Significance
We found that gene mutations/deletions are frequent in mesothelioma and occur through a variety of DNA alterations. We identified genes implicated in malignant mesothelioma: SETD2, SMARCC1, PBRM1. Previous next-generation studies (NGS) underestimated the frequency of genetic alterations in malignant mesothelioma because NGS mainly identifies nucleotide level mutations. Our findings are of general relevance to the field of cancer research that relies almost exclusively on NGS to identify gene alterations in cancer biopsies, and uses this information to design specific molecular therapies. An integrated approach that includes a high-density comparative genomic hybridization array and NGS or targeted-NGS, as conducted here, may reveal additional genes that are inactivated by mechanisms other than point mutations. This information may inform us on how to design more effective molecular therapies.
Keywords: mesothelioma, BAP1, SETD2, SMARCC1, PBRM1
Abstract
We used a custom-made comparative genomic hybridization array (aCGH; average probe interval 254 bp) to screen 33 malignant mesothelioma (MM) biopsies for somatic copy number loss throughout the 3p21 region (10.7 Mb) that harbors 251 genes, including BRCA1 (breast cancer 1)-associated protein 1 (BAP1), the most commonly mutated gene in MM. We identified frequent minute biallelic deletions (<3 kb) in 46 of 251 genes: four were cancer-associated genes: SETD2 (SET domain-containing protein 2) (7 of 33), BAP1 (8 of 33), PBRM1 (polybromo 1) (3 of 33), and SMARCC1 (switch/sucrose nonfermentable- SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily c, member 1) (2 of 33). These four genes were further investigated by targeted next-generation sequencing (tNGS), which revealed sequence-level mutations causing biallelic inactivation. Combined high-density aCGH and tNGS revealed biallelic gene inactivation in SETD2 (9 of 33, 27%), BAP1 (16 of 33, 48%), PBRM1 (5 of 33, 15%), and SMARCC1 (2 of 33, 6%). The incidence of genetic alterations detected is much higher than reported in the literature because minute deletions are not detected by NGS or commercial aCGH. Many of these minute deletions were not contiguous, but rather alternated with segments showing oscillating copy number changes along the 3p21 region. In summary, we found that in MM: (i) multiple minute simultaneous biallelic deletions are frequent in chromosome 3p21, where they occur as distinct events involving multiple genes; (ii) in addition to BAP1, mutations of SETD2, PBRM1, and SMARCC1 are frequent in MM; and (iii) our results suggest that high-density aCGH combined with tNGS provides a more precise estimate of the frequency and types of genes inactivated in human cancer than approaches based exclusively on NGS strategy.
Malignant mesotheliomas (MMs) are highly aggressive adult malignancies that arise from the mesothelial cells covering the pleural, peritoneal, and pericardial cavities. MMs are often caused by exposure to asbestos and other mineral fibers (1). Cofactors and genetic predisposition may also cause MM, especially upon asbestos exposure (2–4). Diagnosis is usually established at a late stage, when MMs are most resistant to medical and surgical therapies. Recent clinical trials targeting molecular pathways have failed to significantly improve MM prognosis (1). Promising biomarkers are being investigated for early detection (5, 6). It is hoped that a better understanding of the molecular pathogenesis of MM will lead to more specific and effective targeted therapies (7).
Using next-generation sequencing (NGS) for exome sequencing involves significant—and at times overlooked—challenges, including DNA quality and quantity, tumor heterogeneity, and the difficulty to detect a wide variety of complex genetic mutations (8). Conventional NGS studies revealed that the most commonly mutated genes in MMs are BRCA1-associated protein 1 (BAP1), CDKN2A (cyclin-dependent kinase inhibitor 2a), and NF2 (neurofibromin 2) (9–12). The reported frequency of BAP1 inactivation, the most commonly mutated gene in MM, using NGS or Sanger sequencing, was between 20% and 30% (9–15).
Overall, NGS studies revealed that each single MM biopsy has its own set of specific mutations, and that driver mutations—except for BAP1—are rare (reviewed in ref. 16). For example, Guo et al. (9) conducted NGS exome sequencing of the whole genome in 22 MM frozen biopsies: they found 490 mutated genes, of which 477 (97%) were mutated only in one biopsy, and found an average of 23 mutations per biopsy (range 2–51). Mutations in human cancers range from as low as one base substitution per exome (<0.1 per megabase) in some pediatric malignancies, to thousands of mutations per exome (∼100 per megabase) in adult malignancies, such as lung cancer and melanoma (17). Thus, the low level of mutations detected by NGS in MM (9, 11, 18) was unexpected and highly unusual. However, past cytogenetic studies revealed that MMs have multiple numerical and structural chromosomal abnormalities (19), findings indicating that genetic alterations in MMs should be frequent, not rare. In support of this hypothesis, comparative genomic hybridization array (aCGH) and multiple ligation-dependent probe amplification studies revealed that several BAP1 deletions were missed by NGS and Sanger sequencing, because of their large size (20, 21).
Large DNA deletions and copy number (CN) changes are best detected by aCGH and SNP arrays. MM CN analyses using whole-genome aCGH and SNP arrays revealed the presence of occasional large genomic deletions (22–26); however, the presence of smaller deletions [i.e., minute deletions (i.e., <3 kb)] has not been specifically investigated. Deletions in that size range fall in a gray area: they are too large to be reliably detected by NGS and Sanger sequencing and too small to be reliably detected by commercial aCGH. We reasoned that such hypothetical minute deletions would not be detected by conventional whole-genome aCGH, as these arrays have only an average of 400,000–1,000,000 probes: because the human genome has over 3 billion base pairs, commercial aCGH provide only one probe every 3,000–8,000 bp (for example, the Agilent CGH array). Whole-exome NGS and targeted NGS (tNGS) sequencing are sensitive to detect minute deletions, but these techniques may produce false-positive results because they are not designed to measure DNA CN (8). Thus, it is not possible to draw definitive conclusions about CN changes relying exclusively on NGS technology, and the NGS results should always be validated by integrated genomic approaches (8).
Here, we hypothesized and tested whether, in addition to the relatively infrequent single-nucleotide mutations detected by NGS studies, larger DNA alterations, including minute deletions, might contribute to inactivate additional tumor-suppressor genes in MM. To screen for the presence of possible minute deletions, we designed a custom-made high-density oligonucleotide aCGH. We selected a specific region of the DNA that we would study in detail: we focused on the 3p21 region, where the BAP1 gene resides. To detect point mutations, we complemented this approach with tNGS (>150 reads) for four tumor-suppressor genes that were found to contain minute deletions by high-density aCGH. tNGS was chosen over conventional NGS exome sequencing of the whole genome, because by targeting a few genes (rather than the over 20,000 genes present in the human genome) it was possible to increase the depth (i.e., number of reads) and the accuracy of the results. We found that this combined approach was more sensitive and specific than NGS-exome–only based approaches to capture the variety of genetic alterations that are present in MM, and likely in other malignancies.
Results
Frequent Biallelic Deletions of Gene Clusters Involving SETD2, SMARCC1, BAP1, and PBRM1 Genes on 3p21.
We built a custom high-density oligonucleotide aCGH with probes designed at an average interval of 254 bp to screen the entire 3p21 region (10.7 Mb) that holds 251 different genes (Table S1). We estimated that we had roughly 30–200 probes per gene and 3–5 probes per each exon region. The density of probes was 12- to 32-times higher than in commercial aCGH. We analyzed a total of 33 MM DNA samples (Table S2). Competitive hybridization was carried out between tumor genome DNA and the matching control normal DNA (Materials and Methods and SI Materials and Methods).
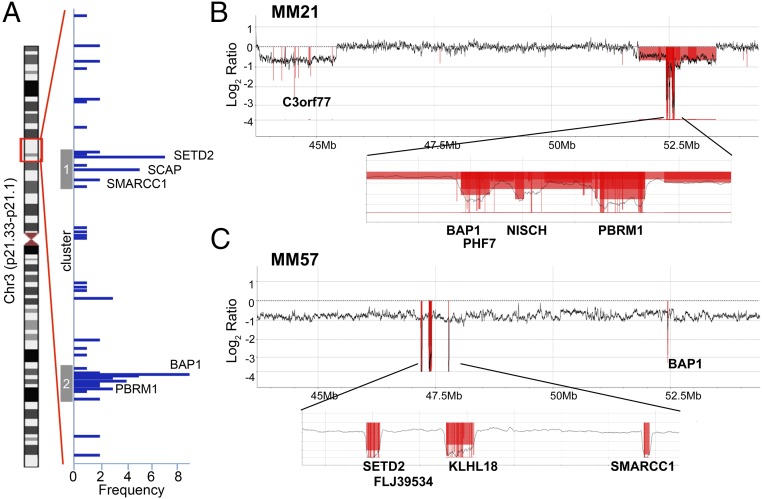
Using our custom-made high-density aCGH, we were able to identify deletions that would have been missed using conventional aCGH analysis (Figs. S1 and S2A). In total, we identified biallelic minute deletions (<3 kb) or larger deletions involving some parts of the 3p21 region in 19 of 33 MMs (Fig. 1A and Table S3). These deletions were concentrated into two DNA regions: a telomeric region at 47–48 Mb (cluster 1) and a centromeric region at 52.2–53.2 Mb (cluster 2) (Fig. 1A). Representative deletions are shown in Fig. 1 B and C and in Fig. S2. Fig. 1B displays sample MM21, showing loss of heterozygosis (LOH) in two regions (43.8–45.4 Mb and 51.9–53.5 Mb). Within the centromeric region, we found biallelic deletions in three noncontiguous regions in gene cluster 2: (i) deletion of the BAP1 gene (BAP1 exons 1–16 and its adjacent PHF7 (PHD finger protein 7) gene; (ii) deletion of the NISCH (nischarin) gene (exons 3–15); and (iii) deletion of the PBRM1 gene (exons 22–30). Fig. 1C shows sample MM57 that contained LOH of the entire 3p21 region. We found biallelic deletions in three noncontiguous regions in telomeric cluster 1: (i) deletion of SETD2 (exon 1) and its adjacent FLJ39534 gene (exons 1–2); (ii) deletion of KLHL18 (kelch like family member 18) (exons 2–10); and (iii) deletion of SMARCC1 (exons 3–5). Moreover, we found biallelic deletions in the centromeric cluster 2, including BAP1 (exons 11–17).
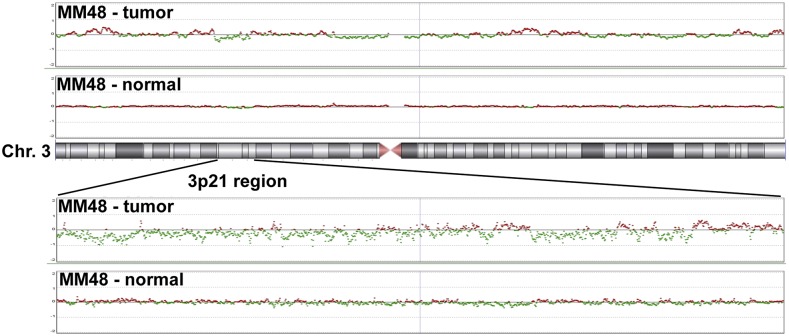
Fig. S1.
High-density aCGH is superior to conventional aCGH: a representative MM sample. MM48 shows monoallelic deleted regions using conventional aCGH.
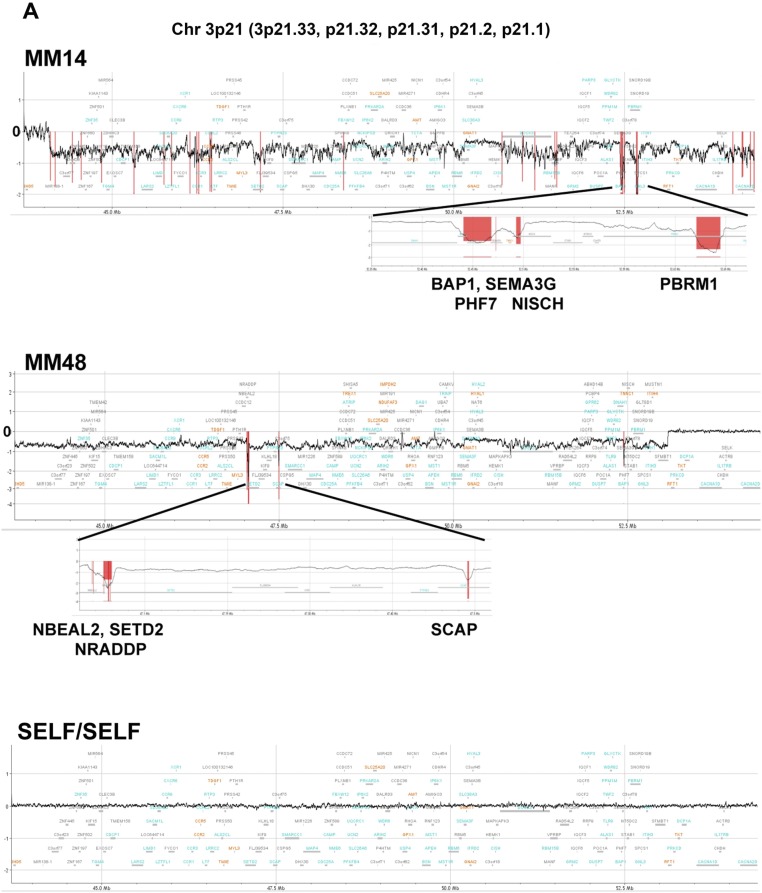
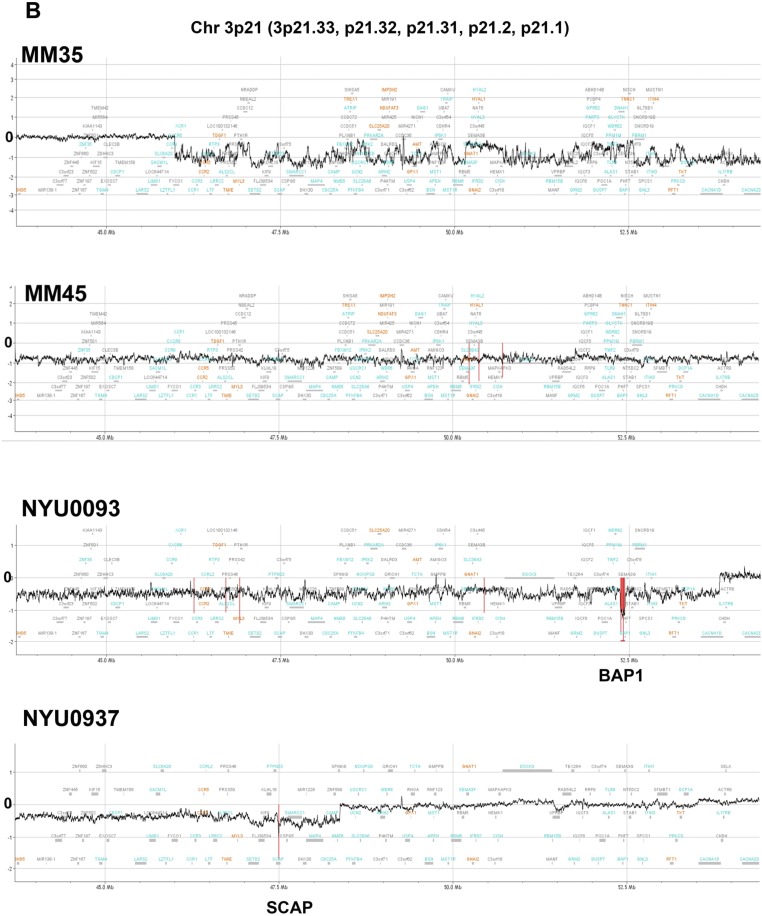
Fig. S2.
CN loss in 3p21 region, detected by high-density aCGH in representative MM samples. The regions judged to be biallelically deleted were marked in a red color and the length of these markers indicates the average log ratio of the consecutive probes in the deleted region. (A) MM14, MM48, SELF/SELF (technical control), which indicates that the same genomic DNA, HapMap Male (NA12891), was used for both Cy5 and Cy3 labeling in the high-density aCGH analysis. (B) MM35, MM45, NYU0093, NYU0937.
Fig. 1.
Segmental CN loss detected in 3p21 by high-density aCGH. (A) Frequency of biallelic deletion detected among 251 genes in 3p21 in 33 MMs. On the vertical-axis, genes are aligned along the genome location. The blue horizontal bars represent each of the 46 genes and the length of the bar represents the case frequency that showed biallelic deletions. (B and C) Representative profiles of CN change in MM21 and MM57, respectively. CN is shown as log2 ratio and the regions showing biallelic deletion are marked in red.
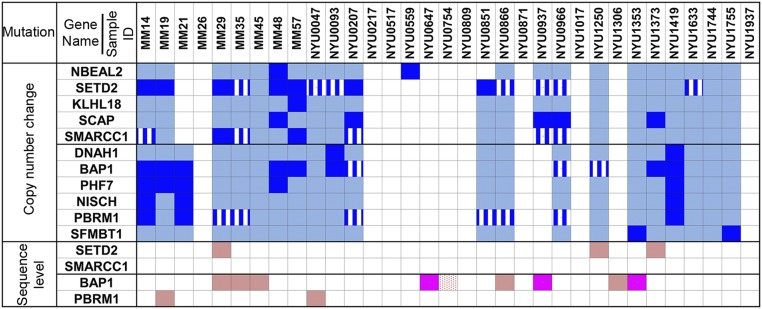
Overall, a total of 46 genes were involved, at least once, in these deletion events (Fig. 1A and Table S3). Most of these 46 deletions displayed complex noncontiguous biallelic deletion patterns, and simultaneous deletions involving two or more genes were observed frequently. The most frequently mutated genes, comprising clusters 1 and 2, are shown in Fig. 2. Among the mutated genes, four genes previously associated with cancer contained frequent biallelic deletions; that is, SETD2 (biallelic deletion frequency: 7 of 33, 21%), SMARCC1 (2 of 33, 6%), BAP1 (8 of 33, 24%), and PBRM1 (3 of 33, 9%) (Figs. 1A and 2). These four genes were further investigated by tNGS, as described below. Among the remaining 42 genes that were found deleted (summarized in Table S3), the most frequently found were: SCAP (SREBP-sterol regulatory element binding transcription factor 1-cleavage activating protein) (5 of 33, 15%), PHF7 (5 of 33), NISCH (3 of 33), SEMA3G (semaphorin 3G) (3 of 33), and FLJ39534 (3 of 33) genes. Several MMs (15 of 33, 45%) contained combined biallelic deletions of two or more genes in 3p21 detectable by aCGH (Table S3). Break points of these genome deletions were different among cases, except for a particular deletion in intron 1 of the SCAP gene, in which the deletion region (chr3: 47.49–47.5 Mb) coincided with the polymorphic CN-variable region in the human population (Database of Genomic Variants); thus, this was no longer investigated (representative samples shown in Fig. 1 and Fig. S2). Finally, the 3p21.3 region containing the tumor-suppressor gene cluster—which includes RBM5, TUSC2, HYAL1, and HYAL2—which is frequently deleted in several human tumors (27), did not show any biallelic deletions in MM.
Fig. 2.
Summary of genomic alterations in 33 malignant mesothelioma cases. Each column indicates genomic alterations in each case. Genes NBEAL2, SETD2, KLHL18, SCAP, and SMARCC1 comprise gene cluster 1; genes DNAH1, BAP1, PHF7, NISCH, PBRM1, and SFMBT1 comprise gene cluster 2. Biallelic deletion detected by both, high-density aCGH array and tNGS, are shown in dark blue. Monoallelic loss detected by both, high-density aCGH array and tNGS, are shown in light blue. Blue with white lines represents biallelic deletion detected by tNGS only. Bright pink: two separate somatic mutations of the same gene detected by tNGS at sequence level; light pink: one somatic mutation detected by tNGS at sequence level; dotted pink: one germ-line and one somatic mutation detected by tNGS at sequence level.
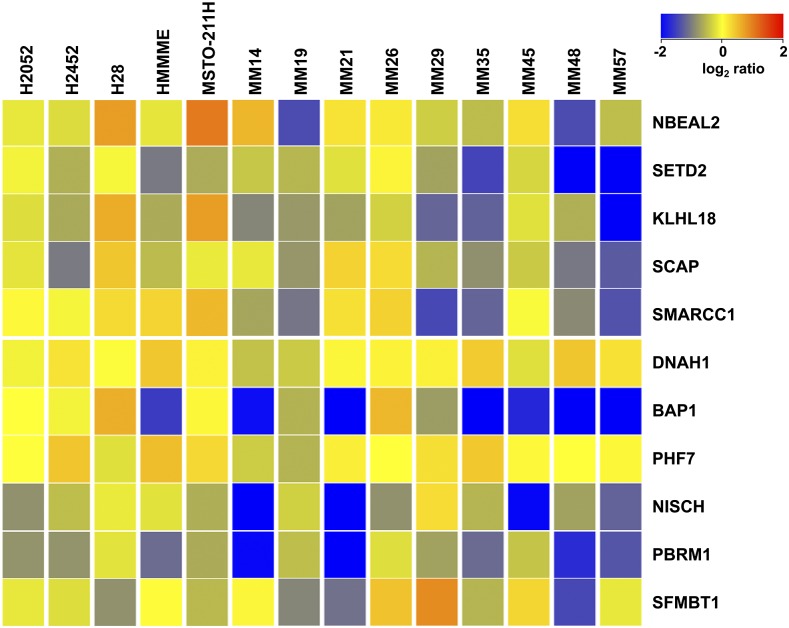
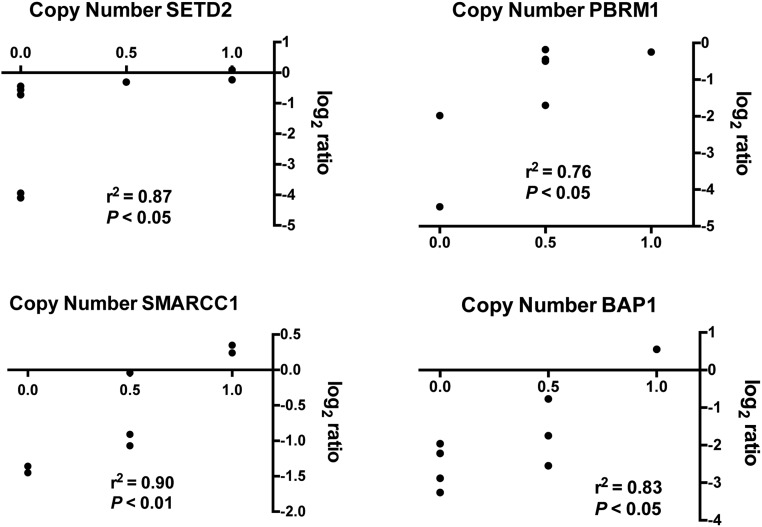
Monoallelic gene loss was also a frequent event: for example, SETD2 (monoallelic deletion frequency: 7 of 33, 21%), SMARCC1 (14 of 33, 42%), BAP1 (10 of 33, 30%), and PBRM1 (12 of 33, 36%) (Fig. 2). Deregulated mRNA expression of these genes was confirmed in a panel of MM cell lines and in a subset of MM samples (Fig. S3); aCGH and mRNA data showed a significant positive correlation, suggesting that monoallelic gene loss was associated with reduced transcript levels (Fig. S4).
Fig. S3.
Heap map showing relative log2 ratio of mRNA expression in five MM cell lines and nine MM clinical samples. Both MM lines (H2052, H2452, H28, HMMME, MSTO-211H) and MM clinical samples (MM14, 19, 21, 26, 29, 35, 45, 48, 57) show deregulated mRNA levels of 3p21 genes.
Fig. S4.
Correlation between high-density aCGH data and mRNA expression. Statistically significant correlation between high-density aCGH data and mRNA expression for SETD2, PBRM1, SMARCC1, and BAP1. For the x axis: 0.0 for biallelic deletion; 0.5 for monoallelic deletion; 1.0 for absence of CN variation. Monoallelic losses often presented intermediate levels of mRNA expression.
Validation of DNA CN Analysis of Selected Genes SETD2, BAP1, PBRM1, and SMARCC1 on 3p21 by tNGS Sequencing.
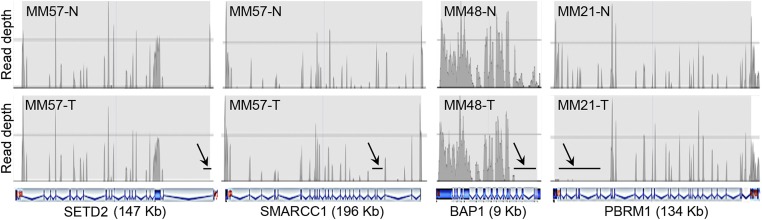
SETD2, BAP1, PBRM1, and SMARCC1 were further investigated by tNGS. In these studies, the read depth was 150–200 for each target gene region, ensuring accuracy of the final sequence. The read depth was measured and compared by pair analysis between tumor DNA and its normal counterpart DNA from the same patient. All of the biallelic deletions detected by aCGH were consistently detected, and thus validated, by tNGS (Fig. 3 and Table S4). Moreover, tNGS analysis identified additional minute biallelic deletions that were not detected by aCGH, at frequencies of 7 of 33 MMs (21%) for SETD2, 3 of 33 (9%) for BAP1, 6 of 33 (18%) for PBRM1, and 5 of 33 (15%) for SMARCC1 (Fig. 2 and Table S4). However, because these deletions were either hidden by nonnegligible noise of aCGH or were very small, they could not be independently validated by high-density aCGH: therefore, we chose a prudent approach and they were not further considered.
Fig. 3.
CN loss detected in SETD2, SMARCC1, BAP1, and PBRM1 by tNGS. The read depth of each target region was measured and compared by paired analysis between tumor DNA and its normal DNA extracted from blood of the same case. (Upper) The read patterns of the normal DNA for the indicated genes; (Lower) read patterns of the tumor DNAs. Arrows and black horizontal bars indicate areas showing no read corresponding to deleted regions of the gene. Schematics of introexon patterns, characteristic of each gene, shown in blue, are beneath the sample profiles. MM57 shows deletions in exon 1 of SETD2 and exons 4–5 of SMARCC1; MM48 shows deletions in exons 1–5 of BAP1 genes; MM21 shows deletions in exons 22–30 of PBRM1 gene. Estimated CNs are reported in Table S4.
Analysis of Sequence-Level Mutations in SETD2, PBRM1, BAP1, and SMARCC1.
tNGS of these same tumors and normal DNAs detected somatic point mutations—which, as expected, were undetectable by aCGH—of these genes. SETD2 point mutations were found in three MMs (3 of 33, 9%): p.S1024fs (MM29), p.K1948X (NYU1250), and p.T1753fs (NYU1373), (Table 1). BAP1 point mutations were found in nine MMs (9 of 33, 27%); that is, p.S460X (MM35), p.W202X (MM45), p.V234- (MM29), c.2278–2283 del/splice site del (NYU0866), p.I72fs (NYU0754), and p.E212X (NYU1306). Three of these nine MMs had two separate inactivating BAP1 somatic sequence-level mutations: p.S341fs and p.O280X (NYU0647), p.I499fs and p.V234fs (NYU0937), and p.Y646X and p.A648fs (NYU1353). PBRM1 point mutations were identified in two MMs (2 of 33, 6%): p.V1582_1582del-stop lost (MM19) and p.Q235X (NYU0047). We did not detect any nucleotide mutation in the SMARCC1 gene. In addition, germ-line variants, each one in these four genes, were found; p.T1033A of SETD2 in MM case NYU0851, p.M1486I of PBRM1 and p.P1075H of SMARCC1 in MM35, and the splice site (NM_004656:c.438-2A > G) of BAP1 in NYU0754. Using tNGS, we were therefore able to identify both the germ-line and somatic mutation present in sample NYU0754, previously published as W-III-04 (14).
Table 1.
Mutations detected by tNGS
| Sample name | Mutation in gDNA (hg19) | cDNA change | Protein change |
| BAP1 (NM_004656) | |||
| MM29 | chr3:52440349, GCAC/G | c.700_702del | p.V234- |
| MM35 | chr3:52437782, G/C | c.C1379G | p.S460X |
| MM45 | chr3:52440899, C/T | c.G605A | p.W202X |
| NYU0647 | chr3:52439218,TG/T and chr3:52439874,G/A | c.1023delC and c.C838T | p.S341fs and p.Q280X |
| NYU0866 | chr3:52436599–52436627, 29 bp del | c.2274–2283del | SPLICE_SITE |
| NYU0937 | chr3:52437663, CG/C and chr3:52440351, AC/A | c.1497delC and c.700delG | p.I499fs and p.V234fs |
| NYU1306 | chr3:52440870, C/A | c.G634T | p.E212X |
| NYU1353 | chr3: 52436840 A/C and chr3: 52436818, 18bp del | c.T2165G and c.2170–2187del | p.Y646X and p.A648fs |
| NYU0754 | chr3:52441334,T/C* and chr3: 52442507–52442531, 25 bp del | intron6:c.438-2A > G and c.214_238del | SPLICE_SITE and p.I72fs |
| SETD2 (NM_014159) | |||
| MM29 | chr3:47163049, TGACCA/T | c.3072_3076del | p.S1024fs |
| NYU1250 | chr3:47125428, T/A | c.A5842T | p.K1948X |
| NYU1373 | chr3:47129622, G/GT | c.5258_5259insA | p.T1753fs |
| NYU0851 | chr3:47163029, T/C* | c.A3097G | p.T1033A |
| PBRM1 (NM_018313) | |||
| MM19 | chr3:52582082, AAC/A | c.4744_4745del | p.V1582_1582del |
| NYU0047 | chr3:52685769, G/A | c.C703T | p.Q235X |
| MM35 | chr3:52584555, C/G* | c.G4458C | p.M1486I |
| SMARCC1 (NM_003074) | |||
| MM35 | chr3:47629793, G/T* | c.C3224A | p.P1075H |
Germ-line variant; italic: point mutation did not cause biallelic deletion of the gene.
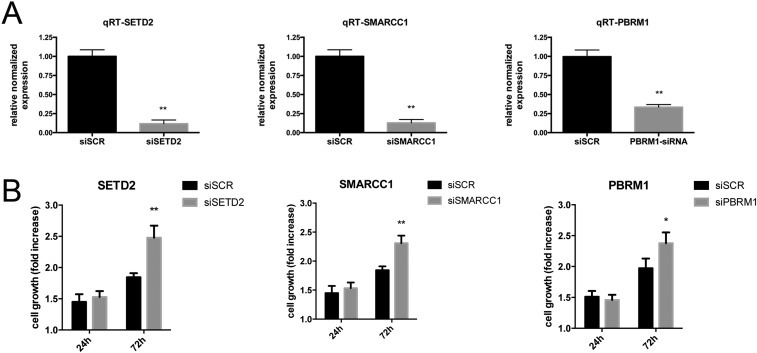
In total, combining all minute alterations and point mutations we confirmed biallelic inactivation of SETD2, BAP1, PBRM1, and SMARCC1 in at least 27% (9 of 33), 48% (16 of 33), 15% (5 of 33), and 6% (2 of 33) of 33 MMs, respectively (Fig. 2). Tissue-culture experiments revealed that silencing of SETD2, SMARCC1, or PBRM1 using siRNAs caused increased proliferation in a MM cell line wild-type for all of these three genes. These findings support the hypothesis that deletions of these genes are biologically relevant (Fig. S5).
Fig. S5.
Silencing of SETD2, SMARCC1, or PBRM1 is associated to increased cell proliferation. (A) PBRM1, SETD2, and SMARCC1 genes were efficiently silenced in PPM-Mill cells at 72 h using siRNAs. (B) At the same time point, viability of silenced cells was significantly higher than control. Data were compared using Mann–Whitney test; *P < 0.05, **P < 0.01.
We also investigated patient information for possible correlations between gene alterations and survival (patients with wild-type or monoallelic deletions vs. patients with biallelic inactivations) using the Kaplan–Meier method stratified by stage. The results were not significant probably because of the small sample size.
SI Materials and Methods
Survival Statistics.
Kaplan–Meier survival estimates were performed using the LIFETEST procedure in the SAS 9.4 software (SAS Institute). We compared patients with at least one wild-type copy of each gene (wild/wild, wild/–, and wild/variant) to patients without any wild-type copy (−/−, –/variant, and variant/variant), stratified by stage (1–2 vs. 3–4).
Data Analysis of High-Density Custom-Made CGH Microarray.
Data analysis was performed using Agilent CytoGenomics 3.0.5.1. Normalization parameters were set on the GC correction (window size = 2 kb) and diploid peak centralization; the aberration detection was using algorithm ADM-2; and for the aberration filter, the minimum number of probes in regions was set to three and the minimum absolute average log-ratio of region was set to each array. The setting of this log ratio was important to judge whether the deletion in the region was monoalleic or biallelic, and the average log ratio per one allele was obtained using the normalized log-ratio distribution.
Cell Line and in Vitro Experiments.
The PPM-Mill cell line was established from a surgically resected human MM specimen, and was characterized and provided by H.I.P. (New York University, New York). This cell line was selected for in vitro experiments because it is wild-type for PBRM1, SETD2, and SMARCC1. Of these PPM-Mill cells, 2 × 103 cells were plated per well in a 96-well plate and grown in DMEM + 1% (vol/vol) FBS. PBRM1 (sc-76075), SETD2 (Dharmacon d-012448-01), SMARCC1 (Dharmacon d-010813-01), or scrambled control siRNAs (sc-37007, d-001810-10-05) were transfected using HiPerfect Transfection Reagent (Qiagen, cat. no.: 301705) according to the manufacturer’s conditions. To evaluate the level of silencing, total RNA was isolated from the cultured cells with TRIzol reagent (Invitrogen) and quantified using microspectrophotometry (Nano-Drop Technologies). Total RNA was reverse-transcribed using the high-capacity cDNA reverse-transcription kit (Invitrogen) according to the manufacturer’s protocols. Quantitative RT-PCR was performed with TaqMan probes and TaqMan universal PCR Master Mix (Invitrogen). The amplification was carried out with an ABI PRISM 7900 RT-PCR System. Triplicate assays were performed with RNA samples isolated from at least two independent experiments. Fold-changes in gene expression were calculated using the ΔΔCt method using β-actin as the normalization control. Results from cells transfected with siRNAs for one of the 3p21 genes were then normalized to those obtained with cells transfected with their respective scrambled control siRNA (siSCR). Growth of siPBRM1-, siSETD2-, siSMARCC1-, and siSCR-transfected cells was determined using the Alamar blue assay according to the manufacturer’s protocol (AlamarBlue, Thermo Fisher). The assay was performed in five replicates from at least two independent experiments.
Gene Microarray Expression.
Gene-expression profiling was performed with Affymetrix Human Genome U133 Plus 2.0 arrays following the manufacturer's instructions. Raw data (CEL files) were summarized using the “plier16” algorithm and quantile normalization was performed by GeneSpring GX11.0.2 (Agilent Technologies). Gene-expression levels were presented compared with the average of three reactive mesothelial cells as log-transformed.
Discussion
We screened for genetic alterations of chromosome 3p21 in 33 MM using a high-density aCGH, followed by tNGS sequencing to validate the alterations detected and to investigate for nucleotide sequence mutations. We found multiple biallelic genome rearrangements involving 46 genes on 3p21. Many of these deletions were not contiguous, but rather they alternated along normal DNA segments, as in chromothripsis (28), although sequential independent deletion events cannot be entirely ruled out. Using high-density custom aCGH, we found frequent minute deletions of BAP1, SETD2, and PBRM1—the latter two genes had previously been found rarely mutated in MM (11, 29)—and of SMARCC1, not previously associated with MM. tNGS independently validated the deletions detected by high-density aCGH. In addition to minute biallelic deletions, tNGS revealed single-nucleotide inactivating mutations (i.e., truncating mutations) of SETD2, PBRM1, and BAP1 in several MMs. Although we detected CN alterations in SMARCC1, no nucleotide level mutations were found in this gene, confirming previous studies based on NGS (9–12) and underscoring the limitations of this technique when used alone. Together, the combination of high-density aCGH and tNGS detected a much higher percentage of genetic alterations in BAP1, SETD2, PBRM1, and SMARCC1 than reported in the MM literature, which is largely based on NGS sequencing (9–12). This is because NGS sequencing is a technique that is effective at identifying nucleotide mutations, but is not optimized for the identification of minute or larger chromosomal deletions (8). Moreover, minute genome changes are unlikely to be detected using commercial SNP arrays or whole-genome array CGH. Thus, an important implication of our study is that conventional NGS approaches are insufficient to identify reliably all of the different genetic alterations that occur in MM, and most likely in other cancers. For example, using a NGS approach, Bueno et al. (11) and Ugurluer et al. (12) reported inactivating nucleotide mutations of BAP1 and SETD2 in 20–30% and 8% of MMs, respectively. Instead, when combining high-density aCGH (to detect minute deletions of <3 kb) and tNGS (to detect nucleotide level mutations), we found that BAP1 and SETD2 were inactivated—either by minute biallelic deletion or LOH combined with point mutations—in 48% and 27% of MMs, respectively. Previous NGS studies underestimated the frequency of SETD2 deletions in MM because minute deletions, which are not reliably detected by NGS, were the most common deletions we found in this gene (i.e., seven of nine MM biopsies). Similarly, we detected biallelic deletions for PBRM1 in 9% of MMs and of SMARCC1 in 6% of MMs that were either underestimated or missed by previous NGS studies. Therefore, our data indicate that an integrated aCGH and tNGS approach is more accurate than NGS based-approaches to identify the different types of gene alterations present in human cancers.
Alterations of SETD2, PBRM1, and SMARCC1 have been linked to several human malignancies (30–39). Briefly, disruptions of the SETD2 gene have been frequently observed in clear cell renal cell carcinomas (ccRCC), gliomas, chronic lymphocytic leukemia, breast fibroepithelial tumors, gastro-intestinal stromal tumors, and melanomas (30–37). PBRM1 is one of the most frequently mutated genes in ccRCC (31), and its alterations were also found in cholangiocarcinomas and liver cancers (38). Moreover, down-regulated PBRM1 expression was positively correlated with tumor stage and the overall survival in breast cancer patients (40). SMARCC1 has been directly shown to be a tumor-suppressor gene in colorectal and ovarian cancer models (41). Additionally, we found that silencing of these genes in a MM cell line results in increased proliferation, as expected for tumor-suppressor genes.
Because NGS and tNGS may overestimate the presence of biallelic deletions (8), only those deletions independently validated by both high-density aCGH and tNGS were included in the final results. Thus, it is possible that our results may underestimate the true frequency of biallelic deletions in BAP1, SETD2, PBRM1, and SMARCC1. Despite our conservative approach, we found a much higher frequency of gene alterations than the frequencies described in the published literature. Thus, our data suggest that we may need to reconsider the hypothesis that MMs are low mutation-frequency malignancies, as we and others had proposed based on NGS studies and Sanger sequencing (9–11).
The implications of our findings may be of general relevance to cancer researchers that currently focus almost exclusively on NGS studies to identify gene mutations in cancer biopsies and then use this information to design specific molecular therapies. A more comprehensive analysis of the whole set of genetic alterations that occur in a cancer, as conducted here, may likely reveal additional mutations in genes that are inactivated by mechanisms other than point mutations, and this information may inform us on how to design more effective molecular therapies.
Our studies revealed multiple biallelic genome alterations of several genes in the 3p21 chromosomal region: 46 genes contained biallelic deletions in at least one MM biopsy. Many of these deletions were not contiguous, but rather alternated with segments showing oscillating CN changes along the 3p21 region. Our findings may be consistent with chromothripsis, a condition caused by catastrophic events, such as aborted apoptosis, which causes DNA fragmentation followed by chromosomal rearrangements and loss of some DNA sequences (28). Alternatively, these segmental losses on 3p21 may have occurred sequentially because of some inherited fragility of this chromosomal region in MM.
Whereas biallelic alterations of tumor-suppressor genes can be easily reconciled with the current models of carcinogenesis, monoallelic alterations might be biologically relevant only for haplo-insufficient genes. The functional characterization of these monoallelic alterations should be the focus of future investigations and is beyond the scope of this current study.
The existing literature, however, supports the hypothesis that monoallelic losses of these genes are of biological relevance: (i) In mice, monoallelic losses of Bap1 alter asbestos-induced peritoneal inflammatory response (42) and induce ccRCC in the presence of a coexisting Vhl deficiency (43). (ii) In chronic lymphocytic leukemia, SETD2 alterations are very often monoallelic, and epigenetic inactivation of the wild-type allele is absent. Thus, this is a typical example of a haplo-insufficient tumor suppressor gene (36). (iii) In the ExAC database, which includes data from more than 60,000 genomes, truncating variants of SETD2, PBRM1, and SMARCC1 are not tolerated in the germ line—probability of loss-of-function intolerance score of 1.0—(44) implying that monoallelic losses of these genes, as observed in our MM samples, have relevant biological consequences. Based on our results and on these observations, we expect that context-specific and co-occurring events might result in haplo-insufficient behavior of at least some of the 3p21 genes that we identified.
BAP1 itself is part of the gene set undergoing biallelic inactivation in the 3p21 region and its loss-of-function might represent a positive feed-back because BAP1 inactivation impairs DNA repair, possibly contributing to the catastrophic massive genomic rearrangement in this particular region of the chromosome. Thus, key events in MM carcinogenesis might include multiple CN losses caused by (i) fragmentation of 3p21 induced by oxidative stress caused by asbestos, the main carcinogen in MM (reviewed in ref. 45), and (ii) aberrant DNA repair because of BAP1 inactivation (reviewed in ref. 46). Similarly, alterations of SETD2, another epigenetic modifier involved in transcription elongation, RNA processing, and DNA repair, plays an important role in maintaining genomic integrity, as observed in ccRCC (35). Therefore, SETD2 alterations may also contribute to the genomic instability of the 3p21 region in MM. Loss of PBRM1 or SMARCC1, as members of the switching/sucrose nonfermenting (SWI/SNF) complex, may also lead to chromosomal instability because of their role in chromatin remodeling and sister chromatid cohesion (41, 47). In other words, each of these four genes that we found to be relatively frequently mutated in 3p21 in MM, is involved in the epigenetic modification of DNA, an unlikely coincidence.
An obvious question that remains to be investigated is whether our observations are specific to the 3p21 region or, more likely, extend to other genome regions in MM. Those DNA regions that include CDKN2A and NF2, which are frequently mutated in MM (9–12), should be studied to investigate the possible presence of similar clusters of gene rearrangements—as observed for 3p21—in the corresponding chromosomal regions (9p21 and 22q21, respectively).
Materials and Methods
DNAs from MM Specimens.
We studied DNAs from 9 MMs surgically resected at the Hyogo College of Medicine, and DNAs from 24 MMs surgically resected at the New York University. For the Japanese samples, DNA was extracted from matching blood samples and from primary tumor cells established in tissue culture, to enrich for tumor cells, from resected MM biopsies using DNeasy Blood & Tissue Kit (Qiagen) or QiAamp DNA Micro Kit (Qiagen 56304) according to the manufacturer’s instructions. For the United States samples, DNA was extracted from blood and from tumor cells obtained by laser-capture microdissection performed on MM biopsies with a MMI CellCut Plus (Molecular Machines & Industries) (21).
This study was approved by the Ethics Committee of Hyogo College of Medicine and by the Institutional Review Board of the New York University and performed in accordance with the Declaration of Helsinki (1995) of the World Medical Association (as revised in 2013 in Fortaleza, Brazil) (48).
Written informed consent was received from all patients. Collection and use of patient information and samples were approved by the Institutional Review Boards of the Hyogo College of Medicine and the New York University.
High-Density Custom-Made CGH Microarray Analysis.
We designed a custom-made microarray containing 42,125 oligonucleotide probes targeting a 10.7-Mb genomic region of 3p21 [Chr 3: 43,700,000–54,400,000 (National Center for Biotechnology Information Build 37, hg19)] at an average spacing of 254 bp within the nonrepeat masked regions of the genome using the Agilent’s database (Agilent Technologies). For quality control and normalization, 10,148 probes covering all chromosomes were also contained in this custom array. MM samples and reference DNAs (each 250 ng) were labeled with Cy5 or Cy3, respectively, using the Agilent DNA labeling kit. Following the manufacturer’s recommendation, hybridization and washes were performed in stringent conditions. Then, the arrays were scanned at 5-μm resolution, using the Agilent microarray scanner and analyzed using Feature Extraction v10.7.3. Data analysis was performed using Agilent CytoGenomics 3.0.5.1; the setting of data analysis is described in SI Materials and Methods. The CN data were plotted after moving average calculated using the linear algorithm, unweighted average, using every probe log-ratio score within 10 kb. The CN alteration of DOCK3 was not verified because this gene was the most frequent outlier for the CN ratio (CN ratio > log2 0.4, < −log2 0.4) by the self/self experiment using the same genomic DNA.
tNGS.
NGS was performed on an Illumina MiSeq using paired-end 150-bp runs. Libraries were prepared from 100 ng of genomic DNA from MM tumors, paired normal and reference DNA, using a Haloplex Custom kit (Agilent Technologies) according to the manufacturer’s instructions. Illumina paired-end reads were each aligned to the human NCBI Build 37 reference sequence using bwa software (bio-bwa.sourceforge.net/, v0.6.0). The aligned sequence files were sorted and merged using SAMtools (samtools.sourceforge.net/, v0.1.18). GATK (https://software.broadinstitute.org/gatk/) was used for realignment, base quality score recalibration, single-nucleotide variant or indel (small insertions and deletions) variant calling, and variant quality recalibration. SnpEff was used to categorize the effects of variants by impact. CN analysis was performed by StrandNGS 2.5, by estimating the contamination rate and ploidy in tumor cells. The estimated CN ratio is listed in Table S4.
Supplementary Material
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research Grants KAKENHI 24590715 and 15K08658 (to T.T.), 25460710 (to M.E.), and 26460689 (to Y.Y.); a Grant-in-Aid for Researchers Grant from Hyogo College of Medicine, 2015 (to Y.Y.); National Cancer Institute Grants NCI-R01 CA198138 (to M.C.) and NCI-R01 CA160715 (to H.Y.); the University of Hawai’i Foundation, which received an unrestricted gift to support malignant mesothelioma research from Honeywell International Inc. (to M.C.); United-4-a-Cure (M.C. and H.Y.); and Belluck and Fox (to H.I.P.).
Footnotes
Conflict of interest statement: M.C. has pending patent applications on BAP1 and provides consultation for malignant mesothelioma expertise and diagnosis.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612074113/-/DCSupplemental.
References
- 1.Carbone M, et al. Consensus report of the 2015 Weinman International Conference on Mesothelioma. J Thorac Oncol. 2016;11(8):1246–62. doi: 10.1016/j.jtho.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carbone M, et al. Combined genetic and genealogic studies uncover a large BAP1 cancer syndrome kindred tracing back nine generations to a common ancestor from the 1700s. PLoS Genet. 2015;11(12):e1005633. doi: 10.1371/journal.pgen.1005633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gazdar AF, Carbone M. Molecular pathogenesis of malignant mesothelioma and its relationship to simian virus 40. Clin Lung Cancer. 2003;5(3):177–181. doi: 10.3816/CLC.2003.n.031. [DOI] [PubMed] [Google Scholar]
- 4.Carbone M, Rizzo P, Pass H. Simian virus 40: The link with human malignant mesothelioma is well established. Anticancer Res. 2000;20(2A):875–877. [PubMed] [Google Scholar]
- 5.Ostroff RM, et al. Early detection of malignant pleural mesothelioma in asbestos-exposed individuals with a noninvasive proteomics-based surveillance tool. PLoS One. 2012;7(10):e46091. doi: 10.1371/journal.pone.0046091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Napolitano A, et al. HMGB1 and its hyperacetylated isoform are sensitive and specific serum biomarkers to detect asbestos exposure and to identify mesothelioma patients. Clin Cancer Res. 2016;22(12):3087–3096. doi: 10.1158/1078-0432.CCR-15-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Napolitano A, Carbone M. Malignant mesothelioma: Time to translate? Trends in Cancer. 2016;2(9):467–474. doi: 10.1016/j.trecan.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray PN, Dunlop CL, Elliott AM. Not all next generation sequencing diagnostics are created equal: Understanding the nuances of solid tumor assay design for somatic mutation detection. Cancers (Basel) 2015;7(3):1313–1332. doi: 10.3390/cancers7030837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo G, et al. Whole-exome sequencing reveals frequent genetic alterations in BAP1, NF2, CDKN2A, and CUL1 in malignant pleural mesothelioma. Cancer Res. 2015;75(2):264–269. doi: 10.1158/0008-5472.CAN-14-1008. [DOI] [PubMed] [Google Scholar]
- 10.Lo Iacono M, et al. Targeted next-generation sequencing of cancer genes in advanced stage malignant pleural mesothelioma: a retrospective study. J Thorac Oncol. 2015;10(3):492–499. doi: 10.1097/JTO.0000000000000436. [DOI] [PubMed] [Google Scholar]
- 11.Bueno R, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48(4):407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 12.Ugurluer G, et al. Genome-based mutational analysis by next generation sequencing in patients with malignant pleural and peritoneal mesothelioma. Anticancer Res. 2016;36(5):2331–2338. [PubMed] [Google Scholar]
- 13.Bott M, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43(7):668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Testa JR, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43(10):1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zauderer MG, et al. Clinical characteristics of patients with malignant pleural mesothelioma harboring somatic BAP1 mutations. J Thorac Oncol. 2013;8(11):1430–1433. doi: 10.1097/JTO.0b013e31829e7ef9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carbone M, Gaudino G, Yang H. Recent insights emerging from malignant mesothelioma genome sequencing. J Thorac Oncol. 2015;10(3):409–411. doi: 10.1097/JTO.0000000000000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153(1):17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Jean D, Daubriac J, Le Pimpec-Barthes F, Galateau-Salle F, Jaurand MC. Molecular changes in mesothelioma with an impact on prognosis and treatment. Arch Pathol Lab Med. 2012;136(3):277–293. doi: 10.5858/arpa.2011-0215-RA. [DOI] [PubMed] [Google Scholar]
- 19.Taguchi T, Jhanwar SC, Siegfried JM, Keller SM, Testa JR. Recurrent deletions of specific chromosomal sites in 1p, 3p, 6q, and 9p in human malignant mesothelioma. Cancer Res. 1993;53(18):4349–4355. [PubMed] [Google Scholar]
- 20.Yoshikawa Y, et al. Frequent inactivation of the BAP1 gene in epithelioid-type malignant mesothelioma. Cancer Sci. 2012;103(5):868–874. doi: 10.1111/j.1349-7006.2012.02223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasu M, et al. High Incidence of Somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol. 2015;10(4):565–576. doi: 10.1097/JTO.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeiger MA, Gnarra JR, Zbar B, Linehan WM, Pass HI. Loss of heterozygosity on the short arm of chromosome 3 in mesothelioma cell lines and solid tumors. Genes Chromosomes Cancer. 1994;11(1):15–20. doi: 10.1002/gcc.2870110104. [DOI] [PubMed] [Google Scholar]
- 23.Lindholm PM, et al. Gene copy number analysis in malignant pleural mesothelioma using oligonucleotide array CGH. Cytogenet Genome Res. 2007;119(1-2):46–52. doi: 10.1159/000109618. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi T, et al. Genomic profiling of malignant pleural mesothelioma with array-based comparative genomic hybridization shows frequent non-random chromosomal alteration regions including JUN amplification on 1p32. Cancer Sci. 2007;98(3):438–446. doi: 10.1111/j.1349-7006.2006.00386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov SV, et al. Genomic events associated with progression of pleural malignant mesothelioma. Int J cancer. 2009;124(3):589–599. doi: 10.1002/ijc.23949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshikawa Y, et al. Frequent deletion of 3p21.1 region carrying semaphorin 3G and aberrant expression of the genes participating in semaphorin signaling in the epithelioid type of malignant mesothelioma cells. Int J Oncol. 2011;39(6):1365–1374. doi: 10.3892/ijo.2011.1158. [DOI] [PubMed] [Google Scholar]
- 27.Hesson LB, Cooper WN, Latif F. Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene. 2007;26(52):7283–7301. doi: 10.1038/sj.onc.1210547. [DOI] [PubMed] [Google Scholar]
- 28.Tubio JM, Estivill X. Cancer: When catastrophe strikes a cell. Nature. 2011;470(7335):476–477. doi: 10.1038/470476a. [DOI] [PubMed] [Google Scholar]
- 29.Yoshikawa Y, et al. Biallelic germline and somatic mutations in malignant mesothelioma: Multiple mutations in transcription regulators including mSWI/SNF genes. Int J Cancer. 2015;136(3):560–571. doi: 10.1002/ijc.29015. [DOI] [PubMed] [Google Scholar]
- 30.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patil V, Pal J, Somasundaram K. Elucidating the cancer-specific genetic alteration spectrum of glioblastoma derived cell lines from whole exome and RNA sequencing. Oncotarget. 2015;6(41):43452–43471. doi: 10.18632/oncotarget.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan J, et al. Genomic landscapes of breast fibroepithelial tumors. Nat Genet. 2015;47(11):1341–1345. doi: 10.1038/ng.3409. [DOI] [PubMed] [Google Scholar]
- 34.Lee JJ, et al. Targeted next-generation sequencing reveals high frequency of mutations in epigenetic regulators across treatment-naïve patient melanomas. Clin Epigenetics. 2015;7:59. doi: 10.1186/s13148-015-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, et al. SETD2: An epigenetic modifier with tumor suppressor functionality. Oncotarget. May 14, 2016 doi: 10.18632/oncotarget.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker H, et al. June 10, 2016. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia.
- 37.Huang KK, et al. SETD2 histone modifier loss in aggressive GI stromal tumours. Gut. September 3, 2015 doi: 10.1136/gutjnl-2015-309482. [DOI] [PubMed] [Google Scholar]
- 38.Jiao Y, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45(12):1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Roberts CW. CARMA: CARM1 methylation of SWI/SNF in breast cancer. Cancer Cell. 2014;25(1):3–4. doi: 10.1016/j.ccr.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mo D, et al. Low PBRM1 identifies tumor progression and poor prognosis in breast cancer. Int J Clin Exp Pathol. 2015;8(8):9307–9313. [PMC free article] [PubMed] [Google Scholar]
- 41.DelBove J, et al. Identification of a core member of the SWI/SNF complex, BAF155/SMARCC1, as a human tumor suppressor gene. Epigenetics. 2011;6(12):1444–1453. doi: 10.4161/epi.6.12.18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Napolitano A, et al. Minimal asbestos exposure in germline BAP1 heterozygous mice is associated with deregulated inflammatory response and increased risk of mesothelioma. Oncogene. 2016;35(15):1996–2002. doi: 10.1038/onc.2015.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang SS, et al. Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proc Natl Acad Sci USA. 2014;111(46):16538–16543. doi: 10.1073/pnas.1414789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lek M, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carbone M, et al. Malignant mesothelioma: Facts, myths, and hypotheses. J Cell Physiol. 2012;227(1):44–58. doi: 10.1002/jcp.22724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carbone M, et al. BAP1 and cancer. Nat Rev Cancer. 2013;13(3):153–159. doi: 10.1038/nrc3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brownlee PM, Chambers AL, Cloney R, Bianchi A, Downs JA. BAF180 promotes cohesion and prevents genome instability and aneuploidy. Cell Reports. 2014;6(6):973–981. doi: 10.1016/j.celrep.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hellmann F, Verdi M, Schlemper BR, Jr, Caponi S. 50th anniversary of the Declaration of Helsinki: The double standard was introduced. Arch Med Res. 2014;45(7):600–601. doi: 10.1016/j.arcmed.2014.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.