Significance
Whereas the role of the ubiquitin system in protein degradation is well established, little is known regarding the regulation of its own components, including its catalytic arm, the 26S proteasome. Here we show that in stressed mammalian cells, the proteasome is targeted by autophagy, which requires site-specific ubiquitination of its ubiquitin receptors. The process is mediated by the p62/SQSTM1 adapter and requires its ubiquitin-associated domain. Independently, p62 serves also as a shuttling protein for ubiquitinated substrates, using its PB1 domain. This places p62 in a pivotal position where under certain conditions it binds to the proteasome as a protease, whereas in other conditions it recognizes the proteasome as a prey. The regulation of this intricate “decision making” process remains elusive.
Keywords: proteasome, autophagy, ubiquitin, degradation
Abstract
The ubiquitin-proteasome system and autophagy are the two main proteolytic systems involved in, among other functions, the maintenance of cell integrity by eliminating misfolded and damaged proteins and organelles. Both systems remove their targets after their conjugation with ubiquitin. An interesting, yet incompletely understood problem relates to the fate of the components of the two systems. Here we provide evidence that amino acid starvation enhances polyubiquitination on specific sites of the proteasome, a modification essential for its targeting to the autophagic machinery. The uptake of the ubiquitinated proteasome is mediated by its interaction with the ubiquitin-associated domain of p62/SQSTM1, a process that also requires interaction with LC3. Importantly, deletion of the PB1 domain of p62, which is important for the targeting of ubiquitinated substrates to the proteasome, has no effect on stress-induced autophagy of this proteolytic machinery, suggesting that the domain of p62 that binds to the proteasome determines the function of p62 in either targeting substrates to the proteasome or targeting the proteasome to autophagy.
The ubiquitin (Ub)-proteasome system (UPS) and autophagy, the two main cellular proteolytic machineries, act in parallel and differentially under changing pathophysiological conditions, thereby maintaining the homeostasis and quality control of the proteome and organelles. An important common regulatory step shared by the two systems is the signaling of their targets for destruction by the covalent attachment of Ub (1, 2).
The UPS is involved in the degradation of the major parts of cytosolic and nuclear proteins, as well as certain membrane proteins (3, 4). The targeting of substrates for proteasomal degradation is mediated by their ubiquitination, and sometimes requires their delivery to the proteasome by shuttling proteins (5). The proteasome, the catalytic arm of the UPS (1, 6), is a large (2.5 MDa) multicatalytic protease complex consisting of the 20S core particle (CP) and the 19S regulatory particles (RPs). The 20S CP is composed of two outer α-rings each containing seven α-subunits (α1–α7) and two inner β-rings each consisting of seven β-subunits (β1–β7). The proteolytic function of the proteasome is mediated by three catalytic β-subunits—β1, β2, and β5—with distinct peptidase activities. The 19S RP, composed of AAA-ATPase (Rpt) and non-ATPase (Rpn) subunits, is responsible for the recognition, unfolding, and subsequent translocation of ubiquitinated substrates into the 20S CP (7). This process is accompanied by enzymatic removal of Ub moieties, which are recycled. The proteasome is widely distributed in the cell, is localized to the cytosol and nucleus, and is also found tethered to several subcellular organelles (8). It is highly abundant, composing ∼1% of the total protein mass of cells (9).
Although the proteasome is responsible mainly for the degradation of single proteins, the autophagy-lysosome system specializes in the degradation of protein aggregates (10) as well as entire organelles, such as mitochondria (11) and pathogens (12). These targets are poorly degraded by the proteasome, and their accumulation underlies the mechanism of many neurodegenerative, liver, muscle, and lung diseases, among others (13). Aggregates and cellular organelles can be eliminated under basal or stress-induced autophagy (14, 15).
Macroautophagy (the main type of stress-induced autophagy) is initiated by the formation of a phagophore, a double-membraned structure that elongates and engulfs a portion of cytoplasmic material. LC3, an autophagic receptor and a key player in the process, is incorporated into the expanding phagophore after its conjugation with phosphatidylethanolamine, at which point it is designated as LC3-II (16). Besides nonselective bulk engulfment of cytosolic material, autophagy also can be a selective process. In this process, phagophore-incorporated LC3-II serves as a docking site for adapter proteins bound to ubiquitinated substrates, in most cases organelles (10). The enclosed vesicle, containing LC3-II, the linking adapter, and the target cargo, is termed an autophagosome. Once it matures, the autophagosome fuses with the lysosome, where subsequent degradation of its contents by lysosomal hydrolysis occurs (16).
An example of a linking adapter is p62/SQSTM-1 (17). This adapter binds ubiquitinated substrates via its Ub-associated (UBA) domain (18) and delivers them to the growing phagophore. There it binds to LC3-II via its LC3-interacting region, and is later degraded along with its substrate (19–21). p62 has another domain, Phox and Bem1p (PB1), through which it binds to the 19S proteasomal subunits Rpn10 and Rpt1. This binding domain allows p62 to function as a shuttling protein, delivering ubiquitinated substrates to the proteasome (22). Thus, p62 plays a pivotal role in targeting Ub-modified proteins to either the proteasome or the autophagic machinery. The “decision making” process of where to deliver the cargo, and under what conditions, is poorly understood.
In the present study we show that the mammalian proteasome is also degraded by selective autophagy. Following amino acid starvation, the proteasome undergoes a notable ubiquitination that appears to be subunit- and site-specific and is essential for its recognition as a substrate. The engulfment of the ubiquitinated proteasome is mediated by its interaction with the UBA domain of p62, which targets the proteasome to the autophagy-lysosome system.
Results
Amino Acid Starvation Induces Autophagy of the 26S Proteasome.
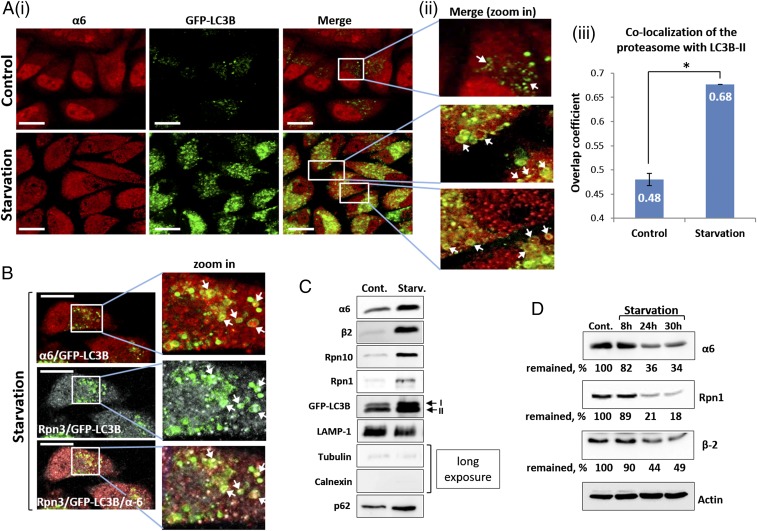
Whereas the proteasome is responsible for the breakdown of numerous cellular proteins, little is known about its own fate. Although unassembled proteasome subunits have been shown to be degraded by mature proteasomes (23), it is reasonable to assume that intact proteasome complexes are also degraded, possibly via autophagy, a destructive system known for its ability to degrade large cargoes (14). To test this hypothesis, we deprived cells of amino acids, a condition known to induce autophagy. As shown in Fig. 1 A and B, after starvation, the proteasome was present within autophagosomal vesicles. Importantly, the subcomplexes of the 26S proteasome—the 20S CP and the 19S RP—also were present in these vesicles (Fig. 1B). The starvation-induced uptake of the proteasome to autophagosomes was also seen in isolated vesicles (Fig. 1C). The purity of the vesicular fraction was confirmed by a complete lack of cytosolic and endoplasmic reticulum markers (Fig. 1C).
Fig. 1.
Autophagic uptake of the proteasome and its subsequent degradation are induced by amino acid starvation. (A) (i) HeLa cells stably expressing GFP-LC3B were incubated for 4 h in the presence of complete (Upper) or amino acid-depleted medium (Lower). Fixed cells were stained with anti-α6 antibody (red). (Scale bars: 20 μm.) (ii) Highly magnified fields. White arrows point to proteasome-containing autophagosomes. (iii) Manders overlap coefficients of LC3B-II (GFP) and α6 (red) colocalization were calculated. *P < 0.0000012. (B) Immunofluorescent staining of GFP-LC3B–transfected HeLa cells with anti-α6 (red; Upper) and Rpn3 (gray; Middle) antibodies following 4 h of amino acid starvation. (Scale bars: 20 μm.) White arrows point to the 20S CP (merge, zoom in; Upper) and 19S RP (merge, zoom in; Middle) within autophagosomal vesicles. (Lower) Merged image of α6, Rpn3, and LC3B-II. (C) Autophagosome-lysosome vesicles were purified from control (Cont.) and 4 h starved cells (Starv.). Isolated vesicles were lysed, resolved via SDS/PAGE, and subjected to immunoblotting with anti-α6 and anti-β2 (20S CP); with anti-Rpn10 and anti-Rpn1 (19S RP); and with anti-LAMP1 (lysosome), anti-tubulin, anti-calnexin and anti-p62 (autophagic machinery). (D) GFP-LC3B–expressing HeLa cells were left untreated (Cont.) or starved for the indicated times. Cell lysates were subjected to immunoblotting after electrophoresis with anti-α6, anti-Rpn1, anti-β2, and anti-actin.
Because the proteasome is a long-lived complex with a half-life of >1 wk (24), we examined whether amino acid starvation affects its stability. As shown in Fig. 1D, depletion of amino acids significantly accelerated degradation of the 26S proteasome. Taken together, the foregoing findings suggest that amino acid starvation leads to increased engulfment of the proteasome by autophagosomes, followed by its subsequent degradation.
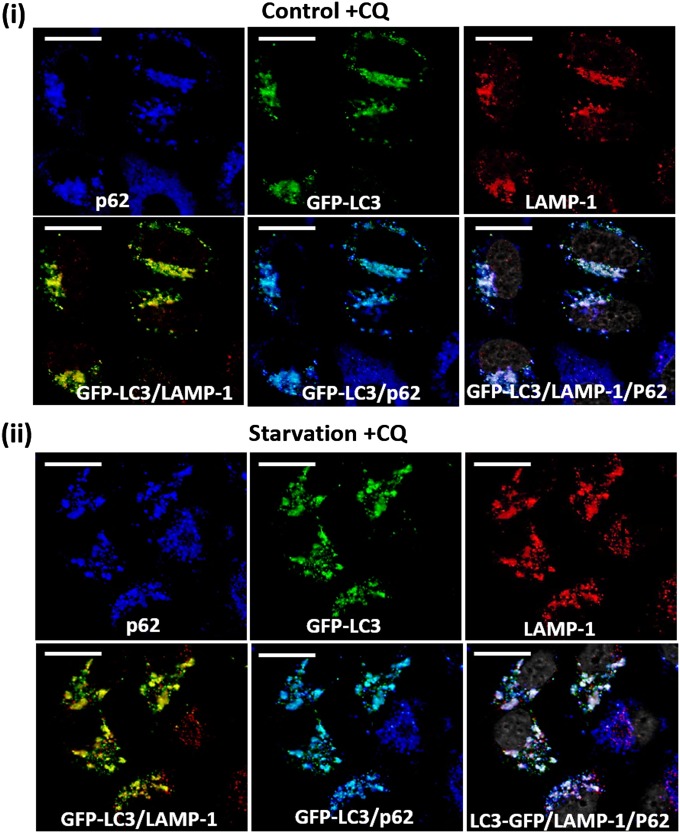
To visualize accumulation of the proteasome in autophagosomal vesicles, we added chloroquine (CQ), a lysosomotropic agent, to all experiments except those in which stability of the proteasome was monitored. To rule out untoward effects of the inhibitor on the autophagic process, we demonstrated that CQ does not affect fusion between the autophagic p62/LC3-containing vesicles and LAMP1-containing lysosomes (Fig. S1).
Fig. S1.
Colocalization of autophagosomes with lysosomes following addition of CQ. Fed (i) and 4-h amino acid-starved (ii) GFP-LC3B–expressing HeLa cells were stained with anti-p62 (blue) and anti-LAMP-1 (red). (Scale bars: 20 μm.)
Amino Acid Starvation Induces Ubiquitination of the Proteasome.
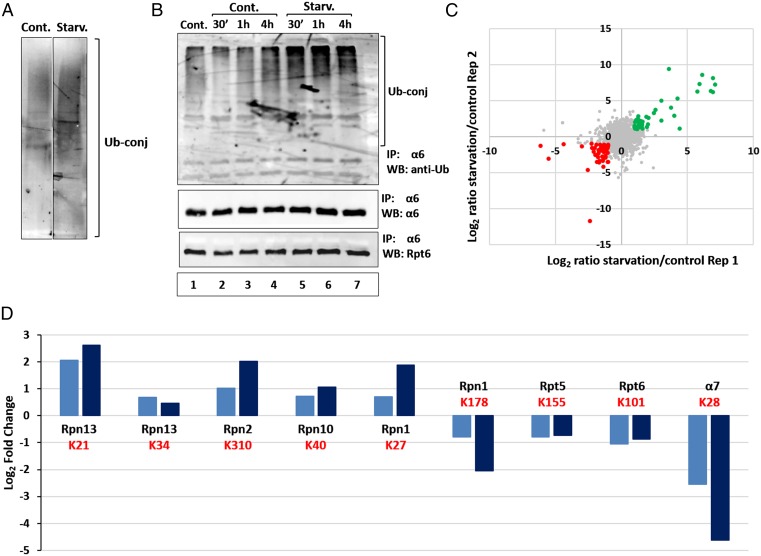
In addition to bulk engulfment of cytoplasmic material, substrates also can be targeted for autophagic degradation selectively, in a process mediated by their ubiquitination and subsequent recognition by autophagic mediators (20, 25). To demonstrate induced ubiquitination of cargo proteins in autophagosomal vesicles following starvation, we probed the resolved isolated vesicular proteins with a Ub conjugate-specific antibody. As shown in Fig. 2A, depletion of amino acids resulted in a significant increase in Ub-conjugated proteins in the purified vesicular fraction.
Fig. 2.
Amino acid deprivation stimulates proteasome ubiquitination. (A) Autophagosomes isolated from control and starved cells (Fig. 1C) were resolved via SDS/PAGE and blotted with anti-Ub conjugates antibody. (B) HeLa cells were incubated for the indicated times in the presence of complete (lanes 1–4; Cont.) or amino acid-depleted (lanes 5–7; Starv.) medium, in the absence (lane 1; Cont.) or presence (lanes 2–7) of CQ. Cell lysates were immunoprecipitated (IP) with anti-α6, and the immunoprecipitates were resolved and subjected to immunoblotting (WB) with anti-Ub, anti-Rpt6, and anti-α6. (C) Log2 ratios of two biological replicates of Ub-anchoring sites (K-ε-GG) enriched from HeLa cells incubated for 1 h under control or starved conditions. Green dots represent ubiquitinated peptides, the level of which increased during starvation, whereas red dots represent those whose level was decreased. (D) Log2 ratios of ubiquitinated sites (K-ε-GG) of specific proteasome subunits obtained in two replicated biological experiments. The Ub-modified lysine (K) is denoted in red.
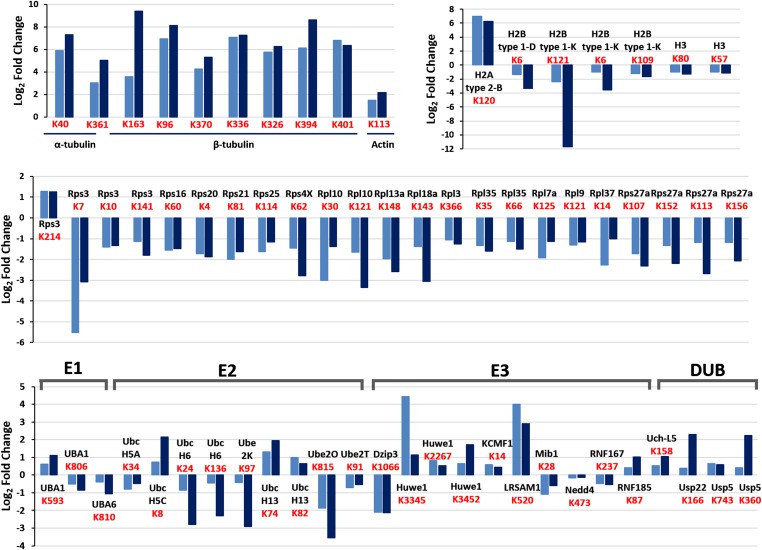
To investigate whether the proteasome is taken up selectively by autophagosomes, we first examined its ubiquitination after amino acid starvation. As shown in Fig. 2B, increased ubiquitination of the 26S proteasome was noted already after 30 min of amino acid deprivation, peaked at 1 h, and decreased slightly after 4 h. To further investigate the effect of amino acid starvation on ubiquitination of the proteasome, we used a specific antibody to the conjugated site, anti–K-ε-GG (26), which allowed us to both quantify the effect and identify the modification sites on the different proteasomal subunits. MS analysis detected 7,808 ubiquitinated peptides, among which were numerous proteasomal subunits, in two independent biological replicates (Fig. 2C). A comparison of control and starved cells revealed a twofold to fivefold increase in ubiquitination of the 19S RP subunits Rpn13, Rpn10, Rpn1, and Rpn2 (Fig. 2D). In addition, we detected a decrease in the ubiquitination of proteasomal subunits Rpn5, Rpn6, α7, and Rpn1. In the case of Rpn1, different ubiquitination sites showed a change in ubiquitination in the opposite direction (Fig. 2D). Thus, it is clear that starvation induces significant changes in the ubiquitination pattern of different 26S proteasomal subunits, which is likely related to the specific recognition of the proteolytic complex by the autophagic machinery. Importantly, as shown in Fig. S2, starvation also affects the ubiquitination state of other proteins, including ribosomal, histone, and cytoskeletal proteins.
Fig. S2.
Amino acid deprivation stimulates the ubiquitination of specific UPS and cellular proteins. Shown are log2 ratios of the ubiquitination sites (K-ε-GG) of components of the UPS and of α- and β-tubulin, actin, histones, and ribosomal proteins that were up-regulated (ratio >1) or down-regulated (ratio <−1) in the two independent biological replicates. Ubiquitination sites are depicted in red.
An interesting question is whether the proteasome is the sole component of the UPS in which ubiquitination is stimulated, or whether there are other components of the system that are also modified under an amino acid shortage, suggesting a coordinated regulation of the system under stress. We found that the ubiquitination state of additional components of the UPS was altered as well (Fig. S2). Importantly, we identified significant stimulation in the ubiquitination of two ligases, Huwe1 and KCMF1, both of which are known as proteasome-associated ligases (27, 28). It is possible that these ligases are activated/stimulated by ubiquitination and in turn modify the proteasome.
Proteasome Ubiquitination Is Essential for Its Autophagosomal Engulfment.
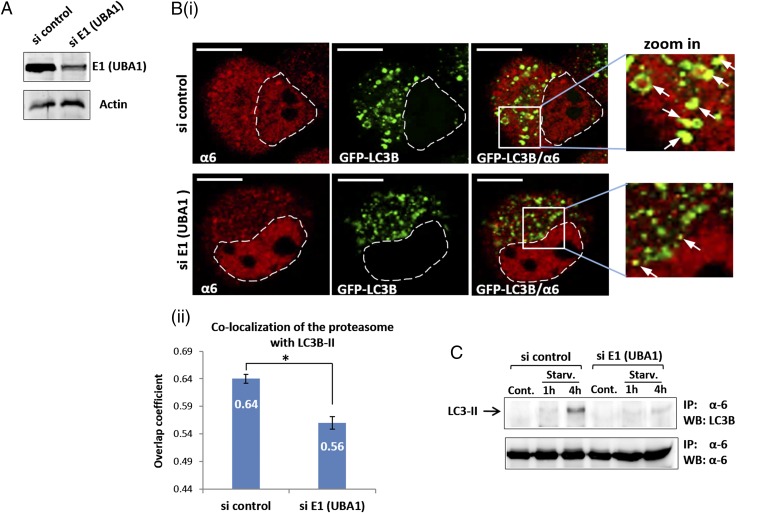
To elucidate the significance of ubiquitination of the proteasome for its autophagosomal uptake, we silenced E1 (UBA1), the enzyme catalyzing the first step in the ubiquitination cascade (4). As shown in Fig. 3A, anti-E1 siRNA effectively inhibited E1 expression. Notably, E1 down-regulation dramatically decreased the amount of proteasome taken up by autophagosomes after starvation (Fig. 3B). We next precipitated the proteasome from cells in which E1 was silenced and found that, compared with si control cells, the amount of LC3B-II coprecipitated with the proteasome after amino acid starvation was decreased significantly (Fig. 3C). These findings point to the importance of proteasome ubiquitination for LC3-dependent autophagosomal engulfment.
Fig. 3.
Autophagosomal uptake of the proteasome depends on Ub conjugation (A) GFP-LC3B–overexpressing HeLa cells were transfected with control or anti-E1 (UBA1) siRNA oligonucleotides. After 2 d, cells were lysed, resolved via SDS/PAGE, and examined for E1 expression. (B) (i) After silencing of E1, cells were starved for amino acids for 4 h and then stained with anti-α6. (Scale bars: 10 μm.) White arrows point to proteasome-containing autophagosomes. (ii) Overlap coefficients of colocalization of the proteasome with LC3B-II were measured according to the method of Manders. *P < 0.000027. (C) HeLa cells were transfected with control or anti-E1 (UBA1) siRNAs. Cells were left untreated (Cont.) or starved to amino acids (Starv.) for the indicated times. Cell lysates were immunoprecipitated (IP) with anti-α6, and the precipitates were resolved, followed by immunoblotting (WB) with anti-LC3B and anti-α6.
Amino Acid Starvation Increases Interaction of the Proteasome with p62 and LC3B-II.
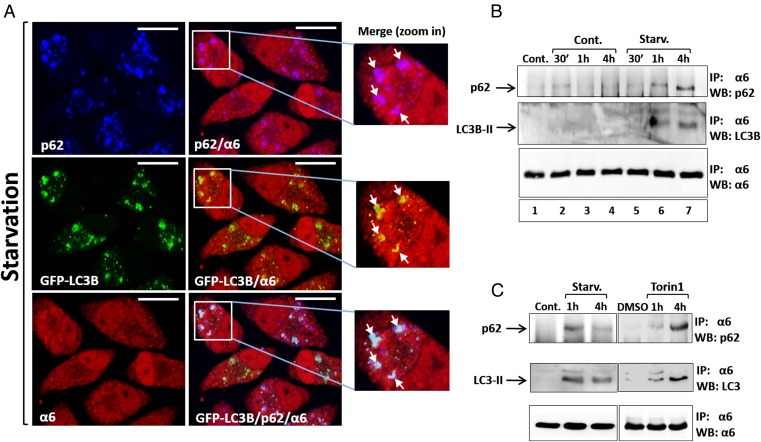
p62 is an autophagic cargo adapter that recruits ubiquitinated proteins and organelles to the autophagosome by its simultaneous interaction with both the modified substrates and the LC3 autophagosomal receptor (10, 17, 29, 30). We next examined whether p62 is also involved in autophagosomal uptake of the proteasome. As shown in Fig. 4A, depriving cells of amino acids resulted in colocalization of p62 with the proteasome and LC3B-II.
Fig. 4.
Amino acid starvation stimulates the interaction of the proteasome with p62 and LC3B. (A) After 4 h of amino acid starvation, GFP-LC3B–expressing HeLa cells were stained with anti-p62 (blue; Upper Left) and anti-α6 (red; Lower Left). (Scale bars: 20 μm.) White arrows point to colocalized proteasome and p62 (zoom in; Upper), LC3B-II (zoom in; Middle), and colocalization of all three (zoom in; Lower). (B) GFP-LC3B–expressing HeLa cells were incubated for the indicated times in the presence of complete (lanes 1–4) or amino acid-depleted (lanes 5–7; Starv.) medium, in the absence (lane 1; Cont.) or presence (lanes 2–7) of CQ. Cell lysates were immunoprecipitated (IP) with anti-α6, and the immunoprecipitates were resolved and blotted (WB) with anti-p62, anti-LC3B, and anti-α6. (C) GFP-LC3B–expressing HeLa cells were left untreated (Cont.) or starved for amino acids (Starv.; Left), or were supplemented with DMSO (DMSO) or Torin1 (0.5 μΜ) for the indicated times (Right). CQ was present in the starved and the Torin1-treated cells. Cell lysates were subjected to immunoprecipitation (IP) and, following resolution, were immunoblotted (WB) with anti-p62, anti-LC3B, and anti-α6.
To further dissect this finding, we isolated the proteasome by immunoprecipitation and looked for its interaction with p62. A notable increase in the proteasome-bound p62 was observed after 1 h and 4 h of starvation (Fig. 4B, Upper). Importantly, this increase was correlated with a parallel and comparable increase in the amount of LC3B-II, which was also associated with the proteasome (Fig. 4B, Middle), indicating selective recognition of the proteasome by the two components of the autophagic machinery. Of note, CQ added to both the control and starved cells did not induce precipitation of either p62 or LC3B-II along with the proteasome (Fig. 4B).
mTORC1 (mechanistic target of rapamycin complex 1) activity is regulated by amino acid levels. Amino acid deprivation results in mTORC1 inhibition and subsequent activation of ULK1, leading to the initiation of autophagy (31, 32). We used Torin1, a potent mTORC1 inhibitor, to check whether increased interaction of the proteasome with p62 and LC3B-II occurs downstream of mTORC1. Similar to amino acid starvation, the Torin1 treatment markedly increased the interaction of the proteasome with p62 (Fig. 4C, Upper). An analogous increase was also noted in the amount of LC3B-II coprecipitated with the proteasome (Fig. 4C, Middle). These findings suggest that the signal for starvation-induced proteasome interaction with p62 and LC3B-II and its concomitant engulfment by the autophagosome is transduced, at least in part, via the mTOR pathway.
p62 Mediates Autophagosomal Uptake of the Proteasome.
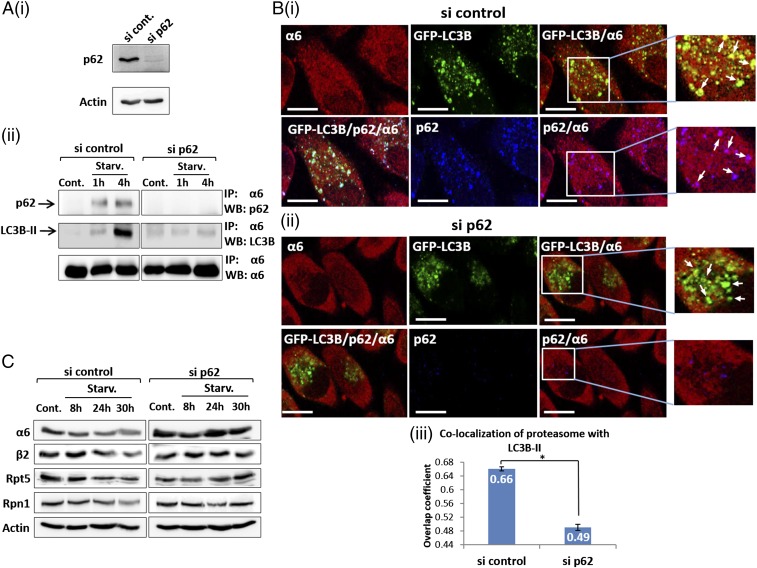
To further study the significance of p62 in autophagosomal uptake of the proteasome, we knocked down p62 expression using siRNA (Fig. 5A, i). The reduction in p62 abolished the interaction of the proteasome with LC3B-II following starvation (Fig. 5A, ii, Middle). Moreover, immunofluorescent staining showed that p62 silencing significantly reduced recruitment of the proteasome to autophagosomes after starvation (Fig. 5B). Importantly, depletion of endogenous p62 impaired degradation of the proteasome after starvation (Fig. 5C), again highlighting the key role of p62 in the recruitment of ubiquitinated proteasomes into autophagosomes.
Fig. 5.
p62/SQSTM1 mediates autophagosomal uptake of the proteasome. (A) (i) GFP-LC3B–expressing HeLa cells were transfected with control or anti-p62 siRNAs. At 2 d after transfection, cells were lysed and p62 expression was detected by Western blot analysis. (ii) si control-transfected and si p62-transfected cells were left untreated (Cont.) or starved for amino acids (Starv.) for the indicated times. Cell lysates were subjected to immunoprecipitation (IP) with anti-α6, followed by electrophoresis and immunoblotting (WB) with anti-p62, anti-LC3B, and anti-α6. (B) After treatment with si control (i) and si p62 (ii), GFP-LC3B–expressing HeLa cells were subjected to amino acid starvation, then stained with anti-p62 and anti-α6. (Scale bars: 20 μm.) (iii) Overlap coefficients of colocalization of the proteasome with LC3B-II in control and p62-silenced cells were measured according to the method of Manders. *P < 0.0000211. (C) Cells were treated as described in A and B, but for different starvation times, and lysates were resolved and blotted with anti-α6, anti-β2, anti-Rpt5, anti-Rpn1, and anti-actin.
The UBA Domain of p62 Mediates Its Interaction with the Proteasome.
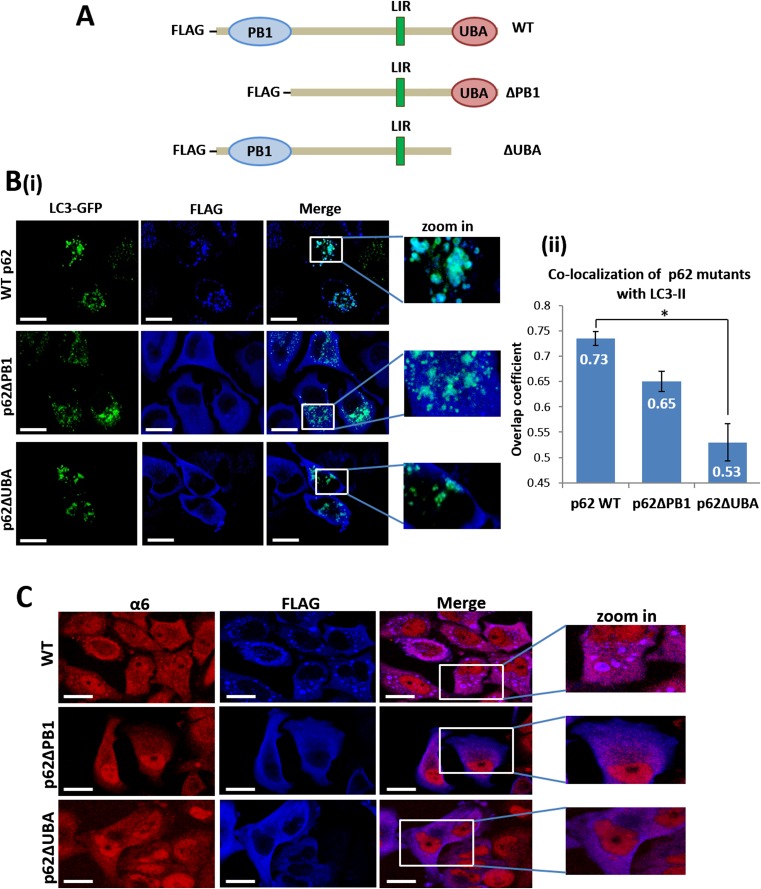
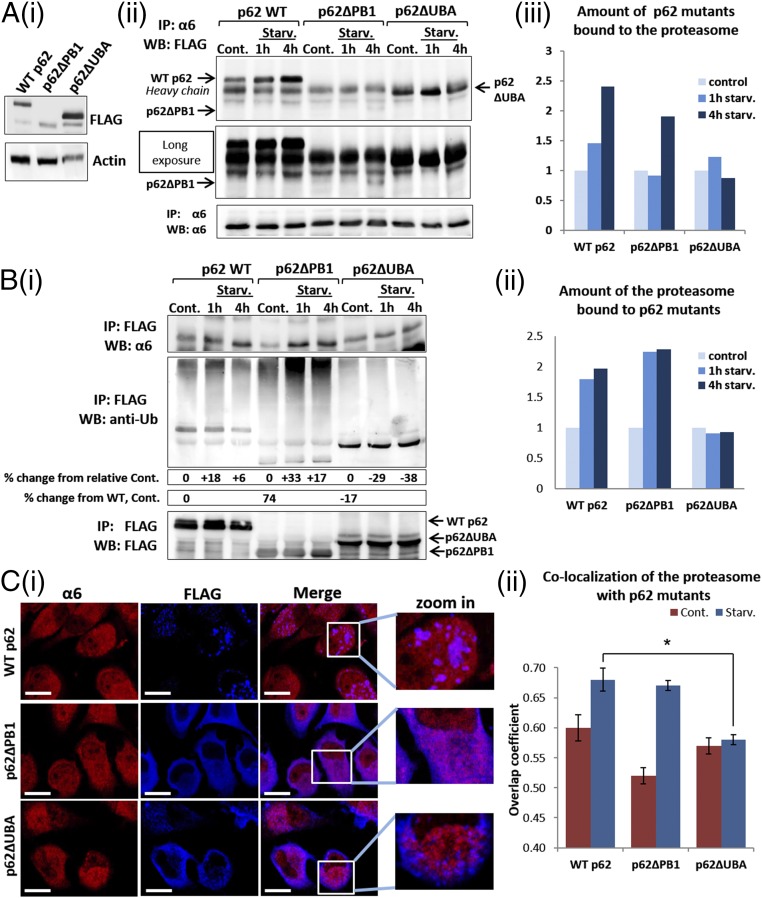
Along with its role as an autophagic mediator, p62 also delivers ubiquitinated substrates for proteasomal degradation by direct interaction of its PB1 domain with the 19S proteasomal subunits (22). To identify the domain of p62 that is important for binding to ubiquitinated proteasome, we constructed two mutated forms of p62, one lacking the PB1 domain and the other lacking the UBA domain (Fig. S3A). GFP-LC3B–transfected HeLa cells transiently expressing WT p62, p62∆PB1, or p62∆UBA were each subjected to amino acid starvation for 1 h and 4 h. This treatment resulted in an increase in the interaction of the proteasome with both WT p62 and p62∆PB1, but not with p62∆UBA (Fig. 6 A, ii and iii and B, i and ii), highlighting the role of the p62 UBA domain in proteasome engulfment by the autophagosome.
Fig. S3.
Deletion of the UBA domain of p62 decreases its colocalization with both the proteasome and LC3B-II. (A) Schematic representation of the p62 constructs. (B) (i) GFP-LC3B–expressing HeLa cells were transiently transfected with FLAG-tagged WT (Upper), ∆PB1 (Middle), and ∆UBA (Lower) p62s. Cells were starved for amino acids for 4 h and then stained with anti-FLAG. (Scale bars: 20 μm.) (ii) Overlap coefficients of LC3B-II colocalization with the p62 constructs were calculated according to the method of Manders. *P < 0.0002. (C) Same as in Fig. 6B, except that the cells were not starved. These cells served as a control for the experiment shown in Fig. 6C, ii. (Scale bars: 20 μm.)
Fig. 6.
The UBA domain of p62 is required for its interaction with the ubiquitinated proteasome following amino acid starvation. (A) (i) Whole-cell lysates of FLAG-tagged WT p62-, p62∆PB1-, and p62∆UBA-expressing HeLa cells were analyzed for expression of the different p62 proteins by immunoblotting with anti-FLAG. (ii) HeLa cells expressing the different p62 species (as described in A, i) were left untreated (Cont.) or starved for amino acids for the indicated times. Cell lysates were immunoprecipitated (IP) with anti-α6, resolved, and immunoblotted (WB) with anti-FLAG and anti-α6. (iii) Densitometric analysis of the p62 proteins depicted in A, ii. The value of p62 in the control lane of each construct was arbitrarily set to 1. (B) (i) The cells and the experimental setup were as described in A, ii. Cell lysates were subjected to immunoprecipitation (IP) with immobilized anti-FLAG, followed by immunoblotting (WB) with anti-α6, anti-FLAG, and anti-Ub conjugates. (ii) Densitometric analysis of the amount of the proteasome coprecipitated with the p62 species after amino acid deprivation. The value of the α6 subunit in the control lane of each p62 construct was arbitrarily set to 1. (C) (i) HeLa cells transiently transfected with FLAG-tagged WT p62, p62∆PB1, and p62∆UBA were starved for amino acids for 4 h and then stained with anti-FLAG and anti-α6. (Scale bars: 20 μm.) (ii) Manders overlap coefficients of colocalization of the different p62 proteins with the proteasome. *P < 0.000313.
It should be noted that p62 can bind to the proteasome via two independent mechanisms. Under basal conditions, p62 binds the proteasome directly via its PB1 domain. In this mechanism, p62 serves as a shuttling protein for ubiquitinated substrates to the proteasome. Under stress, the binding increases through the involvement of the UBA domain, now associating with the proteasome also via its Ub moieties. Here the binding serves to target the proteasome for autophagy.
The low basal amount of ∆PB1-p62 in fed cells that coprecipitated with the proteasome can be explained by the importance of the PB1 domain to the interaction between the two in the context of substrate shuttling to the proteasome, along with its low steady-state level (Fig. 6A, i). Nevertheless, the interaction between p62 and the proteasome is increased in p62∆PB1-expressing cells after starvation (Fig. 6 A and B). Likewise, high expression of p62∆UBA compared with WT p62 (Fig. 6A, i) can explain the large amount of p62∆UBA that coprecipitated with the proteasome under control conditions. Importantly, this relatively high level of p62∆UBA was not elevated further after starvation (Fig. 6 A and B). Taken together, these findings suggest distinct roles for p62 association with the proteasome under basal and stressed conditions, as explained above. As expected, with a lack of the UBA domain, precipitation of ubiquitinated substrates by p62 was dramatically decreased (Fig. 6B, i, Middle), in agreement with the UBA domain’s role in recognition of ubiquitinated cargo (18). On removal of the PB1 domain, more ubiquitinated substrates were precipitated with p62 (Fig. 6B, i, Middle), suggesting compromised degradation of these substrates owing to the inability of p62 to deliver them to the proteasome. Interestingly, we noted an increase in the amount of ubiquitinated proteins bound to WT p62 and even more so to p62∆PB1 after a short starvation, suggesting entry of the UPS as a defense mechanism already in the early stages of stress.
To further confirm the importance of p62’s UBA domain for the interaction between p62 and the ubiquitinated proteasome, we demonstrated that WT p62 and p62∆PB1 were colocalized with the proteasome after starvation (Fig. 6C). In striking contrast, p62∆UBA did not colocalize with the proteasome and actually were clearly separated from one another; p62∆UBA was mostly localized close to the membrane, and the proteasome was observed mainly in the central part of the cytoplasm (Fig. 6C, i, Lower). In addition, the p62 species that lacked the PB1 or UBA domains demonstrated a much lower tendency for self-aggregation following amino acid starvation (in comparison with the WT form of p62) (Fig. 6C, i; compare Upper with Middle and Lower). This observation is in agreement with the known role of these domains in p62 oligomerization (33).
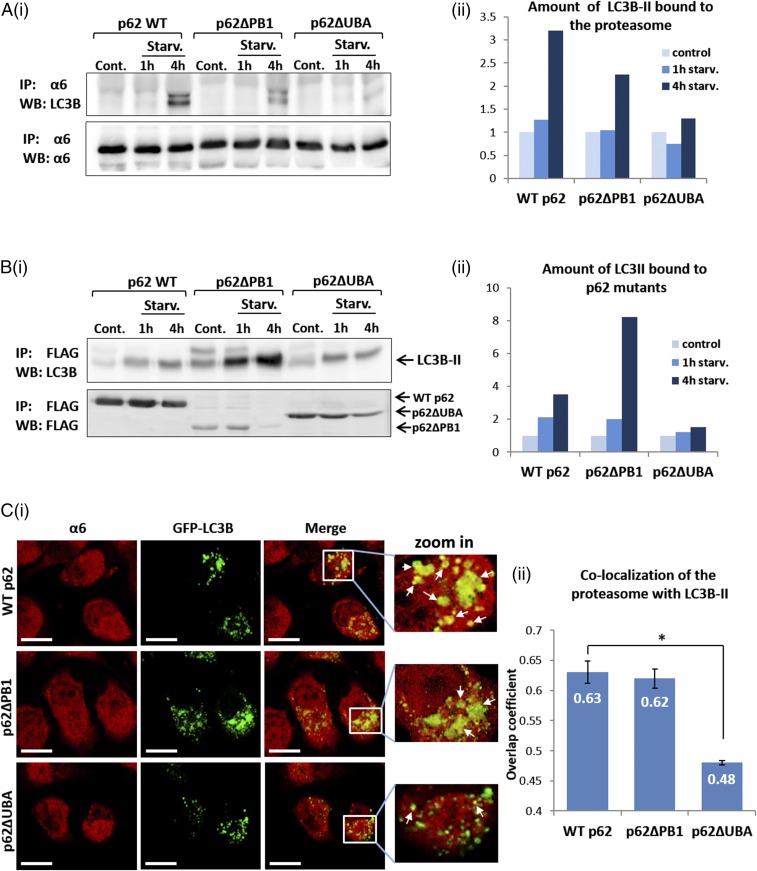
Autophagosomal Uptake of the Proteasome Is Mediated by the UBA Domain of p62 and Its Subsequent Interaction with LC3B-II.
Next, it was important to show that the UBA domain-mediated p62–proteasome interaction plays a role in the targeting of the proteasome to the autophagic machinery. We found that whereas WT p62 and p62∆PB1 facilitated interaction of the proteasome with LC3B-II after amino acid starvation, such an interaction was not evident in the absence of the UBA domain (Fig. 7 A and B). Notably, under basal conditions, this interaction did not occur even in the presence of WT p62 (Fig. 7A). Similarly, immunofluorescence staining of the WT p62- and p62∆PB1-expressing cells revealed increased autophagosomal uptake of the proteasome (Fig. 7C, Upper and Middle), in contrast to a significantly decreased uptake in the presence of p62∆UBA (Fig. 7C, Lower). In addition, deletion of the UBA domain decreased the interaction of p62 with LC3B-II after amino acid starvation (Fig. S3B). This finding may suggest that the lack of binding of p62 to the target ubiquitinated substrate also affects its interaction with the developing autophagosomal vesicle. Taken together, the foregoing results strongly suggest that after amino acid starvation, p62 recognizes the ubiquitinated proteasome via its UBA domain, making it also a connecting “bridge” with LC3B-II and thereby facilitating autophagosomal uptake of the proteasome. In general, this is the case for the uptake and destruction of other cargoes as well (10).
Fig. 7.
The interaction of the ubiquitinated proteasome with the UBA domain of p62 promotes its autophagosomal uptake. (A) (i) FLAG-tagged WT, ∆PB1, and ∆UBA p62s were transiently transfected to GFP-LC3B–expressing HeLa cells and either left untreated or starved to amino acids for the indicated times. Cell lysates were subjected to immunoprecipitation (IP) with anti-α6, resolved, and immunoblotted (WB) with anti-LC3B and anti-α6. (ii) Densitometric analysis of the amount of LC3B-II precipitated with the proteasome. The value of LC3B-II in the control lane of each p62 construct was arbitrarily set to 1. (B) (i) Same as under A, i, except that anti-FLAG was used to immunoprecipitate the different p62 species. (ii) Densitometric analysis of the amount of LC3B-II precipitated with the different p62 proteins. The value of LC3B-II in the control lane of each p62 construct was arbitrarily set to 1. (C) (i) WT, ∆PB1, and ∆UBA p62s were overexpressed in GFP-LC3B–expressing HeLa cells and stained with anti-α6 (red). (Scale bars: 20 μm.) White arrows indicate points of colocalization of the proteasome with LC3B-II. (ii) Coefficients of colocalization of the proteasome with LC3B-II in the different p62 species-expressing cells were calculated according to the method of Manders. *P < 0.000015.
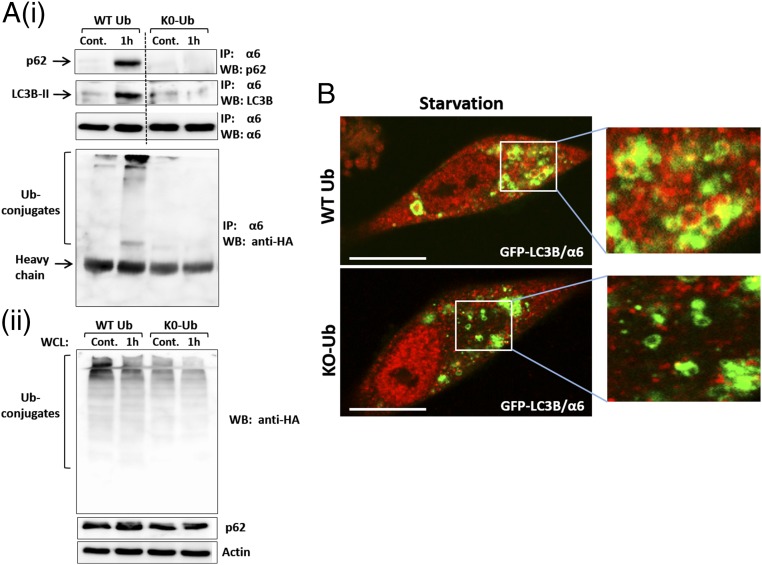
Autophagosomal Engulfment of the Proteasome Requires Its Polyubiquitination.
It has been shown that monoubiquitination can serve as a signal for both proteasomal (26) and autophagic degradation (29). To test the mode of modification required for autophagy of the proteasome, we overexpressed nonpolymerizable Ub (K0-Ub), in which all seven internal lysine residues are replaced with arginines (26). We found that although starvation of cells overexpressing WT Ub resulted in the formation of high molecular mass conjugates of the proteasome and concomitant interaction of the proteasome with the autophagic machinery (p62 and LC3B-II), the expression of the nonpolymerizable Ub inhibited both processes (Fig. 8).
Fig. 8.
Inhibition of polyubiquitination decreases uptake of the proteasome by autophagosomes. (A) (i) GFP-LC3B–expressing HeLa cells were transiently transfected with p62. At 24 h later, the cells were infected with adenoviruses coding for either WT HA-Ub or HA-K0-Ub. Cells were split and were either left untreated or starved for amino acids for 1 h. The proteasome was immunoprecipitated from cell lysates, and the precipitates were resolved and blotted with anti-p62, anti-LC3B, anti-α6, and anti-HA. (ii) Expression of WT Ub and K0-Ub (Upper) and p62 (Middle) was detected in whole-cell lysates (WCL). (B) Immunofluorescent staining with anti-α6 of GFP-LC3B–expressing HeLa cells infected with WT Ub (Upper) or K0-Ub (Lower) was performed after 4 h of amino acid starvation. (Scale bars: 20 μm.)
Discussion
As the catalytic arm of the UPS, the proteasome is involved in the breakdown of a wide spectrum of cellular proteins, making it a key component in the regulatory mechanisms of various vital cellular processes (34–36). The proteasome is stable, with a half-life of ∼1–2 wk (24). Like all proteins, the proteasome turns over not only under basal conditions, but also after several experimental manipulations, including its inhibition (37). Although much is known about the biogenesis of the proteasome, the pathway of its degradation remains poorly understood. It was recently shown that in yeast, nitrogen starvation results in vacuolar localization of the proteasome with its subsequent degradation (38). In addition, it was observed that the proteasome accumulates in rat liver lysosomes after leupeptin treatment or nutrient starvation (39). The mechanism by which the proteasome is delivered to the autophagosome has remained obscure, however.
In this study, we have demonstrated that in mammalian cells, amino acid starvation significantly increases engulfment of the proteasome by autophagosomes (Fig. 1 A–C), resulting in proteasome degradation (Fig. 1D). This process appears to be mediated by the mTOR pathway (Fig. 4C) and is preceded by a starvation-induced increase in ubiquitination of the proteasome, mostly on specific sites in subunits Rpn1, Rpn2, Rpn10, and Rpn13 (Fig. 2). This ubiquitination was found to be essential for the proteasome engulfment (Fig. 3). Furthermore, polyubiquitin chains must be built on the proteasome to allow its autophagosomal uptake to occur; overexpression of a nonpolymerizable Ub abolished the process (Fig. 8). The search for an adapter protein that recruits the proteasome to the autophagosome identified p62 (Figs. 4 and 5). Dissection of the domains in p62 required for delivery demonstrated that the UBA domain is essential (Figs. 6 and 7). This finding underscores the role of ubiquitination in specific targeting of the proteasome to the autophagic machinery.
Of note, although starvation-induced ubiquitination of the proteasome is observed already after a few hours, detecting degradation takes much longer. This is not surprising, given the time needed for the different steps involved in autophagic degradation of an organelle/complex (e.g., assembly, recognition, lysosomal fusion, degradation). A similar time scale has been observed in other cases of selective autophagy; for example, in mitophagy, membrane depolarization following oxidative stress peaks after 6 h and recovers over time although the stress is still present, whereas substantial mitochondrial removal is apparent only later (40). Importantly, the same holds true for selective autophagy of the proteasome in other organisms and experimental systems (37, 38). Along with the requirement for organization of the autophagic machinery, other factors may possibly affect the interval between ubiquitination and detectable degradation of the proteasome. Because the proteasome is an abundant protein, it seems reasonable to assume that only a small fraction of the entire population is undergoing autophagy at any given time, and thus significant degradation may be detected only after a relatively long period. In addition, it has been suggested that the effectiveness of lysosomal degradation varies among substrates. Thus, it has been shown that the degradation rate of proteins depends on their susceptibility to lysosomal proteases, which in turn correlates with their in vivo half-lives, with long-lived proteins being less susceptible to degradation (41). The proteasome is a large complex with a half-life of >1 wk, which may render it resistant to degradation.
Interestingly, inhibition of the proteasome has been reported to increase its ubiquitination on multiple subunits (37, 42), among them Rpn10 and Rpn13 (28). This process is likely mediated by proteasome-associated ligases (28). As noted above, inhibition of the proteasome also accelerates its destruction. Analysis of the ubiquitinated subunits and modification sites after proteasome inhibition revealed a striking similarity with our findings regarding the subunits and sites modified after amino acid starvation. This similarity strongly suggests that the signal that targets the proteasome for autophagy is highly specific and conserved. Moreover, and possibly not surprising, these subunits are localized on the distal part of the 19S RP, rendering them accessible for modification.
We also found a reduction in the ubiquitination of certain subunits, which also may serve as part of the signal, possibly via removal of Ub moieties that serve other regulatory roles and can be “inhibitory” for proteaphagy. In that context, we showed that monoubiquitination is insufficient for signaling the proteasome for autophagy. This finding may be related to the fact that a single Ub moiety or even several single moieties (multiple monoubiquitinations) are sterically hindered when conjugated to such a large complex like the proteasome. Therefore, polyubiquitination is required to provide the flexibility needed to accommodate the proteasome to the engulfing phagophore.
In agreement with this finding, for soluble proteins, a correlation has been found between the size of the protein and its mode of ubiquitination, with monoubiquitination characterizing proteins of low molecular mass (26, 43). The finding that starvation-induced ubiquitination occurs on Rpn1, Rpn10, and Rpn13, the known receptors for ubiquitinated substrates, raises the possibility that this modification inhibits binding of Ub-modified targets, a binding that is futile if the proteasome is defective/inactive and thus destined for destruction.
Another interesting problem relates to the binding of p62 to the proteasome. p62 can bind it via its PB1 domain, in which case it delivers ubiquitinated substrates for degradation. It also can bind it via its UBA domain, which binds to the proteasome Ub moieties, where it targets the proteasome for degradation. The way in which it binds likely depends on the specific mode of ubiquitination of the proteasome (which in turn is determined by the respective pathophysiological condition), as well as on other posttranslational modifications, such as phosphorylation (44).
An interesting question relates to the possible involvement of other mediators in starvation-induced proteaphagy. A study in Arabidopsis thaliana found that Rpn10 in its free state mediates selective proteaphagy after inhibition of the proteasome, but does not appear to play a role in a stress elicited by nitrogen starvation (37). It will be interesting to determine whether p62, which we have shown mediates starvation-induced proteaphagy in mammals, plays such a role in other organisms as well.
After amino acid deprivation, we also noted increased ubiquitination of cytoskeletal components, including α/β-tubulin and actin (Fig. S2). It will be interesting to study whether this modification facilitates the transport of the proteasome as part of its recruitment to the autophagic machinery.
Experimental Procedures
Cell Culture and Amino Acid Starvation.
GFP-LC3B HeLa cervical carcinoma cells were kindly provided by Zvulun Elazar (Weizmann Institute of Science), and nontransfected HeLa cells were purchased from American Type Culture Collection. Cells were cultured in DMEM medium supplemented with antibiotics, pyruvate, glutamine, and 10% FBS in a humidified atmosphere containing 5% CO2 at 37 °C. For amino acid starvation, the growth medium was removed and replaced after two washes with PBS by Earl’s Balanced Salt Solution (EBSS), supplemented with 100 μM CQ.
Cell Lysates, Immunoprecipitation, and Protein Blotting.
Cell cultures were washed twice with ice-cold PBS and scraped into lysis buffer (50 mM Tris⋅HCl pH 7.4, 130 mM NaCl, and 0.5% Nonidet P-40) supplemented with freshly added Protease Inhibitor Mixture (Roche), 5 mM ATP, 10 mM iodoacetamide, and 5 mM NEM. Protein concentration was determined by the BCA assay according to the manufacturer’s instructions (Pierce). Here 30 μg of cellular protein was resolved via SDS/PAGE, transferred to PVDF membrane, and immunoblotted with the appropriate antibodies. For immunoprecipitation, 600 µg of cellular protein was brought to a volume of 1 mL in lysis buffer (supplemented with Protease Inhibitor Mixture, 5 mM ATP, 10 mM iodoacetamide, and 5 mM NEM), incubated with the appropriate antibody for 2 h at 4 °C, and then incubated with protein G-Sepharose for 1 h at 4 °C. Beads were washed five times with the same buffer. Sample buffer was added, and samples were boiled and subjected to gel electrophoresis and immunoblotting as described above.
Immunofluorescence Microscopy.
Untreated or transiently transfected (cDNA or siRNA) cells were split and grown on glass coverslips for 24 h. After a 4-h incubation with Complete Medium or EBSS supplemented with CQ (0.1 mM), cells were fixed with 4% PFA for 20 min. The cells were then washed with PBS and subsequently incubated in PBS containing 10% normal goat serum for 1 h at room temperature, followed by a 2-h incubation with the indicated primary antibody. The cells were then washed extensively with PBS and incubated with the relevant secondary antibodies for 1 h, washed again, and mounted. Staining was analyzed using a Zeiss LSM 700 confocal microscope, and images were acquired under a 63× oil-immersion objective at a definition of 1,024 × 1,024 pixels with the pinhole diameter adjusted to 1 μm. All images were acquired using the same laser parameters and image magnification. Two-channel colocalization analysis and the Manders overlap coefficient of colocalization (45) were determined using ZEN software (Zeiss). Each graph represents the average of three independent experiments, with six fields acquired in each experiment.
SI Experimental Procedures
Antibodies and Reagents.
The following antibodies were purchased from Santa Cruz Biotechnology: anti–SQSTM-1/p62 (sc-25575), anti-LAMP1 (sc-18821), anti-calnexin (sc-11397), anti-20S proteasome β2 (sc-58410), and anti-PSMD2 (sc-271775). A polyclonal antibody to the proteasome 19S ATPase subunit Rpt6/S8 was purchased from Enzo Life Sciences. Anti-proteasome 19S subunit S3 (Rpn3) antibody was purchased from Thermo Fisher Scientific. Anti-S5a (Rpn10) CPTC-PSMD4 [52242.DSHB] and anti-α6 hybridomas were obtained from the Developmental Studies Hybridoma Bank. Anti-actin and anti-tubulin were purchased from Millipore. Anti-Ub conjugates (26) and anti-E1 (46) antibodies have been described previously. Alexa Fluor-conjugated (633 and 568) secondary antibodies were purchased from Invitrogen. VECTASHIELD Mounting Medium with DAPI was purchased from Vector Laboratories. Protein G was purchased from Roche Applied Science. Anti-FLAG M2 affinity gel and anti-FLAG and anti-LC3B antibodies were purchased from Sigma-Aldrich. Anti–K-ε-GG Immunoaffinity Beads were obtained from Cell Signaling Technology. Torin1 was purchased from TOCRIS. N-ethylmaleimide (NEM), iodoacetamide, and chloroquine were obtained from Sigma-Aldrich. Tissue culture media, supplements, and sera were purchased from Biological Industries. EBSS was purchased from Sigma-Aldrich. Peroxidase-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Restriction and modifying enzymes were purchased from New England Biolabs. Oligonucleotides were synthesized by Sigma-Aldrich. Lipofectamine RNAiMAX transfection reagent for siRNA transfection was purchased from Invitrogen. siRNAs were synthesized by Dharmacon. CalFectin reagent for cDNA transfection was purchased from SignaGen. Histodenz (Nycodenz) and OptiPrep for autophagosome purification were purchased from Sigma-Aldrich.
Cell Transfection.
Cells were transfected with cDNAs coding for p62 species using CalFectin transfection reagent. Lipofectamine RNAiMAX was used to transfect siRNA oligonucleotides. Transfections were carried out according to the manufacturers’ instructions. Infection with adenoviruses coding for WT and K0-Ub was conducted as described previously (27).
Plasmids.
Full-length cDNA encoding for SQSTM1/p62 was amplified by PCR with primers harboring FLAG and flanked with EcoRI and XhoI restriction sites (forward: 5′-CCGATCACGAATTCATGGACTACAAAGACGATGACGACAAGGCGTCGCTCACCGTGAAGGCC-3′; reverse: 5′- CGTACCGCTCGAGGATGGTGTCCAGAGCCGCTCC-3′). Subsequently, the purified SQSTM1/p62 PCR products were subcloned into a PCS2 vector. The FLAG-tagged SQSTM1/p62 construct served as a template to generate the SQSTM1 deletion mutants p62∆PB1 (aa 1–123) and p62∆UBA (aa 386–440).
Autophagosome Purification.
Autophagosome purification was carried out essentially as described by Seglen et al. (47), with some modifications. In brief, cells (1 × 108) were harvested with trypsin and washed twice with PBS. The cell pellet was resuspended with 2 mL of homogenization buffer (HB; 10 mM HEPES pH 7.3, 250 mM sucrose, and 1 mM EDTA). Cells were left for 10 min to swell on ice and then homogenized by 10 strokes in a 5-mL glass homogenizer. The cell homogenate was centrifuged at 400 × g for 2 min at 4 °C, and the supernatant was collected and centrifuged again at 1,500 × g. The postnuclear supernatant (PNS) was collected and placed on top of a discontinuous (two-step) Nycodenz gradient to pellet mitochondria [top layer, 2 parts of PNS; middle layer, 2.3 parts of 9.5% (vol/vol) Nicodenz; bottom layer, 1 part 22.5% (vol/vol) of Nycodenz]. The Nycodenz solution was prepared by dissolving Nycodenz in isotonic diluent (5 mM Tris⋅HCl pH 7.5, 217.5 mM sucrose, 3 mM KCL, and 0.3 mM EDTA). The Nycodenz gradient was centrifuged for 1 h at 140,000 × g at 4 °C. The ring (containing autophagosomes and ER) formed between the 9.5% and 22.5% (vol/vol) Nycodenz layers was collected and diluted with HB at a 1:1 ratio, and then layered on top of a Percoll-Nycodenz gradient [top layer, 1.714 parts of diluted ring; middle layer, 3 parts of 33% (vol/vol) Percoll; bottom layer, 1 part of 22.5% (vol/vol) Nycodenz]. The 33% (vol/vol) Percoll solution was prepared by mixing 33 mL of Percoll with 50 mL of double-strength HB (0.5 M sucrose, 20 mM HEPES, and 2 mM EDTA, pH 7.3) and 17 mL of DDW (double-distilled water). The Percoll-Nycodenz gradient was centrifuged at 72,000 × g for 30 min at 4 °C. After centrifugation, the upper part (ring) of the gradient was discarded (ER fraction). The innermost ring (autophagosomal fraction) was collected and diluted with 0.7 volume of 60% (vol/vol) buffered OptiPrep, which was prepared by mixing 100 parts of OptiPrep with 1 part of a homogenization concentrate (1 M HEPES, 100 mM EDTA, pH 7.3). To remove Percoll, 5.66 parts of diluted autophagosomal fraction was overlaid with 1 part of 30% (vol/vol) buffered OptiPrep [which was prepared by mixing 1 part of 60% (vol/vol) buffered OptiPrep with an equal part of HB], followed by centrifugation at 72,000 × g for 30 min at 4 °C. The uppermost ring (autophagosomes) was collected in a minimal volume, mixed with a threefold larger volume of HB, pelleted by centrifugation at 17,000 × g for 10 min at 4 °C, and then washed three times with PBS. The washed pellet was lysed in lysis buffer (50 mM Tris⋅HCl pH 7.4, 130 mM NaCl, and 0.5% Nonidet P-40) supplemented with freshly added Protease Inhibitor Mixture (Roche), 5 mM ATP, 10 mM iodoacetamide, and 5 mM NEM.
Sample Preparation for Mass Spectrometry.
Preparation of cell lysates from two biological replicates was carried out using the PTMScan Ubiquitin Remnant Motif (K-ε-GG) Kit (Cell Signaling Technology) according to the manufacturer's instructions. In brief, cell cultures were washed twice with PBS and scraped into urea lysis buffer supplemented with freshly added 10 mM iodoacetamide. Samples were sonicated on ice using a microtip at 15W output with three bursts of 15 s each. Cell lysates were centrifuged, and 2–3 mg of total protein was incubated with DTT (2.8 mM; 30 min at 60 °C) and iodoacetamide (10 mM; 30 min at room temperature in the dark) and then digested (for 20 h at 37 °C) with trypsin (Promega; 1:50 enzyme:substrate ratio) in 2 M urea and 25 mM ammonium bicarbonate. The tryptic peptides were desalted using Sep-Pak C18 (Waters) and lyophilized using a SpeedVac concentrator (Thermo Fisher Scientific). Proteome analysis was performed by LC-MS/MS using 10 µg of the total protein. Part of the residual material was used for detection of ubiquitination sites, as described by Braten et al. (26).
Mass Spectrometry.
Samples for MS and LC-MS/MS analysis of tryptic peptides were prepared as described elsewhere (26) using a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) fitted with a capillary liquid chromatograph (EASY nLC 1000; Thermo Fisher Scientific).
Data Analysis.
Data analysis was carried out as described elsewhere (26), with the following additions. The starvation:control ratios were normalized based on the measured median ratio for each biological replicate. Peptides with a log2 ratio of <−1 or >1 in both biological replicates were considered differentially regulated. All of the data are available in Dataset S1.
Supplementary Material
Acknowledgments
Research in the laboratory of A.C. is supported by grants from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Israel Science Foundation (ISF), the I-CORE Program of the Planning and Budgeting Committee and the ISF (Grant 1775/12), the Deutsch-Israelische Projektkooperation, and a special fund for research in the Technion established by Albert Sweet. I.L. is supported by the Foulkes Fellowship. A.C. is an Israel Cancer Research Fund USA Professor.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 13266.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615455113/-/DCSupplemental.
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78(1):477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen-Kaplan V, Livneh I, Avni N, Cohen-Rosenzweig C, Ciechanover A. The ubiquitin-proteasome system and autophagy: Coordinated and independent activities. Int J Biochem Cell Biol. 2016 doi: 10.1016/j.biocel.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 3.Ruggiano A, Foresti O, Carvalho P. Quality control. ER-associated degradation: Protein quality control and beyond. J Cell Biol. 2014;204(6):869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 5.Hartmann-Petersen R, Gordon C. Protein degradation: Recognition of ubiquitinylated substrates. Curr Biol. 2004;14(18):R754–R756. doi: 10.1016/j.cub.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Finley D, Chen X, Walters KJ. Gates, channels, and switches: Elements of the proteasome machine. Trends Biochem Sci. 2016;41(1):77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomko RJ, Jr, Hochstrasser M. Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem. 2013;82:415–445. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell SJ, Steger KA, Johnston SA. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J Biol Chem. 1999;274(31):21943–21952. doi: 10.1074/jbc.274.31.21943. [DOI] [PubMed] [Google Scholar]
- 9.Driscoll J. The role of the proteasome in cellular protein degradation. Histol Histopathol. 1994;9(1):197–202. [PubMed] [Google Scholar]
- 10.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 11.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wileman T. Autophagy as a defence against intracellular pathogens. Essays Biochem. 2013;55:153–163. doi: 10.1042/bse0550153. [DOI] [PubMed] [Google Scholar]
- 13.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 15.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4(2):141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 16.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22(1):R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 17.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isogai S, et al. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J Biol Chem. 2011;286(36):31864–31874. doi: 10.1074/jbc.M111.259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjørkøy G, et al. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 20.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Ichimura Y, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283(33):22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 22.Seibenhener ML, et al. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24(18):8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters LZ, et al. The protein quality control machinery regulates its misassembled proteasome subunits. PLoS Genet. 2015;11(4):e1005178. doi: 10.1371/journal.pgen.1005178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka K, Ichihara A. Half-life of proteasomes (multiprotease complexes) in rat liver. Biochem Biophys Res Commun. 1989;159(3):1309–1315. doi: 10.1016/0006-291x(89)92253-5. [DOI] [PubMed] [Google Scholar]
- 25.Reggiori F, Komatsu M, Finley K, Simonsen A. Autophagy: More than a nonselective pathway. Int J Cell Biol. 2012;2012:219625. doi: 10.1155/2012/219625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braten O, et al. Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc Natl Acad Sci USA. 2016;113(32):E4639–E4647. doi: 10.1073/pnas.1608644113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tai H-C, Besche H, Goldberg AL, Schuman EM. Characterization of the brain 26S proteasome and its interacting proteins. Front Mol Neurosci. 2010;3:12. doi: 10.3389/fnmol.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Besche HC, et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 2014;33(10):1159–1176. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008;105(52):20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 31.Jewell JL, Russell RC, Guan K-L. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14(3):133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zientara-Rytter K, Sirko A. Significant role of PB1 and UBA domains in multimerization of Joka2, a selective autophagy cargo receptor from tobacco. Front Plant Sci. 2014;5:13. doi: 10.3389/fpls.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 35.Naujokat C, Hoffmann S. Role and function of the 26S proteasome in proliferation and apoptosis. Lab Invest. 2002;82(8):965–980. doi: 10.1097/01.lab.0000022226.23741.37. [DOI] [PubMed] [Google Scholar]
- 36.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 37.Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell. 2015;58(6):1053–1066. doi: 10.1016/j.molcel.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waite KA, De La Mota-Peynado A, Vontz G, Roelofs J. Starvation induces proteasome autophagy with different pathways for core and regulatory particle. J Biol Chem. 2015;291(7):3239–3253. doi: 10.1074/jbc.M115.699124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995;227(3):792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- 40.Sargsyan A, et al. Rapid parallel measurements of macroautophagy and mitophagy in mammalian cells using a single fluorescent biosensor. Sci Rep. 2015;5:12397. doi: 10.1038/srep12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segal HI, Winkler JR, Miyagi MP. Relationship between degradation rates of proteins in vivo and their susceptibility to lysosomal proteases. J Biol Chem. 1974;249(19):6364–6365. [PubMed] [Google Scholar]
- 42.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shabek N, et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell. 2012;48(1):87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Livneh I, Cohen-Kaplan V, Cohen-Rosenzweig C, Avni N, Ciechanover A. The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res. 2016;26(8):869–885. doi: 10.1038/cr.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. J Microsc. 1993;169(3):375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 46.Mayer A, Siegel NR, Schwartz AL, Ciechanover A. Degradation of proteins with acetylated amino termini by the ubiquitin system. Science. 1989;244(4911):1480–1483. doi: 10.1126/science.2544030. [DOI] [PubMed] [Google Scholar]
- 47.Seglen PO, Brinchmann MF. Purification of autophagosomes from rat hepatocytes. Autophagy. 2010;6(4):542–547. doi: 10.4161/auto.6.4.11272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.