Significance
The ability to engineer specific behaviors into cells would have a significant impact on biomedicine and biotechnology, including applications to regenerative medicine and biofuels production. One way to coax cells to behave in a desired way is to globally modify their gene expression state, making it more like the state of cells with the desired behavior. This paper introduces a broadly applicable algorithm for transcriptome engineering—designing transcription factor deletions or overexpressions to move cells to a gene expression state that is associated with a desired phenotype. This paper also presents an approach to benchmarking and validating such algorithms. The availability of systematic, objective benchmarks for a computational task often stimulates increased effort and rapid progress on that task.
Keywords: gene-regulatory networks, regulatory systems biology, transcriptome, engineering, Saccharomyces cerevisiae
Abstract
The ability to rationally manipulate the transcriptional states of cells would be of great use in medicine and bioengineering. We have developed an algorithm, NetSurgeon, which uses genome-wide gene-regulatory networks to identify interventions that force a cell toward a desired expression state. We first validated NetSurgeon extensively on existing datasets. Next, we used NetSurgeon to select transcription factor deletions aimed at improving ethanol production in Saccharomyces cerevisiae cultures that are catabolizing xylose. We reasoned that interventions that move the transcriptional state of cells using xylose toward that of cells producing large amounts of ethanol from glucose might improve xylose fermentation. Some of the interventions selected by NetSurgeon successfully promoted a fermentative transcriptional state in the absence of glucose, resulting in strains with a 2.7-fold increase in xylose import rates, a 4-fold improvement in xylose integration into central carbon metabolism, or a 1.3-fold increase in ethanol production rate. We conclude by presenting an integrated model of transcriptional regulation and metabolic flux that will enable future efforts aimed at improving xylose fermentation to prioritize functional regulators of central carbon metabolism.
The central premise of regulatory systems biology is that a systematic map of a cell’s regulatory machinery will enable us to understand, predict, and rationally manipulate the cell’s state or behavior. Manipulation of cellular state has many promising applications, including stem cell biology and regenerative medicine, biofuel production, and gene therapy. Progress toward cellular state control has been driven by both the systems biology and the synthetic biology research communities. Systems biology has produced whole-genome regulatory network maps (1), but relatively little research has focused on using these maps for predicting and manipulating cellular behavior (2). Regulatory synthetic biology has focused on creating molecular circuits that can be placed into a cell to control the transcription of a small number of transgenes, but genome-scale engineering of the cell’s native regulatory apparatus is still rare, with most systems restricted to a limited set of controlled targets (3). Here, we demonstrate that transcription factor (TF) network mapping, gene expression profiling, and computational modeling can be integrated to rationally engineer transcriptional state. We call this activity, which bridges the gap between systems biology and synthetic biology, “transcriptome engineering.”
Transcriptome engineering focuses on the manipulation of extant regulatory networks to enforce a state associated with a desired phenotype. The use of native regulatory mechanisms and network models enables designers to access evolutionarily optimized states and to rationally control the expression of hundreds of genes (4). Although there has been previous work on transcriptome engineering via random mutagenesis, we use the term here to refer to rational, design-based approaches (5). Most previous work on transcriptome engineering has taken place in the context of stem cell engineering, with the generation of induced pluripotency being the most prominent example (6). Since then, many transcriptional interventions have been identified that move cells along a specified lineage (7). However, current strategies for lineage conversion are typically guided by human expertise rather than generally applicable analytical methods and often are unable to fully convert cells to the desired fate (8–10).
In the past year, several algorithms have been proposed for recommending TFs to overexpress to convert mammalian cells from one cell type to another (8, 11, 12). These are important contributions to regenerative medicine, but they are not general algorithms for moving cells from any transcriptional state to any other transcriptional state. For example, the CellNet (8) recommendation system requires many expression profiles for the target cell type after many distinct perturbations of that cell type. CellNet is intentionally biased toward TFs with many target genes, and it recommends only activators to overexpress, not activators to knock down or interventions on repressors (13). The Mogrify (11) system requires multiple input databases including a protein–protein interaction network, considers only the activation of genes expressed in the target state (not the repression of genes that are not expressed), and focuses on TFs that are differentially expressed (DE) between the source and target cells (not those that are activated by posttranscriptional mechanisms). Finally, these systems have been tested on only a handful of conversion tasks, perhaps because of the technical challenges of mammalian cell culture, transdifferentiation, and cell type determination.
In this paper, we introduce and systematically evaluate NetSurgeon, a broadly applicable algorithm for recommending TF deletions or overexpressions to move cells from an initial (current) expression state to any desired state associated with any desired phenotype. The only inputs it requires are one expression profile of the initial state, one expression profile of cells in the target state, and an approximate transcriptional regulatory map indicating the direct targets of each TF. The network map can be derived from gene expression data by NetProphet (14) or other network inference algorithms, from TF binding motifs determined in vitro, or from in vivo binding data derived by methods such as chromatin immunoprecipitation sequencing (ChIP-seq) or Calling Cards (15, 16). Unlike the CellNet recommendation system (8), NetSurgeon considers all genes that are DE between the initial and target states, not only those that increase, it considers interventions on all TFs whether or not they are DE, and it has no bias toward TFs with large numbers of targets. Instead, it prefers interventions on TFs most of whose targets are predicted to move in the desired direction upon deletion or overexpression of the TF. Because it considers the predicted direction of change, NetSurgeon has no bias toward activators over repressors or overexpression over knockdown. CellNet, by contrast, considers only interventions in which activators are overexpressed. We also introduce a systematic, genome-scale evaluation procedure for general transcriptome engineering algorithms. Saccharomyces cerevisiae is ideal for evaluating transcriptome engineering because of the ease of experimental testing in yeast and the wealth of data available for TF network mapping and algorithmic validation (17–20).
After evaluation on existing yeast data, we apply NetSurgeon to the goal of improving xylose fermentation in S. cerevisiae. Xylose is a five-carbon sugar that is found, together with glucose and other sugars, in cellulosic biomass. Although S. cerevisiae is the most commonly used organism for industrial ethanol production, its inefficient fermentation of pentoses has hampered the production of ethanol from lignocellulosic biomass (21). Recombinant yeast strains expressing transgenes that facilitate integration of xylose into central carbon metabolism have been constructed (22, 23). However, when they are grown in mixed glucose/xylose cultures, which would be encountered in lysates of cellulosic biomass, they rapidly ferment all available glucose and then shift into a respiratory metabolic state in which little if any ethanol is produced (23). This led us to hypothesize that engineering yeast cells to have a gene expression profile in xylose similar to the profile of wild-type (WT) cells in glucose would increase ethanol yield.
Results
Algorithmic Approach to Transcriptome Engineering.
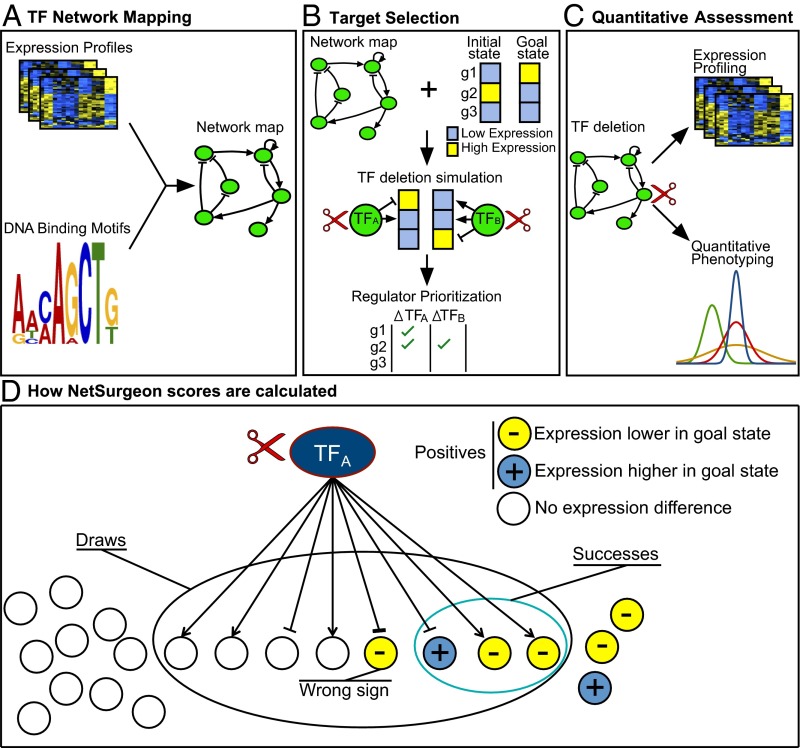
Our approach consists of three steps. First, we build a genome-wide map of the network of direct, functional regulation (Fig. 1A). Each TF–target relationship in this map is labeled as either activating or repressing. Second, we define starting and goal transcriptional states and apply NetSurgeon, which searches through all possible interventions (TF deletions or overexpressions) to identify those that are likely to move the transcriptional state toward the goal state (Fig. 1B). Finally, we construct strains containing the predicted best interventions, profile their transcriptomes, and quantitatively assay the phenotypes of interest—in this study, sugar consumption and the production of biomass and major metabolic products (Fig. 1C).
Fig. 1.
Overview of computational and experimental approach for control of transcriptional state. (A) A gene-regulatory network map is constructed from expression profiles and DNA binding motifs. (B) Given an initial state (gene expression profile), a goal state, and a network map, NetSurgeon simulates the qualitative effects of each intervention (symbolized by the scissors icon) on the expression of each gene. Predicted effects that move the expression of a target gene in the desired direction are indicated by green check marks. Using the results of the simulation, NetSurgeon assigns a priority score to each intervention that reflects its confidence that the intervention will move the cell from the initial state toward the goal state. (C) High-priority interventions are carried out, and their effects on the expression profile and the desired phenotype are assessed quantitatively. (D) NetSurgeon scores are calculated using the hypergeometric probability function with draws, positives, and successes as indicated (see text for details). Arrowheads indicate activation, and T heads indicate repression.
NetSurgeon assigns a score to each possible intervention, representing its confidence that the intervention will yield a substantial shift toward the goal state. The score is loosely based on the cumulative hypergeometric distribution, where the universe is the set of all genes. The “positives” are the genes that are significantly differently expressed between the initial state and the goal state, optionally intersected with a set of genes involved in a process of interest (Fig. 1D). In the hypergeometric enrichment calculation, the “draws” are the target genes of the TF on which the intervention acts, as specified by the input network map. The successes are the positives drawn—that is, the targets of the TF that are also DE between initial and goal states. To count as a success, a gene must not only be a target of the TF, it must also be predicted to move in the direction of the goal state as a result of the intervention (Fig. 1D). Deletion of a TF is predicted to increase the expression of targets it represses and decrease expression of targets it activates. Overexpression of a TF is predicted to have the opposite effects.
The NetSurgeon score is not used for hypothesis testing—that is, rejecting a null hypothesis. It is simply a convenient formula for calculating a score that increases when either of two numbers (the number of targets or the fraction of targets that are DE between the initial and goal states) increases while the other is held constant. The NetSurgeon score differs from the P value of a hypergeometric enrichment test in that different genes among the positives have different weights. This weighting does not change the total number of positives, but it allocates that number unequally among the DE genes. Thus, the number of successes is not just the number of targets that are DE and predicted to move in the right direction, it is the sum of the weights of those targets. To calculate the weight of each gene, we take advantage of the fact that the initial and goal states are both defined by measured expression profiles, so we can calculate a q value (false-discovery rate) for each gene being DE. All genes that pass a q-value threshold for being significantly DE (we used q < 0.05) are weighted by the negative logarithm of the their q values, normalized by the sum of the negative log q values of all positives:
where qi is the false-discovery rate for gene i being DE between the initial and goal states and D is the set of indices of genes whose q value is below the threshold. The sum of Wi over all DE genes is 1. The number of successes for intervention k, Sk, is the total number of DE genes times the sum of the weights of the DE genes that are targets of the TF:
where is the index set of targets that are predicted to move toward the goal state as a result of the intervention. The NetSurgeon score for intervention k given a fixed network is then the hypergeometric probability of obtaining at least Sk positives when drawing a sample of size T from a universe of all genes containing ||D|| positives, where T is the total number of targets of the perturbed TF.
For a given network map in which each gene is categorized as repressed, activated, or unregulated by each TF, the scores for interventions are calculated as above. However, network-mapping algorithms typically return a list of possible TF–target relations ranked from most confident to least confident; setting a confidence threshold is left to the user. NetSurgeon therefore makes the score calculation described above using various thresholds and takes the maximum score over all thresholds considered. It considers a threshold that includes only the top 500 interactions, the top 1,000, the top 1,500, and so on, down to the top 40,000.
Building a TF Network Map.
To test this approach, we first built an integrated gene-regulatory network map by combining separate functional and physical maps. The functional map was built by using NetProphet, an expression-based mapping algorithm that combines a differential expression analysis and a regression analysis (14). Each possible TF–target pair is assigned a differential expression score that is equal to the log odds that the putative target is DE when the TF is perturbed, given the available replicate expression profiles. Each pair is also assigned a regression score based on a regularized regression of the expression level of the target gene (the dependent variable) against the expression levels of all TFs (the predictor variables). The regression score for a given TF–target pair is simply the regression coefficient assigned to the TF’s expression level as a predictor of the target’s expression level. The differential expression and regression scores are combined to provide a final score representing NetProphet’s confidence that the TF binds and regulates the target (see ref. 14 for details). For this paper, we ran NetProphet with 320 potential regulators including proteins with DNA-binding domains, chromatin factors, and other members of DNA-binding protein complexes that regulate gene expression. The input data were gene expression profiles of 269 regulator deletion strains grown in rich medium with glucose (19). For the 51 potential regulators for which there was no expression-profiled deletion, strain 0 was used as the differential expression component of the NetProphet score. The physical map was built by scanning position weight matrix (PWM) models over all yeast promoters (17) to estimate the potential of each TF to bind each promoter (see SI Materials and Methods for details). We integrated the functional and physical network maps by assigning to each TF–target pair a score that was equal to the geometric mean of the scores assigned to it in the functional and physical networks. We refer to this as the NetProphet+PWM network.
Network Models Can Efficiently Guide Transcriptome Engineering.
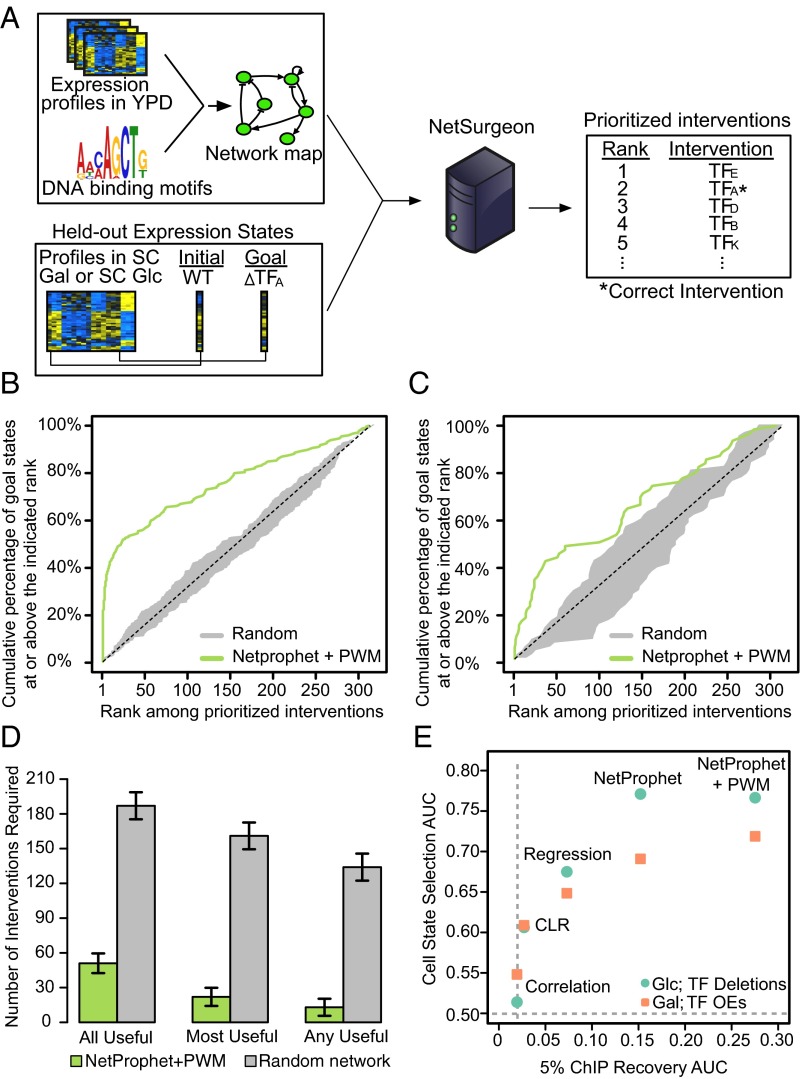
Before embarking on new experiments, we evaluated NetSurgeon using publicly available gene expression datasets from yeast strains in which a single TF had been deleted. The expression profile of WT cells was used as the initial state and the profile of each TF intervention strain was used as a goal state (Fig. 2A). NetSurgeon did not know which TF was deleted or overexpressed to produce the goal state, or even that the goal state was related to a single-TF perturbation. It scored possible TF deletions or overexpressions for moving toward the goal state as described above. We could evaluate the results because we knew the transcriptional profile of each TF-intervention. Thus, we could calculate the Euclidean distance between the expression profile resulting from each intervention and that of the goal state. We could then compare that distance to the distance between the expression profiles of the WT and the goal state to determine whether the intervention reduced the distance to the goal state.
Fig. 2.
Benchmarking NetSurgeon on existing data. (A) Overview of our approach to benchmarking NetSurgeon on existing expression data. (B) Cumulative percentage of correct picks (the TFs that were actually deleted to generate the goal state profile; y axis) prioritized above the rank indicated on the x axis. The goals states were the expression profiles 245 regulator deletion mutants growing in synthetic complete medium (SC) supplemented with 2% glucose. NetSurgeon ranking using the NetProphet+PWM network map (green), NetSurgeon ranking using random permutations of the NetProphet+PWM map (gray), and the expectation for randomly ranked interventions (dotted line). (C) Same as B for goal states defined by the expression profiles of 63 TF overexpression strains growing in SC supplemented with 2% galactose. (D) Median number of top-ranked NetSurgeon interventions required to include all useful interventions, the most useful intervention, or any useful intervention (lower is better). Useful interventions are defined as those that reduce the distance to the goal state by at least 10%. The analogous numbers for randomly ranked networks are shown in gray. (E) NetSurgeon accuracy as a function of the accuracy of the input TF network map for five networks, where NetSurgeon is assessed with goal states defined by TF deletion strains in SC with glucose (green) or TF overexpression strains in SC with galactose (orange). NetSurgeon accuracy is summarized by the area under curves analogous to those shown in B and C. The accuracy of the input GRNs is assessed using ChIP-supported interactions as the gold standard and summarized by the area under the precision recall curve from 0% to 5% recall (SI Materials and Methods).
The first set of goal-state expression profiles were the expression profiles of TF-deletion strains growing on synthetic complete medium with glucose (SC+Glc) (20), whereas the NetProphet+PWM network map was built from profiles of strains growing in rich medium with glucose (YP+Glc). These two media are very different in terms of both gene expression and TF activity (see, e.g., the study of TF Cbf1 in ref. 14). For each of the 245 goal states, NetSurgeon used the NetProphet+PWM network map to assign scores to all 320 possible deletions of regulators in the NetProphet+PWM network. The 320 included 75 “distractor” regulators that were present in the NetProphet+PWM network but did not correspond to goal states. We plotted the cumulative percentage of goal states for which NetSurgeon ranked the best intervention (the one that actually produced the goal state profile) at or above each rank (Fig. 2B, green). We compared NetSurgeon performance to random ranking of TF deletions (Fig. 2B, dotted black) and found that NetSurgeon assigns higher ranks to the correct interventions (Mann–Whitney U test, P < 10−23). NetSurgeon ranks the correct intervention within the top five, a reasonable number to test experimentally, for 91 goal states, which is 24 times better than chance (P < 10−96). We also ran NetSurgeon on 100 random networks with the same topology as the NetProphet+PWM network (Fig. 2B, gray) and observed performance at random chance levels, indicating that an accurate network is critical for NetSurgeon.
We then repeated the analyses described above using goal states defined by cells cultured in conditions even further from those used to construct the input network map. These goal states consisted of 63 expression profiles obtained from regulator overexpression strains grown in SC+Gal (24) (defined medium with galactose), whereas the NetProphet+PWM network was built using expression profiles from regulator deletion strains growing in YP+Glc (rich medium with glucose). As expected, performance on this more challenging task was not as good as when the goals states were from regulator deletion strains grown on SC+Glc. Nonetheless, NetSurgeon assigned higher ranks to the correct interventions compared with randomly assigned ranks (Mann–Whitney U test, P < 10−3) and assigned the best intervention a top five rank for 8 of the 63 goal states (13%), an eightfold improvement over chance (P < 10−5).
To assess the practicality of NetSurgeon-guided transcriptome engineering, we ran NetSurgeon on the NetProphet+PWM network and the SC+Glc goal set (20) and computed the median number of top-ranked interventions that would need to be tested to identify the single most useful intervention. NetSurgeon can identify the single most useful intervention (the one that produced the goal state) in a median of 22 predictions, whereas random guessing would take 161 tries. An intervention was deemed useful if it reduced the distance between the WT cells and the goal by at least 10% (Fig. 2D). NetSurgeon can identify at least one useful intervention in a median of 13 predictions, whereas random guessing would take 134 tries.
NetSurgeon Accuracy Depends on the Accuracy of the TF Network Map.
To evaluate the effect of network accuracy on NetSurgeon performance, we applied NetSurgeon to network maps inferred from the same expression dataset as before but using different algorithms for TF network mapping: CLR (25), LASSO regression (26), NetProphet alone (14), and NetProphet integrated with PWM scores. We first evaluated the structural accuracy of the five maps by their level of support from ChIP data. The results show that NetProphet is more accurate than a simple LASSO regression approach, which is more accurate than CLR, consistent with our previously published comparison of these algorithms (14) (see ref. 27 for a recent review of TF network mapping algorithms.) As expected, scoring potential TF–target relations by the correlation of the TF’s expression level with target’s expression level performed worse than CLR, which postprocesses correlation coefficients. Also as expected, supplementing NetProphet scores with scores reflecting the TF’s potential for binding the sequences in the promoter region of each gene improved accuracy above that of NetProphet alone. We then plotted the accuracy of NetSurgeon when using each of these five input maps on our two goal sets—the TF-deletion strains in SC+Glc and TF-overexpression strains in SC+Galactose—against the accuracy of the input map (Fig. 2E). This shows a clear pattern of improved NetSurgeon performance with more accurate network maps.
Application of Transcriptome Engineering to Ethanol Production.
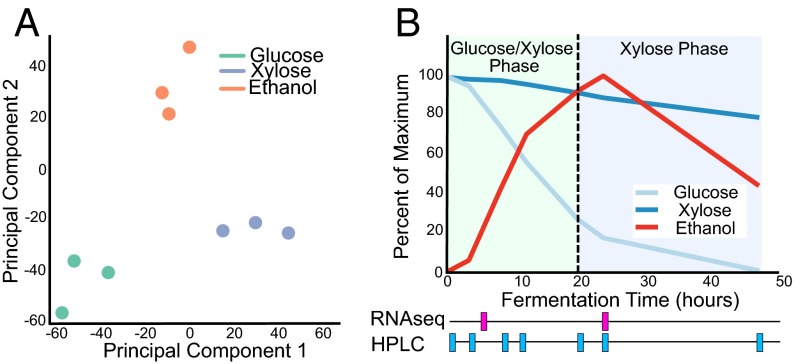
We next applied NetSurgeon to the industrially relevant problem of ethanol production in a mixed glucose–xylose coculture. Principal-components analysis of RNA-seq data from S. cerevisiae cells grown with various carbon sources, including xylose, indicated that cells growing on xylose were in a transcriptional state with some characteristics of cells grown in 5% (wt/vol) glucose, a fermentative state, and some characteristics of cells grown in 1.3% (wt/vol) ethanol, a respiratory state (Fig. 3A). As S. cerevisiae cells do not natively consume xylose, we hypothesized that they were unable to recognize xylose as a fermentable carbon source and therefore entered into a transcriptional state that was nonoptimal for fermentative metabolism. We therefore sought to identify interventions that would shift the cells growing in xylose from the transcriptional state of WT cells growing in xylose (initial state) to the transcriptional state of WT cells growing on glucose (goal state). To apply NetSurgeon to this problem, we generated a network map using all available expression profiles and DNA binding motifs (Full-NetProphet+PWM) and attempted to optimize the expression of 445 genes involved in carbohydrate metabolism. Specifically, we calculated the NetSurgeon score using a subset of the genes that are DE between the initial and goal states—the 445 genes that are annotated in GO or KEGG with the terms Glycolysis, Pentose Phosphate, TCA Cycle, Oxidative Phosphorylation, Carbohydrate Metabolism, Hexose Metabolism, or Pentose Metabolism. Using this network map, NetSurgeon produced a rank-ordered list of regulators whose deletion was predicted to force the system toward the goal state. We tested the top eight interventions, deletion of CAT8 (catabolite repression), HAP4 (heme activator protein), ADR1 (alcohol dehydrogenase II synthesis regulator), MSN2 (multicopy suppressor of SNF1 mutation), MSN4 (multicopy suppressor of SNF1 mutation), GIS1 (GIg1-2 suppressor), AFT2 [activator of Fe (iron) transcription], or USV1 (up in starvation), in the H2217-7 yeast strain (28). As a limited comparison of our algorithmically selected deletions with expert intuition, we also deleted the master regulator SNF1, the yeast ortholog of AMP kinase, which inactivates TFs responsible for glucose repression (29).
Fig. 3.
RNA expression and metabolite analyses reveal a state transition between cells grown on glucose plus xylose and those grown on xylose alone. (A) Principal-components analysis of RNA-seq data indicates that cells growing in xylose are in a transcriptional state with characteristics of both cells growing in high glucose (fermentative state) and those growing in ethanol (respiratory state). (B) Overview of metabolite concentrations (graph at Top) and HPLC/RNA-seq sampling schedule (timeline at Bottom) for aerobic batch fermentations with the WT VTT-C-99318 strain.
The selected interventions looked promising on the basis of existing literature. Cat8 and Hap4 are respiratory factors active in the general cellular response to xylose, and deletion of HAP4 was recently shown to improve cellobiose consumption rates (28, 30). MSN2 and MSN4, encoding stress-associated factors, were observed to be highly up-regulated in xylose and their transcriptional targets misregulated (31). ADR1 is an activator of ADH2, a glucose-repressed gene that encodes an alcohol dehydrogenase that catalyzes a key step in ethanol consumption (32, 33). Usv1, Gis1, and Aft2 have been shown to have clear roles in the yeast transcriptional response to nonfermentable carbon sources and general stress response (34–36).
Aerobic batch fermentations were used to assess the outcome of our interventions at both the transcriptional and the metabolic levels. Cells were inoculated into synthetic complete medium (SC) supplemented with 2% (wt/vol) glucose and 5% (wt/vol) xylose at OD600 = 1.0 ± 0.2 and grown for 48 h. Samples were taken for RNA-seq at 4 and 24 h, representative of the glucose–xylose and xylose phases. Samples were also taken for analyzing output metabolites by HPLC at time points throughout the 48-h fermentation. The results showed a glucose–xylose phase, during which both sugars were consumed and ethanol was produced, followed by a xylose phase during which glucose was largely depleted and ethanol was consumed along with xylose (Fig. 3B).
Transcriptome Engineering Promotes a Fermentative State.
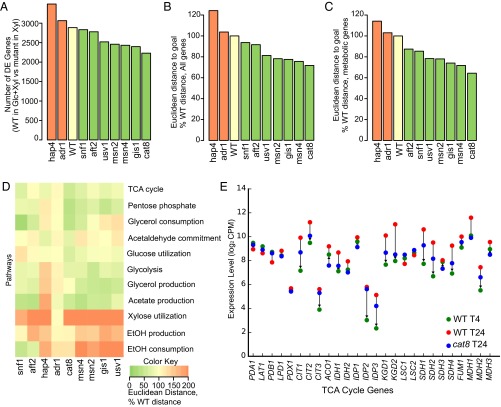
Analysis of our RNA-seq data revealed that 2,887 genes are DE between the 4-h time point, when glucose is abundant, and the 24-h time point, when glucose has been depleted, by at least twofold in WT cells (42% of all genes; Fig. 4A, WT). Six of the eight NetSurgeon-selected interventions lowered the number of DE genes. The best intervention, cat8Δ, reduced the number of DE genes by 22%, performing much better than the choice of our human expert, snf1Δ. To examine this from another perspective, we calculated the Euclidean distance between the goal state (the fermentative state defined by WT expression profile in the glucose–xylose phase) and the state of the engineered strain in the xylose phase (Fig. 4B). Six of eight NetSurgeon interventions lowered the Euclidean distance between the two phases, with an average reduction of 21%. The cat8Δ intervention reduced the genome-wide expression distance by 28%. Among the 445 carbon metabolism genes that NetSurgeon targeted for optimization, cat8Δ reduced the Euclidean distance by 36% (Fig. 4C).
Fig. 4.
NetSurgeon interventions promote a fermentative transcriptional state. (A) Number of twofold or greater DE genes between the WT strain in the glucose-plus-xylose (fermentative) phase and each deletion mutant in the xylose-only (respiratory) phase. (B) Euclidean distance between the full expression profile of WT cells in the fermentative phase and the expression profiles of deletion mutants in the respiratory phase (lower is better). (C) Same as B, restricted to 445 genes encoding metabolically active proteins. (D) Euclidean distance between the expression profiles of selected metabolic pathways in WT cells in the fermentative phase and their profiles in deletion mutants in the respiratory phase (greener is better). (E) The expression of TCA cycle genes in WT cells in the fermentative phase (goal state, red), WT cells in the respiratory phase (initial state, green), and the cat8 deletion mutant in the respiratory phase (blue), on a log scale. Arrowheads indicate the direction the expression of each gene in cat8 (blue) would have to move to get closer to the goal (green). For most TCA cycle genes that show a substantial difference between the initial and goal states, the cat8 deletion moves their expression substantially toward the goal state.
We also evaluated the ability of each TF deletion to promote a fermentative state in 11 pathways of central carbon metabolism (Fig. 4D). Seven of the eight NetSurgeon-selected interventions reduced the Euclidean distance between the initial (T24, respiratory) and goal (T4, fermentative) state expression levels of the genes in at least one of the central carbon metabolism pathways evaluated. Six of these interventions improved the expression of glycolytic genes, and five of them also improved the expression of TCA cycle genes. Deletion of CAT8 promoted a fermentative state in many metabolic pathways essential for xylose fermentation, including genes involved in glucose utilization, the pentose phosphate pathway, glycolysis, the TCA cycle, and acetate/glycerol production. It moved all of the TCA cycle genes toward their expression levels in the goal state (Fig. 4E), although a few genes moved only a small fraction of the way toward their goal state levels (e.g., CIT3, IDP2) and several overshot their goals by a small margin. Considering all genes encoding enzymes in the TCA cycle, deletion of CAT8 reduced the Euclidean distance to the goal state by 60%. These observations highlight the power of transcriptome-level interventions to modulate the expression of many more genes than is feasible by traditional, one-gene-at-a-time genetic engineering.
Identification of Transcriptional States Associated with Improved Fermentation.
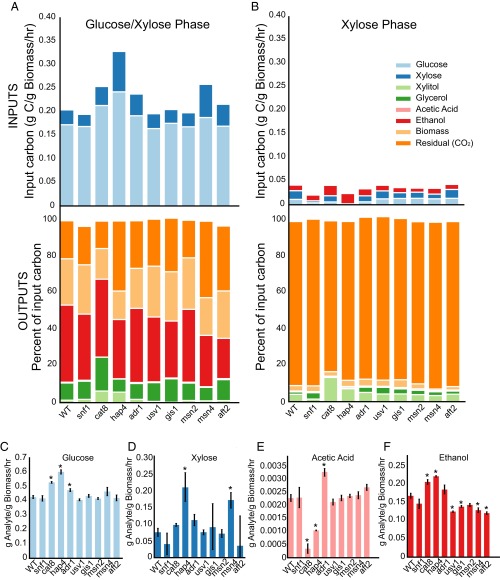
To assess changes in metabolic behavior following our transcriptional interventions, we profiled metabolic intake and output via HPLC. We calculated the carbon import rate per unit biomass for each strain and displayed the percentage of input carbon that is directed to each of the major carbon fates in each growth phase (Fig. 5 A and B). Carbon import rates declined in the xylose phase by 86%, on average, and the cells significantly shifted their carbon commitment from fermentative to respiratory processes in the xylose phase. The proportion of input carbon released as CO2 increased from a mean of 24% in the glucose–xylose phase to 89% in the xylose-only phase.
Fig. 5.
Transcriptome engineering alters carbon uptake rates and commitment ratios but does not prevent the transition to respiratory metabolism. (A) The absolute rate of carbon uptake (Top) and the distribution of carbon fates (Bottom) during the fermentative (glucose-plus-xylose) phase. (B) Same as A for the respiratory (xylose) phase. Red in Top panel indicates net ethanol consumption. (C) Specific rate of glucose consumption (fermentative phase); asterisks indicate a difference from WT with P < 0.05 by Benjamini–Hochberg-corrected t test. (D) Same as C for xylose consumption. (E) Same as C for acetic acid production. (F) Same as C for ethanol production.
Although the interventions we tested did not prevent the transition from fermentative to respiratory metabolism, they did affect carbon fate significantly. Fractional carbon commitment to every measured metabolite was altered by at least one of the TF deletions. The xylitol fraction was significantly increased by interventions affecting TFs associated with respiratory processes, a potential side effect of the respiratory factors regulating the promoters of the XYL1, XYL2, and XKS1 transgenes, which encode enzymes required for integrating xylose into central carbon metabolism. Interestingly, all significant changes in the fraction of input carbon allocated to ethanol or biomass were reductions. The deletions of SNF1, HAP4, USV1, GIS1, MSN4, and AFT1 significantly reduced the fraction of carbon committed to ethanol (average, 26%). Deletions of CAT8 and HAP4 reduced carbon commitment to biomass by 33% and 38%, respectively, in the glucose–xylose phase.
We also analyzed the specific rate (rate per unit biomass) of production of each metabolite by each strain (Fig. 5 C–F). This differs from the previous analysis, which focused on the fraction of carbon input allocated to each carbon fate. Although none of the interventions increased the fraction of input carbon allocated to ethanol, some interventions increased the specific rate of carbon input and hence the specific rate of ethanol production in the glucose–xylose phase. Interventions on respiratory regulators (Cat8, Hap4, and Adr1) improved the specific rate of glucose consumption between 11% and 40% (Fig. 5C). The hap4Δ and msn4Δ mutants improved the specific rate of xylose consumption by 170% and 120%, respectively (Fig. 5D). Acetic acid, a fermentation by-product that has been demonstrated to inhibit glycolysis (37), was produced at lower specific rates in the hap4 and cat8 mutants (53% and 83%; Fig. 5E). Importantly, the specific rate of ethanol production was significantly increased by 22% and 31% in the cat8 and hap4 mutants (Fig. 5F). These strains directed a smaller fraction of the carbon they consumed toward biomass than WT and their peak ethanol levels were significantly higher than those of WT (by 14% and 19%, respectively) and occurred at approximately the same time as WT (24 h). Among the stress-associated factors, we found that the deletion of USV1, MSN2, MSN4, and AFT2 significantly reduced the specific rate of ethanol production (22% on average; Fig. 5F). Taken together, these data demonstrate that transcriptome engineering can generate significant changes in metabolic behavior.
An Integrated Model of Transcriptional Regulation and Metabolic Flux Identifies Direct Regulators of Metabolic Flux.
In the previous sections, we presented both gene expression data and metabolic data on WT yeast and nine TF deletion mutants at multiple time points. We now integrate these datasets to produce a qualitative model linking transcription factors to metabolic fluxes. The purpose of the model is to summarize and integrate the findings reported in this paper and to suggest directions for future work.
As described above, major sources of carbon input and output were measured by HPLC (for soluble metabolites) and optical density (for biomass). These inputs and outputs are shown around the outside of Fig. 6. We then generated a system of linear equations encoding stoichiometric constraints on carbon fluxes across nine metabolic pathways: glucose import, xylose import, the pentose phosphate pathway, glycolysis, the TCA cycle, glycerol production and consumption, acetate production/consumption, ethanol production and consumption, and biomass production (Fig. 6, rectangles and black arrows). These equations, together with our measurements and an approximate partitioning of biomass into components emanating from each pathway, enabled us to calculate fluxes through the pathways. Together, our measurements and the assumption that most of the unaccounted-for carbon was released as CO2 fully constrained the internal fluxes. Unlike many flux balance analyses (e.g., refs. 38 and 39), our fully constrained model did not require the assumption that fluxes are distributed in a way that maximizes biomass production.
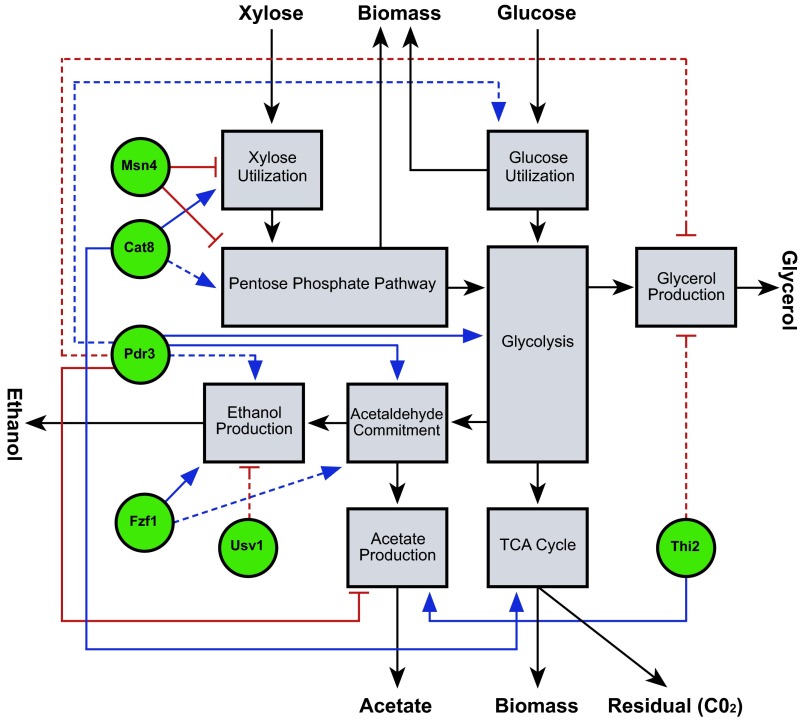
Fig. 6.
An integrated map of transcriptional regulation in central carbon metabolism. Metabolites around the outside are measured, except for the residual, which is carbon consumed minus carbon allocated to all measured carbon fates. Mauve rectangles: selected pathways of central carbon metabolism, including the xylose utilization pathway whose components are encoded by trans genes. Black arrows: connections along which carbon can flow from one pathway to another. Green circles: transcriptional regulators. Blue arrows: inferred transcriptional activation. Red T-head lines: inferred transcriptional repression. Solid lines indicate that the TF directly regulates one or more genes encoding enzymes involved in the pathway, according to the Full-NetProphet+PWM network.
To link transcriptional states with metabolic flux states, we computed the Pearson correlation coefficient between the expression of each TF in the yeast genome and the carbon flux through each pathway. The correlation was calculated across all strains and time points described above. We also generated a null distribution of correlation coefficients by correlating randomly permuted gene expression patterns with the metabolic fluxes. We then identified TFs whose expression is significantly correlated with metabolic flux (false-discovery rate ≤ 0.01) and generated edges (Fig. 6, red and blue lines) linking correlated TFs (Fig. 6, green circles) with each metabolic pathway. Interactions between TFs and pathways were hypothesized to be direct (solid lines) if our TF network map supported the TF as a direct regulator of one or more genes encoding proteins in the pathway. This analysis of our data suggested three transcriptional regulators that may be deeply interconnected with biochemical pathways important for xylose metabolism and fermentation. CAT8 expression was significantly correlated with carbon flux through the xylose utilization, the pentose phosphate, and the TCA cycle. Msn4 was predicted to directly regulate genes involved in xylose utilization and the pentose phosphate pathway, and flux through these pathways was negatively correlated with MSN4 expression. Pdr3 was suggested as a regulator of glycolytic genes, and flux through glycolysis was positively correlated with PDR3 expression. This integrated model suggests important transcriptional regulators that may control both transcriptional and metabolic state and therefore represent high-value targets for future transcriptomic engineering of ethanol productions.
SI Materials and Methods
Saccharomyces cerevisiae Fermentations.
All S. cerevisiae strains were grown aerobically in 60 mL of synthetic complete medium (SC) at 30 °C in 250-mL baffled Erlenmeyer flasks shaken at 225 rpm. Cultures for identification of differential expression associated with carbon sources were grown in triplicate in 50 g/L glucose, 50 g/L xylose, or 1.3 g/L ethanol for 8 h before sampling for RNA-seq. Cultures of cells for evaluating the impact of transcriptional interventions were performed in triplicate and initiated by inoculating overnight cultures into 60 mL of SC supplemented with 20 g/L glucose and 50 g/L xylose resulting in a density of OD600 = 1.0 ± 0.2. Samples taken for RNA-seq were aliquoted from the primary culture, spun down at 3,000 × g, and frozen in liquid nitrogen. The samples for HPLC were supernatants collected after centrifugation of culture samples at 12,000 × g for 3 min before snap freezing for storage in a dry ice/ethanol bath. All samples were stored at −80 °C. At least two independent experiments of three biological replicates were performed for each genotype evaluated by HPLC.
HPLC.
The concentration of input and output soluble metabolites was analyzed by using HPLC. Supernatant solutions were stored at −80 °C and filtered through 0.22-μm syringe filter before HPLC analysis. Metabolites were eluted from an Aminex HPX-87H column maintained at 65 °C and peaks detected by a Waters 2414 refractive index detector. Identified peaks were quantified through integration and interpolated against serial dilutions of standards for glucose, xylose, xylitol, glycerol, acetic acid, and ethanol. Analysis of HPLC data was performed on each biological replicate, with metabolic input/output relationships quantified across each fermentation and pooled into a single distribution based on genotype. Turbidity measurements were converted into units of grams biomass per liter based on the turbidity to biomass conversion factor published (23). To evaluate internal carbon flux, a system of linear stoichiometric constraints was developed for certain branch points in key pathways of S. cerevisiae central carbon metabolism (SI Materials and Methods). This system of equations was fit to experimentally measured parameters of carbon import and export for each genotype across the glucose/xylose and xylose-only phases of each fermentation.
RNA Sequencing.
Total mRNA was isolated using the yeast RiboPure kit (Life Technologies). Libraries for RNA-seq were prepared as in ref. 42 except that the selection of poly(A) RNA from the total RNA isolated was accomplished using an mRNA Catcher Plus kit (Life Technologies) and the resulting poly(A) RNA was subsequently sheared by incubating in TURBO DNA-free buffer at 75 °C for 10 min and purified with a QIAquick PCR Purification kit (Qiagen).
Data Analysis Methods.
HPLC raw data output/structuring.
Analyte detection, peak identification, and peak integration were all performed using the Empower 3 software suite. Following initial labeling by Empower 3, individual peaks were manually confirmed and computationally identified peak boundaries adjusted to minimize error. The area under each labeled peak was calculated as a representation of analyte concentration. Due to the lack of batch data export or API access within Empower 3, tabular data for each HPLC run were manually exported from Empower 3 into a flat text file. Following raw data output, each peak was manually labeled with the molecule responsible for peak generation. Manual labeling was performed through comparison of peak retention time and area against authentic standards for each analyte. This process generated an annotated, comma-separated file suitable for downstream analysis.
Batch effect normalization.
HPLC and refractometry error were controlled for on a per-replicate basis. Data for each replicate were normalized for day effect as follows. All analyte concentrations were divided by the ratio of the HPLC reading for glucose at time 0 (expected to be 20 g/L because that is how the medium was prepared) and an authentic standard of 20 g/L glucose. This reduced the median coefficient of variation among biological replicates by 41%.
Biomass/HPLC data integration, replicate-based analysis.
Measurements of metabolite production from HPLC analysis and turbidity-based biomass measurements collected from spectrophotometry were integrated into a single data structure based on experimental date, strain genotype, and replicate number. All downstream analyses, including stoichiometric flux balance analysis, were performed on a per-replicate basis.
Evaporative normalization.
The presence of a significant evaporative effect on metabolite concentration was observed across the 48-h fermentations. Pilot experiments using standard fermentation media (synthetic complete medium, minus histidine, 20 g/L glucose, 50 g/L xylose) but lacking cells, indicated that all metabolites increased in concentration by ∼20% across a 48-h mock fermentation. To control for this, a quadratic polynomial was fitted to 10 mock fermentations sampled for HPLC analysis across 120 h. For each replicate, across all time points, the predicted evaporative effect was calculated and removed from HPLC and biomass measurements.
Analyte interpolation.
Analyte measurements were interpolated against serial dilutions of purified standards obtained from Sigma-Aldrich. Standards were diluted across physiologically relevant concentrations and then assayed by HPLC. A linear equation was fit to each standard and used to quantify each analyte.
Outlier identification.
Treatment of outliers was performed according to the National Institute of Standards and Technology (NIST) Engineering Statistics Handbook 1.3.5.17 (NIST/SEMATECH, 2012). The generalized extreme Studentized deviate test was used for formal outlier identification, with input parameters of α = 0.05 and 2 for the upper bound of total outliers allowed within a distribution.
Calculation of analyte rates of change, specific rates of change, and carbon commitment ratios.
To examine the effect of transcriptional interventions on cellular metabolism, the analytical process required the identification of biological steady states across which analyte rates of input and output could be calculated and transformed into steady-state models of internal carbon flux. Analysis of ethanol production and consumption indicated that there were three steady states present across the 48-h fermentation. The period from 3 to 12 h was defined as a fermentative metabolic state associated with high external glucose concentrations. A second fermentative phase, associated with low external glucose concentrations, was identified across the 12- to 20-h period. All assayed genotypes transitioned into a respiratory metabolic state during the 24- to 48-h window of the fermentation—this period was therefore used to represent the metabolic state associated with xylose catabolism. A lag phase during the diauxic shift from glucose to xylose and ethanol metabolism was observed from 20 to 24 h, and this time period was excluded from downstream analyses. Analysis of metabolic rates of change, carbon commitment calculations, and stoichiometric flux balance analysis focused on the 3- to 12- and 24- to 48-h fermentative periods for which RNA-seq data were available.
RNA-seq.
Two to three biological replicates of each fermentation strain genotype, growth condition, and time point combination were profiled. For all RNA-seq samples, the mean and median sequencing depth was 7.2 and 6.9 million reads, respectively. The interquartile range of sequencing depth was 4.8–9.1 million reads. Sequenced reads were aligned to the S. cerevisiae S288C reference sequence genome release R63 using TopHat, version 2.0.4 (43), and Bowtie, version 0.12.8 (44). Reads that aligned uniquely to the reference sequence were considered for gene expression quantification with Cufflinks, version 2.0.2 (45). Gene expression was quantitated against known gene boundaries from the gene annotations provided by the SGD S288C reference genome release R63. Gene expression was normalized using the Cufflinks upper-quartile normalization option. After gene expression quantification, samples not passing the following quality control filters were removed from consideration. Each sample was required to be sequenced at a depth of at least 750,000 reads. The expression of the deleted gene in mutant expression profiles was required to be less than 10% of its expression in WT. The mean and median expression of the deleted genes were 1.3% and 0% of the WT levels, respectively. The low levels of expression that remained for some samples were likely the result of errors in the sequencing process itself, either due to contamination between samples or a low level of error in matching barcodes for multiplexed samples to reads. In addition, within each replicate set containing three biological replicates outlier gene expression profiles were detected and removed if the outlier caused the median of all genes’ coefficients of variation (the SD divided by the mean) to be greater than 0.5 for the replicate set or if the outlier caused the Pearson correlation coefficient between replicate gene expression profiles to be less than 0.8. Differential expression analysis comparing initial and goal state expression profiles sets was performed by using LIMMA (46) with the voom transformation applied to read counts (47). The number of reads mapping to each gene was determined by using the htseq-count tool in version 0.5.3 of the HTSeq Python package (48). DE genes responding to the treatment factor were selected based on a q value (false-discovery rate) cutoff of less than or equal to 0.05.
Gene-Regulatory Network Construction.
We took an integrative approach to constructing a gene-regulatory network model by building separate network models from functional and physical data and then combining them. An integrative approach to the network-building process was chosen because integration often leads to more accurate and complete inferred network models (49). The functional gene-regulatory network model is built using NetProphet (14), a method for inferring direct, functional regulation from gene expression data. NetProphet works by combining differential expression analysis with a coexpression analysis. The LASSO regression coexpression component of NetProphet was run on four S. cerevisiae expression profile datasets, three of which are previously published large-scale datasets (19, 24, 50), and the last of the four was a RNA-seq dataset of S. cerevisiae cells grown with various single carbon sources. The four resulting network models were combined into a final coexpression regulatory network model by computing the geometric mean of scores assigned to each TF–target pair. A network model based on differential expression analysis was constructed by using the target differential expression information for all regulators deleted in the Hu et al. (19) dataset supplemented with the target differential expression information for overexpressed regulators in the Chua et al. (24) dataset that were not deleted in the Hu et al. dataset. The NetProphet weighted averaging scheme was used to combine the final coexpression and differential expression networks, producing the fully integrated functional regulatory network. This fully integrated functional regulatory network was normalized to a [0, 1] range by dividing each interaction score by the maximum of all interaction scores.
The physical network model was built using TF binding potential estimates generated by scanning a collection of position weight matrix (PWM) models over all yeast promoters. PWMs for 196 S. cerevisiae TFs were extracted from the ScerTF database (17). To estimate the binding potential of each TF with a PWM on each gene’s promoter, FIMO (51) was used to scan the TF’s PWM over the promoter of each S. cerevisiae gene using the default FIMO background model. Promoters were defined as extending from 800 bases upstream of the transcription start site (excluding sequence from neighboring ORFs) to 200 bases downstream from the transcription start site. Binding sites that were identified by FIMO at a value of P ≤ 0.005 were considered in subsequent analyses. Two models of binding were considered. For each TF, the strong site model ranks promoters containing one or more significant binding sites according to the negative log P value of the most significant site. The weak site model ranks promoters containing one or more significant binding sites by the sum of the negative log P values for all significant sites in a promoter. The final PWM binding potential scores for a TF on each gene’s promoter was computed using the geometric mean of the TF–gene strong and weak site binding potential scores. The final PWM binding potential scores for each TF were normalized into a [0, 1] interaction score range by dividing each binding potential score of a TF by the maximum of all binding potential scores for the TF.
We created our overall gene-regulatory network model by assigning a score to each TF–target gene pair that was equal to the geometric mean of the scores assigned to the pair in the functional and physical networks. This scoring function enforced that high scoring interactions are likely to be direct and functional due to their support by both binding and expression evidence.
NetSurgeon in Silico Validation.
We assessed the ability of NetSurgeon to select regulator interventions (deletions and overexpressions) for moving transcriptional state toward a goal state. To accomplish this, we evaluated NetSurgeon’s ability to identify the correct regulator intervention for goal states defined by postintervention expression profiles from two S. cerevisiae datasets. The first contains 245 regulator deletion expression profiles from cells growing on synthetic complete medium with glucose (SC+Glc) (20). The second contains 63 regulator overexpression expression profiles from cells grown in synthetic complete medium with galactose (SC+Gal) (24). In each case, the initial state was the WT expression profile from the same dataset.
For each expression profiling dataset, we transformed NetSurgeon’s scores for interventions aimed at achieving each goal state into ranks. We then compared NetSurgeon’s accuracy to randomly ranking regulator interventions and to intervention ranks produced by running NetSurgeon on 100 random networks with the same topology as the NetProphet+PWM network. In addition, we also evaluated the number of NetSurgeon selected interventions required to recover all useful interventions, the most useful intervention, and any useful intervention. The most useful intervention was defined as the intervention that decreased the Euclidean distance between the intervention expression state and the goal state by the most. This was, by definition, the intervention that produced the goal state expression profile.
To investigate the relationship between NetSurgeon accuracy and transcriptional network accuracy, we first inferred transcriptional networks using five different inference algorithms: expression correlation, CLR (25), LASSO regression (26), NetProphet alone (14), and NetProphet integrated with PWM scores. Each algorithm was used to infer a transcriptional network from a gene expression dataset consisting of 269 regulator deletions strains grown in YPD (19). The structural accuracy of each resulting network was assessed by comparing the ranked network interaction predictions to the network structures defined by a collection ChIP-seq and ChIP-chip experiments. The ChIP data were compiled as previously described (14). In total, the ChIP data provides 29,260 experimentally validated links between 187 regulators and 5,566 genes, and the ChIP-based network is assigned binary scores based on the presence of ChIP-validated interactions. The area under the precision recall curve at the 5% recall point was used to assess the accuracy of each inferred network compared with the ChIP-defined true network. These inferred networks were also used to make NetSurgeon intervention selections on both goal state expression datasets. NetSurgeon’s accuracy was assessed by the area under the line representing the cumulative percentage of goal states for which NetSurgeon ranked the best intervention (the one that actually produced the goal state profile) at or above each rank. The R glm function was used to investigate the relationship between network accuracy and NetSurgeon accuracy.
RNA-seq Batch Effect Removal.
Before using the RNA-seq data for downstream analysis, we identified technical noise in the expression profiling data that varied with the experimental batches. These batch effects were removed from RNA-seq read counts using the RUVg function in the RUVSeq R package (52). RUVg identifies and removes unwanted variation related to technical effects through factor analysis on a set of negative control genes that were not expected to respond to the biological treatments. We defined a set of 192 negative control genes by identifying genes that are not predicted to be regulated by any of the deleted regulators in the 100,000 highest scoring interactions according to the NetProphet+PWM network scores. Genes were then eliminated from this list if their expression changed significantly (logtwofold change < 1.3 and adjusted P value > 0.1) between any pair of our expression-profiled single carbon growth conditions. Dropping the first factor of unwanted variation, by using RUVg with the parameter k = 1, was sufficient to remove the majority of the batch effect. The batch effect corrected read count expression measurements were further normalized into log2 counts-per-million expression values: log2(cpm + 0.1). We will refer to this as log2 cpm, to be understood as including the pseudocount.
Intervention Strain Fermentative Expression State Convergence.
For each intervention strain we constructed and profiled, we measured the distance between the expression profile of the WT strain at 4 h (fermentative phase with glucose plus xylose) and the intervention strain at 24 h (xylose only). The goal, of course, was to find interventions that reduced this distance. We used two distance metrics. Our first metric quantified the number of twofold DE genes as the distance between two expression profiles. These genes were identified by applying LIMMA (46), with a design matrix considering factors for treatment and batch effects, on voom-corrected raw read counts as described above. We choose to use a twofold-change cutoff, rather than a statistical significance cutoff, because the number of replicate expression profiles (and hence the power to detect statistically significant differences) was not consistent among all intervention strains. Our second metric was Euclidean distance, calculated by using the R function dist on batch-corrected log2 cpm profiles.
Stoichiometric Flux Balance Analysis.
Internal carbon fluxes were described by fitting a system of equations designed to describe the central carbon metabolism of S. cerevisiae. The model was then fitted to measured cellular inputs and outputs using specific rates of analyte change in units of grams carbon per grams cell per hour.
The flux balance analysis equations are as follows:
Integration of Transcriptional Network State with Carbon Fluxes.
To link the transcriptional regulatory network with pathway carbon fluxes, we identified significant correlations between transcription factor expression (TF) levels and calculated metabolic fluxes. For each flux, the Pearson correlation coefficient (PCC) was calculated between the batch-corrected log2 cpm profile of each annotated TF-encoding gene and flux through each pathway. A null distribution of PCCs was generated by computing 10,000 PCCs between the carbon fluxes and randomized gene expression profiles. The random expression profiles were generated by sampling from all genes a log2 cpm expression value once per strain. This null distribution of PCCs was used to calculate the significance of each calculated PCC within the experimental set. In Fig. 6, PCCs with a false-discovery rate of ≤1% between transcription factors and pathway carbon fluxes were plotted.
Discussion
In this paper, we introduced NetSurgeon, an algorithm that uses a map of direct transcriptional regulation to select TFs whose deletion or overexpression will move a cell’s transcriptional state toward a specified goal. The availability of comprehensive TF deletion and overexpression collections in S. cerevisiae enabled us to systematically assess NetSurgeon’s accuracy on a genome-wide scale. Given the expression profile of a cell in which a single TF has been deleted, NetSurgeon can identify that TF in a median of 22 predictions, whereas random guessing would take 161 tries. We observed that network maps built with expression data from one environmental condition can be successfully used to identify interventions for achieving goal states defined by profiles in different environmental conditions. We also demonstrated that TF network maps enriched for direct regulatory relationships are important for this task—network maps generated by NetProphet together with PWM models led to selections that were substantially better than those made by using maps based on expression correlation or CLR (25).
We applied NetSurgeon to optimizing yeast for ethanol production from glucose–xylose coculture. NetSurgeon selected critical regulators supported in the literature without prior knowledge, and six of the eight selected interventions promoted a fermentative transcriptional state when considering the expression of all genes (Fig. 4 A and B) or the expression of 450 genes encoding enzymes involved in carbon metabolism (Fig. 4C). Although the TF deletions were insufficient to entirely prevent a state transition that normally affects 42% of the genes in the yeast genome, they succeeded in significantly changing the allocation of input carbon to various fates and the specific rates of production or consumption of metabolites. We found that regulators associated with respiratory processes had significant metabolic effects in the fermentative phase of the culture. We also found that deletion of stress factors lowers the rate of ethanol production and total ethanol yield.
We noted that five of the six interventions that improved the transcriptional state when considering all genes (Fig. 4 A and B) or 450 genes encoding metabolic enzymes (Fig. 4C) actually reduced the specific rate of ethanol production (Fig. 5F). One possible explanation is that these interventions deleted stress factors, thereby removing tools that S. cerevisiae needs to grow well in stressful conditions, including conditions in which xylose is the sole carbon source. Another possible explanation is that these interventions moved the expression of key ethanol production genes in the wrong direction (Fig. 4D). This cannot be the whole story because hap4Δ, which also moves these ethanol production genes in the wrong direction (Fig. 4D), significantly improves the specific rate of ethanol production (Fig. 5F). Nonetheless, it might be useful to modify NetSurgeon so that it takes in a list of key genes and vetoes any proposed interventions that move any of the key genes in the wrong direction. This rule would veto the hap4Δ mutant, which improved the specific rate of ethanol production, but would have allowed cat8Δ, arguably the best of the interventions tested.
There are several other modifications of NetSurgeon that would be worth exploring in future work. Currently, NetSurgeon weights genes that are DE between the initial and target states by their q value, a measure of confidence that they are in fact DE. The q value increases with both the difference in means between the initial and goal states and the inverse of the pooled variance. This works well in practice, but it is possible that performance would be further improved by considering only the difference in means. NetSurgeon could also be used iteratively to improve a strain by taking the expression profile after a single intervention as the initial state for selecting a second intervention. A more ambitious goal would be to select pairs of interventions a priori, before testing single interventions. One way to approach this would be to take the union of the predicted effects of the two interventions as the set of predicted effects of the pair. However, selecting double interventions might work better in conjunction with a quantitative model of transcriptional regulation that could predict how much the expression of each target gene would change as a result of each intervention.
Transcriptome engineering focuses on attaining naturally evolved transcriptional states under circumstances in which they do not normally occur. This focus contrasts with the more common approach to synthetic biology, in which individual genes encoding enzymes or transporters are knocked out or overexpressed, leading to unnatural transcriptional states. For example, the expression levels of genes encoding proteins in the TCA cycle are highly regulated, maintaining an appropriate ratio of enzymes and thereby avoiding intermediate metabolite accumulation or allosteric inhibition of upstream processes. The engineering of optimal expression levels across entire pathways is a challenging problem that is often addressed through iterative selection strategies (40). However, we found that CAT8 deletion provided a shortcut. The TCA cycle in S. cerevisiae consists of 26 genes, making optimization of this pathway’s expression level a difficult task for one-gene-at-a-time engineering. Deletion of CAT8 moved all 26 genes of the TCA cycle toward the fermentative state, showing that the manipulation of transfactors that have evolved to regulate a naturally occurring transcriptional state can greatly facilitate transcriptome engineering.
Transcriptome engineering is a relatively new endeavor with a bright future. We have demonstrated that transcriptional network maps enriched for direct regulatory interactions are highly valuable for identifying the key regulators that mediate a state transition and prioritizing them for genetic intervention. By exploiting collections of expression profiles from TF deletion or overexpression strains, we have established clear benchmarks by which transcriptome engineering algorithms can be compared with one another, which may accelerate progress (41). We have also demonstrated that transcriptome engineering can be used to move cellular behavior—in this case, metabolic behavior—toward a desired goal. Our dataset of 8,055 metabolic measurements with 73 matched RNA-seq profiles across 14 genotypes will enable future efforts to maximize ethanol production by manipulating key transcriptional regulators. We are optimistic that further progress in transcriptome engineering will lead to improvements in biofuel production, personalized medicine, and stem cell engineering.
Materials and Methods
A software package implementing NetSurgeon is available from the Brent Laboratory web page: mblab.wustl.edu/software.html.
The NetSurgeon scoring algorithm (described at the beginning of Results) can be expressed in the following formula for the score of intervention k:
where h is the hypergeometric probability function, (the positives in the sample) is defined below, G (the population) is the index set of all genes in the genome, (the sample) is the index set of targets of the perturbed regulator among the l most confidently predicted regulatory interactions in the network that are predicted to move toward the goal state as a result of the intervention, and D (the positives) is the set of indices of genes that are DE between the initial and goal state (i.e., those whose q value is below the user-determined threshold).
The number of positives in the sample for intervention k is defined to be the following:
where qi is the false-discovery rate for gene i (and all those q values < qi) being DE between the initial and goal states.
Acknowledgments
We thank Dr. Ron Hector for his gracious provision of plasmids containing the At5g17010 xylose transporter and technical advice on HPLC. We also thank Dr. Laura Salusjarvi and Dr. Laura Ruohonen for providing the H2217 S. cerevisiae strain. D.G.M. was supported by NIH Grant T32HG000045. This work was supported by NIH Grant GM100452.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE69682).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603577113/-/DCSupplemental.
References
- 1.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489(7414):91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chuang HY, Hofree M, Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol. 2010;26:721–744. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat Rev Microbiol. 2014;12(5):381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 4.Cardinale S, Arkin AP. Contextualizing context for synthetic biology—identifying causes of failure of synthetic biological systems. Biotechnol J. 2012;7(7):856–866. doi: 10.1002/biot.201200085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lam FH, Hartner FS, Fink GR, Stephanopoulos G. Enhancing stress resistance and production phenotypes through transcriptome engineering. Methods Enzymol. 2010;470:509–532. doi: 10.1016/S0076-6879(10)70020-3. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Morris SA, Daley GQ. A blueprint for engineering cell fate: Current technologies to reprogram cell identity. Cell Res. 2013;23(1):33–48. doi: 10.1038/cr.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cahan P, et al. CellNet: Network biology applied to stem cell engineering. Cell. 2014;158(4):903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marro S, et al. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9(4):374–382. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng R, et al. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci USA. 2008;105(16):6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rackham OJ, et al. FANTOM Consortium A predictive computational framework for direct reprogramming between human cell types. Nat Genet. 2016;48(3):331–335. doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- 12.D’Alessio AC, et al. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 2015;5(5):763–775. doi: 10.1016/j.stemcr.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris SA, et al. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158(4):889–902. doi: 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haynes BC, et al. Mapping functional transcription factor networks from gene expression data. Genome Res. 2013;23(8):1319–1328. doi: 10.1101/gr.150904.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Mayhew D, Chen X, Johnston M, Mitra RD. “Calling Cards” for DNA-binding proteins in mammalian cells. Genetics. 2012;190(3):941–949. doi: 10.1534/genetics.111.137315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Mayhew D, Chen X, Johnston M, Mitra RD. Calling Cards enable multiplexed identification of the genomic targets of DNA-binding proteins. Genome Res. 2011;21(5):748–755. doi: 10.1101/gr.114850.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spivak AT, Stormo GD. ScerTF: A comprehensive database of benchmarked position weight matrices for Saccharomyces species. Nucleic Acids Res. 2012;40(Database issue):D162–D168. doi: 10.1093/nar/gkr1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158(6):1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet. 2007;39(5):683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 20.Kemmeren P, et al. Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell. 2014;157(3):740–752. doi: 10.1016/j.cell.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 21.Fortman JL, et al. Biofuel alternatives to ethanol: Pumping the microbial well. Trends Biotechnol. 2008;26(7):375–381. doi: 10.1016/j.tibtech.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Hector RE, Dien BS, Cotta MA, Qureshi N. Engineering industrial Saccharomyces cerevisiae strains for xylose fermentation and comparison for switchgrass conversion. J Ind Microbiol Biotechnol. 2010;38(9):1193–1202. doi: 10.1007/s10295-010-0896-1. [DOI] [PubMed] [Google Scholar]
- 23.Hector RE, Qureshi N, Hughes SR, Cotta MA. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption. Appl Microbiol Biotechnol. 2008;80(4):675–684. doi: 10.1007/s00253-008-1583-2. [DOI] [PubMed] [Google Scholar]
- 24.Chua G, et al. Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci USA. 2006;103(32):12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faith JJ, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5(1):e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonneau R, et al. The Inferelator: An algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 2006;7(5):R36. doi: 10.1186/gb-2006-7-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brent MR. Past roadblocks and new opportunities in transcription factor network mapping. Trends Genet. 2016;32(11):736–750. doi: 10.1016/j.tig.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salusjärvi L, et al. Regulation of xylose metabolism in recombinant Saccharomyces cerevisiae. Microb Cell Fact. 2008;7:18. doi: 10.1186/1475-2859-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuttykrishnan S, Sabina J, Langton LL, Johnston M, Brent MR. A quantitative model of glucose signaling in yeast reveals an incoherent feed forward loop leading to a specific, transient pulse of transcription. Proc Natl Acad Sci USA. 2010;107(38):16743–16748. doi: 10.1073/pnas.0912483107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Y, et al. Leveraging transcription factors to speed cellobiose fermentation by Saccharomyces cerevisiae. Biotechnol Biofuels. 2014;7(1):126. doi: 10.1186/s13068-014-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsushika A, Goshima T, Hoshino T. Transcription analysis of recombinant industrial and laboratory Saccharomyces cerevisiae strains reveals the molecular basis for fermentation of glucose and xylose. Microb Cell Fact. 2014;13:16. doi: 10.1186/1475-2859-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tachibana C, et al. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol. 2005;25(6):2138–2146. doi: 10.1128/MCB.25.6.2138-2146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciriacy M. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. II. Two loci controlling synthesis of the glucose-repressible ADH II. Mol Gen Genet. 1975;138(2):157–164. doi: 10.1007/BF02428119. [DOI] [PubMed] [Google Scholar]
- 34.Hlynialuk C, Schierholtz R, Vernooy A, van der Merwe G. Nsf1/Ypl230w participates in transcriptional activation during non-fermentative growth and in response to salt stress in Saccharomyces cerevisiae. Microbiology. 2008;154(Pt 8):2482–2491. doi: 10.1099/mic.0.2008/019976-0. [DOI] [PubMed] [Google Scholar]
- 35.Pedruzzi I, Bürckert N, Egger P, De Virgilio C. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 2000;19(11):2569–2579. doi: 10.1093/emboj/19.11.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaiseau PL, Lesuisse E, Camadro JM. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J Biol Chem. 2001;276(36):34221–34226. doi: 10.1074/jbc.M104987200. [DOI] [PubMed] [Google Scholar]
- 37.Pampulha ME, Loureiro-Dias MC. Activity of glycolytic enzymes of Saccharomyces cerevisiae in the presence of acetic acid. Appl Microbiol Biotechnol. 1990;34(3):375–380. [Google Scholar]
- 38.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat Biotechnol. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plata G, Hsiao TL, Olszewski KL, Llinás M, Vitkup D. Reconstruction and flux-balance analysis of the Plasmodium falciparum metabolic network. Mol Syst Biol. 2010;6:408. doi: 10.1038/msb.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460(7257):894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stolovitzky G, Prill RJ, Califano A. Lessons from the DREAM2 challenges. Ann N Y Acad Sci. 2009;1158:159–195. doi: 10.1111/j.1749-6632.2009.04497.x. [DOI] [PubMed] [Google Scholar]
- 42.Haynes BC, et al. Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS Pathog. 2011;7(12):e1002411. doi: 10.1371/journal.ppat.1002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 47.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marbach D, et al. DREAM5 Consortium Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9(8):796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11(12):4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32(9):896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]