represent an intrinsic process by which both short- and long-range functional connections in the brain are maintained. With this in mind, a growing body of work has begun to show that, via appropriate modeling of MEG data, networks of electrophysiological functional connectivity can be mapped (27–29). The rich temporal complexity of MEG signals means that multiple ways to characterize functional connectivity exist (30). However, one of the most robust methods is amplitude envelope correlation (AEC), which probes temporal relationships between the envelope of oscillations, in a frequency band of interest, at spatially separate brain regions. Our previous work has shown that this intrinsic mechanism of (noninvasively measured) electrophysiological coupling underpins many of the observable resting state networks (RSNs) (27, 31). Given that these measurements represent the principal long-range functional connections in the brain, and given the evidence that electrical activity mediates myelination, we hypothesized that networks of oscillatory envelope correlation, measured between parcellated regions and in multiple frequency bands, would allow prediction of a network of structural covariance representing myeloarchitecture.
Results
Fifty-eight volunteers (39 ± 12 y old, 27 male) took part in the study. Using ultra-high field MRI, we measured magnetization transfer (MT) (32) across the brain, which serves as a marker of myelin. These measurements were parcellated into 64 cortical regions according to the automated anatomical labeling (AAL) atlas (Table S1) and normalized by region volume to give myelin density estimates. Following this, assessment of structural covariance between all possible AAL region pairs allowed derivation of a network matrix showing the degree to which separate (AAL) regions exhibit similarities in their myeloarchitecture (see SI Further Analyses and Fig. S1 for an alternative methodology). The same individuals also underwent resting-state MEG acquisition. MEG data were parcellated according to the same atlas, and RSNs characterizing functional connectivity, between all possible AAL region pairs, were derived using AEC in five frequency bands. With the aid of a recently developed framework (31), we then characterized the relationship between the structural network, representing myelin density, and functional networks, representing the major pathways of electrophysiological communication.
Table S1.
Cortical AAL regions
| Region no. | Description | Region no. | Description |
| 1 | Rectus_L* | 40 | Rectus_R* |
| 2 | Olfactory_L* | 41 | Olfactory_R |
| 3 | Frontal_Sup_Orb_L | 42 | Frontal_Sup_Orb_R |
| 4 | Frontal_Med_Orb_L | 43 | Frontal_Med_Orb_R |
| 5 | Frontal_Mid_Orb_L | 44 | Frontal_Mid_Orb_R |
| 6 | Frontal_Inf_Orb_L | 45 | Frontal_Inf_Orb_R |
| 7 | Frontal_Sup_L | 46 | Frontal_Sup_R |
| 8 | Frontal_Mid_L | 47 | Frontal_Mid_R |
| 9 | Frontal_Inf_Oper_L | 48 | Frontal_Inf_Oper_R |
| 10 | Frontal_Inf_Tri_L | 49 | Frontal_Inf_Tri_R |
| 11 | Frontal_Sup_Medial_L | 50 | Frontal_Sup_Medial_R |
| 12 | Supp_Motor_Area_L | 51 | Supp_Motor_Area_R |
| 13 | Paracentral_Lobule_L | 52 | Paracentral_Lobule_R |
| 14 | Precentral_L | 53 | Precentral_R |
| 15 | Rolandic_Oper_L | 54 | Rolandic_Oper_R |
| 16 | Postcentral_L | 55 | Postcentral_R |
| 17 | Parietal_Sup_L | 56 | Parietal_Sup_R |
| 18 | Parietal_Inf_L | 57 | Parietal_Inf_R |
| 19 | SupraMarginal_L | 58 | SupraMarginal_R |
| 20 | Angular_L | 59 | Angular_R |
| 21 | Precuneus_L | 60 | Precuneus_R |
| 22 | Occipital_Sup_L | 61 | Occipital_Sup_R |
| 23 | Occipital_Mid_L | 62 | Occipital_Mid_R |
| 24 | Occipital_Inf_L* | 63 | Occipital_Inf_R* |
| 25 | Calcarine_L | 64 | Calcarine_R |
| 26 | Cuneus_L | 65 | Cuneus_R |
| 27 | Lingual_L | 66 | Lingual_R |
| 28 | Fusiform_L* | 67 | Fusiform_R* |
| 29 | Heschl_L | 68 | Heschl_R |
| 30 | Temporal_Sup_L | 69 | Temporal_Sup_R |
| 31 | Temporal_Mid_L | 70 | Temporal_Mid_R |
| 32 | Temporal_Inf_L* | 71 | Temporal_Inf_R* |
| 33 | Temporal_Pole_Sup_L | 72 | Temporal_Pole_Sup_R* |
| 34 | Temporal_Pole_Mid_L* | 73 | Temporal_Pole_Mid_R* |
| 35 | ParaHippocampal_L* | 74 | ParaHippocampal_R* |
| 36 | Cingulum_Ant_L | 75 | Cingulum_Ant_R |
| 37 | Cingulum_Mid_L | 76 | Cingulum_Mid_R |
| 38 | Cingulum_Post_L | 77 | Cingulum_Post_R |
| 39 | Insula_L | 78 | Insula_R |
Seventy-eight AAL regions initially used for parcellation of MT and MEG data. Following MT preprocessing, regions (marked with an asterisk) were removed from further analysis.
Fig. S1.
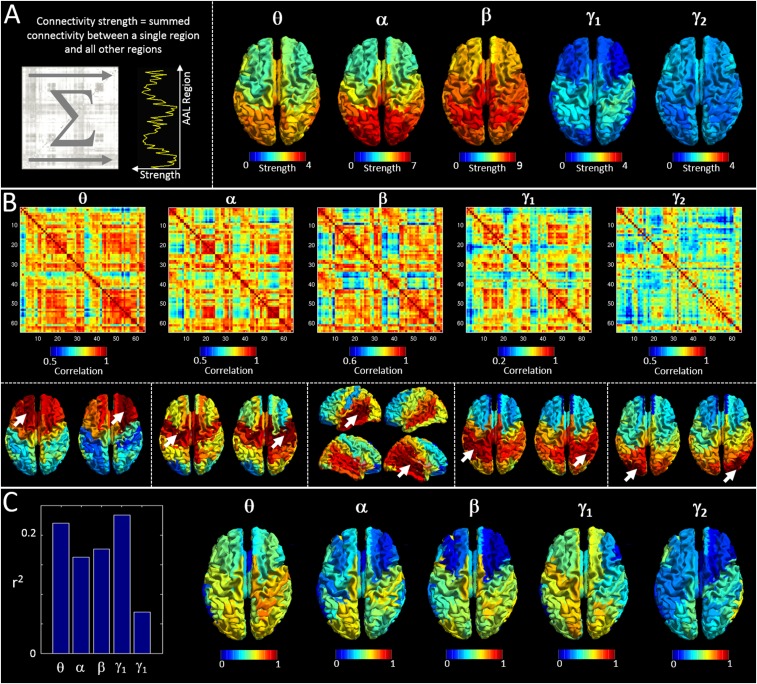
Predicting myelination based on cross-subject covariance. (A) (Left) A schematic representation of a connectivity strength calculation. Connectivity strength is derived as the linear sum of a MEG adjacency matrix in one direction. (Right) Mean (across subjects) connectivity strength, plotted as a function of brain region for all five frequency bands. (B) The matrices show cross-subject correlation of connectivity strength (i.e., high values between two regions, A and B, indicate that subjects with high connectivity strength at region A also indicate high connectivity strength at region B). These matrices thus form a measure of cross-subject covariance in connectivity strength, which we term strength covariance. Note that a dominating feature, in particular in the beta band, is interhemispheric coupling. This is also shown by the inset images in the lower panel. These depict seed-based strength-covariance profiles (single columns in the matrices in the upper panel). Arrows mark the seed AAL region. (C) The link between strength covariance and structural covariance. The bar chart shows correlation between the structural network (Fig. 1D) and the strength-covariance matrices in B. Note correlation is measured over the whole matrix (i.e., for all possible seed regions) and is shown for all frequency bands. The inset images show which seed regions drive the relationship in the bar chart [i.e., red indicates a region whose structural connectivity (MT) profile and strength-covariance profile is highly correlated].
Myelin Maps.
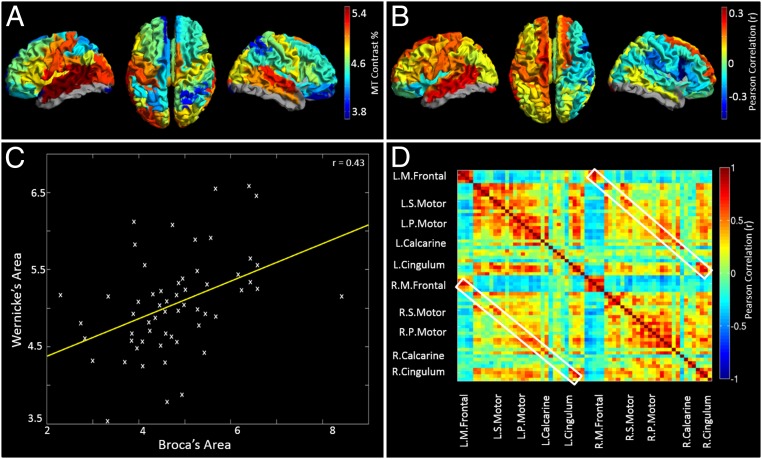
Fig. 1A shows the measured MT, averaged across subjects and plotted for all AAL regions. Red corresponds to high myelin density, blue indicates low myelin density, and gray shows regions where scan coverage was insufficient to gain an accurate estimate. High myelination was observed in primary sensory cortices, with statistical tests showing that this spatial variation was significant; for example, significantly (P < 0.001) higher myelination was found in primary motor cortex compared with the dorsolateral prefrontal cortex. A hemispheric division was also noted with significantly (P < 0.01) higher MT in left compared with right sensorimotor cortex. Fig. 1B maps the cortical variation of correlation between myelin density and handedness (the latter measured using the Edinburgh Handedness Inventory). A positive correlation denotes regions where right handers have more myelin than left handers; a negative correlation denotes regions where left handers have more myelin than right-handed individuals. Note a significant (P < 0.05) split in the polarity of the correlation between hemispheres.
Fig. 1.
In vivo myelination measures and the structural network. (A) Mean MT contrast percentage for all AAL regions. High MT is reflective of high myelination. Note high levels of myelination in primary cortical regions. (B) Correlation between MT and handedness. Positive correlations show regions where myelin is higher in right handers. Negative correlations show regions where myelin is higher in left handers. (C) Example plot showing correlation over subjects between MT measured in the AAL regions capturing Broca’s and Wernicke’s areas. These correlations are the basis of the structural network. (D) The myelin structural network, represented as a matrix. Each element denotes cross-subject correlation in MT between two brain regions.
MT Structural Covariation.
The principle of structural covariance and the structural network are shown in Fig. 1 C and D, respectively. Fig. 1C shows correlation across subjects between myelin concentrations measured in left frontal inferior operculum (capturing Broca’s area) and left supramarginal gyrus (encompassing Wernicke’s area). Results show a significant (P < 0.001) positive correlation across subjects. Equivalent correlations can be derived between all possible AAL region pairs, and results are shown in Fig. 1D. Note that structural covariance between adjacent AAL regions is generally denoted by matrix elements close to the leading diagonal, whereas structural covariance between distal regions is represented far from the diagonal. The white boxes show structural covariance between homologous regions.
MEG Networks.
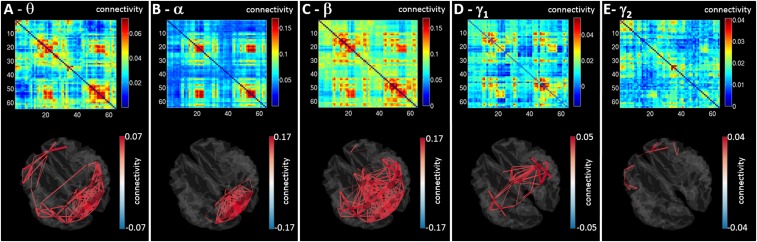
MEG functional connectivity matrices are shown in Fig. 2. All networks are represented by a matrix similar to that in Fig. 1D. The 3D brains display all connections within 5% of the maximum value. It is clear that network structure differs between bands: Theta oscillations support connections in frontal occipital and parietal areas whereas alpha band-mediated connections are predominantly occipital. The beta band shows more widespread connectivity but is dominated by parietal and occipital connections. The low-gamma band was dominated by a sensorimotor network.
Fig. 2.
MEG functional connectivity matrices. Matrices represent AEC in the (A) theta (4–8 Hz), (B) alpha (8–13 Hz), (C) beta (13–30 Hz), (D) low–gamma (30–70 Hz), and (E) high-gamma (70–120 Hz) bands. All matrices show Pearson correlation between AAL region pairs. The 3D plots shown depict all connections within 5% of the maximum value in each band.
The Relationship Between Structural Covariance and MEG.
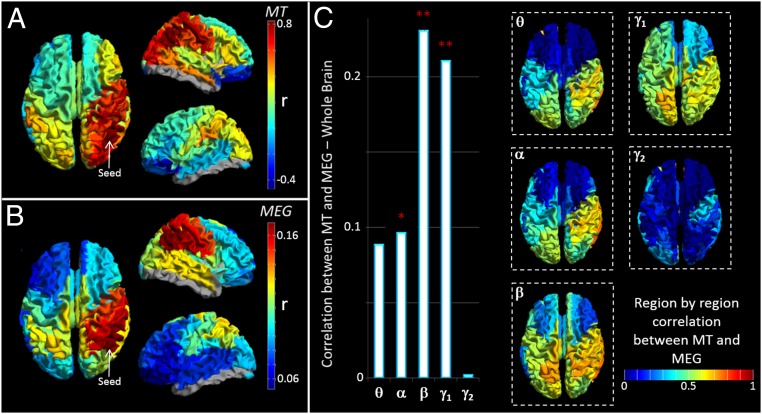
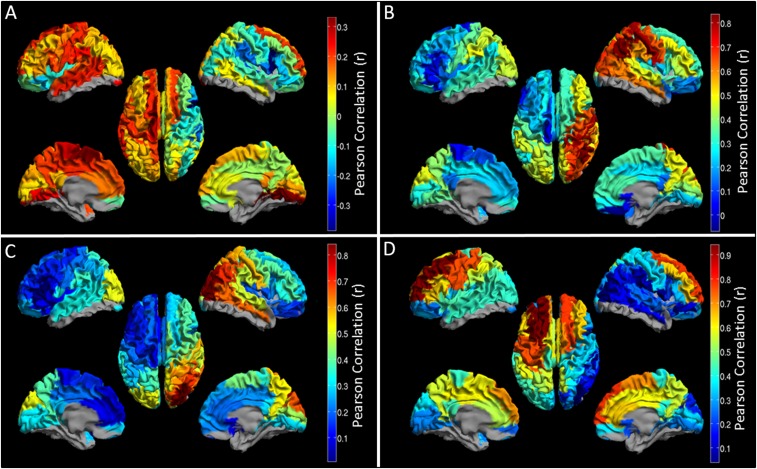
Fig. 3A shows a “seed-based” structural covariance map (i.e., a single column in the matrix in Fig. 1D). Here, a seed region has been placed in right inferior parietal cortex; high values depict brain areas that show high structural covariance to the seed. A cross-hemispheric pattern is observable with high structural covariance between homologous regions. Fig. 3B shows the equivalent seed-based map calculated using beta band MEG data. Note the strong similarity between the functional (Fig. 3B) and the structural (Fig. 3A) networks. To generalize this relationship for all possible seed regions, we tested for correlation between the full MT matrix (representing all seeds; Fig. 1D) and the group-averaged functional connectivity matrices for all bands (Fig. 2). The resulting r2 values (bar chart in Fig. 3C) show that functional networks measured in the beta and low-gamma bands predict significantly the spatial pattern of structural covariance. Neither theta nor high-gamma bands showed a measurable relationship; the alpha band showed a trend. The inset images show seed regions for which structural covariance is best predicted by functional networks. Note that the structure/function relationship is strongest in parietal and occipital areas and weakest in the frontal lobes. It is noteworthy that the MEG-derived functional networks, particularly in the beta band, are driven in part by canonical RSNs (27), and by extension this suggests a significant relationship might also be found between the spatial signature of fMRI-derived RSNs and the structural network. This is indeed the case, and this significant relationship is shown in SI Further Analyses, Functional Connectivity and Myeloarchitecture in RSNs).
Fig. 3.
The relationship between MEG networks and myelination. (A) Structural covariance between a seed region in right inferior parietal cortex and all other brain regions. (B) Seed-based functional connectivity, calculated using MEG in the beta band between the same seed region in inferior parietal cortex and all other regions. Note the similarity between A and B. (C) The bar chart shows correlation between the structural network (Fig. 1D) and the functional networks (Fig. 2). Correlation is measured over the whole matrix (i.e., for all possible seed regions) and is shown for all frequency bands. ** indicates a significant relationship; * indicates a trend. The inset images show which seed regions drive the relationship in the bar chart [i.e., red indicates a region whose structural connectivity (MT) profile and functional connectivity profile are highly correlated].
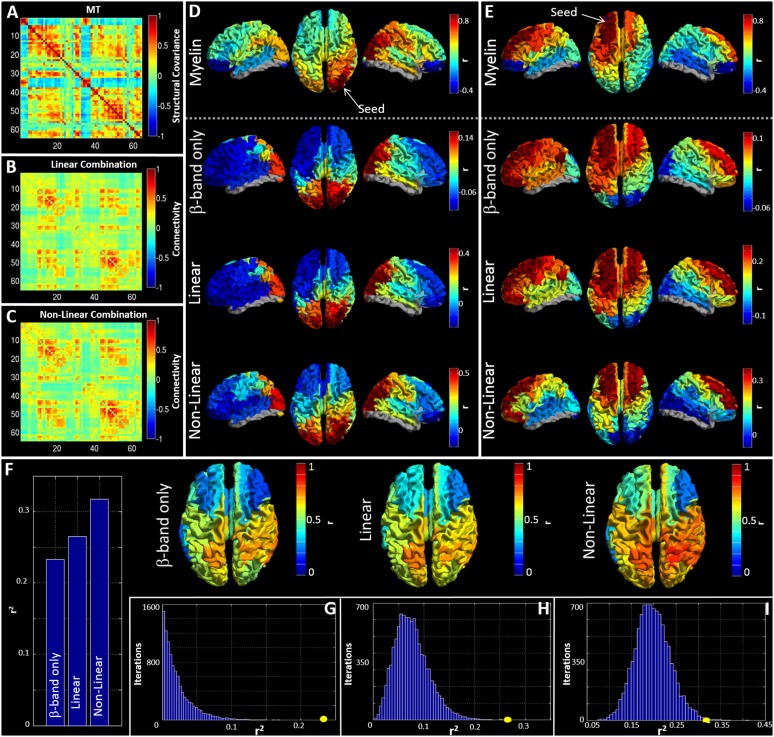
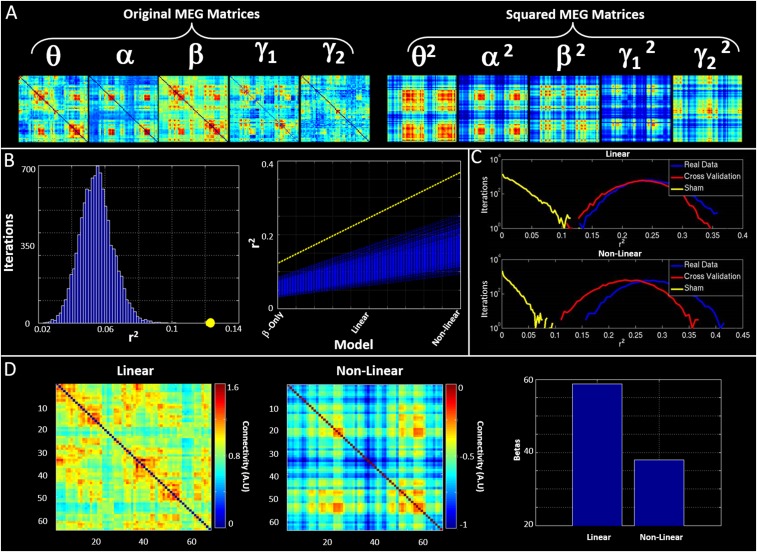
Although functional networks in individual frequency bands show significant correlation, it is likely that the structural network exists to support functional connectivity in all bands. For this reason, we sought to integrate the five MEG networks to test whether such combination could better predict structure than independent frequency bands. Two approaches were used. First, all five MEG matrices were combined in a linear weighted sum. Second, these same matrices were supplemented by nonlinear terms, formed based upon the square of each MEG matrix, and again a weighted sum derived. Importantly, the nonlinear terms have specific meaning: For any squared matrix, a single element, say [1, 2], represents the inner product of the connectivity profile of region 1 and region 2. Because this product is related to covariation, the matrix element will be high if the connectivity profile of region 1 to the rest of the brain overlaps with the equivalent connectivity profile of region 2. In this way the squared terms can be thought of as representing brain regions that share connections to similar areas. Fig. 4 A–C show connectivity matrices representing the structural network (Fig. 4A) and its prediction based upon linear (Fig. 4B) and nonlinear (Fig. 4C) combinations of MEG networks. These relationships, along with that for the best single frequency band, are further visualized in Fig. 4 D and E, which show “seed-based” structural covariance (top row) alongside equivalent maps made using the beta band (upper middle), the best linear combination (lower middle), and the best nonlinear combination (bottom). Seeds were placed in right lateral visual cortex and left superior frontal cortex in Fig. 4 D and E, respectively.
Fig. 4.
Predicting myelination based on integrated MEG networks. (A–C) Matrices representing (A) the MT network, (B) the best linear combination of MEG frequency bands to estimate MT, and (C) the best nonlinear prediction of MT. (D and E) Seed-based visualizations of structural and functional networks with seed regions in right lateral visual cortex (D) and left superior frontal cortex (E). The lower three rows show beta band and the linear and nonlinear predictions of the MT network (Top). (F) r2 values describing goodness-of-fit between structure and function for the best-fitting single frequency band (beta) and predictions based upon linear and nonlinear combinations of MEG frequency bands. The inset images show the seed regions driving these correlations. (G–I) The r2 values (circles) plotted against the null distributions for the beta band (G) linear (H) and nonlinear (I) combinations.
The relationship between structure and integrated functional connectivity is formalized in Fig. 4F, which shows r2 values representing correlation between the MT network and functional networks representing beta band only and linear and nonlinear predictions; the inset images show seed regions for which structural covariance is best predicted by the functional networks. Fig. 4 G–I show the r2 values plotted alongside their representative null distributions. In all cases, correlation between the structural network and MEG networks falls outside the null distribution. Note also that this relationship gets stronger when allowing integration over frequency bands.
Discussion
Although recent years have seen significant progress in mapping the human connectome, the relationship between functional networks and cortical microstructure remains poorly understood. A recent study (24) in animals suggests that electrical activity promotes myelination. We therefore reasoned that, if functional networks represent major pathways of electrophysiological communication, then those pathways should shape myeloarchitecture and a significant relationship between functional connectivity and myelin should be observable. Our results support this, with significant correlation between the structural network and functional networks mediated by neural oscillations in the beta and low-gamma bands. This relationship became stronger when integrating MEG networks across frequency bands, suggesting that myeloarchitecture supports networks at all measurable electrophysiological timescales.
The fact that our functional networks are measured directly using electrophysiological imaging (as distinct from indirectly using hemodynamics) adds an extra (frequency) dimension to our study. In the past, neural oscillations were largely ignored in favor of measurements of evoked responses. However, the last decade has seen a surge of interest, showing these oscillations to be an integral feature of brain function. Recent work suggests that oscillations gate information flow in the cortex (33) and implies that oscillations are an intrinsic form of functional coupling (34). Measurable connectivity depends critically on the frequency band studied; indeed, this is shown in Fig. 2 with marked spatial differences between bands. The fact that the relationship between functional networks and myelin was strongest in the beta band is not surprising given that previous work (27, 31) has shown that beta oscillations mediate long-range connections in a large number of RSNs (see also SI Further Analyses, Functional Connectivity and Myeloarchitecture in RSNs and Fig. S2). The ubiquitous nature of beta-mediated connections is therefore the likely reason why the structural network correlates best with this band. However, it should be pointed out that an inherent problem with MEG is signal-to-noise ratio (SNR), which drops with increasing frequency; it is possible that, at high frequency, the drop in correlation between functional connectivity and myeloarchitecture reflects this SNR limitation. This said, the significant improvement in prediction when combining frequency bands does imply that oscillations across all timescales relate significantly to myelin structure. Interestingly, the addition of nonlinear terms also improved prediction; given that these additional terms represent brain regions that share common connections it is tempting to suggest that shared connectivity also affects myeloarchitecture.
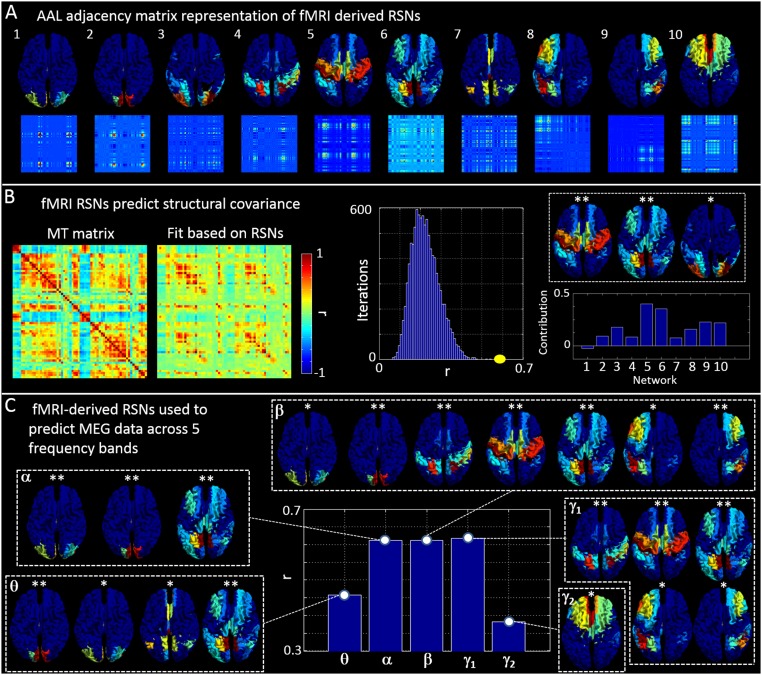
Fig. S2.
Using fMRI-derived RSNs to predict structural covariance and MEG. (A) Vector (Upper) and adjacency matrix (Lower) representations of 10 commonly observed RSNs. (B) (Left) The structural covariance (MT) matrix and a fit to that matrix based on the 10 RSNs in A. (Center) The measured correlation between the MT matrix and the fit (yellow) alongside an empirical null distribution. (Right) The fitted parameters, (bar chart) and the networks with the highest contribution. (C) (Center) Correlation between the five MEG-derived adjacency matrices (Fig. 2) and their best fit based on the 10 fMRI-derived RSNs in A. The inset images show which RSNs contribute maximally to each band.
Our estimation of myelin was based upon measurement of MT. Although serving as an efficient marker of myelination, it is important to note that there is no one-to-one relation between MT and myelin density. Further, if the present method was to be used in pathology care should be taken because the MT/myelin relationship may break down. Nevertheless, our results are in agreement with others, showing that the highest myelin concentration occurs in sensorimotor, auditory, and visual cortices. This finding supports an argument that myelin acts as a means to increase the speed of processing and inhibit plasticity in these primary sensory areas. On average, there was more myelin in left sensorimotor cortex than in right, and this likely reflects the fact that our cohort was biased toward right handers. In agreement with this, the hemispheric split shown in Fig. 1B suggests that this finding is reversed in left handers, implying that brain structure evolves to increase the speed of processing in the dominant hemisphere. Although such asymmetries in myelin have not been previously reported, associated asymmetries in function and structure have been shown previously both in humans (35–37) and animals (38). A potential limitation of our methods is that myelin concentration was parcellated according to the AAL atlas. Although this parcellation allowed derivation of network graphs that could be compared with MEG, the regional parcellation afforded by the AAL atlas does not take into account known differences in myelin signature throughout the cortex. In future studies, a brain parcellation based upon myeloarchitecture (as distinct from cytoarchitecture) would be of high value (12). Although our study is unique in characterizing structural covariance based upon myelin density, previous work has shown that structural covariance networks can also be formed based on macroscopic properties such as cortical thickness (4), and further that such networks correlate with functional connectivity (39). Here, myelin content was normalized by region volume, meaning that our finding of correlation between a structural network and functional connectivity is not related trivially to the previous finding on cortical thickness. However, the question of how myelin density relates to cortical thickness remains, and this should be the topic of future work.
Given that the primary role of myelin is to increase the speed at which nerve impulses travel through neuronal pathways, it is intuitive that cortical myelination is shaped to support functional networks because this will act to maximize the efficiency of their formation. More speculative, however, is the process by which this structural support network evolves. Given that electrical activity on some particular pathway induces myelination, and given that RSNs emerge as early as the third trimester of gestation (40), it is tempting to argue that even before birth myeloarchitecture is being shaped by functional connectivity. Of course, the resulting structural changes would, in turn, refine functional connections and so the likelihood is that changes in myeloarchitecture and functional networks are linked intimately. This notion is supported by the fact that both myelination and functional connectivity change on a similar timescale throughout neurodevelopment. In the present study, our data preclude direct investigation of this interplay between structure and function. However, our study does pave the way for new investigations of this process via longitudinal studies of development. In addition, studies investigating how changing behavior alters both structure and function are becoming popular. This idea is not new (41), and recent evidence (42) suggests that both white-matter and gray-matter structure changes, even on a relatively fast timescale, when learning new skills. This research area would benefit from the use of tools presented here. Perhaps more importantly, our results have implications for future studies of disorders, in particular those involving demyelination or dysconnectivity. For example, gray-matter atrophy, lesions, and demyelination are a better correlate of physical disability and cognitive decline in multiple sclerosis than white-matter lesion load (43). In addition, inefficiency of functional networks has also been reported (39) in this disease. Our method might offer a means to link these findings. Similarly, severe psychosis has been linked with dysconnectivity (44), and recent work has begun to relate this to structural deficiencies (45). Once again, our work might offer a framework to link these abnormalities.
Conclusion
We have probed the relationship between gray-matter myelination and electrophysiological networks, showing a significant correlation. This relationship is strongest for networks mediated by beta oscillations but becomes stronger when integrating across frequency bands, suggesting that myeloarchitecture supports connectivity across all bands. Our study sheds light on the way in which cortical microstructure supports functional networks, the latter being mediated by neural oscillations. Further, it paves the way for future investigations of the structure/function relationship and its breakdown in pathology.
Methods
Myelination and Structural Covariance.
Participants gave written informed consent and ethical approval was granted by the University of Nottingham Medical School Research Ethics Committee. MRI data were collected using a Phillips Achieva 7T system. A phase-sensitive inversion recovery T1 weighted image was acquired and used for MEG coregistration. MT data were obtained from z-spectra acquired using an MT-TFE sequence (46). MT imaging provides contrast based on the exchange of magnetization between free water and protons bound in macromolecules. Although it is likely that no one-to-one relationship exists, experimental and human studies have shown that MT is highly correlated with myelination, which is probably related to the high fraction of water in close proximity to myelin macromolecules. The procedure for extracting MT data from our imaging sequence has been described elsewhere (47). Briefly, z-spectra were corrected for B0 variation and fitted to a database of simulated spectra to extract myelination maps. To investigate structural covariation of myelin within the AAL regions, gray-matter-masked MT data were registered to the AAL atlas and a mean MT value was calculated for each region, for each participant. Due to confounding factors such as scanner drift, the modal value for each individual’s MT data was regressed from that individual’s regional values (7) (Fig. S3). Pearson correlation, measured across subjects, was used to quantify the relationship between MT values measured in AAL region pairs (Fig. 1C). These correlation values form elements of the structural matrix in Fig. 1D. An average MT map (Fig. 1A) was also generated by averaging regional MT values over participants. The relationship between myelination and handedness was probed via correlation between MT and handedness score.
Fig. S3.
Uncorrected MT maps. (A) Correlation between MT and handedness. Positive correlations show regions where myelin is higher in right handers. Negative correlations show regions where myelin is higher in left handers. (B–D) Structural covariance between a seed region and all other brain regions. (B) Parietal seed. (C) Occipital seed. (D) Frontal seed.
Resting-State MEG: Connectivity Analysis.
Three hundred seconds of eyes-open resting-state MEG data were acquired using a 275-channel CTF MEG system operating in third-order synthetic gradiometer configuration (sampling frequency of 1,200 Hz). Three head position indicator coils were placed at fiducial locations on the subject’s head and energized to facilitate continuous tracking of head location. Before acquisition, a 3D head digitization procedure was completed. Coregistration between MEG system geometry and individual brain anatomy was achieved by matching the digitized head surface to the equivalent surface extracted from an anatomical MRI. Functional connectivity was calculated between AAL regions. A scalar beamformer was used to obtain a single MEG signal representative of each region. These regional signals were frequency-filtered to the bands of interest and the confound of signal leakage mitigated using pairwise orthogonalization (48). The magnitude of the analytic signal, computed via a Hilbert transform, was used to generate the amplitude envelope of oscillations, for each regional time course in all frequency bands. Pearson correlation was then computed between envelopes for each region pair. In this way we generated a single adjacency matrix for each subject, and frequency band, representing whole-brain connectivity. These matrices were averaged over subjects.
The Relationship Between MT and MEG.
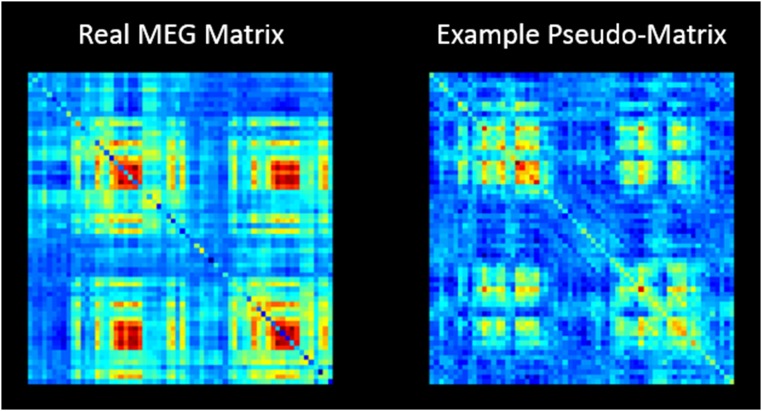
We measured correlation (r2) between the structural covariance matrix (Fig. 1D) and the group-averaged MEG networks (Fig. 2). To test statistically whether these correlation values were significant, we used a permutation test. A set of “pseudo-MEG matrices” were generated, each having spatial properties similar to the real matrices, but crucially they were not based on genuine data (SI Methods and Figs. S4 and S5). This approach accounts for the inherent spatial smoothness in the MEG-derived networks (SI Methods). Correlation between the structural matrix and 10,000 pseudomatrices yielded an empirical null distribution, and comparison of the genuine r2 value with the null distribution allowed computation of a P value.
Fig. S4.
A real MEG matrix (Left) and a single example of a pseudomatrix (Right).
Fig. S5.
Linear and nonlinear fits of MEG to MT data. (A) The five MEG-derived adjacency matrices (Left) and their squares (Right). (B) Results of our rate-of-improvement analysis. (Right) The yellow dashed line represents the improvement in fit with model complexity in real data. The blue lines represent the equivalent analysis applied to pseudomatrices. (Left) The gradient of the dashed line (in yellow) and the equivalent gradients of all of the blue lines in the null distribution (blue). Note that the improvement afforded by increasing model complexity in real data is greater than what would be expected by chance. (C) Results of our cross-validation procedure. (Upper) The case for the linear fit. (Lower) The case for the nonlinear fit. The blue curve shows the estimation of MT data from group A, using linear and nonlinear combinations of MEG matrices, with parameters derived from group A. The red curve shows the estimation of MT data from group B, using linear and nonlinear combinations of MEG matrices, but with parameters derived independently from group A. Yellow curves show the null distributions. (D) The contributions of the original and the squared terms to the nonlinear fit.
Our supplementary information is split into three sections. In SI Further Analyses we describe additional analyses that, due to space constraints, were excluded from the main text. SI Results contains further visualization of primary results. SI Methods describes, in greater detail, our methodology.
SI Further Analyses
Cross-Subject Strength Covariance and Its Relationship with Myeloarchitecture.
Motivation.
In the main text we tested the hypothesis that a network of structural covariance, measured based upon myelination, could be predicted by functional connectivity measured using MEG. To this end, we used functional connectivity matrices averaged over subjects (MEG) to predict a structural covariance matrix formed via assessment of cross-subject correlation in myelin. Although highly instructive, this requires that a structural covariance matrix, based on intersubject variance, be compared with functional connectivity, based on intrasubject variance. A logical extension to this work is therefore to exploit the intersubject variance in MEG and perform a parallel analysis, to assess whether (i) cross-subject correlation in MEG offers a means to generate structured network graphs and (ii) whether such graphs also relate to myeloarchitecture.
Method.
With this in mind, we first reduced the dimensionality of the MEG connectivity matrices by measuring connectivity strength at each of the 64 AAL regions. Connectivity strength was defined as the summed connectivity between a single seed region, and all of the other 63 regions (i.e., connectivity strength is measured as the sum, in one dimension, of the MEG adjacency matrices shown in Fig. 2 – this is sometimes also termed “degree” and is shown schematically in Fig. S1A, Left). These measures generated 64 values of connectivity strength for each subject. Measurement of correlation in strength across subjects for all region pairs then generated a novel MEG metric, somewhat analogous to structural covariance, which we term “cross-subject strength covariance.” This analysis results in a single 64 × 64 adjacency matrix per frequency band, where a high value of strength covariance between regions A and B denotes that subjects with a high connectivity strength in region A also tend to have a high connectivity strength in region B. Note that although the metric is analogous to structural covariance in terms of the way in which it is computed it is based purely on measurement of brain function.
Results.
Fig. S1A shows connectivity strength, calculated for all brain regions and averaged over subjects, for all five frequency bands. Note the clear spatial patterns that differ between frequency bands, alpha and theta being predominantly occipital, beta more widespread, and low gamma dominated by the sensorimotor regions. Note also the significant variation in strength across bands, peaking in the beta band, as would be expected. Fig. S1B shows matrices representing cross-subject strength covariance. Interestingly, a high degree of structure is observed which, although obviously related to the “classical” functional connectivity matrices in Fig. 2, also differs markedly. Here, a clear and dominating feature, in particular in the beta band, is interhemispheric coupling (i.e., subjects with high connectivity strength in one region tend to exhibit similarly high connectivity strength in homologous regions of the opposite hemisphere). This is shown by the matrices and by the inset images. Images depict seed-based strength-covariance profiles (single columns in the matrices in the upper panel). Arrows mark the seed AAL region. Note that for frontal, parietal, temporal, and occipital seeds the highest strength covariance is in homologous areas of the opposite hemisphere. Fig. S1C shows the relationship between strength covariance and structural covariance based upon myelination. Note that this was measured in the same way as was described in SI Methods below. The bar chart shows correlation (r2) between the structural network (Fig. 1D) and the strength covariance adjacency matrices (Fig. S1B). Statistical tests (based on pseudomatrices) show this correlation to be significant. The inset images show the seed regions that drive the relationship in the bar chart (i.e., red indicates a region whose structural connectivity profile and strength covariance profile is highly correlated).
Discussion.
The principal finding in the main text shows that brain areas that are highly functionally connected also exhibit cross-subject covariation in myeloarchitecture. Given this finding, it is perhaps unsurprising that networks representing cross-subject strength covariance also demonstrate a significant relationship with the myelin network (with similar r2 values). To explain this, consider the simplest possible case where a network graph is dominated by a single connection between regions A and B, and that the magnitude of this connectivity differs over individuals. In such a case, strength measures would be dominated by regions A and B, and, further, those strength measures would obviously covary over subjects because they are generated by the same single connection. It follows that strength covariance matrices should bear significant resemblance to standard functional connectivity matrices, and this is indeed the case (e.g., the four patches representing occipital connectivity that dominate the alpha band matrix in Fig. 2 also dominate the alpha band strength covariance matrix in Fig. S1B). This argument suggests that the significant relationship between strength covariance and structural covariance shown in Fig. S1C is secondary to the principal finding of our paper. What is compelling, however, is that as well as obvious similarities between functional connectivity and strength covariance, clear differences are also apparent (e.g., the interhemispheric connectivity in beta band observed in Fig. S1B is much less pronounced in classical functional connectivity measures). The novelty of the strength covariance measure, coupled with this clear demarcation, suggests that these measurements could provide new insights into brain architecture beyond what the widespread AEC (and related) functional connectivity measurements provide.
Functional Connectivity and Myeloarchitecture in RSNs.
Motivation.
The main text shows that a significant relationship exists between MEG-derived functional connectivity and a structural network based upon myeloarchitecture. However, our previous work (27) has shown that similarly derived MEG functional connectivity metrics, in part, represent the electrophysiological basis of RSNs commonly identified by fMRI. It thus follows that these canonical RSNs (e.g., sensorimotor, frontoparietal, default mode, etc.) should predict our network of structural covariance.
Method.
Ten commonly observed canonical RSNs were selected from a previously published fMRI study (49); these included the sensorimotor network, two visual networks, three frontoparietal networks, the default mode network, a bilateral frontal network, a bilateral parietal network, and an occipitoparietal network. To translate these volumetric weighted RSN maps into AAL space, we first multiplied each network map by a set of binary masks depicting each individual AAL region. We then summed (over voxels) the weightings for each RSN within each AAL region and normalized by the number of voxels in each region. This generated a single 1 × 64 vector, per RSN, showing the extent to which each AAL region is involved in that network. These vectors are visualized in Fig. S2A, Upper. Following this, to generate a matrix description of each RSN, the outer product of each 1 × 64 vector with itself was calculated; this generated 10 weighted adjacency matrices (one for each network), which are shown in Fig. S2A, Lower. Finally, to test our hypothesis that canonical networks would predict structural covariance, we constructed a general linear model:
| [S1] |
Here S represents the vectorized structural covariance network (Fig. 1D), G comprised 10 columns representing the vectorized RSN matrices, parameters in determined the contribution of each RSN to the structural network S, and e represents error in the fit. This approach allows a means to explain the structural network as a linear weighted sum of fMRI-derived RSNs. To quantify the success of the fit, Pearson correlation between the model, , and the data, S, was calculated.
To test this GLM fit statistically, a permutation approach was used based on pseudomatrices. Ten thousand iterations of the algorithm were run and, on each iteration, the design matrix, G, was used to predict a new pseudomatrix. Each pseudomatrix was constructed based upon the structural covariance matrix. Again, Pearson correlation between the model and the pseudomatrix was calculated, and these values were used to generate an empirical null distribution. Comparison of the real correlation with the null distribution allowed derivation of a P value to determine the significance of the relationship between fMRI RSNs and structural covariance. In addition to Pearson correlation, for each iteration of the null distribution we measured the magnitude of the fitted parameters, . This generated 10 further statistical null distributions showing chance level contributions of each of the 10 RSNs to the pseudomatrices. These null distributions were compared with the real derived values to determine which RSNs contributed significantly to the fit. Significance was determined at an alpha value of 0.05, but false discovery rate (FDR) correction was applied using the Benjamini–Hochberg procedure to account for multiple comparisons across each of the 10 networks.
Finally, for completeness, this same procedure was used to model each of the five MEG adjacency matrices using the RSNs; this provided a means to replicate previous findings in interrogating which fMRI-derived RSNs are represented by MEG functional connectivity measurements.
Results and discussion.
Fig. S2B shows the results of predicting structural covariance based on RSNs. The left-hand side of the plot shows the structural covariance matrix (left-hand matrix) and the fit based upon a linear combination of RSNs (right-hand matrix). Note the visual similarity. The center panel shows the value of Pearson correlation between the model and the data (in yellow) and the associated empirical null distribution (in blue). Note that the real value of correlation falls outside the null distribution, indicating the significance of the fit and confirming our hypothesis above. The bar chart in the lower right-hand panel shows the parameters, , that represent the contribution of each of the 10 RSNs in Fig. S2A to the structural network; note that all RSNs contribute, with the largest contribution coming from the sensorimotor and frontoparietal networks. Our statistical test also conformed this; the network maps, inset in Fig. S2B, show the three networks that contributed most. ** indicates a significant contribution following correction for multiple comparisons; * indicates a trend (i.e., a P value less than 0.05 but that did not survive FDR correction). The fact that the largest contributions come from networks predominantly centered around the parietal and occipital regions (i.e., visual, sensorimotor, and frontoparietal networks) is in good agreement with results in the main text, which show the strongest relationship between functional connectivity and myeloarchitecture exists in the occipitoparietal regions.
Finally, Fig. S2C shows the equivalent analyses applied to each of the five MEG adjacency matrices (i.e., we test whether the MEG connectivity data can be predicted using RSN network maps). Results show a significant relationship for all five frequency bands. The strongest correlations were observed in the alpha, beta, and low-gamma bands (bar chart in Fig. S2C), and this is in good agreement with previous work (27). The inset images show networks that contribute maximally to each band. As would be expected, most networks are represented in the beta band, including visual, sensorimotor, and frontoparietal.
SI Results
The Effect of Common Mode Rejection.
To protect against confounding factors, such as scanner drift, the common mode was regressed from MT data (7). However, Fig. S3 shows the MT maps from the main text, made using uncorrected data. Fig. S3A shows correlation between MT and handedness. Positive correlations show regions where myelin is higher in right handers. Negative correlations show regions where myelin is higher in left handers. Fig. S3 B–D show seed-based structural covariance maps with seed locations in parietal, occipital, and frontal cortices, respectively.
Linear and Nonlinear Prediction of MT.
Fig. S5 shows validation of our linear and nonlinear fits to the structural covariance matrix. Fig. S5A shows the five MEG matrices (from Fig. 2, Fig. S5A, Left) and their squares (Fig. S5A, Right). Note that for our linear fit only the five original matrices (Fig. S5A, Left) are combined, whereas the nonlinear combination incorporates a sum of all 10 matrices.
Fig. S5B shows our rate-of-improvement analysis. In Fig. S5B, Right, the yellow dashed line represents the improvement in r2 afforded by moving from the beta band only to first linear and then nonlinear fits, in real data. The blue lines show equivalent gradients for the null distribution, which was based upon pseudomatrices. In Fig. S5B, Left, these data are further visualized with the gradient for real data (yellow) plotted against the gradients for pseudodata (blue). Note that the null distribution shows positive gradients, meaning that the introduction of more terms in the model allows a better fit to the data, even in the absence of any genuine effect. However, the gradient for real data falls outside this empirical null, showing clearly that the improvements observed are in excess of what would be expected statistically. Fig. S5C shows the results of our cross-validation procedure for the linear combination (Fig. S5C, Upper) and nonlinear combination (Fig. S5C, Lower). Subjects were split into two groups, A and B; in both cases, the blue curve shows the r2 values for the estimation of MT data from group A, using linear and nonlinear combinations of MEG matrices, with parameters also derived from group A. Conversely, the red curve plots the distribution of the cross-validation r2 values; in other words, it shows the estimation of MT data from group B, using linear and nonlinear combinations of MEG matrices from group B, but with parameters derived independently from group A. The yellow curves show the null distributions (sham r2 values). Note that, as would be expected, a slight drop in r2 is noted when using independent parameters from a different subject group. However, importantly, this drop is slight and the median correlation falls outside the null distribution in both cases. This analysis shows clearly the robustness of the fits. Finally, Fig. S5D shows the relative contributions of the original and the squared terms to the nonlinear fit. The matrix on the left shows the five linear terms combined and the matrix on the right shows the five nonlinear terms combined. The sum of these two matrices generates the result in Fig. 4C. The bar chart shows the matrix norms corresponding to the linear and squared terms. Note that the nonlinear contribution is smaller.
AAL Regions.
Table S1 lists the 78 AAL regions used for analysis in the main text. Following MT preprocessing, 14 regions (marked with an asterisk) were removed from further analysis.
SI Methods
Participants.
Seventy-seven healthy participants were recruited to the study. Nineteen participants were excluded because of either unsuitable MEG or MT data. Following preprocessing, a final cohort of 58 participants were included [mean age of 39 ± 12 y (mean ± SD), 27 male]. Participants completed an online screening form to assess health and lifestyle; this included the Edinburgh Handedness Inventory (50). MRI and MEG data were acquired in all subjects, in two scanning sessions. To avoid the effects of tissue magnetization, MEG data were acquired first.
Resting-State MEG Data Acquisition.
MEG data were acquired using a 275-channel CTF MEG system (MISL) operating in third-order synthetic gradiometer configuration, at a sample frequency of 1,200 Hz. The resting-state paradigm comprised a 5-min recording during which participants fixated on a small centrally positioned red circle presented on a gray background that was back-projected onto a screen placed ∼40 cm from the participant’s face. Subjects were seated in the MEG system. Three head position indicator coils were placed at fiducial locations (nasion, left, and right preauricular points) on the subject’s head. These coils were energized periodically to facilitate continuous tracking of the head location throughout data acquisition. Before MEG acquisition, a 3D head digitization procedure was completed for each individual (Polhemus Inc.), characterizing head shape relative to the fiducial markers. Coregistration between MEG system geometry and individual brain anatomy images was achieved by matching the digitized head surface to the equivalent surface extracted from the participant’s individual anatomical MRI.
Resting-State MEG Connectivity Analysis.
MEG data were down-sampled to 600 Hz and divided into 30 10-s epochs. All epochs were inspected visually and those containing excessive interference or head movement (>5 mm from starting position) were discarded.
Functional connectivity was calculated between 78 discrete cortical regions, defined based on the AAL atlas (51). To assess the time evolution of electrical activity within each region a scalar beamformer was used (52). Each AAL region was split into 4-mm cubic voxels and the beamformer-estimated time course of electrical activity was derived for each voxel. For beamforming, we calculated the data covariance matrix within a 1- to 150-Hz frequency window and a time window spanning the whole experiment (53). Regularization was applied using the Tikhonov method with a regularization parameter equal to 5% of the maximum eigenvalue of the unregularized matrix. The forward model was based upon a dipole approximation (54) and a multiple local sphere head model (55). Dipole orientation was determined using a nonlinear search for optimum SNR. Beamformer time courses for all voxels in any one region were sign flipped where necessary to account for the arbitrary polarity introduced by source orientation estimation. Following this, time courses were combined in a weighted average, where the weighting was defined by a 3D Gaussian function (full width at half maximum of 17 mm) centered on the region’s center of mass (i.e., those voxels further from the center of the AAL region were down-weighted) (56). This procedure allowed a single beamformer-derived electrophysiological time course for each AAL region.
Regional signals were filtered into the theta (4–8 Hz), alpha (8–13 Hz), beta (13–30 Hz), low-gamma (30–70 Hz), and high-gamma (70–100 Hz) frequency bands. Within each band, the confound of signal leakage was mitigated using a pairwise orthogonalization scheme described previously (48, 57). Following leakage correction, a Hilbert transform was used to derive the analytic signal, and the absolute value of the analytic signal was computed to give the amplitude envelope of oscillations, for each time course, in all five frequency bands. Pearson correlation was then computed between the envelopes for each region pair; this was done separately for each 10-s epoch and results averaged over epochs within each participant. In this way we generated a single 78 × 78 adjacency matrix for each subject, and each frequency band, representative of connectivity between all pairs of AAL regions. These connectivity matrices were averaged over subjects within each band to generate five group level connectivity matrices, representing frequency specific amplitude envelope connectivity across the cortex. It should be pointed out that this AEC method has been used extensively in previous work (for reviews see refs. 29 and 58) and represents an intrinsic mode of electrophysiological coupling (34).
MRI Data Acquisition.
MRI data were collected using a Phillips Achieva 7T system, equipped with a whole head volume transmit coil and a 32-channel receive head coil. A high-resolution anatomical phase-sensitive inversion recovery [PSIR; field of view (FOV) = 240 × 216 × 160 mm3, 0.8-mm isotropic resolution] image was acquired and used for MEG coregistration (discussed above). MT data were obtained from z-spectra acquired using an MT-TFE sequence (46). Each point on the z-spectrum was acquired using a saturation-prepared 3D TFE sequence. The saturation consisted of a train of n = 20 Gaussian windowed sinc rf pulses of bandwidth BW = 200 Hz, 30 ms long, repeated every T = 60 ms (50% duty cycle), with a phase increment between each pulse, and a spoiler gradient applied at the end of the train to remove any residual transverse magnetization. Z-spectra were acquired by varying the off-resonance saturation frequency from −5 kHz to 5 kHz, including a scan at 50 kHz off-resonance for normalization. This was repeated for the three nominal B1sat values of 0.33, 0.65, and 1.09 μT (in B1rms). The imaging readout was a volume acquisition with a readout train of 410 gradient echoes, TE/TR/FA = 2.7 ms/5.8 ms/8°, FOV = 192 × 192 × 60 mm3, 1.5-mm isotropic image resolution, low-high k-space acquisition, and a SENSE factor (RL) of 2. The 3D volume acquisition required five repetitions of this cycle. Using this 3D nonsteady-state approach, a 15-point z-spectrum (plus an additional point for normalization) was acquired in 8 min (24 min total for the three powers). The amplitude of readout pulses was modulated to avoid large variations in signal at the start of the TFE train; there were 2 ramped rf pulses before acquisition, followed by 4 ramped rf pulses at the start of the acquisition, with the remaining pulses at constant flip angle alpha = 8° (B1read). Additionally, a B0 map (double echo method, FOV = 252 × 255 × 100 mm3 at 3 × 3 × 2 mm3 voxels) was acquired for B0 correction (1 min), along with a whole-head B1 map (dual TR method, at 20 ms and 120 ms TR, FOV 205 × 180 × 132 mm3 at 3.2 × 4 × 4 mm3 voxels, 3 min) and a T1 map [dual readout PSIR data, FOV = 240 × 216 × 160 mm3 at 0.8-mm isotropic resolution, SENSE factor (RL) of 2.2 and (FH) of 2, 10 min], giving a total scan time of 38 min.
MRI Data Analysis and MT Mapping.
The procedure for extracting high-fidelity MT data from the CEST imaging sequence has been described in detail in a recent paper (47). All data were coregistered onto the MT space using a high contrast-to-noise ratio (hCNR) image created by averaging across z-spectra dynamics acquired at the highest B1 amplitude. Each z-spectra dataset was motion corrected [using MCFLIRT in FSL (59)] and registered onto the hCNR image. Similarly, B0, B1, and PSIR scans were registered onto the hCNR image. All z-spectra were then corrected for B0 fluctuations (commonly observed at ultra-high field strength). The B0 corrected z-spectra were then fitted to a database of simulated spectra to extract myelination (MT) maps using a four-pool model with 15 off-resonance values sampled (47). This method includes B1 and T1 correction, because it is important to obtain unbiased MT maps as described in ref. 47.
To investigate covariation of myelin within the same 78 cortical AAL regions used for MEG, MT data were first segmented to separate gray and white matter. Gray-matter masks were generated from a high-resolution anatomical image (PSIR) using SPM. These were then thresholded at a high probability value. This, in effect, meant a highly conservative gray-matter mask that minimizes any partial volume effects due to the relatively low resolution of the MT data. Gray-matter-masked MT data were then registered to the AAL atlas. A mean MT measure was calculated for each region, for each participant, creating 78 values of MT per person. Due to limited FOV and intersubject variability, 14 AAL regions were either missed or poorly characterized by the MT acquisition and so were removed from any subsequent analyses. The remaining 64 regions were averaged across subjects to give a single map, showing average MT in each region. Due to potentially confounding factors in the MRI such as scanner drift, the common mode for each individual’s MT data was calculated and regressed from that individual’s regional MT values (7). Following this, regional MT values were correlated across subjects to generate a structural matrix. In everything that follows we assume that these MT maps, and the associated structural network, are representative of cortical myeloarchitecture.
To confirm that our myelin maps were related to human physiology we assessed the relationship of myelin with handedness to test a hypothesis that right handers would exhibit greater myelin content in the left hemisphere, whereas left handers would exhibit greater myelin content in their right hemisphere. Given the right-handed bias in our cohort of 58 participants (24 reported being strongly right handed), we first tested statistically for a difference in myelination, across the subject group, between the left and right motor cortices, using a paired t test. Following this, we assessed correlation across subjects between MT and handedness, the latter being assessed by the Edinburgh handedness inventory. The questionnaire was scored such that strongly right-handed individuals have a score of +12, strongly left-handed individuals have a score of −12, and ambidextrous individuals fall between these limits. Thus, for any one brain region, a positive gradient between MT and handedness would indicate more myelin for right handers. Likewise, a negative gradient would indicate more myelin for left handers. We hypothesized that a negative gradient would dominate the right hemisphere whereas a positive gradient would dominate the left hemisphere. To test this statistically, first the difference in mean (across regions) gradient for each hemisphere was measured. This value was then tested against an empirical null distribution. To generate the null, the gradient difference was computed in the same way; however, the gradients for each region were defined following a randomization of the order of the handedness scores across subjects. In other words, we reasoned that, if there was no relationship between handedness and myelination, the handedness scores could be switched randomly between subjects with no effect on the slope difference detected. Hence, the real gradient difference would fall within the null distribution.
The Relationship Between MT and MEG in Individual Frequency Bands.
To assess a relationship between the myelin structural network and the MEG functional networks we probed correlation between network matrices. All matrices were first vectorized and redundancy was removed, so that the leading diagonal (which represents only within-region correlation) did not contribute to the measurement. Following this, linear correlation was measured between the structural network matrix and all five MEG functional connectivity matrices (representing each of the five frequency bands). This yielded five values of correlation (represented by r2). In testing whether these correlation values are statistically significant, a significant problem arises. Specifically, given the logical ordering of brain regions in the AAL atlas and the finite spatial resolution of MEG, functional connectivity matrices exhibit inherent smoothness, meaning that separate connections between pairs of brain regions are not independent. This means that simply assuming that the number of degrees of freedom in the matrix is equivalent to the number of measured connections would vastly inflate the prescribed significance of the correlation. To eliminate this problem we used a permutation test. A set of “pseudo-MEG matrices” was generated, each having spatial properties similar to those of the real matrices but, crucially, not based on genuine data. This procedure has been described recently by Tewarie et al. (31); briefly, to obtain the pseudomatrices we first performed an eigenvalue decomposition of the real MEG-derived matrices. Each eigenvector was then randomized using a phase-based technique (60). Reconstruction of the matrix postrandomization yielded a pseudomatrix similar in mathematical structure to the genuine adjacency matrices but not reflecting genuine MEG-derived functional connectivity (Fig. S4). Correlation between the structural matrix and 10,000 iterations of the pseudomatrix (with a different phase distribution on each iteration) was measured and an empirical null distribution derived. Comparison of the r2 value from real MEG connectivity matrices with the empirical null from the pseudomatrices allowed computation of a P value. Significant correlation was assigned if the P value was less than 0.01 (i.e., a threshold of 0.05 corrected for multiple comparisons across bands).
The Relationship Between MT and MEG in Linear and Nonlinear Combinations.
It is likely that, if our hypothesis that functional connectivity predicts myeloarchitecture is true, then the relationship would not be confined to any single frequency band. Rather, it is likely that the structural network develops based on a combination of electrophysiological activity across all bands. For this reason we used a recently developed mathematical framework (31) to assess whether linear or nonlinear combinations of frequency bands could predict the structural network better than any single MEG band. A three step approach was adopted:
-
•
First, a single-frequency model was used to assess the predictive value of each individual MEG frequency band (as described above).
-
•
Second, all five MEG frequency bands in linear combination were used to predict the structural network.
-
•
Finally, a multivariate truncated Taylor series was adopted allowing for both linear terms (i.e., the regional MEG matrices in Fig. 2) and squared terms (i.e., the square of each of the five MEG matrices in Fig. 2), 10 terms in all.
For all three steps above, a goodness-of-fit measure (r2) was used to assess the efficacy of the model. Importantly, the squared terms have specific meaning because they represent covariance between the neuronal connectivity profiles of separate regions. In other words, for any nonlinear term, a matrix element, say [1, 2], will be high if the connectivity profile of region 1 to the rest of the brain overlaps with the connectivity profile of region 2 to the rest of the brain. This physical interpretation helps explain why their inclusion might improve the prediction of structural matrices; if two regions share electrophysiological interactions with similar areas, then it is likely that their myeloarchitecture may evolve together.
Our hypothesis was that as the model got more complex (i.e., moving from single frequency band to first linear and then nonlinear integration) we would see a better fit to the structural covariance matrix, and that this would be due to the fact that myeloarchitecture supports networks in all frequency bands. However, a confound here is that, when introducing more complexity into a model, the goodness of fit for that model is likely to increase, simply because one tries to fit data using more terms. We therefore tested whether the improvements observed were merely the result of increased model complexity. To do this, the rate of improvement of the fit with increasing model order was calculated and tested statistically. The gradient of the slope representing improvement was first measured using real data. Second, a null distribution of model improvement was constructed. Here, five pseudomatrices generated based upon the five MEG matrices in Fig. 2 were used. These five sham matrices were combined (both linearly and nonlinearly) and a fit to the MT data derived, alongside an associated r2. A gradient, representing improvement in fit to the MT data with increasing model complexity, was then derived. This procedure was repeated 10,000 times (different realizations of the pseudomatrices on each iteration). The 10,000 values of gradient then represented an empirical null distribution; this was compared with the value from real data.
Finally, we performed a cross-validation to test whether the prediction of the structural network based on multiple frequency bands was robust. If the estimated parameters contain meaningful information, then applying the same set of parameters on an independent dataset would result in a significantly better prediction of the structural network than an equivalent prediction based on pseudo MEG networks. To this end, the cohort of 58 subjects was split into two equal groups, A and B (29 subjects in each). For each group a structural network and average MEG networks were estimated. First, the structural network in group A was predicted based on linear and nonlinear combinations of the MEG networks from group A, resulting in an r2 value. Second, parameters from this prediction were used to map the MEG networks to the structural network in group B to obtain a “cross-validation” r2. Third, the same parameters from group A were used to map pseudo MEG networks obtained from group B to the structural network of group B, which yielded in a “sham” r2 value. This procedure was repeated 10,000 times, with the 58 subjects split differently on each iteration. This resulted in three r2 distributions (original, cross-validation, and sham). The parameters of the initial prediction were said to contain meaningful information if the median of the distribution of cross-validation r2 values could be found outside the 95% area under the curve of the sham r2 distribution.
Acknowledgments
This work was funded by Medical Research Council (MRC) New Investigator Research Grant MR/M006301/1, MRC Partnership Grant MR/K005464/1, and MRC Doctoral Training Grant MR/K501086/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: MEG and MT adjacency matrices have been deposited at www.figshare.com and can be found with DOI: 10.6084/m9.figshare.4056441.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608587113/-/DCSupplemental.
References
- 1.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 2.Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360(1457):1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meier J, et al. A mapping between structural and functional brain networks. Brain Connect. 2016;6(4):298–311. doi: 10.1089/brain.2015.0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lerch JP, et al. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage. 2006;31(3):993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 5.Andrews TJ, Halpern SD, Purves D. Correlated size variations in human visual cortex, lateral geniculate nucleus, and optic tract. J Neurosci. 1997;17(8):2859–2868. doi: 10.1523/JNEUROSCI.17-08-02859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White LE, et al. Structure of the human sensorimotor system. II: Lateral symmetry. Cereb Cortex. 1997;7(1):31–47. doi: 10.1093/cercor/7.1.31. [DOI] [PubMed] [Google Scholar]
- 7.He Y, Chen ZJ, Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb Cortex. 2007;17(10):2407–2419. doi: 10.1093/cercor/bhl149. [DOI] [PubMed] [Google Scholar]
- 8.Alexander-Bloch A, Giedd JN, Bullmore E. Imaging structural co-variance between human brain regions. Nat Rev Neurosci. 2013;14(5):322–336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mechelli A, Friston KJ, Frackowiak RS, Price CJ. Structural covariance in the human cortex. J Neurosci. 2005;25(36):8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glasser MF, Goyal MS, Preuss TM, Raichle ME, Van Essen DC. Trends and properties of human cerebral cortex: Correlations with cortical myelin content. Neuroimage. 2014;93(Pt 2):165–175. doi: 10.1016/j.neuroimage.2013.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutti A, Dick F, Sereno MI, Weiskopf N. Using high-resolution quantitative mapping of R1 as an index of cortical myelination. Neuroimage. 2014;93(Pt 2):176–188. doi: 10.1016/j.neuroimage.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Glasser MF, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536(7615):171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J Neurosci. 2011;31(32):11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sánchez-Panchuelo RM, Francis ST, Schluppeck D, Bowtell RW. Correspondence of human visual areas identified using functional and anatomical MRI in vivo at 7 T. J Magn Reson Imaging. 2012;35(2):287–299. doi: 10.1002/jmri.22822. [DOI] [PubMed] [Google Scholar]
- 15.Sereno MI, Lutti A, Weiskopf N, Dick F. Mapping the human cortical surface by combining quantitative T(1) with retinotopy. Cereb Cortex. 2013;23(9):2261–2268. doi: 10.1093/cercor/bhs213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besle J, Sánchez-Panchuelo R-M, Bowtell R, Francis S, Schluppeck D. Event-related fMRI at 7T reveals overlapping cortical representations for adjacent fingertips in S1 of individual subjects. Hum Brain Mapp. 2014;35(5):2027–2043. doi: 10.1002/hbm.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dick F, et al. In vivo functional and myeloarchitectonic mapping of human primary auditory areas. J Neurosci. 2012;32(46):16095–16105. doi: 10.1523/JNEUROSCI.1712-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glasser MF, et al. Improved Cortical Myelin Maps in Humans, Chimpanzees, and Macaques Allow Identification of Putative Areal Homologies. Society for Neuroscience; New Orleans: 2012. [Google Scholar]
- 19.Deoni SCL, Dean DC, 3rd, Remer J, Dirks H, O’Muircheartaigh J. Cortical maturation and myelination in healthy toddlers and young children. Neuroimage. 2015;115:147–161. doi: 10.1016/j.neuroimage.2015.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shafee R, Buckner RL, Fischl B. Gray matter myelination of 1555 human brains using partial volume corrected MRI images. Neuroimage. 2015;105:473–485. doi: 10.1016/j.neuroimage.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fields RD. Nerve impulses regulate myelination through purinergic signalling. Novartis Found Symp. 2006;276:148–158; discussion 158–161, 233–237, 275–281. [PubMed] [Google Scholar]
- 22.Fields RD. A new mechanism of nervous system plasticity: Activity-dependent myelination. Nat Rev Neurosci. 2015;16(12):756–767. doi: 10.1038/nrn4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pajevic S, Basser PJ, Fields RD. Role of myelin plasticity in oscillations and synchrony of neuronal activity. Neuroscience. 2014;276:135–147. doi: 10.1016/j.neuroscience.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson EM, et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science. 2014;344(6183):1252304. doi: 10.1126/science.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helbling S, et al. Structure predicts function: Combining non-invasive electrophysiology with in-vivo histology. Neuroimage. 2015;108:377–385. doi: 10.1016/j.neuroimage.2014.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen D. Magnetoencephalography: Detection of the brain’s electrical activity with a superconducting magnetometer. Science. 1972;175(4022):664–666. doi: 10.1126/science.175.4022.664. [DOI] [PubMed] [Google Scholar]
- 27.Brookes MJ, et al. Investigating the electrophysiological basis of resting state networks using magnetoencephalography. Proc Natl Acad Sci USA. 2011;108(40):16783–16788. doi: 10.1073/pnas.1112685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillebrand A, Barnes GR, Bosboom JL, Berendse HW, Stam CJ. Frequency-dependent functional connectivity within resting-state networks: an atlas-based MEG beamformer solution. Neuroimage. 2012;59(4):3909–3921. doi: 10.1016/j.neuroimage.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Neill GC, Barratt EL, Hunt BAE, Tewarie PK, Brookes MJ. Measuring electrophysiological connectivity by power envelope correlation: A technical review on MEG methods. Phys Med Biol. 2015;60(21):R271–R295. doi: 10.1088/0031-9155/60/21/R271. [DOI] [PubMed] [Google Scholar]
- 30.Schölvinck ML, Leopold DA, Brookes MJ, Khader PH. The contribution of electrophysiology to functional connectivity mapping. Neuroimage. 2013;80:297–306. doi: 10.1016/j.neuroimage.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tewarie P, et al. Predicting haemodynamic networks using electrophysiology: The role of non-linear and cross-frequency interactions. Neuroimage. 2016;130:273–292. doi: 10.1016/j.neuroimage.2016.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolff SD, Balaban RS. Magnetization transfer contrast (MTC) and tissue water proton relaxation in vivo. Magn Reson Med. 1989;10(1):135–144. doi: 10.1002/mrm.1910100113. [DOI] [PubMed] [Google Scholar]
- 33.Zumer JM, Scheeringa R, Schoffelen J-M, Norris DG, Jensen O. Occipital alpha activity during stimulus processing gates the information flow to object-selective cortex. PLoS Biol. 2014;12(10):e1001965. doi: 10.1371/journal.pbio.1001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engel AK, Gerloff C, Hilgetag CC, Nolte G. Intrinsic coupling modes: Multiscale interactions in ongoing brain activity. Neuron. 2013;80(4):867–886. doi: 10.1016/j.neuron.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 35.Amunts K, et al. Asymmetry in the human motor cortex and handedness. Neuroimage. 1996;4(3 Pt 1):216–222. doi: 10.1006/nimg.1996.0073. [DOI] [PubMed] [Google Scholar]
- 36.Driver ID, et al. Hemispheric asymmetry in cerebrovascular reactivity of the human primary motor cortex: an in vivo study at 7 T. NMR Biomed. 2015;28(5):538–545. doi: 10.1002/nbm.3282. [DOI] [PubMed] [Google Scholar]
- 37.Toga AW, Thompson PM. Mapping brain asymmetry. Nat Rev Neurosci. 2003;4(1):37–48. doi: 10.1038/nrn1009. [DOI] [PubMed] [Google Scholar]
- 38.Sun T, Walsh CA. Molecular approaches to brain asymmetry and handedness. Nat Rev Neurosci. 2006;7(8):655–662. doi: 10.1038/nrn1930. [DOI] [PubMed] [Google Scholar]
- 39.Tewarie P, et al. Disruption of structural and functional networks in long-standing multiple sclerosis. Hum Brain Mapp. 2014;35(12):5946–5961. doi: 10.1002/hbm.22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doria V, et al. Emergence of resting state networks in the preterm human brain. Proc Natl Acad Sci USA. 2010;107(46):20015–20020. doi: 10.1073/pnas.1007921107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hebb DO. The Organization of Behavior: A Neuropsychological Theory. Taylor & Francis; Abingdon, UK: 1949. [Google Scholar]
- 42.Draganski B, May A. Training-induced structural changes in the adult human brain. Behav Brain Res. 2008;192(1):137–142. doi: 10.1016/j.bbr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Calabrese M, et al. Exploring the origins of grey matter damage in multiple sclerosis. Nat Rev Neurosci. 2015;16(3):147–158. doi: 10.1038/nrn3900. [DOI] [PubMed] [Google Scholar]
- 44.Friston KJ, Frith CD. Schizophrenia: A disconnection syndrome? Clin Neurosci. 1995;3(2):89–97. [PubMed] [Google Scholar]
- 45.Iwabuchi SJ, Liddle PF, Palaniyappan L. Structural connectivity of the salience-executive loop in schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2015;265(2):163–166. doi: 10.1007/s00406-014-0547-z. [DOI] [PubMed] [Google Scholar]
- 46.Mougin O, Clemence M, Peters A, Pitiot A, Gowland P. High-resolution imaging of magnetisation transfer and nuclear Overhauser effect in the human visual cortex at 7 T. NMR Biomed. 2013;26(11):1508–1517. doi: 10.1002/nbm.2984. [DOI] [PubMed] [Google Scholar]
- 47.Geades N, et al. October 17, 2016. Quantitative analysis of the z-spectrum using a numerically simulated look-up table: Application to the healthy human brain at 7T. Magn Reson Med 10.1002/mrm.26459.
- 48.Brookes MJ, Woolrich MW, Barnes GR. Measuring functional connectivity in MEG: A multivariate approach insensitive to linear source leakage. Neuroimage. 2012;63(2):910–920. doi: 10.1016/j.neuroimage.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Filippini N, et al. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci USA. 2009;106(17):7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oldfield RC. The assessment and analysis of handedness: The Edinburgh inventory. Neuropsychologia. 1971;9(1):97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- 51.Tzourio-Mazoyer N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 52.Robinson S, Vrba J. Functional neuroimaging by synthetic aperture magnetometry (SAM) In: Yoshimoto T, Kotani M, Kuriki S, Karibe H, Nakasato N, editors. Recent Advances in Biomagnetism. Tohoko Univ Press; Sendai, Japan: 1998. pp. 302–305. [Google Scholar]
- 53.Brookes MJ, et al. Optimising experimental design for MEG beamformer imaging. Neuroimage. 2008;39(4):1788–1802. doi: 10.1016/j.neuroimage.2007.09.050. [DOI] [PubMed] [Google Scholar]
- 54.Sarvas J. Basic mathematical and electromagnetic concepts of the biomagnetic inverse problem. Phys Med Biol. 1987;32(1):11–22. doi: 10.1088/0031-9155/32/1/004. [DOI] [PubMed] [Google Scholar]
- 55.Huang MX, Mosher JC, Leahy RM. A sensor-weighted overlapping-sphere head model and exhaustive head model comparison for MEG. Phys Med Biol. 1999;44(2):423–440. doi: 10.1088/0031-9155/44/2/010. [DOI] [PubMed] [Google Scholar]
- 56.Brookes MJ, et al. A multi-layer network approach to MEG connectivity analysis. Neuroimage. 2016;132:425–438. doi: 10.1016/j.neuroimage.2016.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hipp JF, Hawellek DJ, Corbetta M, Siegel M, Engel AK. Large-scale cortical correlation structure of spontaneous oscillatory activity. Nat Neurosci. 2012;15(6):884–890. doi: 10.1038/nn.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hall EL, Robson SE, Morris PG, Brookes MJ. The relationship between MEG and fMRI. Neuroimage. 2014;102(Pt 1):80–91. doi: 10.1016/j.neuroimage.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 59.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 60.Prichard D, Theiler J. Generating surrogate data for time series with several simultaneously measured variables. Phys Rev Lett. 1994;73(7):951–954. doi: 10.1103/PhysRevLett.73.951. [DOI] [PubMed] [Google Scholar]