Significance
A fundamental question in biology is how the biophysical parameters describing protein folding in vitro are altered during cotranslational folding. Because protein synthesis is slow, nascent chains can fold cotranslationally while still tethered to the ribosome and before the entire protein has emerged. Here we examine the effects of ribosomal tethering on protein stability and how that stability is modulated by the distance away from the ribosome’s active site. We develop a quantitative assay for measuring protein stability on stalled nascent chains and find that the ribosome destabilizes nascent chains. This destabilization may play a key role in ensuring folding fidelity by helping the nascent chain avoid partially folded kinetic traps before the entire chain has been synthesized.
Keywords: protein folding, cotranslational folding, pulse proteolysis, protein stability
Abstract
Accurate protein folding is essential for proper cellular and organismal function. In the cell, protein folding is carefully regulated; changes in folding homeostasis (proteostasis) can disrupt many cellular processes and have been implicated in various neurodegenerative diseases and other pathologies. For many proteins, the initial folding process begins during translation while the protein is still tethered to the ribosome; however, most biophysical studies of a protein’s energy landscape are carried out in isolation under idealized, dilute conditions and may not accurately report on the energy landscape in vivo. Thus, the energy landscape of ribosome nascent chains and the effect of the tethered ribosome on nascent chain folding remain unclear. Here we have developed a general assay for quantitatively measuring the folding stability of ribosome nascent chains, and find that the ribosome exerts a destabilizing effect on the polypeptide chain. This destabilization decreases as a function of the distance away from the peptidyl transferase center. Thus, the ribosome may add an additional layer of robustness to the protein-folding process by avoiding the formation of stable partially folded states before the protein has completely emerged from the ribosome.
Proper protein folding is necessary for the function of all cells, and changes in cellular protein folding capacity can lead to cell death and disease (1). For more than 50 years, experimental studies have probed the physical properties of proteins, paving the way for incredible advances in protein design and protein structure prediction, as well as for understanding the first principles of protein folding (2, 3). These in vitro studies, however, do not necessarily recapitulate the folding process in vivo, including the constraints that the ribosome imposes on the emerging nascent chain during translation (4).
In the cell, proteins are synthesized by the ribosome one amino acid at a time on a time scale that is slower than most in vitro protein folding rates (5). Thus, the emerging chain has time to explore conformational space and adopt structured conformations before its entire sequence has been synthesized (6). This vectorial process, and the proximity of the ribosome itself, can modulate the emerging chain’s energy landscape in ways that are just beginning to be appreciated. Translation can affect folding efficiency, enable the population of intermediates that are not revealed during off-ribosome studies, and even determine the final conformational fate of the nascent protein (6–10). To understand the folding process in vivo, general rules and biophysical mechanisms for these modulations of the emerging chain’s energy landscape need to be elucidated.
To unravel the in vivo folding landscape, we need to interrogate the energetics and dynamics of ribosome-bound nascent chains in a manner analogous to studies of the in vitro folding process. The challenge, however, is that standard methods used to probe protein energy landscapes in vitro—such as circular dichroism and intrinsic fluorescence— demand large quantities (milligrams) of extremely pure protein in conditions that differ significantly from those found in the cytosol. Therefore, these experimental approaches are not amenable to buffers that more closely replicate in vivo conditions or to heterogeneous protein complexes, such as ribosome nascent chains (RNCs). Recently, approaches such as single-molecule optical trapping and multidimensional NMR have revealed interesting effects of the folding of a few different RNCs, but these studies are technically very challenging and thus are not practical for characterizing many different proteins (11, 12). Furthermore, these approaches do not provide the most basic features of a protein’s energy landscape, such as the global stability of the nascent chain.
Here we present an approach that we believe takes a first step toward quantitatively measuring the energy landscape of RNCs. Our technique uses in vitro translation (IVT) and an easily accessible cotranslational labeling scheme in combination with a gel-based method to measure the protein stability of RNCs. We measure the stability of three proteins both on and off the ribosome and demonstrate that the ribosome destabilizes the nascent chain in a manner dependent on the distance away from the ribosome's peptidyl-transferase center (PTC). This ribosome-mediated destabilization may safeguard the emerging chain by preventing the formation of stable off-pathway intermediates that lead to misfolding.
Results
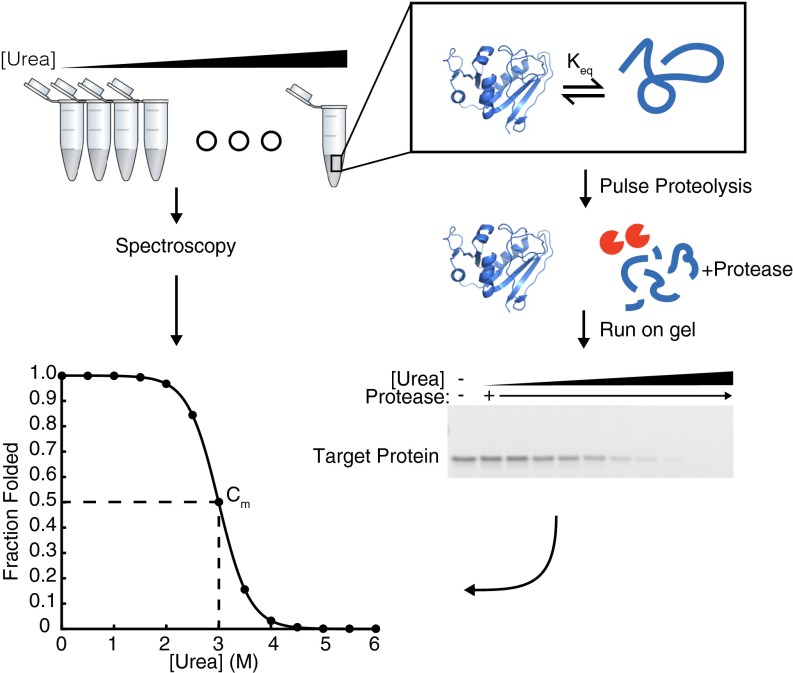
The simple, first-order description of any protein’s energy landscape is its global stability, ∆Gunfolding, the relative population of the unfolded and native conformers. This simple parameter is a key determinant of in vivo protein lifetimes, and many disease-causing SNPs are associated with changes in protein stability (13–15). In vitro, ∆Gunfolding is easily determined by equilibrium denaturation—monitoring the spectroscopic signal from the folded protein as a function of chemical perturbant, such as urea or guanidinium chloride (Fig. 1); however, in the presence of the ribosome, these simple measurements are not feasible, because the ribosome itself will overwhelm the spectroscopic signal of the nascent chain.
Fig. 1.
Comparison of spectroscopy and pulse proteolysis for determining protein stability. Samples are equilibrated in urea overnight for both spectroscopic methods (Left) and pulse proteolysis (Right). For spectroscopic methods, equilibrated samples are read directly in a spectrophotometer to determine the signal difference between the folded and unfolded states. Alternatively, for pulse proteolysis, a protease is added to digest all unfolded protein at a given urea concentration. Samples are analyzed by SDS/PAGE, and the remaining band represents the amount of folded protein at each urea concentration. The intensity of each band is plotted against the urea concentration and then fit to determine the concentration of urea where half of the folded protein remains, or the Cm. The Cm is multiplied by the protein’s m-value to determine the stability, ∆Gunfolding.
To overcome this, we use a gel-based method, pulse proteolysis, which measures protein stability by taking advantage of the fact that unfolded proteins are more susceptible to proteolysis than folded proteins (16). In pulse proteolysis, an equilibrium mixture of folded and unfolded proteins is subjected to a short pulse of protease, which digests the unfolded proteins and leaves the folded proteins intact. This is then repeated at increasing urea concentrations to alter the equilibrium ratio of folded and unfolded proteins. Finally, the fraction of folded protein is determined by SDS/PAGE (Fig. 1). As long as the protein-unfolding rate is slow compared with the duration of the proteolysis pulse (1 min), the intensity of the full-length band on SDS/PAGE is directly proportional to the equilibrium fraction of native protein before digestion. No protease is added to the zero molar urea sample as a control for any proteolysis of folded protein. The intensities of the full length bands at each urea concentration are then fit to determine the denaturant midpoint, or Cm, which when multiplied by a protein’s m-value (slope, or denaturant dependence, of stability) yields the protein’s stability in zero denaturant. The m-values are not determined by pulse proteolysis but are either calculated (as here) or determined using another experimental method (16, 17).
Pulse Proteolysis of in Vitro Translated and Labeled Proteins.
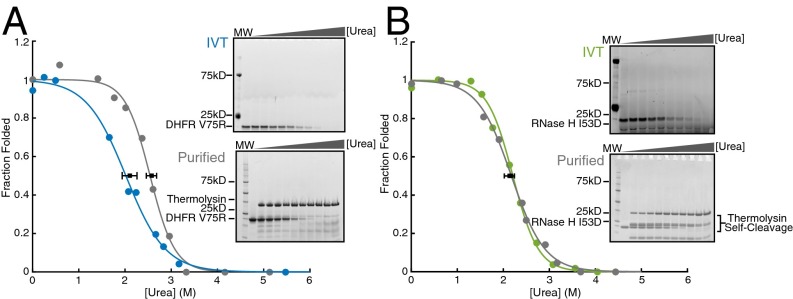
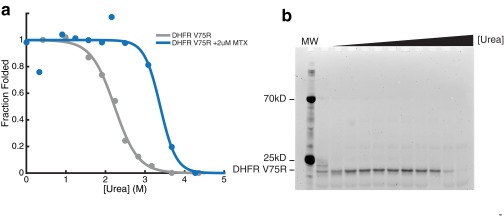
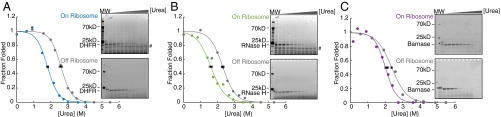
We performed pulse proteolysis on RNCs by combining a commercially available in vitro coupled transcription/translation system (PURExpress; New England Biolabs) with commercially available BODIPY-FL-LysineAAA-tRNA (Promega). This pulse proteolysis/labeling approach can successfully determine the stability of in vitro translated proteins that contain a single lysine (18). To generalize this method, we first compared the stability of proteins containing multiple lysines produced via the same IVT strategy or purified recombinantly from Escherichia coli. We measured the stabilities of two model proteins—RNase H I53D (11 lysines) and DHFR V75R (six lysines)— by pulse proteolysis. Because lysines are predominately surface-exposed, we expected BODIPY-FL-lysine incorporation to minimally perturb each protein’s native state stability. We observed a single band for each protein assayed (top gels in Fig. 2 A and B) and thus assume that only a single BODIPY-FL-lysine is incorporated per protein. Fig. 2A shows that for DHFR V75R, the stability of the in vitro translated, labeled protein is slightly lower than that of the purified recombinant protein. For RNase H I53D, the stability of the in vitro translated, labeled protein matches that of the unlabeled recombinant protein purified from E. coli (Fig. 2B and Table 1). Slight destabilization of DHFR may be due to the incorporation of the fluorophore or from slight differences in the buffer used in the IVT reaction compared with those used on the purified protein, because DHFR is highly sensitive to changes in salt concentrations. It should be noted, however, that incorporation of BODIPY-FL-lysine does not affect the ability of DHFR V75R to bind one of its inhibitors, methotrexate, suggesting that although the stability of the protein is somewhat decreased, the native conformation of DHFR V75R is not disturbed (Fig. S1). Although this could pose a problem when comparing purified protein with in vitro translated protein, it should not affect a direct comparison of IVT-produced protein on and off the ribosome.
Fig. 2.
Stability of proteins purified from E. coli and made using IVT by pulse proteolysis. (A) DHFR V75R purified (in gray and upper gel) and in vitro translated (in blue and lower gel). (B) RNase H I53D purified (in gray and upper gel) and in vitro translated (in green and lower gel). Note that the concentration of urea is different for each lane between gels. Each gel and trace shown is representative of three separate experiments. Error bars represent the SD of the Cm for each curve plotted, determined by three separate experiments.
Table 1.
Comparison of IVT and recombinant protein
| Protein | Cm, purified (urea, M) | ∆Gunf, purified (kcal mol−1) | Cm, IVT (urea, M) | ∆Gunf, IVT (kcal mol−1) |
| DHFR V75R | 2.50 ± 0.18 | 4.50 ± 0.32 | 2.10 ± 0.16 | 3.78 ± 0.29 |
| RNase H I53D | 2.21 ± 0.09 | 4.41 ± 0.18 | 2.16 ± 0.10 | 4.31 ± 0.19 |
Fig. S1.
Methotrexate (MTX) binds in vitro translated DHFR V75R. (A) Gray indicates the fraction folded as a function of urea for in vitro translated DHFR V75R. Blue indicates the fraction folded as a function of urea for in vitro translated DHFR V75R in the presence of 2 μM MTX. (B) Gel of MTX used for quantitation in A.
Urea Sensitivity of 70S Ribosome and RNCs.
Determining protein stability (ΔGunfolding) requires modulating and measuring the population of both the folded and unfolded states; this is usually done using thermal or chemical denaturation. Urea is the preferred denaturant for pulse proteolysis, because it does not alter the samples’ ionic strength and can be used in combination with SDS/PAGE. Considering its ubiquity in protein-folding experiments, it is surprising that, to our knowledge, no group has determined the sensitivity of RNCs to urea denaturation. To probe this, we used both sucrose gradient ultracentrifugation and fluorescence correlation spectroscopy (FCS).
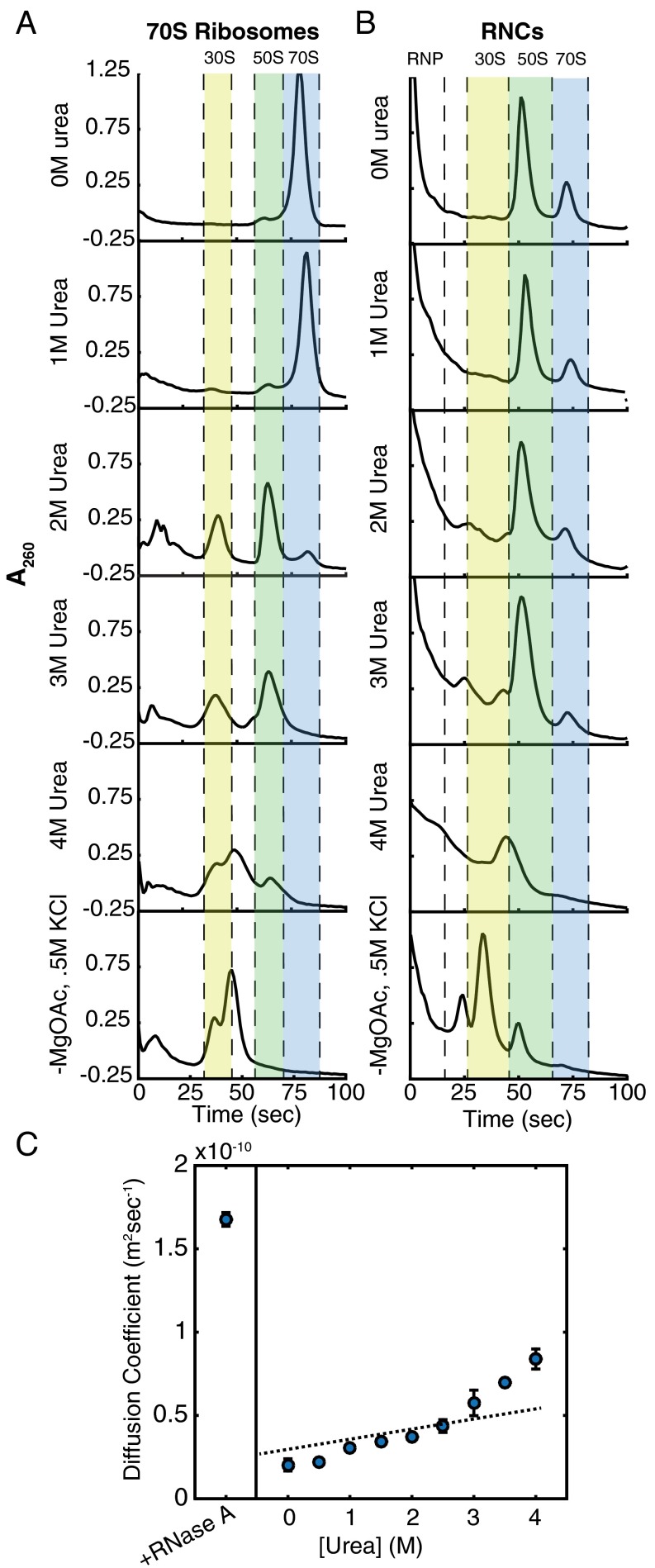
We first performed sucrose gradient ultracentrifugation on purified 70S ribosomes after incubation in varying urea concentrations (Fig. 3A). The 70S ribosomes appear stable in 1.0 M urea, although the peak shifts down the gradient marginally, perhaps signifying slight expansion or partial unfolding of ribosomal proteins or rRNA. At 2.0 M urea, however, the 70S peak decreases concurrent with an increase in the heights of both the 30S and 50S peaks. The 70S peak then completely disappears at 3.0 M urea, and the 50S peak broadens and simultaneously decreases in intensity, signifying denaturation of 50S subunits.
Fig. 3.
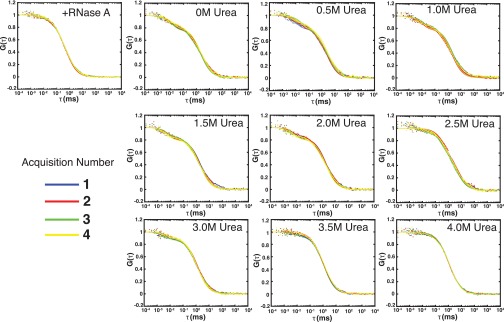
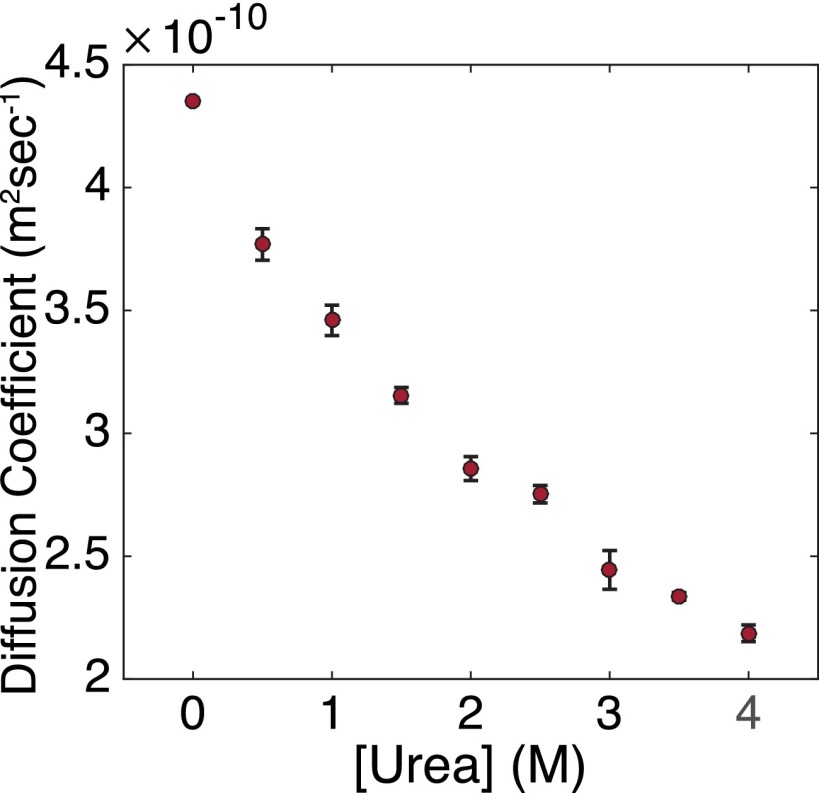
Urea sensitivity of 70S ribosomes and RNCs. (A and B) Sucrose gradient ultracentrifugation of 70S ribosomes (A) and RNCs (B). Highlighted in blue is the peak corresponding to 70S ribosomes. The 50S peak is highlighted in green; the 30S peak, in yellow. The 70S ribosomes and RNCs were also run in gradients containing no magnesium and 0.5 M KCl as a negative control. (C) Diffusion coefficients of RNCs plotted as a function of urea concentration as determined by FCS. Error bars represent the SD of at least 10 experiments. Fits are shown in Fig. S5.
Fig. S5.
Sample FCS runs with fits for each urea concentration. Colors represent four different acquisitions. Diffusion coefficients of RNCs are plotted as a function of urea concentration. Error bars represent the SD of at least 10 experiments.
SecM-stalled ribosomes should be more stable than reconstituted 70S ribosomes because the SecM stalling sequence is known to make extensive contacts with the ribosome exit tunnel and to strengthen 30S–50S contacts (19). To test the urea sensitivity of SecM-stalled RNCs, we translated DHFR V75R-(GS)5-SecM for 1 h, halted translation with 2 mM chloramphenicol, incubated the IVT reactions in urea overnight, and subjected these samples to sucrose-gradient ultracentrifugation. At 0 M and 1.0 M urea, as observed for reconstituted 70S ribosomes, the height of the 70S peak changes little for SecM-stalled RNCs. Unlike reconstituted 70S ribosomes, however, a significant portion of the 70S peak remains for SecM-stalled RNCs incubated at 2.0 M and 3.0 M urea (compare Fig. 3 A and B). It is possible that a small population of the 70S ribosomes is not properly engaged with SecM, and this could account for the small decrease in 70S peak intensity between 1.0 M and 2.0 M urea. At 4.0 M urea, there is no apparent 70S peak. Overall, these data suggest that the SecM-stalled RNCs are stable to >3.0 M urea. To confirm this observation, we turned to FCS to follow RNCs specifically, rather than the entire 70S ribosome population.
We carried out FCS studies on RNCs labeled with Atto647N via unnatural amino acid incorporation (Fig. 3C) and were incubated overnight in urea as described above. From 0 M urea to 3.0 M urea, the small increase in the measured diffusion coefficient is likely due to a small population of free, dye-labeled nascent chains or a small proportion of RNCs improperly engaged with SecM, as was seen in the previously mentioned sucrose gradient experiments. Because we use a single-species fit (Materials and Methods), a small proportion of free nascent chains would appear as an increase in the observed diffusion coefficient. The diffusion coefficient measured in low concentrations of urea is ∼2.0 × 10−11 m2s−1, close to the diffusion coefficient measured for 70S ribosomes using laser light-scattering microscopy (20), although in different conditions. Above 3.0 M urea, there is a dramatic increase in the observed diffusion coefficient, likely as a result of 70S dissociation into 30S and 50S subunits. At 4.0 M urea, the diffusion coefficient measured corresponds to that of sphere with mass ∼40 kD (approximately the molecular weight of the protein and attached tRNA) and remains smaller than that of nascent chains released by RNase A digestion. These data support those seen in the sucrose gradient ultracentrifugation experiments. SecM-stalled RNCs are stable to >3.0 M but <4.0 M urea.
Pulse Proteolysis of Stalled Ribosome Nascent Chains.
To determine whether the ribosome can alter the stability of the emerging chain, we applied pulse proteolysis to stalled RNCs. We used protein variants with a Cm < 2.5 M urea off the ribosome—RNase H I53D, DHFR V75R, and Barnase W35F/W94F/H102A—to ensure that the unfolding transition occurred before RNC dissociation. All three of these variants have been well characterized in vitro and display two-state equilibrium unfolding (21, 22). For each protein tested, RNCs are destabilized compared with the same protein free from the ribosome (Fig. 4 A–C and Table 2).
Fig. 4.
Determination of RNC stability by pulse proteolysis. (A) DHFR V75R RNCs. Blue indicates protein on the ribosome; gray, protein off the ribosome. (B) RNase H I53D RNCs. Green indicates on the ribosome; gray, off the ribosome. (C) Barnase W35F/W94F/H102M RNCs. Purple indicates on the ribosome; gray, off the ribosome. Each gel and trace shown is representative of three separate experiments. *Incomplete tRNA digestion. #RNase A. Error bars represent the SD of the Cm for each curve determined by three separate experiments, except for Barnase, for which the Cm is the average of two experiments.
Table 2.
Determination of RNC stability
| Protein | Cm, on (urea, M) | Cm, off (urea, M) | ∆Gunf, on (kcal mol−1) | ∆Gunf, off (kcal mol−1) | ∆∆Gunf, on-off (kcal mol−1) |
| DHFR V75R-(GS)5-SecM | 1.81 ± 0.05 | 2.58 ± 0.11 | 4.57 ± 0.14 | 6.50 ± 0.25 | −1.93 ± 0.29 |
| RNase H I53D-(GS)5-SecM | 1.65 ± 0.10 | 2.14 ± 0.13 | 4.07 ± 0.23 | 5.29 ± 0.33 | −1.22 ± 0.16 |
| Barnase W35F/W94F/H102M-(GS)5-SecM | 2.09 ± 0.01 | 2.31 ± 0.04 | 3.90 ± 0.02 | 4.32 ± 0.08 | −0.42 ± 0.08 |
In addition, the magnitude of destabilization is anticorrelated to the isoelectric point of the protein. DHFR V75R is destabilized by 1.93 ± 0.29 kcal/mol and has an isoelectric point of 4.94, whereas RNase H I53D and Barnase W35F/W94F/H102M, with respective isoelectric points of 7.97 and 8.86, are destabilized by 1.63 ± 0.36 kcal/mol and 0.42 ± 0.08 kcal/mol, respectively. These data are consistent with previous studies on ribosome-stalled intrinsically disordered proteins (IDPs) demonstrating that more negatively charged IDPs have increased dynamic motions when stalled on the ribosome compared with when free in solution (23). Similarly, NMR data show increased dynamics of the nascent chain when proximal to the ribosome (12, 24). Both of these results can be explained by a general destabilization of the nascent chain owing to proximity to the ribosome.
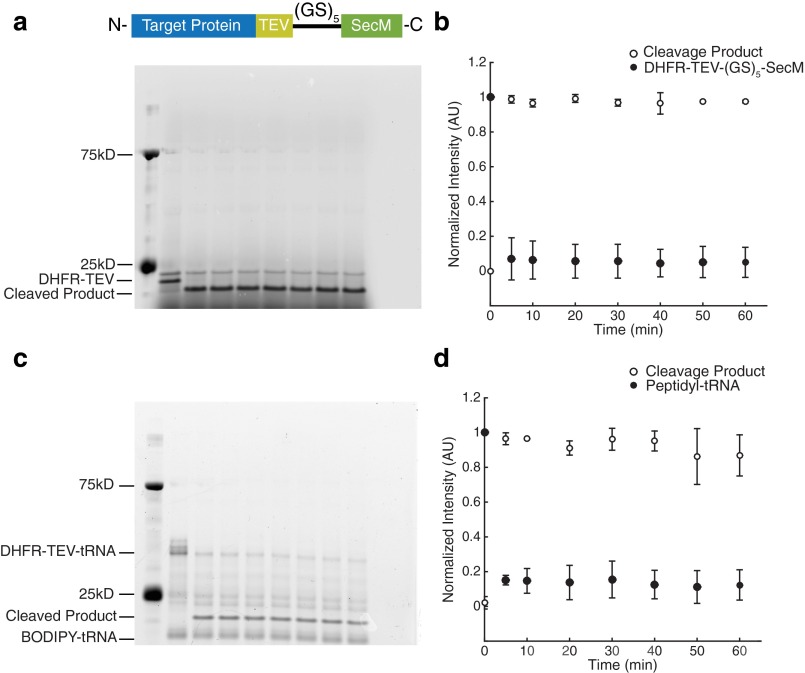
To ensure that our proteolysis measurements are not biased due to steric occlusion of the protease by the ribosome, we appended a TEV cleavage site between the natural C terminus of the protein and the glycine–serine linker. We monitored proteolysis kinetics using TEV protease on RNCs both before and after RNaseA treatment. In both cases, the protein is fully cleaved within 5 min of TEV addition (Fig. S2). Thus, even the C termini of the RNCs are fully accessible to proteolysis.
Fig. S2.
The presence of the ribosome does not inhibit protease accessibility. (A) TEV cleavage of DHFR V75R-TEV-(GS)5-SecM as a function of time after RNase A digestion overnight. (B) Quantification of gel band intensities normalized to the highest band intensity on the gel. The original band is shown as filled circles; the band corresponding to cleavage product, as open circles. (C and D) Same as A and B, respectively, but as RNCs. Each gel and point shown is representative of three separate experiments. Error bars represent the SD of the normalized band intensity for each time point.
Reversible Folding of Stalled Nascent Chains.
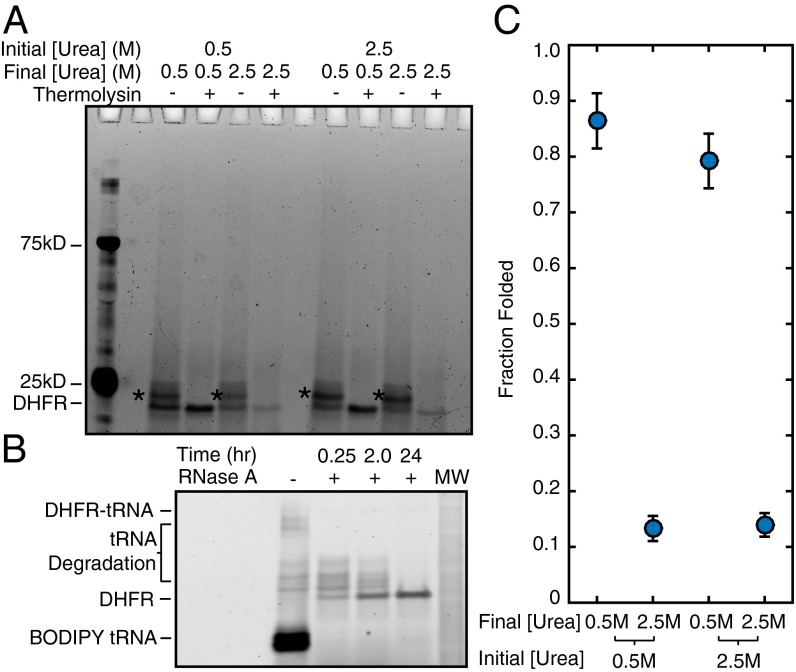
Measuring thermodynamic stability requires that the folding process be reversible and that the proteolysis-resistant conformation not represent a kinetic trap. To test for reversibility, after purification through a sucrose cushion, we resuspended stalled DHFR V75R-(GS)5-SecM RNCs in both 0.5 M and 2.5 M urea and allowed them to reach equilibrium overnight. Each RNC/urea sample was then divided and diluted into either 2.5 M urea or 0.5 M urea (Fig. 5). After a second overnight equilibration, the fraction of folded protein was measured by pulse proteolysis. Regardless of the initial urea concentration, the fraction folded of RNC at 0.5 M urea remained the same whether or not the sample had been unfolded in 2.5 M urea (Fig. 5 A and C); therefore, stalled nascent chains fold reversibly.
Fig. 5.
Folding on the ribosome is reversible. DHFR V75R RNCs reached equilibrium in either 0.5 M or 2.5 M urea. Samples were then split in half and diluted to 0.5 M urea or 2.5 M urea. (A) After another equilibration step, each sample was again split in half and either treated or not treated with thermolysin to assess the amount of folded protein remaining, and then run on a gel. Starred bands are due to incomplete RNase A digestion of attached tRNA. (B) Gel showing complete cleavage of peptidyl-tRNA as a function of time. (C) Quantitation of the data shown in A. Because there is the same amount of folded protein independent of the initial concentration of urea, folding is reversible. Error bars represent the SD of duplicate experiments.
Nascent Chain Destabilization Is Distance-Dependent.
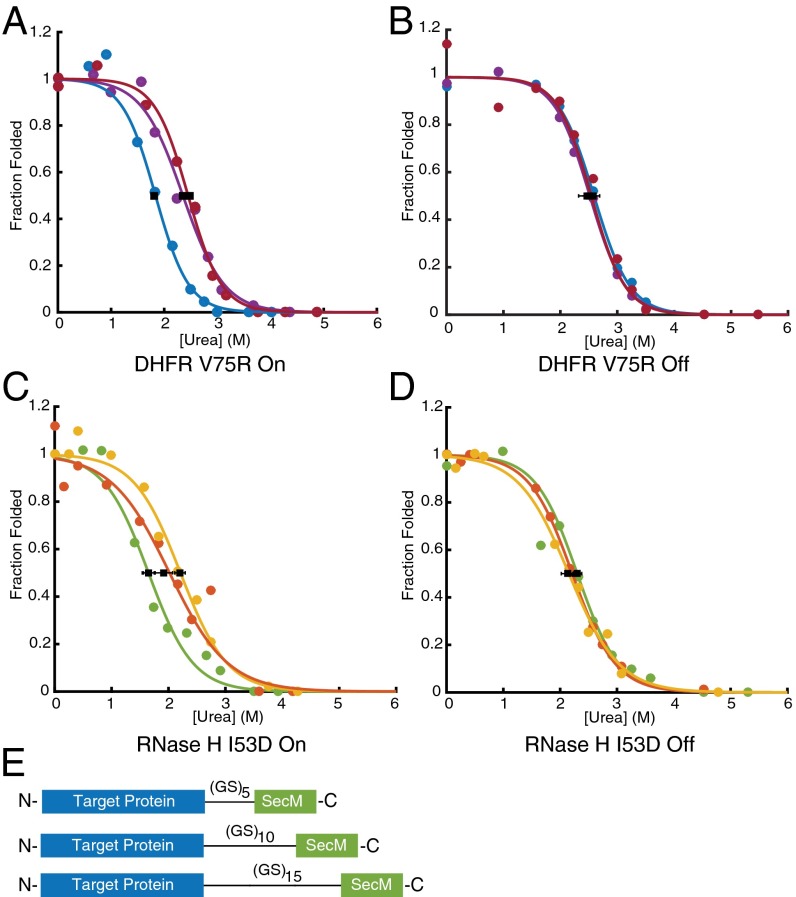
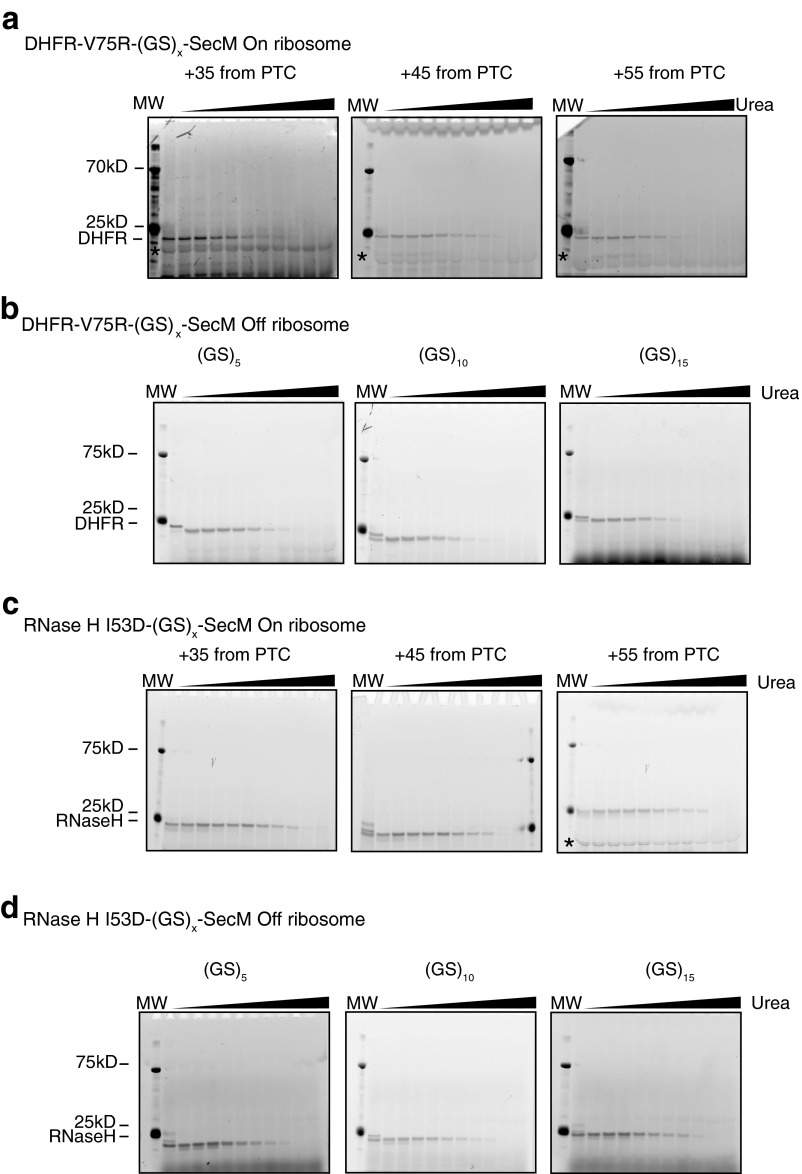
Several previous studies have suggested a distance dependence of the effects of ribosome-mediated changes in nascent chain behavior (6, 11, 12). To investigate the distance dependence of the stability of RNCs, we used both DHFR V75R and RNase H I53D to measure protein stability as a function of amino acid distance from the PTC by increasing the linker length between the SecM stalling sequence and the natural C terminus of the protein (Fig. 6E). The linker length was extended by increasing the number of glycine-serine repeats in 10-aa steps, resulting in distances of 35, 45, and 55 aa between the natural C terminus of the target protein and the PTC. For both proteins, nascent chain stability increased as a function of linker length, approaching the stability of the free protein at a distance of 55 residues away from the PTC (Fig. 6 A–D and Table 3). These results can explain previous observations of both increased protection from limited proteolysis and increased peak dispersion as the distance from the PTC increases (12, 25–27), suggesting that the increased protection is likely due to changes in global stability and not to interactions with the ribosome or changes in native state dynamics.
Fig. 6.
RNC stability increases as the distance to the PTC increases, as determined by pulse proteolysis. (A) DHFR V75R RNC stability as a function of linker length. Blue, 35 aa from PTC; purple, 45 aa; red, 55 aa. (B) Same as A but with a stalling-deficient SecM mutant, 1FSTPVWISQAQGIAAGA17. (C) RNase H I53D RNC stability as a function of linker length. Green, 35 aa from PTC; orange, 45 aa; yellow 55 aa. (D) Same as C except after RNase A digestion overnight. Each trace shown is representative of three separate experiments. Error bars represent the SD of the Cm determined by three separate experiments. (E) Constructs used in this study. The sequence coding for each target protein was appended with a variable size glycine-serine linker, (GS)x, followed by the SecM stalling sequence at its C terminus. Gels of traces plotted here are present in Fig. S6.
Table 3.
Ribosome-mediated destabilization is dependent on distance from the PTC
| Protein | Distance from PTC, aa | Cm, on (urea, M) | Cm, off (urea, M) | ∆Gunf, on (kcal mol−1) | ∆Gunf, off (kcal mol−1) |
| DHFR V75R-(GS)x-SecM | 35 | 1.81 ± 0.05 | 2.58 ± 0.11 | 4.57 ± 0.14 | 6.50 ± 0.25 |
| 45 | 2.47 ± 0.01 | 2.60 ± 0.15 | 6.24 ± 0.02 | 6.55 ± 0.39 | |
| 55 | 2.75 ± 0.02 | 2.72 ± 0.12 | 6.93 ± 0.05 | 6.85 ± 0.30 | |
| RNase H-I53D-(GS)x-SecM | 35 | 1.65 ± 0.10 | 2.14 ± 0.13 | 4.07 ± 0.23 | 5.29 ± 0.33 |
| 45 | 1.92 ± 0.15 | 2.23 ± 0.11 | 4.99 ± 0.39 | 5.80 ± 0.27 | |
| 55 | 2.13 ± 0.11 | 2.18 ± 0.09 | 5.96 ± 0.24 | 5.96 ± 0.30 |
Fig. S6.
Gels used in Fig. 6. (A) DHFR V75R-(GS)x-SecM on ribosome gels. (B) DHFR V75R-(GS)x-SecM off ribosome gels. (C) RNase H I53D-(GS)x-SecM on ribosome gels. (D) RNase H I53D-(GS)x-SecM off ribosome gels.
Discussion
Here we have developed a simple gel-based assay for measuring the stability of RNCs and compared the global stability of three proteins both on the ribosome and free in solution. We provide quantitative measurements of a protein’s global stability on the ribosome and find that RNCs are destabilized relative to the same protein off the ribosome. This ribosome-dependent modulation of the energy landscape is dependent on the amino acid distance from the PTC. By the time the nascent chain is 55 residues away from the PTC (a spacer of 20–30 residues from the end of the exit tunnel), the global stability of the protein is no longer modulated by the presence of the ribosome. These results are consistent with, and provide a quantitative explanation for, several other observations of RNC behavior (6, 11, 12, 28). For instance, the point at which the nascent chain acquires its native stability as measured here is very similar to the distance from the PTC required for full acquisition of folded peaks by NMR (12). Furthermore, work on stalled IDPs suggests that the ribosome acts comparably to a denaturant on the emerging chain, which again is consistent with what we observe here for the folding energetics of RNCs (23).
It is particularly interesting to compare our results with those obtained by von Heijne and colleagues using an arrest peptide-mediated assay to assess the force generated by the nascent chain during translation (28, 29). For DHFR, they measured the maximum force generated by the emerging chain at approximately 45 aa away from the PTC, close to where we see a return to off-ribosome–like stability. How the force applied to the arrest peptide is related to the protein’s stability, folding trajectory, or folding rates, however, remains unclear. Our approach, which can be applied under various conditions and at a range of nascent chain lengths, should help shed light on the biophysical effects driving the peptide arrest assay.
Several other studies have implicated electrostatic forces in modulating nascent chain energy landscapes (11, 12, 23). The distance dependence of the stability changes that we observe, combined with the fact that the magnitude of destabilization is inversely correlated with the isoelectric point of the proteins that we measure, are consistent with this idea; however, further studies are needed to determine the physical basis of the destabilization observed here.
What role could general ribosomal destabilization play in ensuring folding fidelity in vivo? For the emerging chain, accessing the native state is not an option, simply because the full sequence of the protein is yet to be synthesized. However, hydrophobic collapse or other partially folded states are accessible to the incomplete nascent chain, and translation is certainly slow enough such that the incomplete nascent chain has sufficient time for the formation of such potential native and nonnative intermediates. Although some experiments have shown cotranslational folding to increase folding efficiency and speed, it is possible that intermediates that form cotranslationally may be off-pathway and result in nonnative, toxic species (7, 30, 31). To avoid such conformations, cells may have evolved chaperones, such as trigger factor, to “hold and unfold” proteins as they emerge. Indeed, trigger factor is known to bind to proteins approximately 60 aa away from the PTC, the same distance at which the destabilization that we observe is abrogated (26, 27). Perhaps the observed destabilization allows the nascent chain to fold more efficiently and avoid off-pathway kinetic traps. Therefore, destabilizing collapsed states that form as the nascent peptide emerges but before its entire sequence is accessible may help ensure that the protein forms a stable structure only after completely emerging from the ribosome.
Finally, our approach is adaptable as a tool to probe other differences between protein energy landscapes in vitro and in vivo. Although we believe this method to be widely applicable, when applying it to new systems, it is important to take into account the limitations of pulse proteolysis, including possible steric hindrance because of biomolecular interactions (16, 32). Nevertheless, it should be possible to extend our approach to determine how the ribosome modulates protein folding kinetics by monitoring unfolding rates via pulse proteolysis (33). In addition, given the seemingly important role of quality control at the ribosome in general cellular proteostasis (34), it will be important to know how RNC stability is altered in the presence of ribosome-associated chaperones such as trigger factor (in bacteria) or the ribosome quality control complex (in eukaryotes). Importantly, moving from more descriptive studies of RNCs to measuring their biophysical properties will enable both validation and extension of recent computational studies (35–37), as well as uncover general biophysical principles governing cotranslational folding.
Materials and Methods
Sample Preparation.
Fifty-microliter IVT reactions (PURExpress; New England Biolabs) were initiated by the addition of 500 ng of plasmid DNA in the presence of 2.5 µL of Flourotect Greenlys tRNA (Promega) and 2 μL of RNase inhibitor, murine (New England Biolabs). For pulse proteolysis off the ribosome or without a SecM sequence, samples were incubated for 1 h at 37 °C, followed by the addition of chloramphenicol to 2 mM and RNase A to a final concentration of 0.1 mg/mL These samples were incubated at room temperature overnight and then spun at 21,000 × g for 30 min at 4 °C. The resulting supernatant was used for pulse proteolysis.
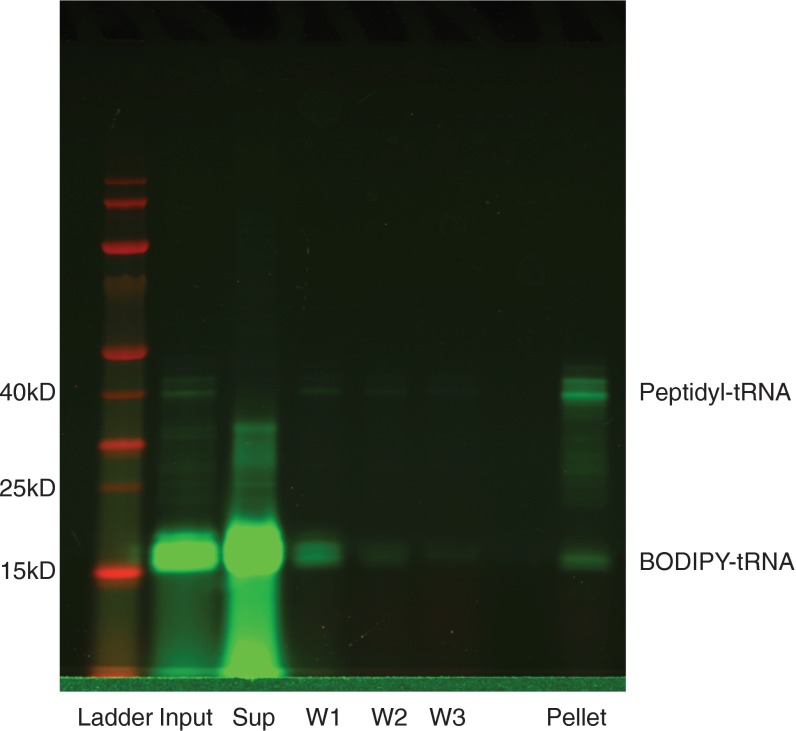
For RNCs, after incubation for 30 min at 37 °C, IVT reactions without release factors were loaded onto a 125-µL 1 M sucrose cushion in 25 mM HEPES pH 7.5, 15 mM MgOAc, 150 mM KCl, and 2 mM DTT (HKM+DTT), and centrifuged at 200,000 × g for 40 min at 4 °C. Supernatant was aspirated, and ribosome pellets were washed three times with 200 µL of HKM+DTT, then resuspended in 35 µL of HKM+DTT (Fig. S3).
Fig. S3.
Purification of labeled RNCs. IVT reactions (input) were loaded onto a sucrose cushion and centrifuged as described in Materials and Methods. Supernatant (sup) was aspirated, and the pellets were washed three times (W1, W2, and W3) with 200 μL of ice-cold HKM+DTT. Pellets were then resuspended, mixed with SDS/PAGE loading dye, and loaded on a gel.
Pulse Proteolysis.
For protein purified from E. coli, pulse proteolysis was conducted as described previously (16, 32) in HKM+DTT. For released or stalled nascent chains, 3 µL of halted IVT reactions or RNCs, respectively, were diluted into 7 µL of HKM+DTT and urea to the desired urea concentration. After incubation for at least 12 h, 1 µL of 6.8 mg/mL thermolysin was added to each 10 µL of reaction and 8 µL was quenched into 3 µL of 500 mM EDTA, pH 8.5. After pulse proteolysis, RNase A was added to 1 mg/mL to each reaction, followed by incubation at 37 °C overnight to digest any remaining peptidyl-tRNA. For IVT reactions off the ribosome, RNase A was added to a final concentration of 1 mg/mL, followed by incubation for 15 min at 37 °C. Samples were then mixed with SDS/PAGE loading dye and loaded onto 4–12% Bis-Tris gels (Thermo Fisher Scientific). Gels were run in MES buffer and imaged with a Typhoon laser scanner (GE Healthcare) using a 488-nm laser and 520BP filter. Analysis and quantification of gels was done using ImageJ as described previously (32). Urea concentrations were measured using a refractometer as described previously (32).
FCS.
RNCs with fluorescently labeled nascent chains were a gift from Madeleine Jensen. For experiments, they were diluted into appropriate urea concentrations and allowed to reach equilibrium overnight at room temperature in 1× HKM+DTT. FCS measurements and analysis were performed as described previously (38) fitting to a single species, using an additional term to correct for the triplet state. To control for effects of urea on optics and viscosity, diffusion of free Alexa Fluor 488 was measured at the same urea concentrations as the RNCs (Fig. S4). The measured Alexa Fluor 488 diffusion coefficients were then normalized to the 0 M urea coefficient to determine the viscosity. These values were used to calculate RNC diffusion coefficients.
Fig. S4.
Diffusion of Alexa Fluor 488 as a function of urea concentration. The diffusion of Alexa Fluor 488 was used to determine the viscosity of the solution when calculating diffusion coefficients of RNCs. Shown here is the apparent diffusion coefficient without correcting for changes in viscosity due to the presence of high concentrations of urea. Error bars are SDs of 15 experiments.
Sucrose Gradients.
The 70S ribosomes with no nascent chain were a gift from Jonas Noeske, University of California, Berkeley, and prepared as described previously (39). For RNCs, IVT reactions were quenched with chloramphenicol to a final concentration of 2 mM after preparation as described above, diluted to 100 µL to the desired urea concentrations, and incubated for at least 12 h at room temperature. Samples were then layered on top of 10–50% sucrose gradients and centrifuged in a SW41Ti rotor at 40,000 rpm for 3 h.
Acknowledgments
We thank K. Hamadani and the entire staff of the S.M. laboratory for advice throughout the project. We thank J. S. Fraser for comments on the manuscript and D. Weiner and C. Reidel for help with FCS data collection and analysis. This work was funded by the National Institutes of Health (Grants GM05945, to S.M., and R01-GM065050, to J.H.D.C.) and SynBERC (a National Science Foundation-funded engineering research center). M.K.J. is supported through National Science Foundation (NSF) Graduate Research Fellow Program (GRFP) Grant DGE1106400.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610272113/-/DCSupplemental.
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Clark PL. Protein folding in the cell: Reshaping the folding funnel. Trends Biochem Sci. 2004;29(10):527–534. doi: 10.1016/j.tibs.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16(6):574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 4.Braselmann E, Chaney JL, Clark PL. Folding the proteome. Trends Biochem Sci. 2013;38(7):337–344. doi: 10.1016/j.tibs.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabrita LD, Dobson CM, Christodoulou J. Protein folding on the ribosome. Curr Opin Struct Biol. 2010;20(1):33–45. doi: 10.1016/j.sbi.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Holtkamp W, et al. Cotranslational protein folding on the ribosome monitored in real time. Science. 2015;350(6264):1104–1107. doi: 10.1126/science.aad0344. [DOI] [PubMed] [Google Scholar]
- 7.Ugrinov KG, Clark PL. Cotranslational folding increases GFP folding yield. Biophys J. 2010;98(7):1312–1320. doi: 10.1016/j.bpj.2009.12.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sander IM, Chaney JL, Clark PL. Expanding Anfinsen’s principle: Contributions of synonymous codon selection to rational protein design. J Am Chem Soc. 2014;136(3):858–861. doi: 10.1021/ja411302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16(3):274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 10.Hsu S-TD, et al. Structure and dynamics of a ribosome-bound nascent chain by NMR spectroscopy. Proc Natl Acad Sci USA. 2007;104(42):16516–16521. doi: 10.1073/pnas.0704664104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser CM, Goldman DH, Chodera JD, Tinoco I, Jr, Bustamante C. The ribosome modulates nascent protein folding. Science. 2011;334(6063):1723–1727. doi: 10.1126/science.1209740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabrita LD, et al. A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding. Nat Struct Mol Biol. 2016;23(4):278–285. doi: 10.1038/nsmb.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17(4):263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- 14.Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353(2):459–473. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Parsell DA, Sauer RT. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J Biol Chem. 1989;264(13):7590–7595. [PubMed] [Google Scholar]
- 16.Park C, Marqusee S. Pulse proteolysis: A simple method for quantitative determination of protein stability and ligand binding. Nat Methods. 2005;2(3):207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 17.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4(10):2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallam AL, Jackson SE. Knot formation in newly translated proteins is spontaneous and accelerated by chaperonins. Nat Chem Biol. 2011;8(2):147–153. doi: 10.1038/nchembio.742. [DOI] [PubMed] [Google Scholar]
- 19.Mitra K, et al. Elongation arrest by SecM via a cascade of ribosomal RNA rearrangements. Mol Cell. 2006;22(4):533–543. doi: 10.1016/j.molcel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Gabler R, Westhead EW, Ford NC. Studies of ribosomal diffusion coefficients using laser light-scattering spectroscopy. Biophys J. 1974;14(7):528–545. doi: 10.1016/S0006-3495(74)85933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garvey EP, Matthews CR. Effects of multiple replacements at a single position on the folding and stability of dihydrofolate reductase from Escherichia coli. Biochemistry. 1989;28(5):2083–2093. doi: 10.1021/bi00431a018. [DOI] [PubMed] [Google Scholar]
- 22.Spudich GM, Miller EJ, Marqusee S. Destabilization of the Escherichia coli RNase H kinetic intermediate: Switching between a two-state and three-state folding mechanism. J Mol Biol. 2004;335(2):609–618. doi: 10.1016/j.jmb.2003.10.052. [DOI] [PubMed] [Google Scholar]
- 23.Knight AM, et al. Electrostatic effect of the ribosomal surface on nascent polypeptide dynamics. ACS Chem Biol. 2013;8(6):1195–1204. doi: 10.1021/cb400030n. [DOI] [PubMed] [Google Scholar]
- 24.Eichmann C, Preissler S, Riek R, Deuerling E. Cotranslational structure acquisition of nascent polypeptides monitored by NMR spectroscopy. Proc Natl Acad Sci USA. 2010;107(20):9111–9116. doi: 10.1073/pnas.0914300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomic S, Johnson AE, Hartl FU, Etchells SA. Exploring the capacity of trigger factor to function as a shield for ribosome bound polypeptide chains. FEBS Lett. 2006;580(1):72–76. doi: 10.1016/j.febslet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann A, et al. Trigger factor forms a protective shield for nascent polypeptides at the ribosome. J Biol Chem. 2006;281(10):6539–6545. doi: 10.1074/jbc.M512345200. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann A, et al. Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding. Mol Cell. 2012;48(1):63–74. doi: 10.1016/j.molcel.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Nilsson OB, et al. Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep. 2015;12(10):1533–1540. doi: 10.1016/j.celrep.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson OB, Müller-Lucks A, Kramer G, Bukau B, von Heijne G. Trigger factor reduces the force exerted on the nascent chain by a cotranslationally folding protein. J Mol Biol. 2016;428(6):1356–1364. doi: 10.1016/j.jmb.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 30.Frydman J, Erdjument-Bromage H, Tempst P, Hartl FU. Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat Struct Biol. 1999;6(7):697–705. doi: 10.1038/10754. [DOI] [PubMed] [Google Scholar]
- 31.Deckert A, et al. Structural characterization of the interaction of α-synuclein nascent chains with the ribosomal surface and trigger factor. Proc Natl Acad Sci USA. 2016;113(18):5012–5017. doi: 10.1073/pnas.1519124113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park C, Marqusee S. 2006. Quantitative determination of protein stability and ligand binding by pulse proteolysis. Curr Protoc Protein Sci Chapter 20:Unit 20.11.
- 33.Na Y-R, Park C. Investigating protein unfolding kinetics by pulse proteolysis. Protein Sci. 2009;18(2):268–276. doi: 10.1002/pro.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choe Y-J, et al. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature. 2016;531(7593):191–195. doi: 10.1038/nature16973. [DOI] [PubMed] [Google Scholar]
- 35.O’Brien EP, Vendruscolo M, Dobson CM. Prediction of variable translation rate effects on cotranslational protein folding. Nat Commun. 2012;3:868. doi: 10.1038/ncomms1850. [DOI] [PubMed] [Google Scholar]
- 36.Sharma AK, Bukau B, O’Brien EP. Physical origins of codon positions that strongly influence cotranslational folding: A framework for controlling nascent-protein folding. J Am Chem Soc. 2016;138(4):1180–1195. doi: 10.1021/jacs.5b08145. [DOI] [PubMed] [Google Scholar]
- 37.O’Brien EP, Christodoulou J, Vendruscolo M, Dobson CM. New scenarios of protein folding can occur on the ribosome. J Am Chem Soc. 2011;133(3):513–526. doi: 10.1021/ja107863z. [DOI] [PubMed] [Google Scholar]
- 38.Riedel C, et al. The heat released during catalytic turnover enhances the diffusion of an enzyme. Nature. 2015;517(7533):227–230. doi: 10.1038/nature14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noeske J, et al. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 2015;22(4):336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]