Significance
We identified the biosynthetic pathway for the nonsugar sweetener mogroside V, a noncaloric with a sweetening strength 250-fold that of sucrose. This compound is produced by the fruit of the endemic Chinese cucurbit Siraitia grosvenoriii, also known as monk fruit and luo-han-guo. The metabolic pathway was identified using a combination of genomic and transcriptomic databases of the Siraitia plant, together with a large-scale functional expression of candidate genes. The novelty of the pathway could be attributed to a highly coordinated gene expression pattern responsible for the unique epoxidations, hydroxylations, and glucosylations leading to the sweet mogrosides. These discoveries will facilitate the development of alternative natural sweeteners.
Keywords: mogrosides, metabolic pathway discovery, functional genomics
Abstract
The consumption of sweeteners, natural as well as synthetic sugars, is implicated in an array of modern-day health problems. Therefore, natural nonsugar sweeteners are of increasing interest. We identify here the biosynthetic pathway of the sweet triterpenoid glycoside mogroside V, which has a sweetening strength of 250 times that of sucrose and is derived from mature fruit of luo-han-guo (Siraitia grosvenorii, monk fruit). A whole-genome sequencing of Siraitia, leading to a preliminary draft of the genome, was combined with an extensive transcriptomic analysis of developing fruit. A functional expression survey of nearly 200 candidate genes identified the members of the five enzyme families responsible for the synthesis of mogroside V: squalene epoxidases, triterpenoid synthases, epoxide hydrolases, cytochrome P450s, and UDP-glucosyltransferases. Protein modeling and docking studies corroborated the experimentally proven functional enzyme activities and indicated the order of the metabolic steps in the pathway. A comparison of the genomic organization and expression patterns of these Siraitia genes with the orthologs of other Cucurbitaceae implicates a strikingly coordinated expression of the pathway in the evolution of this species-specific and valuable metabolic pathway. The genomic organization of the pathway genes, syntenously preserved among the Cucurbitaceae, indicates, on the other hand, that gene clustering cannot account for this novel secondary metabolic pathway.
Sweetness is one of the fundamental human hedonic pleasures (1), even more reinforcing and attractive than drugs such as heroin and cocaine (2). However, in satisfying this desire, sugar consumption has risen exponentially from nearly 250 y ago and meta-analyses implicate sugar consumption in the development of obesity, diabetes, metabolic syndrome, and cardiovascular diseases (3). Noncaloric artificial sweeteners offer hope for calorie reduction and, although there has been a general acceptance of many of them as safe for consumption (4), recent research has pointed to effects of synthetic sweeteners on the intestinal microbiome, ironically leading to metabolic syndrome and glucose intolerance (5, 6). In light of the problems associated with both natural sugar and synthetic noncaloric sweeteners, there is great interest in developing alternative natural nonsugar sweeteners to satisfy the human “need” for sweet.
The natural compounds with strong sweetening capacity belong to numerous chemical families, including proteins, flavonoids, and terpenoids (7). The mogroside family of triterpenoids, derived from the ripe fruit of the Chinese cucurbit, Siraitia grosvenorii [Cucurbitaceae, luo-han-guo or monk fruit, discovered and classified initially in the 1930s (8)], is used as a natural sweetener in China, having a sweetening strength of 250 times that of sucrose (9). The mogrosides are derived from the cucurbitane skeleton of triterpenoids, one of hundreds of possible cyclic triterpenoid backbones (10, 11), which is ubiquitous throughout the Cucurbitaceae. The intensely bitter cucurbitacins are derivatives of the same skeleton, but differ from the intensely sweet mogrosides primarily in the oxygenated decorations on the cucurbitane backbone.
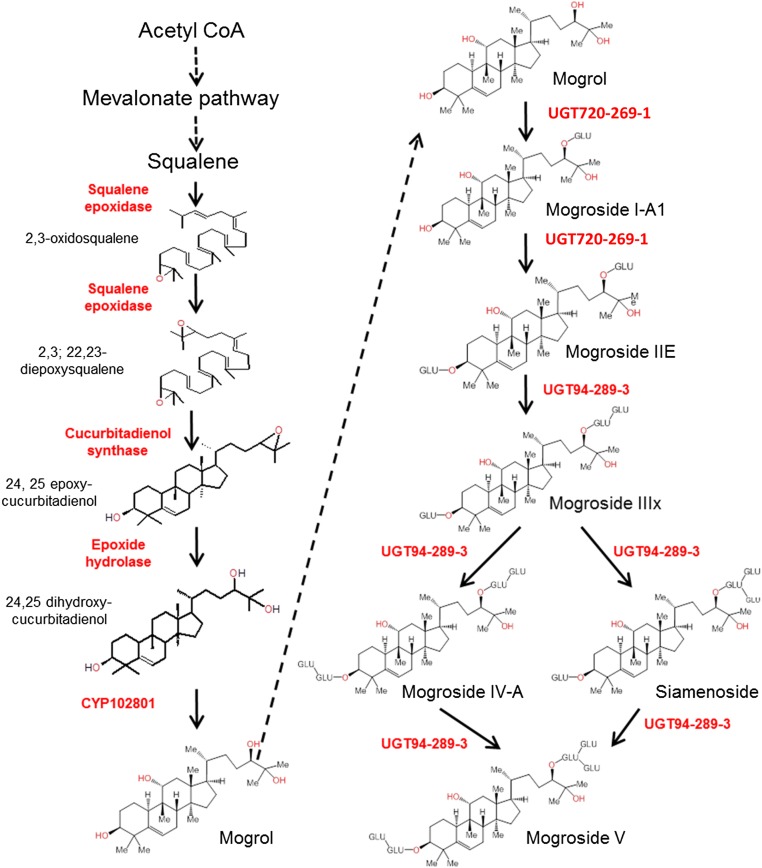
The novelty of the mogrosides among the cucurbitane triterpenoids are their four regio-specific oxygenations, at C3, C11, C24, and C25, forming the tetra-hydroxylated cucurbitane, mogrol (Fig. 1). Hydroxylation of triterpenoids at the C3 position is ubiquitous because it is inherent in the cyclization of the squalene monoxygenase substrate, and hydroxylations of triterpenoids at C11 are fairly common. The C24 and C25 trans-hydroxylations in the cucurbitanes are rare, reported in only a few instances in the Cucurbitaceae (12).
Fig. 1.
Schematic diagram of the proposed pathway for mogroside biosynthesis in fruit of S. grosvenorii. The left portion of the schematic represents the steps leading to the nonglycosylated tetra-hydroxycucurbitane, mogrol. The right side indicates the successive glucosylations. Enzyme names and numbers are described in the text.
The enzymes responsible for these oxygenations are not known and could be members of numerous enzyme families from multiple groups of monooxygenases, dioxygenases, and epoxidases (cytochrome P450-, Fe-, α-ketoglutaric acid–, FAD-, or NAD-linked). Because these enzyme families each comprise multiple members, with the cytochrome P450s alone comprising the largest family of plant enzymes with more than 100 representatives in species studied (13, 14), a purely biochemical search for the enzymes responsible for the oxygenations would be Sisyphean.
Following the oxygenation reactions, the mogrosides are successively glucosylated, at positions C3 and C24, to yield from one to six glucosyl groups, and up to three at each of the two positions (15) (Fig. 1). These include the primary glucosylation events at each of the C3 and C24 positions, as well as branched glucosylations on the two primary glucosyl moieties. The perceived taste of the mogrosides is dependent on the number of glucosylations. Mogrosides with four glucosyl groups or more are intensely sweet and the penta-glucosylated M5 makes up the major component of the commercial sweet powder derived from the mature monk fruit. Similar to the oxygenation reactions, the identification of the glycosylation enzymes is also a challenge because they are expected to be members of the large family 1 UGTs, the second largest family of plant enzymes, comprising over 100 members in plant species studied (16).
A paradigm breakthrough of the past decade in the area of novel secondary plant metabolism has been the discovery of “operon-like” or regulon gene clustering (17, 18), reported so far in over a dozen biosynthetic pathways including those encoding, for example, cucumber bitter triterpenoid cucurbitacins (19) and tomato tomatine glycoalkaloids (20). Wada et al. (21) estimated about 100 such gene clusters for secondary metabolism in the Arabidopsis genome, and other plant species are likely not to be different. We therefore initially hypothesized that the novel secondary metabolic pathway of sweet mogroside synthesis might be similarly clustered, which could expedite the discovery of the pathway.
To uncover the metabolic pathway leading to sweet mogroside accumulation, we prepared a draft genome of S. grosvenorii, combined with a transcriptomic analysis of the fruit through development and supported by large-scale functional analysis of candidate genes. Time-resolved comparative transcriptomic and metabolomics databases are a useful strategy for the elucidation of biochemical pathways in plant secondary metabolism (20, 22). An earlier analysis of the transcriptome of three stages of Siraitia fruit development (23) proposed a limited number of candidate genes. Following the identification of the pathway, the comparison of our Siraitia genomic and transcriptomic database with those of other Cucurbitaceae allowed us to shed light on the evolution and novelty of the mogroside pathway within this plant family and to offer perspective to the clustering paradigm of plant secondary metabolic pathways.
Results
Developmental Accumulation of Mogrosides.
We determined the mogroside levels in developing fruit and vegetative tissues of the Siraitia plant to correlate the spatial and temporal metabolite levels with the transcription patterns of the candidate genes. Mogrosides were limited to the developing fruit and were not observed in the root, stem, or leaf tissue. The developmental pattern of mogroside accumulation in the fruit clearly indicated progressive glycosylations to the mogrol moiety (SI Appendix, Fig. S1A and Table S1). At the youngest stage of immature fruit, at 15 days after anthesis (DAA), the majority of the mogrosides were present in the di-glucosylated form in which both the C3 and C24 carbons were monoglucosylated. Nonglucosylated or alternative M2 compounds, in which the second glucosyl moiety was present as a branched glucose on one of the primary glucose moieties, were not observed, indicating that the initial metabolic steps of mogroside glucosylations are the two primary glucosylations and that these occur early in fruit development.
Of particular significance is the net conservation of mogroside content in the developing fruitlet. The total mogroside levels in the fruit remained similar throughout development, and there was certainly no indication of a net increase in mogroside levels with development (SI Appendix, Fig. S1B). This observation coincides with data presented in an earlier study of Siraitia fruit that also showed that the total mogroside content did not increase during development (24). Rather, the existing mogroside pool in the immature fruit was progressively glucosylated to the penta-glucosylated form. These results indicate a strong temporal division of mogroside metabolism and that the early steps of the synthesis of the aglycone mogrol and the initial primary glucosylations are limited to very early fruit development, preparing a reservoir of mogrosides for subsequent glucosylations later in development.
Following the synthesis of M2, there was an additional branched 1–6 glycosylation at the C24 position leading to the accumulation of M3X. During the later stages (77 and 90 DAA) a number of M4 compounds appeared, primarily siamenoside, which was confirmed by NMR as containing a third, branched glucosylation at the C24 position (SI Appendix, Fig. S1A). Alternative tetra-glucosylated mogrosides, such as M4A, were also present but in low amounts. M5, with a second glucosylation at the C3 position, began to accumulate at the expense of the M4 compounds at 77 DAA and increased sharply during the final stages of ripening. The major fruit mogroside component in the ripe 103-DAA fruit was M5, along with small traces of isomogroside V, IM5.
Siraitia Genome Assembly and Developmental Transcriptome.
The genome assembly of S. grosvenorii was developed largely based on the Illumina TSLR platform (TruSeq Synthetic Long Reads, formerly known as Moleculo) in combination with additional libraries (SI Appendix, Table S2). The preliminary assembly, based on 15.5 Gb of mixed library reads, including over 400,000 long reads of an average 6,000 bp from the TSLR, comprises 12,772 scaffolds with an N50 of over 100 kb. The estimated total genome size, following filtering and removal of organellar genome sequences, of ∼420 MB (SI Appendix, Table S3) is similar to that of three other cucurbits, Citrullus lanatus (watermelon), Cucumis melo (melon), and Cucumis sativus (cucumber) (https://genomevolution.org/wiki/index.php/Sequenced_plant_genomes). Although not complete, this assembly served the purpose of identifying the genomic organization of the gene families of interest, including gene duplications and clustering characteristics.
In combination with the genome, we performed a transcriptome assembly and expression analysis (based on an Illumina RNA-seq of over 300 million reads) of the developing Siraitia fruit samples, together with those of the root, leaf, and stem portions (SI Appendix, Tables S4 and S5) and ∼78% of the transcript reads mapped to the genome assembly. The reads were de novo assembled using the CLC-BIO software, resulting in a total of ∼111,000 Siraitia contigs, representing potential RNA units. Of these, over 43,856 were homologous to genes in the melon genome database (Dataset S1).
Based on this Siraitia genomic and transcriptomic bioinformatics infrastructure, combined with BLAST searches against known catalogs of the gene families of interest, we collected a complete set of the Siraitia members of the large gene families of Cytochrome P450-dependent monooxygenase (CYP) 450s and UGTs, as well as of the smaller gene families of squalene epoxidases (SQE), triterpene synthases, and epoxide hydrolases (EPH), and characterized their expression patterns based on the RNA-seq data by remapping these members to the genome assembly (Dataset S2). Candidate genes for functional expression were selected largely based on these expression patterns in light of the mogroside accumulation patterns. We posited that the early metabolic stages leading to mogrol synthesis, together with the two primary glycosylations, would be highly expressed in the young fruit stage and that the branching enzyme genes would be up-regulated during the later stages of fruit development.
Functional Expression.
We began our systematic functional expression studies from the steps following the synthesis of squalene, carried out by squalene synthase (Fig. 1). There is only a single copy of a squalene synthase gene in the Siraitia genome, as is also the case in other cucurbit genomes (SI Appendix, Table S6A). The expression pattern of this gene (SI Appendix, Table S6B) supports a ubiquitous role for the enzyme in general plant metabolism, as squalene serves as the precursor for all triterpenoid and sterol biosynthesis.
SQE.
The Siraitia genome harbors five genes encoding squalene epoxidases (SI Appendix, Fig. S2A). Of these, two showed high expression in the 15-d fruit (SI Appendix, Fig. S2B). Squalene epoxidase can carry out two successive epoxidations, performing the mirror image epoxidations of both the 2,3 and 22,23 end positions of the squalene molecule. Di-epoxidation of squalene by squalene epoxidase has been reported in numerous triterpenoid synthase systems (25–29), and plant squalene epoxidases have been functionally expressed and shown to yield both mono- and di-oxidosqualene (30, 31). Modeling of the SgSQE protein supports di-epoxidation and indicates that the presence of the first epoxy oxygen does not hinder the docking for the second epoxidation (SI Appendix, Fig. S3). The erg7− yeast line GIL77 (32), null for the triterpene cyclase lanosterol synthase, which we used for functional expression studies, accumulates di-oxidosqualene together with mono-oxidosqualene, due to endogenous squalene epoxidase (Fig. 2A), allowing for the identification of the di-oxidosqualene-based biosynthesis pathway of mogrol.
Fig. 2.
Cucurbitadienol synthase yields both cucurbitadienol and epoxycucurbitadienol. (A) Extracted ion chromatogram of CDS activity. The control panel shows the yeast GIL77 erg7− accumulation of both 2,3 oxidosqualene (peak 6) and 2,3;22,23-diepoxysqualene (peak 4) in the absence of lanosterol synthase activity. In the presence of SgCDS (Lower) both cucurbitadienol (peak 7) and 24,25-epoxycucurbitadienol (peak 3) are accumulated. Corroborative results with transgenic tobacco plants expressing SgCDS are presented in SI Appendix, Fig. S4. Mass spectra of compounds are presented in SI Appendix, Fig. S5, and NMR results for cucurbitadienol and 24,25-epoxycucurbitadienol are presented in SI Appendix, Table S8. (B) Modeling of CDS with epoxycucurbitadienol. Calculated affinities for cucurbitadienol and epoxycucurbitadienol are, respectively, −12.3 and −12.5 kcal/mol. Detailed docking model is presented in SI Appendix, Fig. S6.
Triterpene cyclases: cucurbitadienol synthase.
The single cucurbitadienol synthase (CDS) gene in the Siraitia genome was functionally expressed in the di-oxidosqualene producing the erg7− yeast line and indeed produced both cucurbitadienol and 24,25-monoepoxycucurbitadienol (Fig. 2A). We substantiated this finding by expressing the SgCDS in tobacco plants as well, and leaf tissue of transgenic tobacco plants also synthesized 24,25-monoepoxycucurbitadienol (SI Appendix, Fig. S4). Homologous CDS genes from various Cucurbitaceae have been functionally implicated in the synthesis of cucurbitadienol since its initial identification in Cucurbita (19, 32–34), but 24,25-monoepoxycucurbitadienol, derived from the cyclization of the 2,3;22,23 di-oxidosqualene, has not previously been reported as a product. Modeling of the SgCDS protein indicated that both 2,3 mono-oxidosqualene and 2,3;22,23 di-oxidosqualene can be cyclized and that the additional epoxy group at the 22,23 position does not interfere with the docking (Fig. 2B).
Other triterpene cyclases have also been reported to cyclicize di-oxidosqualene, leading to cyclic triterpenoids with an epoxy group remaining at the 24,25 position. This has been most clearly shown with regard to cholesterol metabolism in which the di-oxidosqualene is actually the preferred substrate for lanosterol synthase in the synthesis of the 24,25-epoxycholesterol, which inhibits the reaction with mono-oxidosqualene and serves as part of a feedback control in cholesterol biosynthesis (27, 35, 36).
EPH.
The major challenge in identifying the novel steps of mogrol synthesis is its unique hydroxylations, specifically the trans-24,25 hydroxyl pair. We initially hypothesized that cytochrome P450 enzymes would be responsible for all of the mogrol hydroxylations. Of a total of 191 Siraitia CYP450 genes (described in following section), over 40 that showed expression in the young fruit stage were functionally expressed in the cucurbitadienol-producing yeast line. However, none were observed to carry out either C24 or C25 cucurbitadienol hydroxylations. The absence of expressed CYP450 genes of the immature Siraitia fruit capable of C24 and C25 hydroxylations led us to examine members of the epoxide hydrolase family (EC 3.3.2.9) as candidates for the synthesis of the trans-24,25-dihydroxycucurbitadienol from the 24,25-epoxycucurbitadienol formed in the CDS reaction.
The Siraitia genome contains eight genes encoding annotated epoxide hydrolases (Fig. 3A), three of which showed high expression in the 15-DAA fruit. The three—SgEPH1, SgEPH2, and SgEPH3—were individually expressed in the GIL77 yeast containing SgCDS (with endogenous yeast squalene epoxidase and epoxide hydrolase activities), and all three yielded approximately threefold increases in levels of 24,25-dihydroxycucurbitadienol, compared with the yeast containing SgCDS alone (Fig. 3B; SI Appendix, Fig. S7). Docking modeling of the active SgEPH proteins with 24(R),25-epoxycucurbitadienol indeed showed strong affinities and perfect positioning of the 24,25 epoxide oxygen with the established EPH catalytic residues (Fig. 3C; SI Appendix, Fig. S8).
Fig. 3.
EPH expression, activity, and protein-docking model. (A) Hierarchial cluster heat map of expression patterns of the eight epoxide hydrolase genes expressed in the developing Siraitia fruit. The five stages of fruit development presented are 15, 34, 51, 77, and 103 DAA. Genes EPH1, EPH 2, and EPH 3 that showed high expression in young fruit were functionally expressed in yeast (chromatograms presented in SI Appendix, Fig. S7). (B) Relative levels of di-hydroxycucurbitadienol and epoxycucurbitadienol in the control and EPH-expressing yeast lines. Metabolites were identified by LC-MS as described in SI Appendix, Methods, and quantification is presented as peak area of the chromatograms in SI Appendix, Fig. S7. Three independent cultures were analyzed, and results are presented as means ± SD. (C) Docking modeling of SgEPH protein with 24(R),25-epoxycucurbitadienol showing strong affinities and perfect matches with the epoxide oxygen positioned just between the two tyrosine residues and the nucleophile asp-101 in close proximity (3.0 Å) to the C24 and C25 positions. Detailed description of the docking model is provided in SI Appendix, Fig. S8.
CYP 450.
The genomic and transcriptomic databases identified 191 members of the family (Fig. 4A; Dataset S2). Numerous studies of CYP function have identified members of multiple families with hydroxylation activity with triterpenoids (SI Appendix, Table S7, CYP families 51, 705, 710, 716, 72, 73, 81, 85, 88, 90, and 93), thereby necessitating functional expression of ∼40 CYP450 genes expressed in young Siraitia fruit (SI Appendix, Fig. S10). These were expressed in yeast (together with the Arabidopsis CYP450 reductase, AtCPR), and two were identified that produced hydroxylated cucurbitadienol products. One catalyzed the mogrol-type C11 hydroxylation (Fig. 4B). The C11 hydroxylase phylogenetically classifies as a member of CYP87 family: CYP87D18 of clan 85. During the preparation of this article the CYP87D18 was reported by Zhang et al. (37) The second active CYP, CYP88L4 of clan 85, yielded a C19 hydroxylation product (Fig. 4B). The cucumber ortholog of the latter, Csa3G903540 (58% identity), was recently reported to also carry out the C19 hydroxylation in cucumber cucurbitacin biosynthesis (19). In the same study of cucumber cucurbitacin metabolism, a CYP450 that carried out C25 hydroxylation was functionally identified (19) (Csa6G088160), but the closest Siraitia ortholog to this gene (contig 24468, listed as 9654_1, CYP81Q58, 74% identity) is not expressed in the developing Siraitia fruit but only in the root and thus cannot be responsible for mogroside C25 hydroxylation.
Fig. 4.
CYP450 family: expression and activity with cucurbitadienol. (A) Phylogenetic tree of the cytochrome P450 genes (expandable version in SI Appendix, Fig. S9). Protein sequences used are presented in Dataset S2. (B) LC-MS analysis of extracts of yeast coexpressing SgCDS with CYPs showing cucurbitadienol-hydroxylating activity. The extracted ion chromatogram (m/z = 407–444) represents relevant triterpenoid compounds and derivatives accumulated in the yeast. Yeast coexpressing SgCDS with CYP87D18 (Middle chromatogram) produced mainly 11-hydroxy cucrbitadienol (peak 5), which coeluted with 11-oxo-cucurbitadienol. Yeast coexpressing SgCDS with CYP88L4 (Bottom chromatogram) produced mainly 19-hydroxycucurbitadienol (peak 11). A chromatogram from yeast harboring SgCDS alone is illustrated in the Upper chromatogram as negative control. (C) Production of mogrol in yeast extracts expressing SgCDS, SgEPH3, SgCYP87D18, and AtCPR in presence (+) and absence (−) of lanosterol synthase inhibitor R0 48–8072 at 24 and 48 h after addition of the inhibitor. Lower chromatogram shows the mogrol standard (peak 1), and the mogrol can be identified at 48 h after inhibitor addition (48 h+). Peak numbers are as follows: 1—mogrol; 2—24,25-dihydroxycucurbitadienol; 3—unidentified C30H48O3; 4—unidentified C30H52O3; 5—11-hydroxycucurbitadienol; 6—24,25-epoxycucurbitadienol; 7—2,3;22,23-diepoxysqualene; 8—2,3-oxidosqualene; 9—cucurbitadienol; 10—lanosterol; and 11—19-hydroxycucurbitadienol. MS spectra and NMR results are represented in SI Appendix, Fig. S5 and Table S8, respectively.
To determine whether the coexpression of SgCDS, SgEPH3, SgCYP87D18, and AtCPR leads to mogrol, we used the yeast strain BY4743_YHR072, which could accommodate multiple expression constructs. To reduce the flux through the native lanosterol synthase reaction in this strain, we incorporated the lanosterol synthase inhibitor R0 48–8072 in the medium (SI Appendix, Methods). The results indicated that, although in the absence of the lanosterol synthase inhibitor there accumulated only cucurbitadienol and 11-OH cucurbitadienol, the addition of the inhibitor led to the accumulation of the epoxide and 11-OH intermediates at 24 h and to the synthesis of mogrol after 48 h. (Fig. 4C).
Order of the preglucosylation reactions.
Protein-modeling and docking studies support the following order of reactions in the synthesis of mogrol. Our first conclusion is that 2,3;22,23-diepoxysqualene, rather than 2,3-monoepoxysqualene, is the substrate for the cyclization reaction by CDS. This is due to the narrow pocket size of the squalene epoxidase enzyme, making the accommodation of a cyclized substrate unlikely for a second epoxidation (SI Appendix, Fig. S3). Second, the epoxide hydrolase reaction likely follows the CDS cyclization because the EPH protein prefers the cyclic epoxycucurbitadienol over the noncyclic 2,3;22,23-diepoxysqualene (SI Appendix, Fig. S11). Finally, the C11 hydroxylation takes place after the EPH reaction because the additional hydrophilic OH at C11 would preclude docking in the hydrophobic pocket of the hydrolase (SI Appendix, Fig. S12). Accordingly, the proposed pathway of mogrol biosynthesis is as presented in Fig. 1: squalene to mono-oxidosqualene, to di-oxidosqualene, to 24,25-epoxycucurbitadienol, to 24,25-dihydroxycucurbitadienol, to 11,24,25-trihydroxycucurbitadienol (mogrol).
Primary glucosylations.
Previous studies of UGT function have identified members of multiple UGT families (71, 73, 74, 91, and 94) as active with various triterpenoid acceptors (SI Appendix, Table S9). To identify the UGT enzymes responsible for mogrol glucosylation, of the total ∼131 UGTs in the Siraitia genome (Fig. 5A; SI Appendix, Fig. S13; Dataset S2), we functionally expressed nearly 100 genes that showed expression in the developing fruit (SI Appendix, Fig. S14). These were expressed in Escherichia coli and tested for activity with each of the possible mogroside substrates, ranging from M to M5 (SI Appendix, Table S1).
Fig. 5.
Siraitia UGTs involved in mogroside glucosylations. (A) Phylogenetic tree of Siraitia UGTs (expandable version in SI Appendix, Fig. S13). UGTs referred to in this study are boxed in red. Protein sequences used are presented in Dataset S2. (B) Schematic summary of primary glucosylation reactions using the various substrate precursors, as described in SI Appendix, Fig. S1 and Methods. The schematic representation of the mogroside compounds comprises two blue ovals, representing two cyclic rings each of the tetracyclic cucurbitane skeleton; red circles represent glucosyl moieties in the substrate, and green circles represent the newly glycosylated positions due to the reaction. In the case of branched glycosylations, a 1–6 glucosyl arrangement is represented with the attached circle pointing upwards, while a 1–2 arrangement is represented by the attached circle pointing downwards. (C) Chromatograms showing the single glucosylation performed by UGT720-269–4 (Upper chromatogram) and the double glucosylation performed by UGT720-269–1 (Bottom chromatogram). (D) Schematic summary of branched glucosylations using various substrates, described in SI Appendix, Table S1. Chromatograms and MS data are presented in SI Appendix, Fig. S15.
The results identified three genes that supported C3 glucosylations: members of UGT families 74, 75, and 85 (reclassified recently as UGT720). A fourth gene, also from the UGT720 family (UGT720-269-1), was the only UGT identified capable of the C24 primary glucosylation (Fig. 5B; SI Appendix, Fig. S15). Additional enzymes from the UGT73 family produced mixtures of C24 and C25 glucosylations (SI Appendix, Fig. S16), as determined by NMR of the products (SI Appendix, Table S8), and this C24/C25 regio-specificity could be explained by protein modeling and docking analysis (SI Appendix, Fig. S17).
Most interesting, however, was the observation that UGT720-269–1 was not only capable of carrying out the primary C24 glucosylation of mogrol, but also subsequently performed the C3 primary glucosylation of C24-glucosylated M1A1, thus accounting for the synthesis of the di-glucosylated M2-E (Fig. 5C). Modeling of the UGT720-269–1 protein with its substrates showed that the first glucosylation at C24 increases the affinity for the C3 glucosylation (SI Appendix, Fig. S18). In accordance with the docking model results, the UGT720-269–1 enzyme, when incubated with mogrol, yielded both M1A1 (C24 glucosylation) and M2E (C3 and C24 glucosylations), but not M1E1 (C3 glucosylation) (Fig. 5C).
Branched glucosylations.
The subsequent secondary glucosylations were carried out by three members of a single UGT family, UGT94, which were specific for branching and did not perform primary glucosylations (Fig. 5D; SI Appendix, Fig. S19). The three UGT94 enzymes share between 89% and 93% identity (SI Appendix, Fig. S20) and showed differences in in vitro substrate specificity (Fig. 5D). UGT94-289–1 and -289–3 appear to be the most versatile, each leading to the penta-glucosylated mogroside (M5), whereas UGT94-289–2 appears to be most limited in its substrate specificity. We observed an M6 product (m/z 1,642.5) in reactions of UGT94-289–3 with M5 as substrate, but this was not always reproducible (SI Appendix, Fig. S21). Interestingly, UGT720-269–1 also showed branching activity, specifically on the C3 primary glucose (Fig. 5D), and it too may contribute to the branching portion of the pathway.
Protein modeling identified the structural novelty of the branching UGT94 family as due to an expanded pocket size in the active site of the glycosylation reaction, which accommodates the enlarged glycosylated substrates (Fig. 6A). This accommodation of the glucosyl moiety is made possible by a hydrophilic wall of polar amino acids in the enlarged pocket, specifically conserved in this group of branching enzymes (Fig. 6B).
Fig. 6.
Modeling of the mogroside branching UGTs. (A) Docking model of UGT94-289–3 with a glucosylated mogroside residing in the substrate pocket, shown in blue shading. The model shows a deep pocket behind the catalytic His14 (stick figure) that easily accommodates one or two glucose moieties. The wall pocket is created by two helices (colored in yellow) that create a polar interface suitable for glucose binding. The Glu193 is positioned behind the space-filled mogroside. The UDP-glu donor molecule is represented as a stick and ball model. (B) Alignment of the region containing the three characteristic polar residues of the branching UGT94 enzymes (listed as 289–1,2,3 and colored in blue) of representative UGT proteins listed in Dataset S2. Complete protein alignment is presented in SI Appendix, Fig. S22.
In summary, based on the combined metabolic profiling, functional expression and protein-modeling results, we can propose the following metabolic pathway for mogroside biosynthesis. During the initial stage of fruit development, squalene is metabolized to the di-glucosylated, tetra-hydroxycucurbitadienol, via the progressive actions of squalene synthase, squalene epoxidase, cucurbitadienol synthase, epoxide hydrolase, CYP87D18, and UGT720. During fruit maturation, there is a progressive addition of branched glucosyl groups, catalyzed by the UGT94 members, and perhaps also the UGT720-269–1, leading to the sweet M4, M5, and M6 compounds. It might be of significance that our study was performed on parthenocarpic Siraitia fruit that developed in response to hormonal treatment of the nonpollinated female flowers. This may have been fortuitous for the identification of a strong coordinated expression pattern by delineating and separating the developmental stages of the fruit pericarp.
Novelty of the Siraitia Mogroside Pathway.
Following the elucidation of the metabolic pathway, we investigated the molecular evolutionary novelty of mogroside synthesis in Siraitia fruit, taking into consideration the Siraitia genomic assembly and the analysis of Siraitia gene expression during fruit development, contrasted with the genome and transcriptome of other Cucurbitaceae representatives.
Considering the recently well-established phenomenon of genomic clustering of secondary metabolic pathways (17, 18, 21), particularly the clustering of terpene synthases with CYP450s (38), and further supported by the report of clustering in cucurbitacin synthesis in the related cucurbit cucumber (19), we hypothesized that the Siraitia mogroside pathway would similarly be accounted for by novel genomic clustering. Thus, we searched the assembled Siraitia scaffolds for combinations of the identified genes. Surprisingly, the mogroside pathway genes did not show any indication of clustering among the members of the five enzyme families involved. In fact, in striking contrast to the hypothesis of novel mogroside metabolism clustering in Siraitia, the genomic organization of the identified mogroside pathway genes is syntenously preserved by their closest homologs in genomes of Cucurbitaceae that do not accumulate mogrosides, i.e., melon, cucumber, and watermelon (SI Appendix, Table S10). For example, the cluster harboring the CDS gene, described by Shang et al. (19), is syntenously preserved in Siraitia, melon, watermelon, and cucumber (SI Appendix, Table S11), as is the cluster surrounding the CYP performing the C11 hydroxylase reaction (SI Appendix, Table S12). Similarly, there is no indication of novel tandem duplications that can account for the pathway genes. Although the EPH and UGT genes identified are indeed members of tandem duplicated genes, these duplications are also preserved in the other cucurbit species (SI Appendix, Fig. S23).
This absence of gene clustering of the pathway is, at first glance, in dissonance with the numerous cases of clustered novel secondary metabolic pathways recently reported (17, 18, 38). For example, Chae et al. (39) compared the clustering of secondary metabolism among 16 unrelated plant genomes and recognized a significant presence of clustering among secondary metabolism pathways, especially of terpenoid, phenylpropanoid, and nitrogen-containing compounds, depending on the plant family member analyzed.
However, upon reconsideration, the existence of individual examples of clustering in secondary metabolism at the plant family level cannot be expected to account for the wide diversity in a particular secondary metabolism pathway within a single plant family. Perhaps the breadth of the triterpene cucurbitane compounds present in the Cucurbitaceae is exemplary of the inability of clustering to account for this variation. A single CDS terpene synthase is the first and singular committed step in the synthesis of the large family of cucurbitacins and other cucurbitane metabolites. Chen et al. (12) classified this chemical family and reported structures of over 200 (!) different cucurbitanes, primarily from members of the Cucurbitaceae, all presumably based on the single CDS. This considerable variety of related compounds is due to numerous combinatorial variations in chemical decorations, including various and varied oxygenation, reduction, methylation, and acetylation reactions. The particular characteristic decorations are species-, tissue-, and temporal-specific within the plant family, and it would be metabolically inconceivable to exert control over particular cucurbitane pathways merely via genomic organization. The highly conserved clustering of CDS with orthologous oxygenation and acetylation genes among the different cucurbit genomes shows that clustering cannot account for more than one, or a few, of the multiple cucurbitane metabolism pathways. The conserved clustering of some members of the cucurbitacin biosynthetic pathway with the CDS gene throughout the distantly related cucurbits may be useful in revealing the ancestral function of the clustered CDS pathway; however, the control of the wide variety of other branches of the pathway must be looked for elsewhere.
In light of the failure to attribute the Siraitia mogroside pathway to clustering or novel duplications, we investigated the evidence for a coordinated control of transcriptional regulation (40) of the mogroside pathway in S. grosvenorii, uniquely characterizing this species in contrast to other cucurbit fruit. We performed RNA-seq transcriptomic analyses on developing melon and watermelon fruit and compared the expression patterns of the pathway members of Siraitia fruit with their respective orthologs in melon and watermelon fruit.
Strikingly, the temporal expression of the complete early pathway leading to mogrol (SQE, CDS, EPH, and CYP87D18) is strictly coordinated. A hierarchical tree based on the expression patterns of all of the Siraitia SQE, CDS, EPH, CYPs, and UGTs indicates strong hierarchical clustering of the mogroside biosynthetic genes. Fig. 7 shows the single CDS, one of the SQEs, two of the EPHs, and the CYP87D18 (listed as “s623.1” based on its scaffold position) clustered together, all characterized by the developmental pattern of strong expression in the youngest fruit followed by a sharp decline in expression. SQE2 and EPH2 also show a very strong young fruit expression but, due to their high expression in stems and leaves, clustered separately (SI Appendix, Fig. S24). Interestingly, the CDS and CYP87D18 cluster independently from the SQE1 and EPH1 and EPH3 based on their expression in the root whereas CDS and CYP87D18 are strongly expressed in the roots, which have high levels of bitter cucurbitacins; the SQE1 and especially the EPH1 and EPH3 genes are more fruit-specific in their expression (Fig. 7; SI Appendix, Fig. S24). No such coordination of the mogroside genes identified is evident in either developing melon fruit or watermelon fruit (SI Appendix, Table S13). In these, the SQE and EPH genes are expressed in a noncoordinated manner, whereas the CDS and C11 hydroxylase genes are nearly silent in their developing fruit.
Fig. 7.
Hierarchical clustering of the expression patterns of the members of the five gene families responsible for mogroside biosynthesis: SE, EPH, CDS, CYPs, and UGTs. The cluster containing the genes of the pathway is outlined in blue and is expanded to the right. The genes shown to be involved in mogroside biosynthesis are marked by an asterisk. An expandable version of the entire heat map is presented in SI Appendix, Fig. S24 in which the additional genes identified in this paper are marked. Genes with numbers containing decimal points are derived from the UGT-containing scaffolds, and the number following the decimal indicates its position in the tandem arrangement. Genes with numbers containing a lower hyphen are derived from the CYP-containing scaffolds, and the last number indicates its position in the tandem arrangement. Genes beginning with the letter “S” are derived from the genomic scaffolds. All genes are described in Dataset S2. The CYP87D18 coding for the C11-hydroxylating enzyme is listed as S623.1, closely clustering with the CDS gene.
The role of UGT720-269–1 in mogroside synthesis is also supported by its gene expression pattern, which is broadly coordinated with those of the enzymes of mogrol synthesis. It is most highly expressed in the young fruit (15 d and 34 d) and declines with fruit maturity, although not as strikingly as the strict hierarchical cluster of early fruit expression (SI Appendix, Figs. S14 and S24). This correlates as well with the developmental pattern of mogroside accumulation that indicated that the M2 product was already at its peak concentration in the early fruit. The expression patterns of the other C3 glucosylation genes of UGT74 and UGT75 did not show the pattern of early fruit expression, nor did the members of the UGT73 family (SI Appendix, Figs. S14 and S24). Nevertheless, these functionally active genes were expressed in the young Siraitia fruit, and their involvement in mogroside synthesis cannot be ruled out.
Only the branching enzyme family UGT94 exhibited a strikingly different expression pattern, strongly up-regulated in the latter stages of development (SI Appendix, Figs. S14 and S24) in agreement with the accumulation of the branched mogrosides in the maturing fruit. This expression pattern is also novel to Siraitia fruit; however, the evolutionary changes in the expression of this enzyme family are unlikely to be causal to mogroside metabolism because it is temporally distinct from the committed early stages of mogroside M2 biosynthesis, when the novelty of mogroside accumulation is already determined.
The possibility exists that, together with the coordinated expression of the mogroside pathway in the fruit, the defining characteristic of which is the trans-dihydroxy decorations at the 24,25 position, there is additional metabolic control on the flux of the pathway, perhaps specifically through an enhanced level of diepoxysqualene. The novelty of Siraitia fruit mogroside synthesis may also lie with metabolic compartmentation characteristics of the fruit. The Siraitia squalene epoxidase reaction drives the pathway to diepoxysqualene, perhaps by involving novel metabolic channeling (41) or perhaps by the low levels of competing pathways for the initial squalene monoepoxide product. In support of the latter, the expression pattern of the single cycloartenol synthase (CAS) gene is temporally asynchronous with early mogroside accumulation, compared with CDS. CAS expression is about 20% the expression as that of CDS at the earliest stage of fruit development and increases dramatically only upon the sharp decline of CDS expression in the 34-DAA fruit (SI Appendix, Table S14). Alternatively, the compartmentalization/subcellular localization of the pathway enzymes may contribute to the flux control, as it appears from analysis of predicted enzyme localization using numerous targeting programs that all of the enzymes may be localized to organelles, either ER or peroxisomes (SI Appendix, Table S15). Further research is required to account for these components of control of the flux and accumulation of mogrosides.
In conclusion, we have identified the and strikingly coordinated metabolic pathway for the natural, noncaloric sweetener mogroside. This insight is expected to facilitate the production of low-cost, high-intensity natural sweeteners.
Methods
Plant Material.
Seeds of S. grosvenorii were purchased from B & T World Seeds, germinated under sterile conditions, and grown trellised under shade netting in Bet Dagan, Israel. Of nearly 30 plants, a single female plant produced a flush of flowers in May 2011 and, in the absence of male flowers, the newly opened female flowers were treated with a hormone spray (20 ppm naphthalenacetic acid, commercial formulation from Alphatop, Perelman Co.) to induce a parthenocarpic fruit set. Flowers were tagged and fruit was sampled over the following 103-d period, when the fruits had turned a ripe beige. In the laboratory, plant tissue was stored at −80 °C. Samples of at least three fruit at each stage were harvested at 15, 34, 55, 77, 90, and 103 DAA. Samples of mature leaves (most recent full-sized), stems, and root tissue were similarly prepared.
Chromatographic Separations and Identifications.
Details of metabolite extractions, chromatographic separations, standards preparation, and NMR analysis are provided in SI Appendix, Methods.
DNA Isolation, RNA Isolation, Library Preparation, and Sequencing.
See SI Appendix, Methods.
Genome Assembly.
Initial assembly of short 100-bp PE reads into 2,000-bp “super-reads”’ was carried out using the MaSuRCA‐2.1.0 Genome Assembler (42). Following this, the super-reads were combined with the TLSR reads using Celera Assembler 8.2 (43) (assembly parameters and assembly statistics in SI Appendix, Table S2). Scaffolding was done with SSPACE (44), combining the two additional mate-pair libraries (SI Appendix, Table S3). Due to the single-strand origin of the TLSR sequences it is possible that alleles for the same locus are represented on different scaffolds, accounting for the high similarities especially among the CYP and UGT collections of genes. Genome-size estimation (420 Mb) was based on the total size of assembled scaffolds and corresponded to the value derived from the k-mer distribution of Illumina 100-bp PE reads (445 Mb) using Jellyfish (45). The organellar (mitochondrial and chloroplastic) genomes were filtered based on BLAST analysis against the melon organellar genome database and were excluded from the genome assembly. To collect the complete genomic catalog of the five enzyme families (squalene epoxidases, epoxide hydrolases, triterpene synthases, CYP450, and UGT), irrespective of their expression in the tissues studied, the genome assembly was used to identify all of the members of the families.
Data Availability.
The raw sequencing data were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive database as Bioproject PRJNA339375.
Specific Gene Family Expression Analysis.
The transcriptome assembly contigs annotated as CYP450, together with annotated CYP450 sequences from the melonomics database (https://melonomics.net/; version 3.5), combined with a comprehensive list of plant Cytochrome P450-dependent monooxygenase (CYP) sequences (38, 46), were combined. This collection was used to search against the Moleculo long-reads using Basic Local Alignment Search Tool (BLASTN) (47) to extract “CYP reads” (E-value cutoff of 0.001). The CYP reads were de novo-assembled using Celera Assembler 8.2 (43) using default parameters and manually corrected as necessary into a Siraitia genome CYP450 database. These are presented in Dataset S2.
Similarly, 158 UDP-glucosyltransferase sequences from the melon database (https://melonomics.net/; version 3.5) combined with the annotated UGT transcript contigs from the Siraitia transcript de novo assembly were searched against the TLSR library (via BLASTN, E-value< 0.001). The “UGT reads” were also assembled using Celera Assembler 8.2 using default parameters and manually corrected as necessary into a Siraitia genome UGT database. These are presented in Dataset S2.
In a similar manner the smaller gene families were also collected, searching the annotated melon database combined with the annotated de novo transcriptome assembly. These are presented in Dataset S2.
GeneScan software (48) was used for predicting the locations and exon–intron structures of genes in the CYP and UGT genomic assemblies. These were corrected manually, based on BLASTX comparisons of the genomic scaffolds, against the NCBI nr database. The predicted genes were annotated against the melon protein database (https://melonomics.net/; version 3.5; BLASTX) for extricating the genes of the five families.
The expression estimation of the five family genes was carried out using the RNA-seq libraries. The nine single-end 100-bp libraries of 15-DAA, 34-DAA, 50-DAA, 77-DAA, 90-DAA, and 103-DAA fruit, stems, leaves, and roots) were mapped on the reference five-family genes using version 2.1.0 of bowtie2 (49), and soap.coverage (version 2.7.7) was used for depth estimation of each gene in each sample. The expression data are presented in Dataset S2.
Phylogenetic and Cluster Analysis.
Alignments of protein sequences for the large phylogenetic analyses (UGT and CYP) were performed using the mafft program using default parameters (mafft.cbrc.jp/alignment/server/). Phylogenetic trees were constructed based on a maximum likelihood framework using phyml software (50) based on the LG matrix-based model (51). The trees were graphically designed using Figtree version 1.4 (tree.bio.ed.ac.uk/software/figtree/). For the smaller phylogenetic analyses (SQE, EPH) the one-click phylogeny.fr program (phylogeny.lirmm.fr/phylo_cgi/index.cgi) was used (52). Hierarchical clustering of genes was performed based on the expression values using Expander 7 software (53) and the CLICK and Kmeans algorithms (54).
Gene Expression and Functional Analysis.
Candidate genes were cloned using standard laboratory methods, or synthetic genes were prepared by Gen9Bio based on identified sequences.
Cucurbitadienol synthase.
The coding region fragment for CDS was synthesized by Gen9Bio to contain additional restriction sites (NotI and Cla) to allow its cloning in MCS1 of the yeast expression vector pESC-URA (Agilent) under the control of the GAL10 promoter and designated as pESC-CDS. The constructs were transformed into DH5α competent cells, and plasmid DNAs were prepared and used to transform the yeast strain GIL77 (gal2 hem3-6 erg7 ura3-167), which was kindly provided by M. Shibuya and Y. Ebizuka, Graduate School of Pharmaceutical Sciences, University of Tokyo, Tokyo (32). Yeast GIL77 was maintained on YEPD medium supplemented with ergosterol (20 mg/mL) and Tween 80 (5 mg/mL). Transgenic tobacco plants expressing CDS were produced as follows. The full-length CDS was used as a template for a PCR with the forward and reverse primers (F-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGTGGAGGTTAAAGGTCGGA and R-GGGGACCACTTTGTACAAGAAAGCTGGGTTTATTCAGTCAAAACCCGATGG), rendering it suitable for the Gateway system (Invitrogen). The resulting amplicon was cloned into the pENTR vector and subsequently cloned into the plant expression vector Pk7wg2.0. The resulting plasmid, Pk7wg2.0-CDS, was expressed in tobacco plants as described in Cohen et al. (55).
Epoxide hydrolase.
The coding-region fragments for the candidate EPHs were synthesized by Gen9Bio to contain additional restriction sites (SalI and NheI) for their cloning into the MSC2 site of the pESC-CDS vector under the control of the GAL1 promoter. The resulting plasmids were designated pESC-CDS_EPH1, -2, or -3 and were then transformed in the yeast strain GIL77. Relative activity of EPH was calculated from the integrated peak area of di-hydroxycucurbitadienol and 24,25-epoxycucurbitadienol in the control and EPH-expressing yeast lines, as calculated by the MassHunter Qualitative analysis version B.06.00 using Find by Molecular Formula algorithm (Agilent Technologies, Inc.).
CYP450s.
The coding-region fragments for the candidate CYPs were synthesized by Gen9Bio, as above. The resulting plasmids were designated pESC-CDS_CYPXXX and were then transformed in the yeast strain BY4743_YHR072 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 kanMax::erg7/ERG7; Thermo). For the analysis of CYPs, yeast was transformed with pESC-HIS vector harboring the Arabidopsis thaliana NADPH-CYP reductase under GAL1 induction (56). For the additional expression of SgEPH3 in BY4743_YHR072 yeast expressing SgCYP87D18, AtCPR, and SgCDS, the SgEPH3 was cloned in pESC-Leu. Yeast for this experiment were grown in the presence of the lanosterol synthase inhibitor R0 48–8072 (57) (50 µg/mL) (Cayman Chemicals), added 4–10 h following galactose induction.
UGTs.
UGT sequences were designed to contain the NheI restriction site at 5′ to create fusion with His-tag and the NotI restriction site at 3′ following the stop codon. Synthetic DNA was cut with NheI and Not enzymes, subcloned into pET28a expression vector (Novagen), and sequenced for verification. For UGT functional expression, Arctic Express (Agilent Technologies, Inc.) bacteria, containing UGT genes in pET28a (Novagen) vector, were grown overnight in 5 mL of LB at 37 °C at 200 × g. Then, 20 mL of LB was inoculated with 0.8 mL of overnight culture and grown until OD600 = 0.4. Next they were induced in 4 mM IPTG overnight at room temperature, cells were collected (5-min centrifugation at 10,000 × g), and the pellet was resuspended in 1.5 mL of 50 mM Tris⋅HCl, pH 7.0, 15% (vol/vol) glycerol, 0.1 mM EDTA, and 5 mM β-mercaptoethanol. After breaking the cells by five cycles of half-hour sonication/freeze in liquid nitrogen, insoluble material was removed by centrifugation for 1 h at 20,000 × g, and the soluble fractions were used for characterization of the enzymes. Proteins were stored at −20 °C until further analysis.
Heterologous Expression in Recombinant Yeast and Analysis.
Single transformed yeast colonies were incubated overnight in 3 mL synthetic complete medium without the appropriate selection markers at 30 °C and 220 × g and were then used for the inoculation of 20 mL medium. After 2% (wt/vol) galactose induction for 2 d, cells were collected by centrifugation, and the pellet was disrupted with 2.5 mL of hot 20% (wt/vol) potassium hydroxide and 50% (vol/vol) ethanol and extracted twice with a similar volume of n-hexane. The hexane extract was evaporated and resuspended in 1 mL methanol for the analysis of the cucurbitadienol product using a liquid chromatography-mass spectrometry (LC-MS) (SI Appendix, Methods). In the case of hydroxylation products, the extraction protocol was modified, and we used the commercial yeast lysis buffer Yeast Buster (Merck) for cell disruption and ethyl acetate for extraction.
Heterologous Expression in E. coli, UGT Enzyme Assays.
The mogroside substrates were dissolved to 1 mM in 50% DMSO. Enzyme assays were carried out in 50 mM Tris⋅HCl, pH 7.0, containing 5 mM β-mercaptoethanol, using up to 25 µL crude extract, 8 mM UDP-glucose, and 0.1 mM substrate in a final reaction volume of 100 µL. After overnight incubation at 37 °C, reactions were stopped by addition of 300 µL methanol, followed by brief vortexing. Subsequently, the extracts were centrifuged for 10 min at 20,000 × g and analyzed initially by LC-DAD (liquid chromatography with diode array detector); reactions showing evidence of new products were further analyzed by LC-MS (SI Appendix, Methods). The amount of product was measured by the peak area in the LC-MS chromatogram and compared with the control with an enzyme preparation of E. coli harboring an empty vector.
Protein Modeling.
See SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Prof. Shibuta and Prof. Ebizuka for the generous gift of the yeast strain GIL77; Prof. Michael Court for the classification of the relevant UGTs; and Dr. Sariel Hubner for advice regarding the genome assembly. The mogroside standards of M4, M5, IM5, and M6 were supplied by Dr. Indra Prakash, Dr. Venkata Sai Prakash Chaturvedula, and Dr. Michael Brown from The Coca Cola Co. The research was partially supported by The Coca Cola Co.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw sequencing data were deposited in the National Center for Biotechnology Information Sequence Read Archive database as Bioproject PRJNA339375.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604828113/-/DCSupplemental.
References
- 1.Castro DC, Berridge KC. Opioid hedonic hotspot in nucleus accumbens shell: Mu, delta, and kappa maps for enhancement of sweetness “liking” and “wanting.”. J Neurosci. 2014;34(12):4239–4250. doi: 10.1523/JNEUROSCI.4458-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madsen HB, Ahmed SH. Drug versus sweet reward: Greater attraction to and preference for sweet versus drug cues. Addict Biol. 2015;20(3):433–444. doi: 10.1111/adb.12134. [DOI] [PubMed] [Google Scholar]
- 3.Bray GA, Popkin BM. Dietary sugar and body weight: Have we reached a crisis in the epidemic of obesity and diabetes?: Health be damned! Pour on the sugar. Diabetes Care. 2014;37(4):950–956. doi: 10.2337/dc13-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroger M, Meister K, Kava R. Low-calorie sweeteners and other sugar substitutes: A review of the safety issues. Compr Rev Food Sci Food Saf. 2006;5(2):35–47. [Google Scholar]
- 5.Suez J, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514(7521):181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 6.Pepino MY. Metabolic effects of non-nutritive sweeteners. Physiol Behav. 2015;152(Part B):450–455. doi: 10.1016/j.physbeh.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim N-C, Kinghorn AD. Highly sweet compounds of plant origin. Arch Pharm Res. 2002;25(6):725–746. doi: 10.1007/BF02976987. [DOI] [PubMed] [Google Scholar]
- 8.Swingle WT. Momordica grosvenori sp. nov. The source of the Chinese Lo Han Kuo. J Arnold Arbor. 1941;22:197–203. [Google Scholar]
- 9.Kasai R, et al. Sweet cucurbitane glycosides from fruits of Siraitia siamensis (chi-zi luo-han-guo), a Chinese folk medicine. Agric Biol Chem. 1989;53(12):3347–3349. [Google Scholar]
- 10.Xu R, Fazio GC, Matsuda SP. On the origins of triterpenoid skeletal diversity. Phytochemistry. 2004;65(3):261–291. doi: 10.1016/j.phytochem.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Thimmappa R, Geisler K, Louveau T, O’Maille P, Osbourn A. Triterpene biosynthesis in plants. Annu Rev Plant Biol. 2014;65:225–257. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- 12.Chen JC, Chiu MH, Nie RL, Cordell GA, Qiu SX. Cucurbitacins and cucurbitane glycosides: Structures and biological activities. Nat Prod Rep. 2005;22(3):386–399. doi: 10.1039/b418841c. [DOI] [PubMed] [Google Scholar]
- 13.Nelson D, Werck-Reichhart D. A P450-centric view of plant evolution. Plant J. 2011;66(1):194–211. doi: 10.1111/j.1365-313X.2011.04529.x. [DOI] [PubMed] [Google Scholar]
- 14.Hamberger B, Bak S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos Trans R Soc Lond B Biol Sci. 2013;368(1612):20120426. doi: 10.1098/rstb.2012.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, et al. Chemistry and pharmacology of Siraitia grosvenorii: A review. Chin J Nat Med. 2014;12(2):89–102. doi: 10.1016/S1875-5364(14)60015-7. [DOI] [PubMed] [Google Scholar]
- 16.Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012;69(6):1030–1042. doi: 10.1111/j.1365-313X.2011.04853.x. [DOI] [PubMed] [Google Scholar]
- 17.Boycheva S, Daviet L, Wolfender JL, Fitzpatrick TB. The rise of operon-like gene clusters in plants. Trends Plant Sci. 2014;19(7):447–459. doi: 10.1016/j.tplants.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Nützmann HW, Osbourn A. Gene clustering in plant specialized metabolism. Curr Opin Biotechnol. 2014;26:91–99. doi: 10.1016/j.copbio.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Shang Y, et al. Plant science. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science. 2014;346(6213):1084–1088. doi: 10.1126/science.1259215. [DOI] [PubMed] [Google Scholar]
- 20.Itkin M, et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science. 2013;341(6142):175–179. doi: 10.1126/science.1240230. [DOI] [PubMed] [Google Scholar]
- 21.Wada M, et al. Prediction of operon-like gene clusters in the Arabidopsis thaliana genome based on co-expression analysis of neighboring genes. Gene. 2012;503(1):56–64. doi: 10.1016/j.gene.2012.04.043. [DOI] [PubMed] [Google Scholar]
- 22.Frusciante S, et al. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci USA. 2014;111(33):12246–12251. doi: 10.1073/pnas.1404629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Q, et al. An efficient approach to finding Siraitia grosvenorii triterpene biosynthetic genes by RNA-seq and digital gene expression analysis. BMC Genomics. 2011;12:343. doi: 10.1186/1471-2164-12-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D, et al. Seasonal variation of mogrosides in Lo Han Kuo (Siraitia grosvenori) fruits. J Nat Med. 2007;61(3):307–312. [Google Scholar]
- 25.Corey EJ, Gross SK. Formation of sterols by the action of 2,3-oxidosqualene-sterol cyclase on the factitious substrates 2,3:22,23-dioxidosqualene and 2,3-oxido-22,23-dihydrosqualene. J Am Chem Soc. 1967;89(17):4561–4562. doi: 10.1021/ja00993a079. [DOI] [PubMed] [Google Scholar]
- 26.Rowan MG, Dean PD, Goodwin TW. The enzymic conversion of squalene, 2(3),22(23)-diepoxide to alpha-onocerin by a cell-free extract of Ononis spinosa. FEBS Lett. 1971;12(4):229–232. doi: 10.1016/0014-5793(71)80027-3. [DOI] [PubMed] [Google Scholar]
- 27.Nelson JA, Steckbeck SR, Spencer TA. Biosynthesis of 24,25-epoxycholesterol from squalene 2,3;22,23-dioxide. J Biol Chem. 1981;256(3):1067–1068. [PubMed] [Google Scholar]
- 28.Boutaud O, Dolis D, Schuber F. Preferential cyclization of 2,3(S):22(S),23-dioxidosqualene by mammalian 2,3-oxidosqualene-lanosterol cyclase. Biochem Biophys Res Commun. 1992;188(2):898–904. doi: 10.1016/0006-291x(92)91140-l. [DOI] [PubMed] [Google Scholar]
- 29.Godio RP, Fouces R, Martín JF. A squalene epoxidase is involved in biosynthesis of both the antitumor compound clavaric acid and sterols in the basidiomycete H. sublateritium. Chem Biol. 2007;14(12):1334–1346. doi: 10.1016/j.chembiol.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki H, Achnine L, Xu R, Matsuda SP, Dixon RA. A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J. 2002;32(6):1033–1048. doi: 10.1046/j.1365-313x.2002.01497.x. [DOI] [PubMed] [Google Scholar]
- 31.Rasbery JM, et al. Arabidopsis thaliana squalene epoxidase 1 is essential for root and seed development. J Biol Chem. 2007;282(23):17002–17013. doi: 10.1074/jbc.M611831200. [DOI] [PubMed] [Google Scholar]
- 32.Shibuya M, Adachi S, Ebizuka Y. Cucurbitadienol synthase, the first committed enzyme for cucurbitacin biosynthesis, is a distinct enzyme from cycloartenol synthase for phytosterol biosynthesis. Tetrahedron. 2004;60:6995–7003. [Google Scholar]
- 33.Davidovich-Rikanati R, et al. Recombinant yeast as a functional tool for understanding bitterness and cucurbitacin biosynthesis in watermelon (Citrullus spp.) Yeast. 2015;32(1):103–114. doi: 10.1002/yea.3049. [DOI] [PubMed] [Google Scholar]
- 34.Dai L, et al. Functional characterization of cucurbitadienol synthase and triterpene glycosyltransferase involved in biosynthesis of mogrosides from Siraitia grosvenorii. Plant Cell Physiol. 2015;56(6):1172–1182. doi: 10.1093/pcp/pcv043. [DOI] [PubMed] [Google Scholar]
- 35.Wong J, Quinn CM, Brown AJ. Synthesis of the oxysterol, 24(S), 25-epoxycholesterol, parallels cholesterol production and may protect against cellular accumulation of newly-synthesized cholesterol. Lipids Health Dis. 2007;6:10. doi: 10.1186/1476-511X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinto JT, Cooper AJ. From cholesterogenesis to steroidogenesis: Role of riboflavin and flavoenzymes in the biosynthesis of vitamin D. Adv Nutr. 2014;5(2):144–163. doi: 10.3945/an.113.005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, et al. Oxidation of cucurbitadienol catalyzed by CYP87D18 in the biosynthesis of mogrosides from Siraitia grosvenorii. Plant Cell Physiol. 2016;57(5):1000–1007. doi: 10.1093/pcp/pcw038. [DOI] [PubMed] [Google Scholar]
- 38.Boutanaev AM, et al. Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci USA. 2015;112(1):E81–E88. doi: 10.1073/pnas.1419547112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chae L, Kim T, Nilo-Poyanco R, Rhee SY. Genomic signatures of specialized metabolism in plants. Science. 2014;344(6183):510–513. doi: 10.1126/science.1252076. [DOI] [PubMed] [Google Scholar]
- 40.Patra B, Schluttenhofer C, Wu Y, Pattanaik S, Yuan L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim Biophys Acta. 2013;1829(11):1236–1247. doi: 10.1016/j.bbagrm.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Jørgensen K, et al. Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr Opin Plant Biol. 2005;8(3):280–291. doi: 10.1016/j.pbi.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 42.Zimin AV, et al. The MaSuRCA genome assembler. Bioinformatics. 2013;29(21):2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers EW, et al. A whole-genome assembly of Drosophila. Science. 2000;287(5461):2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 44.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 45.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson DR. A world of cytochrome P450s. Philos Trans R Soc Lond B Biol Sci. 2013;368(1612):20120430. doi: 10.1098/rstb.2012.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268(1):78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 50.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 51.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 52.Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ulitsky I, et al. Expander: From expression microarrays to networks and functions. Nat Protoc. 2010;5(2):303–322. doi: 10.1038/nprot.2009.230. [DOI] [PubMed] [Google Scholar]
- 54.Shamir R. Algorithmic approaches to clustering gene expression data. In: Jiang T, Smith T, Xu Y, Zhang MQ, editors. Current Topics in Computational Biology. MIT Press; Cambridge, MA: 2002. pp. 269–299. [Google Scholar]
- 55.Cohen S, et al. The PH gene determines fruit acidity and contributes to the evolution of sweet melons. Nat Commun. 2014;5:4026. doi: 10.1038/ncomms5026. [DOI] [PubMed] [Google Scholar]
- 56.Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D. Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem. 1997;272(31):19176–19186. doi: 10.1074/jbc.272.31.19176. [DOI] [PubMed] [Google Scholar]
- 57.Morand OH, et al. Ro 48-8.071, a new 2,3-oxidosqualene:lanosterol cyclase inhibitor lowering plasma cholesterol in hamsters, squirrel monkeys, and minipigs: Comparison to simvastatin. J Lipid Res. 1997;38(2):373–390. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive database as Bioproject PRJNA339375.